Supplementary materials. Computational study of β-n-acetylhexosaminidase from. Talaromyces flavus, a glycosidase with high substrate flexibility
|
|
|
- Patrick Sullivan
- 6 years ago
- Views:
Transcription
1 Supplementary materials Computational study of β-n-acetylhexosaminidase from Talaromyces flavus, a glycosidase with high substrate flexibility Natallia Kulik 1,*, Kristýna Slámová 2, Rüdiger Ettrich 1,3, Vladimír Křen 2 1 Department of Structure and Function of Proteins, Institute of Nanobiology and Structural Biology of GCRC, Academy of Sciences of the Czech Republic, Zamek 136, Nove Hrady, Czech Republic 2 Laboratory of Biotransformation, Institute of Microbiology, Academy of Sciences of the Czech Republic, Videnska 1083, Praha 4, Czech Republic 3 Faculty of Sciences, University of South Bohemia in Ceske Budejovice, Zamek 136, Nove Hrady, Czech Republic *Corresponding author: Kulik Natallia, Institute of Nanobiology and Structural Biology of GCRC, Academy of Sciences of the Czech Republic, Zamek 136, Nove Hrady, Czech Republic, phone: , kulik@nh.cas.cz. 1
2 Figure S1. Part of multiple sequence alignment containing loop 1 used for phylogenetic analysis. Amino acid composition of loop 1 in plants and fungi is similar and rich in aromatic residues and proline, so we could expect that they will have also similar conformation. In the hexosaminidases from Aspergillus oryzae and Penicillium oxalicum this loop has the same size and similar amino acid composition as the hexosaminidase from Talaromyces flavus (TfHex). 2
3 Figure S2. Part of multiple sequence alignment containing loop 2 used for phylogenetic analysis. Loops 1 and 2 are longest in plant (especially in Arabidopsis lyrata and Prunus persica) and in fungal sequences. N-terminal and C-terminal ends of loop 1 and loop 2 have similar amino acid composition in animals, fungi and plants. 3
4 Figure S3. Statistics of the refinement of TfHex model. A. Root means square deviation (RMSD) of Cα atoms of monomers. B. Distance between monomers in dimer. C. Ramachandran plot. The red, yellow and green regions correspond to the favored, allowed, and "generously allowed" regions, grey color fields are assumed as disallowed for phi/psy backbone angles. D. Protein quality analyzed by ProSA. ProSA z-score is -9.06, that is fitted in region of values found for structures solved by X-ray crystallography. Knowledge-based potential of mean force averaged over 40 residues is negative for all residues, except for the region of loop 2, which could be connected to its high flexibility and also absence of a suitable template. 4
5 Figure S4. Glycosylated TfHex. A. Part of model of hexosaminidase from T. flavus with shown glycosyl antennae at Asn 378, partly covering loop 1 (green) and interacting with loop 2 (blue). One monomer is shown by yellow color, another is magenta, glycan chain is shown in stick representation, hydrogen bonds are marked by yellow dotted lines. B. and C. Root means square deviation of residues of loop 1 (B) and loop 2 (C) in the vicinity of the glycan at Asn 378 (threshold was 0.3 nm for distance from carbohydrates). 5
6 Figure S5. Overlay of hexosaminidases with docked product GlcNAc. Position of product (2-acetamido-2-deoxy-D-glucopyranose) in the active sites of hexosaminidase from T. flavus before molecular dynamics simulation (green) and after (red) and hexosaminidase from S. plicatus after molecular dynamics (blue) is shown extracted from representative molecular dynamics run. Initial position of catalytic residues in S. plicatus hexosaminidase is similar to T. flavus enzyme and hence not shown. Change in the position of the corresponding atoms of catalytic residues is highlighted by arrows with distances in nanometers, calculated from independent molecular dynamis run for a stable period. 6
7 Figure S6. Change in the hydrogen bonding interaction of catalytic glutamic residue. A. Complex of pnp-glcnac with S. plicatus hexosaminidase in the beginning of simulation is green, snapshot of this complex after 10 ns molecular dynamics is magenta. Loops close to the active site are shown like tubes, Glu 314 and 4-nitrophenyl 2-acetamido-2-deoxy-β-D-glucopyranoside as sticks, the rest of the protein and hydrogen atoms are hidden. B. and C. Formation of hydrogen bonds during molecular dynamics: 0 means absence of interaction, 1 means presence of hydrogen bond. 7
Computational study of β-n-acetylhexosaminidase from Talaromyces flavus, a glycosidase with high substrate flexibility
 Kulik et al. BMC Bioinformatics (2015) 16:28 DOI 10.1186/s12859-015-0465-8 RESEARCH ARTICLE Open Access Computational study of β-n-acetylhexosaminidase from Talaromyces flavus, a glycosidase with high
Kulik et al. BMC Bioinformatics (2015) 16:28 DOI 10.1186/s12859-015-0465-8 RESEARCH ARTICLE Open Access Computational study of β-n-acetylhexosaminidase from Talaromyces flavus, a glycosidase with high
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis
 Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein
 Paper Presentation PLoS ONE 2011 Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein Debanu Das, Mireille Herve, Julie Feuerhelm, etc. and Dominique
Paper Presentation PLoS ONE 2011 Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein Debanu Das, Mireille Herve, Julie Feuerhelm, etc. and Dominique
Comparative Modeling Part 1. Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center
 Comparative Modeling Part 1 Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center Function is the most important feature of a protein Function is related to structure Structure is
Comparative Modeling Part 1 Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center Function is the most important feature of a protein Function is related to structure Structure is
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I
 BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
Conformational changes in IgE contribute to its. uniquely slow dissociation rate from receptor FcεRI
 Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Bioinformatics & Protein Structural Analysis. Bioinformatics & Protein Structural Analysis. Learning Objective. Proteomics
 The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Information. Structural basis for duplex RNA recognition and cleavage by A.
 Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial:
 Name (s): Swiss PDB viewer assignment chapter 4. If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial: http://spdbv.vital-it.ch/themolecularlevel/spvtut/index.html
Name (s): Swiss PDB viewer assignment chapter 4. If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial: http://spdbv.vital-it.ch/themolecularlevel/spvtut/index.html
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
Protein Folding Problem I400: Introduction to Bioinformatics
 Protein Folding Problem I400: Introduction to Bioinformatics November 29, 2004 Protein biomolecule, macromolecule more than 50% of the dry weight of cells is proteins polymer of amino acids connected into
Protein Folding Problem I400: Introduction to Bioinformatics November 29, 2004 Protein biomolecule, macromolecule more than 50% of the dry weight of cells is proteins polymer of amino acids connected into
Supplementary Information Titles
 Supplementary Information Titles Please list each supplementary item and its title or caption, in the order shown below. Note that we do NOT copy edit or otherwise change supplementary information, and
Supplementary Information Titles Please list each supplementary item and its title or caption, in the order shown below. Note that we do NOT copy edit or otherwise change supplementary information, and
Molecular Modeling 9. Protein structure prediction, part 2: Homology modeling, fold recognition & threading
 Molecular Modeling 9 Protein structure prediction, part 2: Homology modeling, fold recognition & threading The project... Remember: You are smarter than the program. Inspecting the model: Are amino acids
Molecular Modeling 9 Protein structure prediction, part 2: Homology modeling, fold recognition & threading The project... Remember: You are smarter than the program. Inspecting the model: Are amino acids
Protein Structure Prediction
 Homology Modeling Protein Structure Prediction Ingo Ruczinski M T S K G G G Y F F Y D E L Y G V V V V L I V L S D E S Department of Biostatistics, Johns Hopkins University Fold Recognition b Initio Structure
Homology Modeling Protein Structure Prediction Ingo Ruczinski M T S K G G G Y F F Y D E L Y G V V V V L I V L S D E S Department of Biostatistics, Johns Hopkins University Fold Recognition b Initio Structure
LIST OF ACRONYMS & ABBREVIATIONS
 LIST OF ACRONYMS & ABBREVIATIONS ALA ARG ASN ATD CRD CYS GLN GLU GLY GPCR HIS hstr ILE LEU LYS MET mglur1 NHDC PDB PHE PRO SER T1R2 T1R3 TMD TRP TYR THR 7-TM VFTM ZnSO 4 Alanine, A Arginine, R Asparagines,
LIST OF ACRONYMS & ABBREVIATIONS ALA ARG ASN ATD CRD CYS GLN GLU GLY GPCR HIS hstr ILE LEU LYS MET mglur1 NHDC PDB PHE PRO SER T1R2 T1R3 TMD TRP TYR THR 7-TM VFTM ZnSO 4 Alanine, A Arginine, R Asparagines,
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Electronic Supplementary Information Unraveling the effects of amino acid substitutions
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Electronic Supplementary Information Unraveling the effects of amino acid substitutions
Structural bioinformatics
 Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
6-Foot Mini Toober Activity
 Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
CSE : Computational Issues in Molecular Biology. Lecture 19. Spring 2004
 CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
SUPPLEMENTARY INFORMATION. Determinants of laminin polymerization revealed. by the structure of the α5 chain amino-terminal region
 SUPPLEMENTARY INFORMATION Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region Sadaf-Ahmahni Hussain 1, Federico Carafoli 1 & Erhard Hohenester 1 1 Department
SUPPLEMENTARY INFORMATION Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region Sadaf-Ahmahni Hussain 1, Federico Carafoli 1 & Erhard Hohenester 1 1 Department
Workunit: P Title:MT
 Workunit: P000003 Title:MT Model Summary: Model information: Modelled residue range: 48 to 340 Quaternary structure information: Based on template: 3aptA (1.85 A) Template (3aptA): Unknown Sequence Identity
Workunit: P000003 Title:MT Model Summary: Model information: Modelled residue range: 48 to 340 Quaternary structure information: Based on template: 3aptA (1.85 A) Template (3aptA): Unknown Sequence Identity
CAVER Analyst 1.0: Graphic tool for interactive visualization and analysis of tunnels in protein structures
 Supplementary data CAVER Analyst 1.0: Graphic tool for interactive visualization and analysis of tunnels in protein structures Barbora Kozlikova 1,#, Eva Sebestova 2,# Vilem Sustr 1, Jan Brezovsky 2, Ondrej
Supplementary data CAVER Analyst 1.0: Graphic tool for interactive visualization and analysis of tunnels in protein structures Barbora Kozlikova 1,#, Eva Sebestova 2,# Vilem Sustr 1, Jan Brezovsky 2, Ondrej
University of Groningen
 University of Groningen Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp lactis Bl-04 Ejby, Morten; Fredslund, Folmer; Vujicic - Zagar, Andreja;
University of Groningen Structural basis for arabinoxylo-oligosaccharide capture by the probiotic Bifidobacterium animalis subsp lactis Bl-04 Ejby, Morten; Fredslund, Folmer; Vujicic - Zagar, Andreja;
Supplementary Information for. Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction
 Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Homology modeling and structural validation of tissue factor pathway inhibitor
 www.bioinformation.net Hypothesis Volume 9(16) Homology modeling and structural validation of tissue factor pathway inhibitor Piyush Agrawal, Zoozeal Thakur & Mahesh Kulharia* Centre for Bioinformatics,
www.bioinformation.net Hypothesis Volume 9(16) Homology modeling and structural validation of tissue factor pathway inhibitor Piyush Agrawal, Zoozeal Thakur & Mahesh Kulharia* Centre for Bioinformatics,
Supporting Information
 Supporting Information Peroxidase vs. Peroxygenase Activity: Substrate Substituent Effects as Modulators of Enzyme Function in the Multifunctional Catalytic Globin Dehaloperoxidase Ashlyn H. McGuire, Leiah
Supporting Information Peroxidase vs. Peroxygenase Activity: Substrate Substituent Effects as Modulators of Enzyme Function in the Multifunctional Catalytic Globin Dehaloperoxidase Ashlyn H. McGuire, Leiah
Supplementary Information
 Supplementary Information Probing Medin Monomer Structure and its Amyloid Nucleation Using 13 C-Direct Detection NMR in Combination with Structural Bioinformatics Hannah A. Davies, Daniel J. Rigden, Marie
Supplementary Information Probing Medin Monomer Structure and its Amyloid Nucleation Using 13 C-Direct Detection NMR in Combination with Structural Bioinformatics Hannah A. Davies, Daniel J. Rigden, Marie
Fundamentals of Biochemistry
 Donald Voet Judith G. Voet Charlotte W. Pratt Fundamentals of Biochemistry Second Edition Chapter 6: Proteins: Three-Dimensional Structure Copyright 2006 by John Wiley & Sons, Inc. 1958, John Kendrew Any
Donald Voet Judith G. Voet Charlotte W. Pratt Fundamentals of Biochemistry Second Edition Chapter 6: Proteins: Three-Dimensional Structure Copyright 2006 by John Wiley & Sons, Inc. 1958, John Kendrew Any
Exploring Suboptimal Sequence Alignments and Scoring Functions in Comparative Protein Structural Modeling
 Exploring Suboptimal Sequence Alignments and Scoring Functions in Comparative Protein Structural Modeling Presented by Kate Stafford 1,2 Research Mentor: Troy Wymore 3 1 Bioengineering and Bioinformatics
Exploring Suboptimal Sequence Alignments and Scoring Functions in Comparative Protein Structural Modeling Presented by Kate Stafford 1,2 Research Mentor: Troy Wymore 3 1 Bioengineering and Bioinformatics
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Institute of Physics, Academy of Sciences of the Czech Republic, v.v.i., Na Slovance 2, CZ , Prague 8, Czech Republic 2
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Theoretical and experimental study of antifreeze protein AFP752, trehalose and dimethyl sulfoxide
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Theoretical and experimental study of antifreeze protein AFP752, trehalose and dimethyl sulfoxide
In silico measurements of twist and bend. moduli for beta solenoid protein self-
 In silico measurements of twist and bend moduli for beta solenoid protein self- assembly units Leonard P. Heinz, Krishnakumar M. Ravikumar, and Daniel L. Cox Department of Physics and Institute for Complex
In silico measurements of twist and bend moduli for beta solenoid protein self- assembly units Leonard P. Heinz, Krishnakumar M. Ravikumar, and Daniel L. Cox Department of Physics and Institute for Complex
BRITISH BIOMEDICAL BULLETIN
 Journal Home Page www.bbbulletin.org BRITISH BIOMEDICAL BULLETIN Original Bioinformatics Analysis of α-amylase Three-Dimensional Structure in Aspergillus oryzae Javad Sharifi-Rad,2, Fatemeh Taktaz* 3 and
Journal Home Page www.bbbulletin.org BRITISH BIOMEDICAL BULLETIN Original Bioinformatics Analysis of α-amylase Three-Dimensional Structure in Aspergillus oryzae Javad Sharifi-Rad,2, Fatemeh Taktaz* 3 and
Supporting Information
 Supporting Information Shuffling Active Site Substate Populations Affects Catalytic Activity: The Case of Glucose Oxidase Dušan Petrović, 1 David Frank, 2, Shina Caroline Lynn Kamerlin, 3 Kurt Hoffmann,
Supporting Information Shuffling Active Site Substate Populations Affects Catalytic Activity: The Case of Glucose Oxidase Dušan Petrović, 1 David Frank, 2, Shina Caroline Lynn Kamerlin, 3 Kurt Hoffmann,
Biochemistry Proteins
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Recitation Section 3 February 9-10, 2005 A. CBS proteins, 3D representation Biochemistry Proteins The protein we will consider today and
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Recitation Section 3 February 9-10, 2005 A. CBS proteins, 3D representation Biochemistry Proteins The protein we will consider today and
Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction
 Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction Conformational Analysis 2 Conformational Analysis Properties of molecules depend on their three-dimensional
Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction Conformational Analysis 2 Conformational Analysis Properties of molecules depend on their three-dimensional
SUPPLEMENTARY INFORMATION Figures. Supplementary Figure 1 a. Page 1 of 30. Nature Chemical Biology: doi: /nchembio.2528
 SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
Heat and SDS insensitive NDK dimers are largely stabilised by hydrophobic interaction to form functional hexamer in Mycobacterium smegmatis
 Suplementary Materials Vol. 60, No 2/2013 199 208 on-line at: www.actabp.pl Regular paper Heat and SDS insensitive NDK dimers are largely stabilised by hydrophobic interaction to form functional hexamer
Suplementary Materials Vol. 60, No 2/2013 199 208 on-line at: www.actabp.pl Regular paper Heat and SDS insensitive NDK dimers are largely stabilised by hydrophobic interaction to form functional hexamer
Minicollagen cysteine-rich domains encode distinct modes of. Anja Tursch, Davide Mercadante, Jutta Tennigkeit, Frauke Gräter and Suat
 Supplementary Figure S1. Conformational dynamics of partially reduced CRDs. (A) Root mean square deviation (RMSD) and (B) radius of gyration (R G ) distributions for the N-CRD (green) and C-CRD (red) domains,
Supplementary Figure S1. Conformational dynamics of partially reduced CRDs. (A) Root mean square deviation (RMSD) and (B) radius of gyration (R G ) distributions for the N-CRD (green) and C-CRD (red) domains,
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein.
 Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Sequence Analysis '17 -- lecture Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction
 Sequence Analysis '17 -- lecture 16 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction Alpha helix Right-handed helix. H-bond is from the oxygen at i to the nitrogen
Sequence Analysis '17 -- lecture 16 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction Alpha helix Right-handed helix. H-bond is from the oxygen at i to the nitrogen
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
Globins. The Backbone structure of Myoglobin 2. The Heme complex in myoglobin. Lecture 10/01/2009. Role of the Globin.
 Globins Lecture 10/01/009 The Backbone structure of Myoglobin Myoglobin: 44 x 44 x 5 Å single subunit 153 amino acid residues 11 residues are in an a helix. Helices are named A, B, C, F. The heme pocket
Globins Lecture 10/01/009 The Backbone structure of Myoglobin Myoglobin: 44 x 44 x 5 Å single subunit 153 amino acid residues 11 residues are in an a helix. Helices are named A, B, C, F. The heme pocket
GPCR structure modeling and ligand docking: GPCR Dock2008
 KO18/KP32 第 37 回構造活性相関シンポジウム平成 21 年 11 月 20 日北里大学薬学部コンベンションホール GPCR structure modeling and ligand docking: GPCR Dock2008 Kazuhiko Kanou 1, Daisuke Takaya 2, Genki Terashi 1, Mayuko Takeda Shitaka 1,2 and
KO18/KP32 第 37 回構造活性相関シンポジウム平成 21 年 11 月 20 日北里大学薬学部コンベンションホール GPCR structure modeling and ligand docking: GPCR Dock2008 Kazuhiko Kanou 1, Daisuke Takaya 2, Genki Terashi 1, Mayuko Takeda Shitaka 1,2 and
Enzymatic properties and subcellular localization of Arabidopsis -N-acetylhexosaminidases (correction)
 Enzymatic properties and subcellular localization of Arabidopsis β-n-acetylhexosaminidases (correction) Richard Strasser, Jayakumar Singh Bondili, Jennifer Schoberer, Barbara Svoboda, Eva Liebminger, Josef
Enzymatic properties and subcellular localization of Arabidopsis β-n-acetylhexosaminidases (correction) Richard Strasser, Jayakumar Singh Bondili, Jennifer Schoberer, Barbara Svoboda, Eva Liebminger, Josef
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
SUPPLEMENTARY INFORMATION
 Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Introduction to Proteins
 Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport
 Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
SUPPLEMENTARY INFORMATION
 This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Textbook Reading Guidelines
 Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: May 1, 2009 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: May 1, 2009 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Protein Structure. Protein Structure Tertiary & Quaternary
 Lecture 4 Protein Structure Protein Structure Tertiary & Quaternary Dr. Sameh Sarray Hlaoui Primary structure: The linear sequence of amino acids held together by peptide bonds. Secondary structure: The
Lecture 4 Protein Structure Protein Structure Tertiary & Quaternary Dr. Sameh Sarray Hlaoui Primary structure: The linear sequence of amino acids held together by peptide bonds. Secondary structure: The
Programme Good morning and summary of last week Levels of Protein Structure - I Levels of Protein Structure - II
 Programme 8.00-8.10 Good morning and summary of last week 8.10-8.30 Levels of Protein Structure - I 8.30-9.00 Levels of Protein Structure - II 9.00-9.15 Break 9.15-11.15 Exercise: Building a protein model
Programme 8.00-8.10 Good morning and summary of last week 8.10-8.30 Levels of Protein Structure - I 8.30-9.00 Levels of Protein Structure - II 9.00-9.15 Break 9.15-11.15 Exercise: Building a protein model
Figure S1
 Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
vector AvrRpt2 vector AvrRpt2 AvrRpt2 3.5 hours 6 hours
 A 30 kd RIN4 ACP2 17 kd vector AvrRpt2 vector AvrRpt2 AvrRpt2 3.5 hours 6 hours B ACP3 7 kd vector AvrRpt2 vector AvrRpt2 total membrane Figure S1. Accumulation of ACP2 and ACP3 upon cleavage of native
A 30 kd RIN4 ACP2 17 kd vector AvrRpt2 vector AvrRpt2 AvrRpt2 3.5 hours 6 hours B ACP3 7 kd vector AvrRpt2 vector AvrRpt2 total membrane Figure S1. Accumulation of ACP2 and ACP3 upon cleavage of native
Acceleration of protein folding by four orders of magnitude through a single amino acid substitution
 Acceleration of protein folding by four orders of magnitude through a single amino acid substitution Daniel J. A. Roderer 1, Martin A. Schärer 1, Marina Rubini 2 * and Rudi Glockshuber 1 AUTHOR ADDRESS
Acceleration of protein folding by four orders of magnitude through a single amino acid substitution Daniel J. A. Roderer 1, Martin A. Schärer 1, Marina Rubini 2 * and Rudi Glockshuber 1 AUTHOR ADDRESS
Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1
 Supplemental information for: Structure determination and activity manipulation of the turfgrass AA receptor FePYR1 Zhizhong Ren 1, 2,#, Zhen Wang 1, 2,#, X Edward Zhou 3, Huazhong Shi 4, Yechun Hong 1,
Supplemental information for: Structure determination and activity manipulation of the turfgrass AA receptor FePYR1 Zhizhong Ren 1, 2,#, Zhen Wang 1, 2,#, X Edward Zhou 3, Huazhong Shi 4, Yechun Hong 1,
Molecules VII. Structures and Functions of Proteins
 Molecules VII Structures and Functions of Proteins The 20 Amino Acids Sequences from Protein Data Bank (PDB) The primary sequence can be indicated very compactly using the one letter abbreviations. This
Molecules VII Structures and Functions of Proteins The 20 Amino Acids Sequences from Protein Data Bank (PDB) The primary sequence can be indicated very compactly using the one letter abbreviations. This
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Building an Antibody Homology Model
 Application Guide Antibody Modeling: A quick reference guide to building Antibody homology models, and graphical identification and refinement of CDR loops in Discovery Studio. Francisco G. Hernandez-Guzman,
Application Guide Antibody Modeling: A quick reference guide to building Antibody homology models, and graphical identification and refinement of CDR loops in Discovery Studio. Francisco G. Hernandez-Guzman,
Molecular Modeling Lecture 8. Local structure Database search Multiple alignment Automated homology modeling
 Molecular Modeling 2018 -- Lecture 8 Local structure Database search Multiple alignment Automated homology modeling An exception to the no-insertions-in-helix rule Actual structures (myosin)! prolines
Molecular Modeling 2018 -- Lecture 8 Local structure Database search Multiple alignment Automated homology modeling An exception to the no-insertions-in-helix rule Actual structures (myosin)! prolines
Protein Structure/Function Relationships
 Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
CS273: Algorithms for Structure Handout # 5 and Motion in Biology Stanford University Tuesday, 13 April 2004
 CS273: Algorithms for Structure Handout # 5 and Motion in Biology Stanford University Tuesday, 13 April 2004 Lecture #5: 13 April 2004 Topics: Sequence motif identification Scribe: Samantha Chui 1 Introduction
CS273: Algorithms for Structure Handout # 5 and Motion in Biology Stanford University Tuesday, 13 April 2004 Lecture #5: 13 April 2004 Topics: Sequence motif identification Scribe: Samantha Chui 1 Introduction
3D Structure Prediction and Validation of Novel Exo-beta-1, 3-Glucanase from Psychroflexus torquis ATCC700755/ACAM623
 Available online at www.ijpab.com Isa et al DOI: http://dx.doi.org/10.18782/2320-7051.2206 Research Article 3D Structure Prediction and Validation of Novel Exo-beta-1, 3-Glucanase from Psychroflexus torquis
Available online at www.ijpab.com Isa et al DOI: http://dx.doi.org/10.18782/2320-7051.2206 Research Article 3D Structure Prediction and Validation of Novel Exo-beta-1, 3-Glucanase from Psychroflexus torquis
Structure. Structural Components of Nucleotides Base. Sugar. Introduction Nucleotide to Cells & Microscopy and Nucleic Acid. Phosphate Glycosidic bond
 11 Structural Components of Nucleotides Base Sugar Introduction Nucleotide to Cells & Microscopy and Nucleic Acid Structure Phosphate Glycosidic bond H NUCLEOTIDE H Nucleic acid polymer of nucleotides
11 Structural Components of Nucleotides Base Sugar Introduction Nucleotide to Cells & Microscopy and Nucleic Acid Structure Phosphate Glycosidic bond H NUCLEOTIDE H Nucleic acid polymer of nucleotides
Supporting Information Contents
 Supporting Information Choy Theng Loh, Kiyoshi Ozawa, Kellie L. Tuck, Nicholas Barlow, Thomas Huber, Gottfried Otting, and Bim Graham Lanthanide tags for site-specific ligation to an unnatural amino acid
Supporting Information Choy Theng Loh, Kiyoshi Ozawa, Kellie L. Tuck, Nicholas Barlow, Thomas Huber, Gottfried Otting, and Bim Graham Lanthanide tags for site-specific ligation to an unnatural amino acid
SUPPLEMENTARY INFORMATION
 Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
KEMM15 Lecture note in structural bioinformatics: A practical guide. S Al-Karadaghi, Biochemistry & Structural Biology, Lund University
 KEMM15 Lecture note in structural bioinformatics: A practical guide S Al-Karadaghi, Biochemistry & Structural Biology, Lund University 1 BASICS OF PROTEIN STRUCTURE 3 SOME DEFINITIONS 3 THE 20 AMINO ACIDS
KEMM15 Lecture note in structural bioinformatics: A practical guide S Al-Karadaghi, Biochemistry & Structural Biology, Lund University 1 BASICS OF PROTEIN STRUCTURE 3 SOME DEFINITIONS 3 THE 20 AMINO ACIDS
A hybrid structural approach to analyze ligand binding by the 5-HT4. receptor
 Supplement A hybrid structural approach to analyze ligand binding by the 5-HT4 receptor Pius S. Padayatti 1**, Liwen Wang 2**, Sayan Gupta 3, Tivadar Orban 4, Wenyu Sun 1, David Salom 1, Steve Jordan 5,
Supplement A hybrid structural approach to analyze ligand binding by the 5-HT4 receptor Pius S. Padayatti 1**, Liwen Wang 2**, Sayan Gupta 3, Tivadar Orban 4, Wenyu Sun 1, David Salom 1, Steve Jordan 5,
Super Models. Deoxyribonucleic Acid (DNA) Molecular Model Kit. Copyright 2015 Ryler Enterprises, Inc. Recommended for ages 10-adult
 Super Models Deoxyribonucleic Acid (DNA) Molecular Model Kit Copyright 2015 Ryler Enterprises, Inc. Recommended for ages 10-adult! Caution: Atom centers and vinyl tubing are a choking hazard. Do not eat
Super Models Deoxyribonucleic Acid (DNA) Molecular Model Kit Copyright 2015 Ryler Enterprises, Inc. Recommended for ages 10-adult! Caution: Atom centers and vinyl tubing are a choking hazard. Do not eat
STAAR Biology: Assessment Activities. Biomolecules. Cell Structure and Function. The Charles A. Dana Center at The University of Texas at Austin
 STAAR Biology: Assessment Activities ell Structure and Function The harles A. Dana enter at The University of Texas at Austin 115 STAAR Biology: Assessment Activities 116 The harles A. Dana enter at The
STAAR Biology: Assessment Activities ell Structure and Function The harles A. Dana enter at The University of Texas at Austin 115 STAAR Biology: Assessment Activities 116 The harles A. Dana enter at The
Nucleic Acids and the RNA World. Pages Chapter 4
 Nucleic Acids and the RNA World Pages 74-89 Chapter 4 RNA vs. Protein Chemical Evolution stated that life evolved from a polymer called a protein. HOWEVER, now many scientists question this. There is currently
Nucleic Acids and the RNA World Pages 74-89 Chapter 4 RNA vs. Protein Chemical Evolution stated that life evolved from a polymer called a protein. HOWEVER, now many scientists question this. There is currently
Figure A summary of spontaneous alterations likely to require DNA repair.
 DNA Damage Figure 5-46. A summary of spontaneous alterations likely to require DNA repair. The sites on each nucleotide that are known to be modified by spontaneous oxidative damage (red arrows), hydrolytic
DNA Damage Figure 5-46. A summary of spontaneous alterations likely to require DNA repair. The sites on each nucleotide that are known to be modified by spontaneous oxidative damage (red arrows), hydrolytic
i. Monomers for which class(s) of macromolecules always have phosphorous? Circle all that apply. Carbohydrates Proteins Lipids DNA RNA
 Question 1 (25 points) a) There are four major classes of biological macromolecules: carbohydrates, proteins, lipids and nucleic acids (deoxyribonucleic acid or DNA and ribonucleic acid or RNA). i. Monomers
Question 1 (25 points) a) There are four major classes of biological macromolecules: carbohydrates, proteins, lipids and nucleic acids (deoxyribonucleic acid or DNA and ribonucleic acid or RNA). i. Monomers
Hmwk # 8 : DNA-Binding Proteins : Part II
 The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
Introduction to BioLuminate
 Introduction to BioLuminate Janet Paulsen Stanford University 10/16/17 BioLuminate has Broad Functionality Antibody design Structure prediction from sequence, humanization Protein design Residue scanning
Introduction to BioLuminate Janet Paulsen Stanford University 10/16/17 BioLuminate has Broad Functionality Antibody design Structure prediction from sequence, humanization Protein design Residue scanning
What s New in Discovery Studio 2.5.5
 What s New in Discovery Studio 2.5.5 Discovery Studio takes modeling and simulations to the next level. It brings together the power of validated science on a customizable platform for drug discovery research.
What s New in Discovery Studio 2.5.5 Discovery Studio takes modeling and simulations to the next level. It brings together the power of validated science on a customizable platform for drug discovery research.
DNA Structure, Function, and Engineering Page /14/2018 Dr. Amjid Iqbal 1
 DNA Structure, Function, and Engineering Page 40-51 2/14/2018 Dr. Amjid Iqbal 1 Background Organisms exhibit striking similarity at the molecular level. The structures and metabolic activities of all cells
DNA Structure, Function, and Engineering Page 40-51 2/14/2018 Dr. Amjid Iqbal 1 Background Organisms exhibit striking similarity at the molecular level. The structures and metabolic activities of all cells
PeppyChains: Simplifying the assembly of 3D- printed generic protein models
 PeppyChains: Simplifying the assembly of 3D- printed generic protein models Promita Chakraborty (QuezyLab) Peppytides is a coarse- grained, accurate, physical model of the polypeptide chain [1-4]. I have
PeppyChains: Simplifying the assembly of 3D- printed generic protein models Promita Chakraborty (QuezyLab) Peppytides is a coarse- grained, accurate, physical model of the polypeptide chain [1-4]. I have
Q22M, T44W, R81G, H83G, T84M, N130G, N172M, A234S, T236L, C9G, V48G, L50W, T78S, S101E, E237M, T265S, W267F 7, 11, , 239,
 111 Table 4-1. Summary of design calculations for Kemp elimination enzymes. The residues allowed for each required catalytic contact are indicated along with the actual catalytic residue chosen from the
111 Table 4-1. Summary of design calculations for Kemp elimination enzymes. The residues allowed for each required catalytic contact are indicated along with the actual catalytic residue chosen from the
An insilico Approach: Homology Modelling and Characterization of HSP90 alpha Sangeeta Supehia
 259 Journal of Pharmaceutical, Chemical and Biological Sciences ISSN: 2348-7658 Impact Factor (SJIF): 2.092 December 2014-February 2015; 2(4):259-264 Available online at http://www.jpcbs.info Online published
259 Journal of Pharmaceutical, Chemical and Biological Sciences ISSN: 2348-7658 Impact Factor (SJIF): 2.092 December 2014-February 2015; 2(4):259-264 Available online at http://www.jpcbs.info Online published
SUPPLEMENTARY INFORMATION
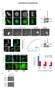 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
Virtual bond representation
 Today s subjects: Virtual bond representation Coordination number Contact maps Sidechain packing: is it an instrumental way of selecting and consolidating a fold? ASA of proteins Interatomic distances
Today s subjects: Virtual bond representation Coordination number Contact maps Sidechain packing: is it an instrumental way of selecting and consolidating a fold? ASA of proteins Interatomic distances
Homology Modeling of the Chimeric Human Sweet Taste Receptors Using Multi Templates
 2013 International Conference on Food and Agricultural Sciences IPCBEE vol.55 (2013) (2013) IACSIT Press, Singapore DOI: 10.7763/IPCBEE. 2013. V55. 1 Homology Modeling of the Chimeric Human Sweet Taste
2013 International Conference on Food and Agricultural Sciences IPCBEE vol.55 (2013) (2013) IACSIT Press, Singapore DOI: 10.7763/IPCBEE. 2013. V55. 1 Homology Modeling of the Chimeric Human Sweet Taste
RNA World Hypothesis and RNA folding. By Lixin Dai
 RNA World Hypothesis and RNA folding By Lixin Dai Outline: RNA World Hypothesis RNA structure primary secondary tertiary Folding of RNA structure Problems with the folding Solutions RNA World Hypothesis:
RNA World Hypothesis and RNA folding By Lixin Dai Outline: RNA World Hypothesis RNA structure primary secondary tertiary Folding of RNA structure Problems with the folding Solutions RNA World Hypothesis:
Thr Gly Tyr. Gly Lys Asn
 Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Bioinformation Volume 5 open access
 Molecular docking analysis of new generation cephalosporins interactions with recently known SHV-variants Asad Ullah Khan 1 *, Mohd Hassan Baig 1, Gulshan Wadhwa 2 1 Interdisciplinary Biotechnology Unit,
Molecular docking analysis of new generation cephalosporins interactions with recently known SHV-variants Asad Ullah Khan 1 *, Mohd Hassan Baig 1, Gulshan Wadhwa 2 1 Interdisciplinary Biotechnology Unit,
Fundamentals of Protein Structure
 Outline Fundamentals of Protein Structure Yu (Julie) Chen and Thomas Funkhouser Princeton University CS597A, Fall 2005 Protein structure Primary Secondary Tertiary Quaternary Forces and factors Levels
Outline Fundamentals of Protein Structure Yu (Julie) Chen and Thomas Funkhouser Princeton University CS597A, Fall 2005 Protein structure Primary Secondary Tertiary Quaternary Forces and factors Levels
