SUPPLEMENTARY INFORMATION
|
|
|
- Christal Harrell
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid data are available for unique sirnas. Worksheet 2, List of 153 genes selected as candidates for positive ciliogenesis modulators. Worksheet 3, List of 79 genes selected as candidates for negative ciliogenesis modulators. To minimize false negative, MAX NPI and MIN NPI values from the duplicates were used for the selection of positive modulators and negative modulators, respectively. Supplementary Table 2. An excel file containing confirmation screen data. Worksheet 1, Measurement for ciliogenesis inhibition resulted from silencing of 153 positive modulator candidates. Data (percentage of ciliated cells) was converted to Z score for plate normalization using median and MAD values from 48 non-targeting sirnas included to each plate. Criteria for hit selection were at least 2 sirnas with (-) Z score 2.0. Worksheet 2, List of 40 genes selected as positive ciliogenesis modulators. Annotation of the 40 genes was obtained from ENTREZ gene database. Worksheet 3, Measurement for an increase in average cilium size resulted from silencing of 79 negative modulator candidates. Criteria for hit selection were at least 2 sirnas with Z score 1.2 (the variability in non-targeting controls was mainly ascribed to a decrease in cilium size; Supplementary Fig. 2b). Worksheet 4, List of 13 genes selected as negative ciliogenesis modulators. Annotation of the 13 genes was obtained from ENTREZ gene database. Supplementary Table 3. An excel file summarizing hit classification study. Worksheet 1, Investigation for ciliogenesis defect caused by silencing of confirmed positive modulators using anti-acetylated tubulin immunofluorescence staining. Worksheet 2, Validation experiment for the result shown in worksheet 1. Worksheet 3, Test for serum starvation independent ciliogenesis in cells depleted with confirmed negative modulators. Worksheet 4, Validation experiment for the result shown in worksheet 3. Supplementary Table 4. An excel file containing the sequences of sirnas for selected hits.
2 Supplementary Figure 1. Assay development. a, Serum starvation for 48 hrs efficiently induced ciliogenesis in htrpe-smoegfp cells. b, The specificity of SmoEGFP as a cilium maker was confirmed by co-labeling with anti-acetylated-tubulin antibody, a commonly used cilium marker. c, Image processing to detect cilia per cell. A.1 Merged, B.1 GFP and C.1 DAPI channel images are shown. Respective object masks for the cilia detected from the GFP image are shown in B.2 and nuclei masks detected from the DAPI image in C.2. Tessellation lines marking cell areas associated with each nucleus are shown in yellow in C.3, while nuclei outlines are shown in blue and nuclei centroids in red. A.4 shows the final segmented cell objects with nuclei outlines in blue, cell object ID in red, detected cilia in green and cell regions in yellow. d, Confirmation of the sensitivity of the screen strategy using a positive control sirna. Cells were serum-starved 24 hrs after transfection and ciliated cells were counted 48 hrs later. e, Effect of plating density on ciliogenesis. f, Schematic of the primary and confirmation screen. Values, mean ± SD (n = 4 experiments). Scale bars, 10 μm.
3 Supplementary Figure 2. Histogram and scatter plots displaying screen results. a, Histogram showing primary screen results. For normalization of plate-to-plate variability, percentages of ciliated cells were converted to normalized percent inhibition (NPI) values. Two NPI values (from duplication) for each sirna were averaged. Overall NPI mean was 11.5, and SD was Threshold for active sirnas in the primary screen was set to ± 1.5 SDs. b, Scatter plots showing the results of cilium length measurement for 48 distinct non-targeting sirnas tested. Only 1 non-targeting sirna increased cilium length over Z score 1.2, indicating that cilium length increase is a highly specific phenotype.
4 Supplementary Figure 3. CytochalasinD dose-response. a,b,c, Cells were cultured for 16 hrs in serumfree medium containing cytochalasind. b, Ciliated cell numbers were increased by the treatment of cytochalasind at 50 nm or higher. c, Cilium lengths were increased in cells treated with cytochalasind at 100 nm or higher. Cell viability was affected in the presence of higher concentrations ( 1 um) of cytochalasind. Arrow, fragmented nuclei. Scale bar, 10 μm.
5 Supplementary Figure 4. CytochalasinD treatment enhances ciliogenesis without global rearrangement of actin cytoskeleton. a,b, Enhanced ciliogenesis in cytochalasind treated HEK293T cells. HEK293T cells were serum starved for 12 hrs, and incubated for additional 12 hrs with 1 μm cytochalasind. Cilia were labeled by Arl13b immunofluorescence (Caspary et al., 2007, Dev Cell 12:762), and the basal bodies were stained by γ-tubulin. Values, mean ± SD (n = 3 experiments). Student s t-test: **P < c, htrpe cells treated with 0.5 μm cytochalasind did not show noticeable changes in the morphology of stress fibers (labeled by phalloidin-alexa594). Scale bars, 5 μm (a); 20 μm (c).
6 Supplementary Figure 5. PARVA is involved in negative regulation of ciliogenesis. a,b, PARVA knockdown significantly increased the numbers of cells with longer cilium (> 6 μm). c, Correlation of knockdown phenotype with protein levels was confirmed by Western blotting. Values, mean ± SD (n = 4 experiments). Student s t-test: *P < 0.05, **P < Scale bar, 10 μm.
7 Supplementary Figure 6. Live-imaging of htrpe-smoegfp cells transfected with ACTR3 sirnas. a, htrpe-smoegfp cells were transfected with indicated sirnas for 2.5 days in the presence of 10 % serum and then non-ciliated cells were live-imaged for 6 hrs in serum-free medium. b, Magnified view of selected images. Note that cells transfected with ACTR3 sirna#1 displayed pronounced PPC at the base of newly forming cilia. Scale bars, 2.5 μm.
8 Supplementary Figure 7. PPC is a temporary structure. a, A subset of cells with premature cilia displays PPC after 4 hr serum starvation. In contrast, PPC was undetectable from most of both control and ACTR3-depleted cells with mature cilia after 24 hr serum starvation. s.s., serum starvation. Arrow, a premature cilium. Arrowhead, PPC. b, IMCD cells overexpressing SmoEGFP also temporarily displayed PPC at the base of nascent cilia. Scale bars, 10 μm (a); 5 μm (b).
9 Supplementary Figure 8. CytochalasinD stabilizes PPC. a, Live imaging revealed that PPC in DMSO treated htrpe-smoegfp cells undergoes dynamic morphological changes. b, In the presence of 0.2 μm cytochalasind, morphological changes of PPC were noticeably decreased. Right panels are magnified view of PPC. Scale bars, 10 μm (left panels); 2.5 μm (right panels).
10 Supplementary Figure 9. PPC overlaps with pericentrosomal recycling endosomes. PPC was identified by SmoEGFP fluorescence and the pericentrosomal recycling endosome (PRE) was labeled by endocytosed transferrin-alexa594. A subset of interphase cells cultured in normal medicum (containing 10 % FBS) displayed prominent PRE. The number of cells exhibiting PRE decreased after 24 hr serum starvation. Before serum starvation, 68 % of PRE was labeled by SmoEGFP, while all SmoEGFP positive PPC was labeled by transferrin-alexa594 fluorescence, suggesting that PPC is a specific stage of PRE. Scale bar, 5 μm.
11 Supplementary Figure 10. Confirmation of PTPN23 knockdown phenotype. a, Multiple distinct sirnas targeting PTPN23 significantly reduced ciliated cell numbers. Results from sirna#1-4 are also presented in Fig. 3b. Values, mean ± SD [n = 4 (sirna #1-4) and 2 (sirna #5-7) experiments]. Student s t-test: *P < 0.05, **P < b, Western blotting showing correlation of knockdown phenotype with protein levels. c, Specificity of anti-ptpn23 antibody staining was confirmed by PTPN23 knockdown. Anti-γ-tubulin antibody was used as a marker for the centrosome. Scale bar, 10 μm.
12 Supplementary Figure 11. PTPN23 knockdown interferes with PPC formation. a, PPC/pericentrosomal recycling endosomes (identified by SmoEGFP/endocytosed transferrin-alexa594) were not observed in cells displaying punctuate SmoEGFP accumulation after PTPN23 knockdown. Arrow indicates cells displaying PPC/pericentrosomal recycling endosomes. b, In contrast to PTPN23 knockdown, GSN knockdown that inhibited ciliogenesis (Fig. 2a,b) did not cause an accumulation of SmoEGFP on EEA1 positive early endosomes. Scale bars, 20 μm (a); 5 μm(b).
13 Supplementary Figure 12. Recycling route of the endocytic pathway is specifically involved in ciliogenesis. a, Immunofluorescence staining showing pericentrosomal enrichment of ASAP1, a component of the endocytic recycling component. b,c, ASAP1 knockdown caused a reduction of ciliated cell numbers in htrpe cells after 48 hr serum starvation. Values, mean ± SD (n = 2 experiments). Student s t-test: *P < d, Blocking endocytic degradation pathway by concanamycina did not cause early endosomal SmoEGFP accumulation, and did not interfere with ciliogenesis or PPC formation. Scale bars, 5 μm (a,d); 15 μm (b).
14 Supplementary Figure 13. Confirmation of PLA2G3 knockdown phenotype. a, Multiple distinct sirnas targeting PLA2G3 significantly increased the number of cells with longer cilium (> 6 μm). Results from sirna#1-4 are also presented in Fig. 3e. Values, mean ± SD [n = 4 (sirna #1-4) and 2 (sirna #5-7) experiments]. Student s t-test: *P < 0.05, **P < b, Western blotting showing correlation of knockdown phenotype with protein levels. c, Specificity of anti-pla2g3 antibody staining was confirmed by PLA2G3 knockdown. Anti-γ-tubulin antibody was used as a marker for the centrosome. d, Western blotting showing overexpressed full-length human PLA2G3 proteins with c-myc epitope at the C-terminus. e, Extensive colocalization of overexpressed PLA2G3myc and endocytosed Transferrin-Alexa594 was observed (arrows), supporting PLA2G3 s involvement in the endocytic pathway. f, PLA2G3myc overexpression for 36 hrs in the absence of serum decreased the number of ciliated cells. Values are mean ± SD (more than 100 PLA2G3myc positive cells from two experiments were counted). Scale bars, 10 μm (c); 5 μm (e).
15 Supplementary Figure 14. Inhibition of PLA2G3 enhances ciliogenesis and ciliary SmoEGFP targeting. a, Oleyloxyethyl phosphorylcholine (OPC), an inhibitor of secreted phospholipases, caused an increase in ciliated cell number in the presence of 10 % serum. Values, mean ± SD (n = 5 experiments). Student s t-test: **P < b, Images of representative ciliary EGFP fluorescence were captured 3 days after sirna transfection (cells were cultured in serum-free medium for final 2 days). c, Histogram showing quantification data of the images. Fluorescent intensity is an arbitrary unit. Note that cilia from cells depleted with PLA2G3 display higher levels of SmoEGFP fluorescence. Scale bars, 2.5 μm.
16 Supplementary Figure 15. CytochalasinD enhanced both axoneme assembly and ciliary membrane biogenesis. Cells were cultured in serum free medium for 24 hrs before fixation (final 16 hr with 0.2 μm cytochalasind). a, Axoneme was labeled by both anti-acetylated tubulin antibody and anti-glutamylated tubulin antibody, and ciliary membrane was identified by SmoEGFP fluorescence. b, Quantification data of the length measurement for 25 representative cilia. Values, mean ± SD. Scale bar, 5 μm.
Xu et al., Supplementary Figures 1-7
 Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Xu et al., Supplementary Figures 1-7 Supplementary Figure 1. PIPKI is required for ciliogenesis. (a) PIPKI localizes at the basal body of primary cilium. RPE-1 cells treated with two sirnas targeting to
Supplemental Material
 Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplemental Material 1 Figure S1. Phylogenetic analysis of Cep72 and Lrrc36, comparative localization of Cep72 and Lrrc36 and Cep72 antibody characterization (A) Phylogenetic alignment of Cep72 and Lrrc36
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
NIH Public Access Author Manuscript Nature. Author manuscript; available in PMC 2010 October 1.
 NIH Public Access Author Manuscript Published in final edited form as: Nature. 2010 April 15; 464(7291): 1048 1051. doi:10.1038/nature08895. Functional genomic screen for modulators of ciliogenesis and
NIH Public Access Author Manuscript Published in final edited form as: Nature. 2010 April 15; 464(7291): 1048 1051. doi:10.1038/nature08895. Functional genomic screen for modulators of ciliogenesis and
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr
 Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
Supplemental figure legends Supplemental Fig. 1: PEA-15 knockdown efficiency assessed by immunohistochemistry and qpcr A, LβT2 cells were transfected with either scrambled or PEA-15 sirna. Cells were then
SUPPLEMENTARY INFORMATION
 DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
DOI: 1.1/ncb2918 Supplementary Figure 1 (a) Biotin-dUTP labelling does not affect S phase progression. Cells synchronized in mid-s phase were labelled with biotindutp during a 5 min hypotonic shift (red)
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information Table S1. Definition of quantitative cellular features
T H E J O U R N A L O F C E L L B I O L O G Y
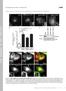 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Allison et al., http://www.jcb.org/cgi/content/full/jcb.201211045/dc1 Figure S1. Spastin depletion causes increased endosomal tubulation.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
doi: 10.1038/nature06721 SUPPLEMENTARY INFORMATION. Supplemental Figure Legends Supplemental Figure 1 The distribution of hatx-1[82q] in Cos7 cells. Cos7 cells are co-transfected with hatx-1[82q]-gfp (green)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Supplemental material JCB. Pitaval et al., Cell shape and ciliogenesis Pitaval et al.
 Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplemental material THE JOURNAL OF CELL BIOLOGY Pitaval et al., http://www.jcb.org/cgi/content/full/jcb.201004003/dc1 S1 Figure S1. Cell cycle exit analysis. (A) Experimental procedure. RPE1 cells were
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various
 Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
Supplementary Figure 1. APP cleavage assay. HEK293 cells were transfected with various GST-tagged N-terminal truncated APP fragments including GST-APP full-length (FL), APP (123-695), APP (189-695), or
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
1 Supplementary Figure 1. Kif13b expression and localization in serum starved
 1 2 3 4 5 6 Supplementary Figure 1. Kif13b expression and localization in serum starved cells. (a) Immunoblot of NIH3T3 cell lysates after 24 hrs of serum deprivation, using rabbit polyclonal KIF13B antiserum.
1 2 3 4 5 6 Supplementary Figure 1. Kif13b expression and localization in serum starved cells. (a) Immunoblot of NIH3T3 cell lysates after 24 hrs of serum deprivation, using rabbit polyclonal KIF13B antiserum.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
SUPPLEMENTARY INFORMATION doi:10.1038/nature12138 Supplementary Figure 1. Knockdown of KRAS leads to a reduction in macropinocytosis. (a) KRAS knockdown in MIA PaCa-2 cells expressing KRASspecific shrnas
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
SUPPLEMENTARY INFORMATION
 a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
a before amputation regeneration regenerated limb DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS DERMIS SKELETON MUSCLE SCHWANN CELLS EPIDERMIS developmental origin: lateral plate mesoderm presomitic mesoderm
Journal of Cell Science Supplementary Material
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 SUPPLEMENTARY FIGURE LEGENDS Figure S1: Eps8 is localized at focal adhesions and binds directly to FAK (A) Focal
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Dual PI3K/ERK inhibition induces necroptotic cell death of Hodgkin Lymphoma cells through IER3 downregulation
 Dual PI3K/ERK inhibition induces necroptotic cell death of Hodgkin Lymphoma cells through IER3 downregulation *Silvia Laura Locatelli, 1 Giuseppa Careddu, 1 Giuliano Giuseppe Stirparo, 1 Luca Castagna,
Dual PI3K/ERK inhibition induces necroptotic cell death of Hodgkin Lymphoma cells through IER3 downregulation *Silvia Laura Locatelli, 1 Giuseppa Careddu, 1 Giuliano Giuseppe Stirparo, 1 Luca Castagna,
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
supplementary information
 DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure 1. Drawing of spinal cord open-book preparations and DiI tracing. Nature Neuroscience: doi: /nn.3893
 Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Supplementary Figure 1 Drawing of spinal cord open-book preparations and DiI tracing. Supplementary Figure 2 In ovo electroporation of dominant-negative PlexinA1 in commissural neurons induces midline
Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking
 Human Molecular Genetics, 2009, Vol. 18, No. 20 3926 3941 doi:10.1093/hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a
Human Molecular Genetics, 2009, Vol. 18, No. 20 3926 3941 doi:10.1093/hmg/ddp335 Advance Access published on July 22, 2009 Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
DOI: 10.1038/ncb2880 Supplementary Figure 1 Sequence alignment of Deup1 and Cep63. The protein sequence alignment was generated by the Clustal X 2.0 multiple sequence alignment program using default parameters.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
www.sciencesignaling.org/cgi/content/full/8/404/ra120/dc1 Supplementary Materials for The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1 Akihiro
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in
 Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
Supplementary Figure 1 Pfn1, but not other Pfn isoforms are expressed in platelets. (a) RT-PCR of Pfn isoforms in control mouse platelets, Pfn1 -/- platelets and control heart. Expected band size for Pfn1
J. Cell Sci. 128: doi: /jcs : Supplementary Material. Supplemental Figures. Journal of Cell Science Supplementary Material
 Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
Supplemental Figures Figure S1. Trio controls endothelial barrier function. (A) TagRFP-shTrio constructs were expressed in ECs. Western blot shows efficient Trio knockdown in TagRFP-expressing ECs. (B)
J. Cell Sci. 130: doi: /jcs : Supplementary information. Supplementary figure. Journal of Cell Science Supplementary information
 Supplementary figure Fig.S1 Normal rhabdomere biogenesis and structure during development in dpip4k 29 and dpip4k over expressing PRs. (A) dpip4k 29 PRs showing immunogold labeling for Rh1 in MVBs (i)
Supplementary figure Fig.S1 Normal rhabdomere biogenesis and structure during development in dpip4k 29 and dpip4k over expressing PRs. (A) dpip4k 29 PRs showing immunogold labeling for Rh1 in MVBs (i)
Supplementary Figure 1. Additional RNAi screen data
 Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin
 Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
Fig. S1. Endocytosis of extracellular cargo does not depend on dia. (A) To visualize endocytosis, fluorescently labelled wheat germ agglutinin (WGA-Alexa555) was injected into the extracellular perivitteline
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Paul et al.,
 Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplemental material JCB Paul et al., http://www.jcb.org/cgi/content/full/jcb.201502040/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Mutant p53-expressing cells display limited retrograde actin flow at
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Table, Figures and Videos
 Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Table, Figures and Videos Table S1. Oligonucleotides used for different approaches. (A) RT-qPCR study. (B) qpcr study after ChIP assay. (C) Probes used for EMSA. Figure S1. Notch activation
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and
 Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Determining positive selection gates for LRCs and nonlrcs Positive selection gates for the collection of LRCs or nonlrcs had to be drawn based on the location and shape of the Gaussian distributions. For
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
THE JOURNAL OF CELL BIOLOGY
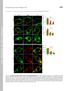 Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Supplementary Data
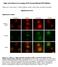 Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
Single cell resolution in vivo imaging of DNA damage following PARP inhibition Katherine S. Yang, Rainer H. Kohler, Matthieu Landon, Randy Giedt, and Ralph Weissleder Supplementary Data Supplementary Figures
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Flag-Rac Vector V12 V12 N17 C40. Vector C40 pakt (T308) Akt1. Myc-DN-PAK1 (N-SP)
 a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
a b FlagRac FlagRac V2 V2 N7 C4 V2 V2 N7 C4 p (T38) p (S99, S24) p Flag (Rac) NIH 3T3 COS c +Serum p (T38) MycDN (NSP) Mycp27 3 6 2 3 6 2 3 6 2 min p Myc ( or p27) Figure S (a) Effects of Rac mutants on
Assay Name: Antibody-Dependent Drug Uptake Assay
 Assay Name: Antibody-Dependent Drug Uptake Assay Assay ID: Celigo_02_0019 Table of Contents Experiment: Antibody-Dependent Drug Uptake Assay... 2 Celigo Setup...2 Assay Protocol and Plate Setup...3 Results...5
Assay Name: Antibody-Dependent Drug Uptake Assay Assay ID: Celigo_02_0019 Table of Contents Experiment: Antibody-Dependent Drug Uptake Assay... 2 Celigo Setup...2 Assay Protocol and Plate Setup...3 Results...5
Supplementary Information
 Supplementary Information stability is regulated by CK2-dependent interaction with R2TP complex Patrick von Morgen 1,2, Kamila Burdova 1, Thomas G. Flower 3, Nicola J. O'Reilly 4, Simon J. Boulton 5, Stephen
Supplementary Information stability is regulated by CK2-dependent interaction with R2TP complex Patrick von Morgen 1,2, Kamila Burdova 1, Thomas G. Flower 3, Nicola J. O'Reilly 4, Simon J. Boulton 5, Stephen
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Prospéri et al.,
 Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Supplemental material JCB Prospéri et al., http://www.jcb.org/cgi/content/full/jcb.201501018/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Myo1b Tail interacts with YFP-EphB2 coated beads and genistein inhibits
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
3 P p25. p43 p41 28 FADD. cflips. PE-Cy5 [Fluorescence intensity]
![3 P p25. p43 p41 28 FADD. cflips. PE-Cy5 [Fluorescence intensity] 3 P p25. p43 p41 28 FADD. cflips. PE-Cy5 [Fluorescence intensity]](/thumbs/87/97394430.jpg) L S p4 3 D3 76 N S L Ve ct or p4 3 D3 76 N S L Ve ct or A aspase 8 FADD TRAF2 D95-R - + Vector D95L TL I S L D376N T RAIL-R1 T RAIL-R2 D95-R E-y5 [Fluorescence intensity] Supplemental Fig. 1 Different
L S p4 3 D3 76 N S L Ve ct or p4 3 D3 76 N S L Ve ct or A aspase 8 FADD TRAF2 D95-R - + Vector D95L TL I S L D376N T RAIL-R1 T RAIL-R2 D95-R E-y5 [Fluorescence intensity] Supplemental Fig. 1 Different
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas.
 Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
Supplementary Table 1. Sequences for BTG2 and BRCA1 sirnas. Target Gene Non-target / Control BTG2 BRCA1 NFE2L2 Target Sequence ON-TARGET plus Non-targeting sirna # 1 (Cat# D-001810-01-05) sirna1: GAACCGACAUGCUCCCGGA
The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from
 Materials and Methods Antibodies: The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from D. A. Compton); Dynein Intermediate Chain - 74.1 (Chemicon, Temecula, CA); Dynein
Materials and Methods Antibodies: The following antibodies were used in this study: NuMA - rabbit-anti NuMA (gift from D. A. Compton); Dynein Intermediate Chain - 74.1 (Chemicon, Temecula, CA); Dynein
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
DOI: 10.1038/ncb2271 Supplementary Figure a! WM266.4 mock WM266.4 #7 sirna WM266.4 #10 sirna SKMEL28 mock SKMEL28 #7 sirna SKMEL28 #10 sirna WM1361 mock WM1361 #7 sirna WM1361 #10 sirna 9 WM266. WM136
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive
 Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
Supplementary Figure 1 Structural modeling and purification of V. cholerae ABH. (a) The migration of the purified rabh and catalytically inactive variants rabhs, rabhd, and rabhh are shown on 12% SDS-PAGE
Supplementary Figure 1. Related to Figure 1. Characterization of centrosome fragmentation in mitotically delayed RPE1 cells. (a) Cells transiently
 Supplementary Figure 1. Related to Figure 1. Characterization of centrosome fragmentation in mitotically delayed RPE1 cells. (a) Cells transiently transfected with EGFP centrin-2 (Green), synchronized
Supplementary Figure 1. Related to Figure 1. Characterization of centrosome fragmentation in mitotically delayed RPE1 cells. (a) Cells transiently transfected with EGFP centrin-2 (Green), synchronized
T H E J O U R N A L O F C E L L B I O L O G Y
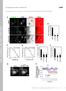 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Yamaguchi et al., http://www.jcb.org/cgi/content/full/jcb.201009126/dc1 S1 Figure S2. The expression levels of GFP-Akt1-PH and localization
hnrnp C promotes APP translation by competing with FMRP for APP mrna recruitment to P bodies
 hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
hnrnp C promotes APP translation by competing with for APP mrna recruitment to P bodies Eun Kyung Lee 1, Hyeon Ho Kim 1, Yuki Kuwano 1, Kotb Abdelmohsen 1, Subramanya Srikantan 1, Sarah S. Subaran 2, Marc
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration
 /, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
/, Supplementary Advance Publications Materials 2016 CD93 and dystroglycan cooperation in human endothelial cell adhesion and migration Supplementary Materials Supplementary Figure S1: In ECs CD93 silencing
(B) Comparable expression of major integrin subunits and glycoproteins on the surface of resting WT and Lnk -/- platelets.
 Supplemental Figure S1. Characteristics of Lnk -/- platelets. (A) Electron micrographs of resting platelets showing the normal intracellular structure of Lnk -/ platelets. Samples were fixed with 4% PFA
Supplemental Figure S1. Characteristics of Lnk -/- platelets. (A) Electron micrographs of resting platelets showing the normal intracellular structure of Lnk -/ platelets. Samples were fixed with 4% PFA
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
High content imaging and applications
 High content imaging and applications Nick Dolman Ph.D. Senior Staff Scientist Biosciences Division- Thermo Fisher Scientific For Research Use Only. Not for use in diagnostic procedures. The world leader
High content imaging and applications Nick Dolman Ph.D. Senior Staff Scientist Biosciences Division- Thermo Fisher Scientific For Research Use Only. Not for use in diagnostic procedures. The world leader
Description: Nuclear morphology and dynamics in nontargeting sirna transfected cells. HeLa Kyoto
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tables Title of file for HTML: Supplementary Movie 1 Description: Nuclear morphology and dynamics
Supplementary Infomation
 Supplementary Infomation Mycobacterium avium MAV2054 protein induces macrophage apoptosis through targeting to mitochondria and reduces intracellular growth of the bacteria. Kang-In Lee 1,2, Jake Whang
Supplementary Infomation Mycobacterium avium MAV2054 protein induces macrophage apoptosis through targeting to mitochondria and reduces intracellular growth of the bacteria. Kang-In Lee 1,2, Jake Whang
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
(A) Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site.
 SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
SUPPLEMENTRY INFORMTION SUPPLEMENTL FIGURES Figure S1. () Schematic illustration of sciatic nerve ligation. P, proximal; D, distal to the ligation site. () Western blot of ligated and unligated sciatic
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells.
 Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
Supplementary Fig. 1 Supplementary Fig. 1 Identification of Nedd4 as an IRS-2-associated protein in camp-treated FRTL-5 cells. (a) FRTL-5 cells were treated with 1 mm dibutyryl camp for 24 h, and the lysates
A Tumor Necrosis Factor-Alpha-Mediated Pathway Promoting Autosomal Dominant Polycystic Kidney Disease.
 A Tumor Necrosis Factor-Alpha-Mediated Pathway Promoting Autosomal Dominant Polycystic Kidney Disease. Xiaogang Li, Brenda S. Magenheimer, Sheng Xia, Teri Johnson, Darren P. Wallace, James P. Calvet and
A Tumor Necrosis Factor-Alpha-Mediated Pathway Promoting Autosomal Dominant Polycystic Kidney Disease. Xiaogang Li, Brenda S. Magenheimer, Sheng Xia, Teri Johnson, Darren P. Wallace, James P. Calvet and
Supplementary Figure 1
 Supplementary Figure 1 Cell cycle distribution (%) 1 8 6 4 2 Cell Cycle G1 S G2 Viability (%) 5 4 3 2 1 Viability Supplementary Figure 1. Cell cycle distribution and viability during DSB repair measurements.
Supplementary Figure 1 Cell cycle distribution (%) 1 8 6 4 2 Cell Cycle G1 S G2 Viability (%) 5 4 3 2 1 Viability Supplementary Figure 1. Cell cycle distribution and viability during DSB repair measurements.
Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of
 Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
Supplementary Information Cytotoxicity of Botulinum Neurotoxins Reveals a Direct Role of Syntaxin 1 and SNAP-25 in Neuron Survival Lisheng Peng, Huisheng Liu, Hongyu Ruan, William H. Tepp, William H. Stoothoff,
