Proteomics. Proteomics is the study of all proteins within organism. Challenges
|
|
|
- Clyde Taylor
- 6 years ago
- Views:
Transcription
1 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we need to know 2. All protein-protein interactions. 3. One protein or peptide may have multiple functions depending on context. 4. Regulation of protein function. 5. Modification 6. Location Location will help us to understand the proteins role in the cell, what its function is, and what controls its function. 7. Detection and quantitation The concentration of all proteins changes by 10 orders of magnitude within the cell. Currently there are no easy methods for determining the concentrations over this large of a concentration range. 1
2 What areas are being studied in Proteomics? The areas being studied under the name of proteomics is changing and often growing as the field develops. I will present a few of the areas in this class. Some areas in proteomics are: 1. Mass spectrometer based proteomics This area is most commonly associated with proteomics and is a method to determine which proteins are expressed and the amounts of those proteins. 2. Array based proteomics This method uses various arrays to try and define the function of the proteins, regulation levels and interacting partners within the cell. 3. Informatics This is trying to define what information will be needed, stored, accessed and how it can be used to study the proteome of the cell. 4. Clinical 5. Orthagonalomics 2
3 MASS SPECTROMETER BASED PROTEOMICS Principles Mass spectrometry is based on the fact that ions of differing charge and mass will move differently in a magnetic field. In proteomics the proteins are first separated by some means and then analyzed with a mass spectrometer. Let s begin by discussing various means for separating proteins to prepare them for mass spectrometry and then discuss mass spectrometry and how it is used. Separating the genome The protein genome is separated by several different methods. Many researchers are first separating portions of the genome, such as isolating organelles, and then analyzing that portion. This is because often proteins of interest, regulatory proteins are in low abundance. The most commonly used method is 2-dimensional gel electrophoresis. First I will explain the individual methods that are coupled to create the two dimensions. 3
4 Iso-electric focusing - This separates proteins based on isoelectric point - The isoelectric point is the ph at which the protein has no net charge. - Typically this is done with a tube gel containing a low concentration of poly-acrylamide. - Ampholytes are added to create a ph gradient in an electric field and the proteins are loaded. - The tube gel is placed in and electrophoresis system for up to 24 hours and the proteins form tight bands at their iso-electric point. - The tube gels are now ready for the second method. 4
5 - - - Figure is from Lehninger fourth edition Principles of Biochemistry 5
6 - SDS Poly-Acrylamide Gel Electrophoresis - The second dimension separates the proteins based on size. - There are two parts, the stacking gel which concentrates the sample and the running gel that is used to separate the proteins. - The tube gel is soaked in a solution containing chemical to denature the proteins including sodium dodecyl sulfate a detergent which gives the proteins a net negative charge. This means that all proteins will move in one direction. - The tube gel is then put in the one long well in the stacking gel, sealed in place with agarose, and the proteins subjected to an electric field to separate. - The larger proteins are found at the top and the smaller ones are found at the bottom of the gel. - In a 2D gel the proteins appear as spots on the gel rather than bands. These spots can then be further processed or used for mass spectrometry directly. - Further processing usually include trypsin digestion 6
7 - - Figure is from Lehninger fourth edition Principles of Biochemistry 7
8 - Figure is from Lehninger fourth edition Principles of Biochemistry 8
9 9
10 Alternate methods of separation. Recently there has been a change in the way people are approaching separating the proteins. The first dimension is run in larger agarose tube gels with ampholytes. This has less resolution than polyacrylamide gels. The tubes are sliced and the proteins are allowed to diffuse out. The diffused proteins are then separated by capillary electrophoresis. Capillary electrophoresis has a much greater resolution for the proteins mass. Proteins are eluted from the capillary in the process and can be collected. They are readily available for mass spectrometry. 10
11 Mass Spectrometery 1. Separates ions based on mass to charge ratio. Charges are placed on the protein or the peptide by ionization. 2. Two most common types of ionization are: a. Electrospray ionization (ESI) ESI can give whole protein masses as well as complex masses. If the proteins is first separated by reverse phase HPLC before injection only the subunits masses will be known. - Figure is from Lehninger fourth edition Principles of Biochemistry 11
12 b. Matrix-assisted laser desorption Ionization. MALDI causes fragmentation of the protein during ionization. Can be used to get more information about the fragments. Easier to do than ESI. 3. Important parameters a. sensitivity b. resolution c. mass accuracy 4. Mass Analyzers d. Quadrapole i. Sensitive, acceptable mass accuracy and ii. e. Time of Flight resolution Easily coupled to chromatography. i. Sensitive, high mass accuracy, high resolution ii. Limited to small m/z ratios iii. Not easily coupled to chromatography f. Ion Trap i. Sensitive ii. Low mass accuracy g. Fourier Transform ion cyclotron i. High sensitivity, mass accuracy, resolution, ii. dynamic range Expensive, difficult to operate, low fragmentation efficiency 12
13 4. Instruments are often coupled a. MS/MS b. Collider - forms collision induced spectra (CID) 13
14 Protein identification and Quatitation 1. Protein separation by 2 dimensional electrophoresis or similar methods 2. Limited protein purification and automated MS. Limited purification and MS 1. multiple chromatography stages a. ion exchange, reverse phase, affinity b. electrophoresis 2. To quantitate a. add stable isotopes b. post separation modification i. SH, NH 2, N-linked carbohydrates c. incorporation of isotopes in culture Protein Identification 1. Use collision induced spectra i. provides sequence information ii. provides unique m/z spectra for each peptide 2. Problem is large number of CID information i. need methods for searching ii. Filtering has been tried (limited success) iii. Most successful with human intervention. iv. Improving 14
Proteomics and Cancer
 Proteomics and Cancer Japan Society for the Promotion of Science (JSPS) Science Dialogue Program at Niitsu Senior High School Niitsu, Niigata September 4th 2006 Vladimir Valera, M.D, PhD JSPS Postdoctoral
Proteomics and Cancer Japan Society for the Promotion of Science (JSPS) Science Dialogue Program at Niitsu Senior High School Niitsu, Niigata September 4th 2006 Vladimir Valera, M.D, PhD JSPS Postdoctoral
SGN-6106 Computational Systems Biology I
 SGN-6106 Computational Systems Biology I A View of Modern Measurement Systems in Cell Biology Kaisa-Leena Taattola 21.2.2007 1 The cell a complex system (Source: Lehninger Principles of Biochemistry 4th
SGN-6106 Computational Systems Biology I A View of Modern Measurement Systems in Cell Biology Kaisa-Leena Taattola 21.2.2007 1 The cell a complex system (Source: Lehninger Principles of Biochemistry 4th
Lecture 5: 8/31. CHAPTER 5 Techniques in Protein Biochemistry
 Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Computing with large data sets
 Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Proteomics and Vaccine Potency Testing
 Proteomics and Vaccine Potency Testing Louisa B. Tabatabai, PhD Respiratory Diseases of Livestock Research Unit National Animal Disease Center, ARS, USDA, Ames, Iowa What is proteomics Methodologies of
Proteomics and Vaccine Potency Testing Louisa B. Tabatabai, PhD Respiratory Diseases of Livestock Research Unit National Animal Disease Center, ARS, USDA, Ames, Iowa What is proteomics Methodologies of
Proteomics and some of its Mass Spectrometric Applications
 Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Advances in analytical biochemistry and systems biology: Proteomics
 Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis
 Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Strategies for Quantitative Proteomics. Atelier "Protéomique Quantitative" La Grande Motte, France - June 26, 2007
 Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Mass Spectrometry-based Methods of Proteome Analysis
 1 Mass Spectrometry-based Methods of Proteome Analysis Boris L. Zybailov and Michael P. Washburn Stowers Institute for Medical Research, Kansas City, MO, USA 1 Principles and Instrumentation 3 1.1 Proteome
1 Mass Spectrometry-based Methods of Proteome Analysis Boris L. Zybailov and Michael P. Washburn Stowers Institute for Medical Research, Kansas City, MO, USA 1 Principles and Instrumentation 3 1.1 Proteome
Bioinformatics (Lec 19) Picture Copyright: the National Museum of Health
 3/29/05 1 Picture Copyright: AccessExcellence @ the National Museum of Health PCR 3/29/05 2 Schematic outline of a typical PCR cycle Target DNA Primers dntps DNA polymerase 3/29/05 3 Gel Electrophoresis
3/29/05 1 Picture Copyright: AccessExcellence @ the National Museum of Health PCR 3/29/05 2 Schematic outline of a typical PCR cycle Target DNA Primers dntps DNA polymerase 3/29/05 3 Gel Electrophoresis
Mass Spectrometry. Kyle Chau and Andrew Gioe
 Mass Spectrometry Kyle Chau and Andrew Gioe Computation of Molecular Mass - Mass Spectrum is a plot of intensity as a function of masscharge ratio, m/z. - Mass-charge ratio can be determined through accelerating
Mass Spectrometry Kyle Chau and Andrew Gioe Computation of Molecular Mass - Mass Spectrum is a plot of intensity as a function of masscharge ratio, m/z. - Mass-charge ratio can be determined through accelerating
11/22/13. Proteomics, functional genomics, and systems biology. Biosciences 741: Genomics Fall, 2013 Week 11
 Proteomics, functional genomics, and systems biology Biosciences 741: Genomics Fall, 2013 Week 11 1 Figure 6.1 The future of genomics Functional Genomics The field of functional genomics represents the
Proteomics, functional genomics, and systems biology Biosciences 741: Genomics Fall, 2013 Week 11 1 Figure 6.1 The future of genomics Functional Genomics The field of functional genomics represents the
Protein analysis. Dr. Mamoun Ahram Summer semester, Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5
 Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Proteomics: A Challenge for Technology and Information Science. What is proteomics?
 Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
LECTURE-3. Protein Chemistry to proteomics HANDOUT. Proteins are the most dynamic and versatile macromolecules in a living cell, which
 LECTURE-3 Protein Chemistry to proteomics HANDOUT PREAMBLE Proteins are the most dynamic and versatile macromolecules in a living cell, which regulates essential activities of the cell. The classical protein
LECTURE-3 Protein Chemistry to proteomics HANDOUT PREAMBLE Proteins are the most dynamic and versatile macromolecules in a living cell, which regulates essential activities of the cell. The classical protein
Extracting Pure Proteins from Cells
 Extracting Pure Proteins from Cells 0 Purification techniques focus mainly on size & charge 0 The first step is homogenization (grinding, Potter Elvejhem homogenizer, sonication, freezing and thawing,
Extracting Pure Proteins from Cells 0 Purification techniques focus mainly on size & charge 0 The first step is homogenization (grinding, Potter Elvejhem homogenizer, sonication, freezing and thawing,
What are proteomics? And what can they tell us about seed maturation and germination?
 Danseed Symposium Kobæk Strand, Skelskør 14 March, 2017 What are proteomics? And what can they tell us about seed maturation and germination? Ian Max Møller Department of Molecular Biology and Genetics
Danseed Symposium Kobæk Strand, Skelskør 14 March, 2017 What are proteomics? And what can they tell us about seed maturation and germination? Ian Max Møller Department of Molecular Biology and Genetics
Chapter 3. Exploring Proteins and Proteomes. Dr. Jaroslava Miksovska
 Chapter 3 Exploring Proteins and Proteomes Dr. Jaroslava Miksovska Chapter objectives: Protein purification and isolation techniques Protein sequencing Immunology for protein identification Protein identification
Chapter 3 Exploring Proteins and Proteomes Dr. Jaroslava Miksovska Chapter objectives: Protein purification and isolation techniques Protein sequencing Immunology for protein identification Protein identification
Chapter 6. Techniques of Protein and Nucleic Acid Purification
 Chapter 6 Techniques of Protein and Nucleic Acid Purification Considerations in protein expression and purification Protein source Natural sources Recombinant sources Methods of lysis and solubilization
Chapter 6 Techniques of Protein and Nucleic Acid Purification Considerations in protein expression and purification Protein source Natural sources Recombinant sources Methods of lysis and solubilization
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Proteomics 6/4/2009 WESTERN BLOT ANALYSIS
 SDS-PAGE (PolyAcrylamide Gel Electrophoresis) Proteomics WESTERN BLOT ANALYSIS Presented by: Nuvee Prapasarakul Veterinary Microbiology Chulalongkorn University Proteomics has been said to be the next
SDS-PAGE (PolyAcrylamide Gel Electrophoresis) Proteomics WESTERN BLOT ANALYSIS Presented by: Nuvee Prapasarakul Veterinary Microbiology Chulalongkorn University Proteomics has been said to be the next
The Role of Chemistry Consumables in Biopharmaceutical Product Development
 Technology & Services The Role of Chemistry Consumables in Biopharmaceutical Product Development a report by John C Gebler, Jeff R Mazzeo and William J Warren Director, Life Sciences Reasearch and Development,
Technology & Services The Role of Chemistry Consumables in Biopharmaceutical Product Development a report by John C Gebler, Jeff R Mazzeo and William J Warren Director, Life Sciences Reasearch and Development,
Chapter 9 Proteomics. From genomics to proteomics
 Chapter 9 Proteomics From genomics to proteomics From structural genomics, functional genomics to proteomics http://www.nature.com/scitable/topicpage/the-proteome-discovering-the-structure-and-function-613
Chapter 9 Proteomics From genomics to proteomics From structural genomics, functional genomics to proteomics http://www.nature.com/scitable/topicpage/the-proteome-discovering-the-structure-and-function-613
NPTEL VIDEO COURSE PROTEOMICS PROF. SANJEEVA SRIVASTAVA
 LECTURE-06 PROTEIN PURIFICATION AND PEPTIDE ISOLATION USING CHROMATOGRAPHY TRANSCRIPT Welcome to the proteomics course. Today, we will talk about protein purification and peptide isolation using chromatography
LECTURE-06 PROTEIN PURIFICATION AND PEPTIDE ISOLATION USING CHROMATOGRAPHY TRANSCRIPT Welcome to the proteomics course. Today, we will talk about protein purification and peptide isolation using chromatography
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham. Bioscience) were washed four times with 1 mm HCl and twice with coupling buffer
 Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS
 Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS Sensitive workflow for the detection of PTMs in biological samples from small sample volumes Klaus Faserl, 1 Bettina Sarg, 1
Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS Sensitive workflow for the detection of PTMs in biological samples from small sample volumes Klaus Faserl, 1 Bettina Sarg, 1
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH)
 User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
New workflows for protein expression analysis ICAT. Reagent Technology
 New workflows for protein expression analysis ICAT Reagent Technology Breakthrough technology for protein expression analysis accelerate In protein-expression analysis studies, ICAT reagent technology
New workflows for protein expression analysis ICAT Reagent Technology Breakthrough technology for protein expression analysis accelerate In protein-expression analysis studies, ICAT reagent technology
Protein Purification and Characterization Techniques. Nafith Abu Tarboush, DDS, MSc, PhD
 Protein Purification and Characterization Techniques Nafith Abu Tarboush, DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Extracting Pure Proteins from Cells Purification techniques focus
Protein Purification and Characterization Techniques Nafith Abu Tarboush, DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Extracting Pure Proteins from Cells Purification techniques focus
PROTEOMICS. Timothy Palzkill Baylor College of Medicine. KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow
 PROTEOMICS PROTEOMICS by Timothy Palzkill Baylor College of Medicine KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow ebook ISBN: 0-306-46895-6 Print ISBN: 0-792-37565-3 2002
PROTEOMICS PROTEOMICS by Timothy Palzkill Baylor College of Medicine KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow ebook ISBN: 0-306-46895-6 Print ISBN: 0-792-37565-3 2002
Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Quantitation techniques Label-free TMT SILAC Peptide identification and FDR 194 Lecture
Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Quantitation techniques Label-free TMT SILAC Peptide identification and FDR 194 Lecture
Medicinal Chemistry of Modern Antibiotics
 Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2012 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2012 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Medicinal Chemistry of Modern Antibiotics
 Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2008 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2008 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
timstof Innovation with Integrity Powered by PASEF TIMS-QTOF MS
 timstof Powered by PASEF Innovation with Integrity TIMS-QTOF MS timstof The new standard for high speed, high sensitivity shotgun proteomics The timstof Pro with PASEF technology delivers revolutionary
timstof Powered by PASEF Innovation with Integrity TIMS-QTOF MS timstof The new standard for high speed, high sensitivity shotgun proteomics The timstof Pro with PASEF technology delivers revolutionary
Nanoflow LC Q-TOF MS for De Novo Peptide Sequencing in Microbial Proteomics
 COUPLING MATTERS Nanoflow LC Q-TOF MS for De Novo Peptide Sequencing in Microbial Proteomics Bart Devreese and Jozef Van Beeumen, Laboratory of Protein Biochemistry and Protein Engineering, Ghent University,
COUPLING MATTERS Nanoflow LC Q-TOF MS for De Novo Peptide Sequencing in Microbial Proteomics Bart Devreese and Jozef Van Beeumen, Laboratory of Protein Biochemistry and Protein Engineering, Ghent University,
Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research
 PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
(Refer Slide Time: 00:16)
 (Refer Slide Time: 00:16) Proteins and Gel-Based Proteomics Professor Sanjeeva Srivastava Department of Biosciences and Bioengineering Indian Institute of Technology, Bombay Mod 02 Lecture Number 5 Welcome
(Refer Slide Time: 00:16) Proteins and Gel-Based Proteomics Professor Sanjeeva Srivastava Department of Biosciences and Bioengineering Indian Institute of Technology, Bombay Mod 02 Lecture Number 5 Welcome
Faster, easier, flexible proteomics solutions
 Agilent HPLC-Chip LC/MS Faster, easier, flexible proteomics solutions Our measure is your success. products applications soft ware services Phospho- Anaysis Intact Glycan & Glycoprotein Agilent s HPLC-Chip
Agilent HPLC-Chip LC/MS Faster, easier, flexible proteomics solutions Our measure is your success. products applications soft ware services Phospho- Anaysis Intact Glycan & Glycoprotein Agilent s HPLC-Chip
Lecture 23: SDS Gel Electrophoresis and Stacking Gels
 Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2018 Lecture 23: SDS Gel Electrophoresis and Stacking Gels 3 April 2018 c David P. Goldenberg University of Utah goldenberg@biology.utah.edu
Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2018 Lecture 23: SDS Gel Electrophoresis and Stacking Gels 3 April 2018 c David P. Goldenberg University of Utah goldenberg@biology.utah.edu
So.. Let us say you have an impure solution containing a protein of interest. Q: How do you (a) analyze what you have and (b) purify what you want?
 So.. Let us say you have an impure solution containing a protein of interest. Q: How do you (a) analyze what you have and (b) purify what you want? Polyacrylamide Gel Electrophoresis (PAGE) Note: proteins
So.. Let us say you have an impure solution containing a protein of interest. Q: How do you (a) analyze what you have and (b) purify what you want? Polyacrylamide Gel Electrophoresis (PAGE) Note: proteins
SUPPLEMENTARY INFORMATION
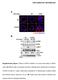 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
Comparability Analysis of Protein Therapeutics by Bottom-Up LC-MS with Stable Isotope-Tagged Reference Standards
 Comparability Analysis of Protein Therapeutics by Bottom-Up LC-MS with Stable Isotope-Tagged Reference Standards 16 September 2011 Abbott Bioresearch Center, Worcester, MA USA Manuilov, A. V., C. H. Radziejewski
Comparability Analysis of Protein Therapeutics by Bottom-Up LC-MS with Stable Isotope-Tagged Reference Standards 16 September 2011 Abbott Bioresearch Center, Worcester, MA USA Manuilov, A. V., C. H. Radziejewski
Improving Productivity with Applied Biosystems GPS Explorer
 Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Application Note TOF/MS
 Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
NPTEL
 NPTEL Syllabus Proteomics: Principles and Techniques - Video course COURSE OUTLINE An introduction to proteomics: Basics of protein structure and function, An overview of systems biology, Evolution from
NPTEL Syllabus Proteomics: Principles and Techniques - Video course COURSE OUTLINE An introduction to proteomics: Basics of protein structure and function, An overview of systems biology, Evolution from
FACTORS THAT AFFECT PROTEIN IDENTIFICATION BY MASS SPECTROMETRY HAOFEI TIFFANY WANG. (Under the Direction of Ron Orlando) ABSTRACT
 FACTORS THAT AFFECT PROTEIN IDENTIFICATION BY MASS SPECTROMETRY by HAOFEI TIFFANY WANG (Under the Direction of Ron Orlando) ABSTRACT Mass spectrometry combined with database search utilities is a valuable
FACTORS THAT AFFECT PROTEIN IDENTIFICATION BY MASS SPECTROMETRY by HAOFEI TIFFANY WANG (Under the Direction of Ron Orlando) ABSTRACT Mass spectrometry combined with database search utilities is a valuable
PROTEIN AND PROTEOME ANALYTICS IDENTIFICATION, CHARACTERIZATION AND EXPRESSION ANALYSIS OF PROTEINS
 FRAUNHOFER INSTITUTE FOR INTERFACIAL ENGINEERING AND BIOTECHNOLOGY PROTEIN AND PROTEOME ANALYTICS IDENTIFICATION, CHARACTERIZATION AND EXPRESSION ANALYSIS OF PROTEINS 1 1 2 2 PROTEINS PROTEOMICS Proteins,
FRAUNHOFER INSTITUTE FOR INTERFACIAL ENGINEERING AND BIOTECHNOLOGY PROTEIN AND PROTEOME ANALYTICS IDENTIFICATION, CHARACTERIZATION AND EXPRESSION ANALYSIS OF PROTEINS 1 1 2 2 PROTEINS PROTEOMICS Proteins,
timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics
 timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics The timstof Pro powered by PASEF enables sequencing speed of > 2 Hz, high sensitivity
timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics The timstof Pro powered by PASEF enables sequencing speed of > 2 Hz, high sensitivity
1-DE 2-DE??? PCPPAU 04/2011
 1-DE 2-DE??? PCPPAU 04/2011 THEORY Any charged ion or group of ions will migrate when placed in an electric field. ph -----IEP (pi) Migration is dependent on charge density Charge/Mass + - PCPPAU 04/2011
1-DE 2-DE??? PCPPAU 04/2011 THEORY Any charged ion or group of ions will migrate when placed in an electric field. ph -----IEP (pi) Migration is dependent on charge density Charge/Mass + - PCPPAU 04/2011
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements
 Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Zakopane, April, 2011
 Clinical Proteomics: A Technology to shape the future? Zakopane, April, 2011 Protein Chemistry/Proteomics/Peptide Synthesis and Array Unit Institute of Biomedicine/Anatomy Biomedicum Helsinki, 00014 University
Clinical Proteomics: A Technology to shape the future? Zakopane, April, 2011 Protein Chemistry/Proteomics/Peptide Synthesis and Array Unit Institute of Biomedicine/Anatomy Biomedicum Helsinki, 00014 University
QSTAR XL Hybrid LC/MS/MS System. Extended Performance Plus Exceptional Flexibility. QSTAR XL. Hybrid LC/MS/MS System
 QSTAR XL Hybrid LC/MS/MS System Extended Performance Plus Exceptional Flexibility. QSTAR XL Hybrid LC/MS/MS System Extended performance and flexibility for your most challenging analyses. Applied Biosystems/MDS
QSTAR XL Hybrid LC/MS/MS System Extended Performance Plus Exceptional Flexibility. QSTAR XL Hybrid LC/MS/MS System Extended performance and flexibility for your most challenging analyses. Applied Biosystems/MDS
Principles of Proteomic Methods
 Principles of Proteomic Methods Marc Wilkins School of Biotechnology and Biomolecular Sciences What is Proteomics? - proteomics is the study of proteomes - proteomics aims to: separate, identify and characterise
Principles of Proteomic Methods Marc Wilkins School of Biotechnology and Biomolecular Sciences What is Proteomics? - proteomics is the study of proteomes - proteomics aims to: separate, identify and characterise
Protein Microarrays?
 Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
PROTEOMICS TODAY Protein Assessment and Biomarkers Using Mass Spectrometry, 2D Electrophoresis, and Microarray Technology
 PROTEOMICS TODAY Protein Assessment and Biomarkers Using Mass Spectrometry, 2D Electrophoresis, and Microarray Technology MAHMOUD HAMDAN GlaxoSmithKline PIER GIORGIO RIGHETTI University of Verona, Italy
PROTEOMICS TODAY Protein Assessment and Biomarkers Using Mass Spectrometry, 2D Electrophoresis, and Microarray Technology MAHMOUD HAMDAN GlaxoSmithKline PIER GIORGIO RIGHETTI University of Verona, Italy
Protein Techniques 1 APPENDIX TO CHAPTER 5
 Protein Techniques 1 APPENDIX T CHAPTER 5 Dialysis and Ultrafiltration If a solution of protein is separated from a bathing solution by a semipermeable membrane, small molecules and ions can pass through
Protein Techniques 1 APPENDIX T CHAPTER 5 Dialysis and Ultrafiltration If a solution of protein is separated from a bathing solution by a semipermeable membrane, small molecules and ions can pass through
Identification of human serum proteins detectable after Albumin removal with Vivapure Anti-HSA Kit
 Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Determination of Isoelectric Point (pi) By Whole-Column Detection cief
 Determination of Isoelectric Point (pi) By Whole-Column Detection cief Tiemin Huang and Jiaqi Wu CONVERGENT BIOSCIENCE Determination of Isoelectric Point (pi) by Whole Column Detection cief Definition
Determination of Isoelectric Point (pi) By Whole-Column Detection cief Tiemin Huang and Jiaqi Wu CONVERGENT BIOSCIENCE Determination of Isoelectric Point (pi) by Whole Column Detection cief Definition
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS
 Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
2D difference gel electrophoresis (2D-DIGE) Spot picking and in-gel digestion
 2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the
2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the
Embryology in the era of proteomics
 New approaches for non-invasive embryo quality assessment Tours, France April 11-12 2008 Embryology in the era of proteomics Roger Sturmey Department of Biology rgs102@york.ac.uk Background Definitions
New approaches for non-invasive embryo quality assessment Tours, France April 11-12 2008 Embryology in the era of proteomics Roger Sturmey Department of Biology rgs102@york.ac.uk Background Definitions
Mass Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System
 GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes
 Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes The PRIME lab and AMS Importance of protein complexes in biology Methods for isolation of protein complexes In
Really high sensitivity mass spectrometry and Discovery and analysis of protein complexes The PRIME lab and AMS Importance of protein complexes in biology Methods for isolation of protein complexes In
Thermo Scientific Peptide Mapping Workflows. Upgrade Your Maps. Fast, confident and more reliable peptide mapping.
 Thermo Scientific Peptide Mapping Workflows Upgrade Your Maps Fast, confident and more reliable peptide mapping. Smarter Navigation... Peptide mapping is a core analytic in biotherapeutic development.
Thermo Scientific Peptide Mapping Workflows Upgrade Your Maps Fast, confident and more reliable peptide mapping. Smarter Navigation... Peptide mapping is a core analytic in biotherapeutic development.
2-Dimensional Electrophoresis
 2-Dimensional Electrophoresis Proteomics Pathway Proteomics Pathway History of 2-DE 1956 -paper & starch gel 1975 -Coupling of IEF & SDS-PAGE IPG-IEF 2DE-Techniques Immobilized ph Gradient IsoElectric
2-Dimensional Electrophoresis Proteomics Pathway Proteomics Pathway History of 2-DE 1956 -paper & starch gel 1975 -Coupling of IEF & SDS-PAGE IPG-IEF 2DE-Techniques Immobilized ph Gradient IsoElectric
CaptiveSpray nanobooster
 CaptiveSpray nanobooster The Revolution in Proteomics Ionization Innovation with Integrity LC-MS Source The Key to Performance and Reliability for nano flow MS Bruker CaptiveSpray principle: Stable and
CaptiveSpray nanobooster The Revolution in Proteomics Ionization Innovation with Integrity LC-MS Source The Key to Performance and Reliability for nano flow MS Bruker CaptiveSpray principle: Stable and
METHODS IN CELL BIOLOGY EXAM II, MARCH 26, 2008
 NAME KEY METHODS IN CELL BIOLOGY EXAM II, MARCH 26, 2008 1. DEFINITIONS (30 points). Briefly (1-3 sentences, phrases, word, etc.) define the following terms or answer question. A. depot effect refers to
NAME KEY METHODS IN CELL BIOLOGY EXAM II, MARCH 26, 2008 1. DEFINITIONS (30 points). Briefly (1-3 sentences, phrases, word, etc.) define the following terms or answer question. A. depot effect refers to
Protein Methods. Second Edition. DANIEL M. BOLLAG Merck Research Laboratories West Point, Pennsylvania
 Protein Methods Second Edition DANIEL M. BOLLAG Merck Research Laboratories West Point, Pennsylvania MICHAEL D. ROZYCKI Department of Chemistry The Henry H. Hoyt Laboratory Princeton University Princeton,
Protein Methods Second Edition DANIEL M. BOLLAG Merck Research Laboratories West Point, Pennsylvania MICHAEL D. ROZYCKI Department of Chemistry The Henry H. Hoyt Laboratory Princeton University Princeton,
Overview. Tools for Protein Sample Preparation, 2-D Electrophoresis, and Imaging and Analysis
 Expression Proteomics // Tools for Protein Separation and Analysis www.expressionproteomics.com 1 2 3 4 Overview Tools for Protein Sample Preparation, 2-D Electrophoresis, and Imaging and Analysis overview
Expression Proteomics // Tools for Protein Separation and Analysis www.expressionproteomics.com 1 2 3 4 Overview Tools for Protein Sample Preparation, 2-D Electrophoresis, and Imaging and Analysis overview
Biotherapeutic Non-Reduced Peptide Mapping
 Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
Quantitative mass spec based proteomics
 Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
Spectrum Mill MS Proteomics Workbench. Comprehensive tools for MS proteomics
 Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
Assessment of Active Biopharmaceutical Ingredients Prior To and Following Removal of Interfering Excipients
 Assessment of Active Biopharmaceutical Ingredients Prior To and Following Removal of Interfering Excipients Andrew J Reason and Howard R Morris BioPharmaSpec Ltd, and BioPharmaSpec Inc, The EMA guideline
Assessment of Active Biopharmaceutical Ingredients Prior To and Following Removal of Interfering Excipients Andrew J Reason and Howard R Morris BioPharmaSpec Ltd, and BioPharmaSpec Inc, The EMA guideline
Chapter 3-II Protein Structure and Function
 Chapter 3-II Protein Structure and Function GBME, SKKU Molecular & Cell Biology H.F.K. Active site of the enzyme trypsin. Enzymes (proteins or RNAs) catalyze making or breaking substrate covalent bonds.
Chapter 3-II Protein Structure and Function GBME, SKKU Molecular & Cell Biology H.F.K. Active site of the enzyme trypsin. Enzymes (proteins or RNAs) catalyze making or breaking substrate covalent bonds.
Proteomics software at MSI. Pratik Jagtap Minnesota Supercomputing institute
 Proteomics software at MSI. Pratik Jagtap Minnesota Supercomputing institute http://www.mass.msi.umn.edu/ Proteomics software at MSI. proteomics : emerging technology proteomics workflow search algorithms
Proteomics software at MSI. Pratik Jagtap Minnesota Supercomputing institute http://www.mass.msi.umn.edu/ Proteomics software at MSI. proteomics : emerging technology proteomics workflow search algorithms
PROTEOMICS IN DRUG TARGET DISCOVERY
 : 19/4/07 19:13 Page 43 PROTEOMICS IN DRUG TARGET DISCOVERY High-throughput meets high-efficiency Biochemistry is enjoying a renaissance under the guise of proteomics due to the availability of sequenced
: 19/4/07 19:13 Page 43 PROTEOMICS IN DRUG TARGET DISCOVERY High-throughput meets high-efficiency Biochemistry is enjoying a renaissance under the guise of proteomics due to the availability of sequenced
BIL 256 Cell and Molecular Biology Lab Spring, Molecular Weight Determination: SDS Electrophoresis
 BIL 256 Cell and Molecular Biology Lab Spring, 2007 Molecular Weight Determination: SDS Electrophoresis Separation of Proteins by Electrophoresis A. Separation by Charge All polypeptide chains contain
BIL 256 Cell and Molecular Biology Lab Spring, 2007 Molecular Weight Determination: SDS Electrophoresis Separation of Proteins by Electrophoresis A. Separation by Charge All polypeptide chains contain
ARE YOU LOOKING FOR 1-D OR 2-D HIGH RESOLOTION GEL ELECTROPHORESIS SYSTEM, WE HAVE A SOLUTION
 ARE YOU LOOKING FOR 1-D OR 2-D HIGH RESOLOTION GEL ELECTROPHORESIS SYSTEM, WE HAVE A SOLUTION We are pleased to inform that Unigenetics Instruments Pvt. Ltd. now is a distributors of WITA GmbH, Germany
ARE YOU LOOKING FOR 1-D OR 2-D HIGH RESOLOTION GEL ELECTROPHORESIS SYSTEM, WE HAVE A SOLUTION We are pleased to inform that Unigenetics Instruments Pvt. Ltd. now is a distributors of WITA GmbH, Germany
Peptide enrichment and fractionation
 Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Quantification of Host Cell Protein Impurities Using the Agilent 1290 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System
 Application Note Biotherapeutics Quantification of Host Cell Protein Impurities Using the Agilent 9 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System Authors Linfeng Wu and Yanan Yang
Application Note Biotherapeutics Quantification of Host Cell Protein Impurities Using the Agilent 9 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System Authors Linfeng Wu and Yanan Yang
392 Index. peptide N-glycosidase F. treatment, 331
 Index 391 Index A Albumins, seed protein extraction, 20, 21, 23 Antibody screening, see Protein microarray Arabidopsis thaliana, see Chloroplast; Plasma membrane B Blue-native polyacrylamide gel (BN-PAGE),
Index 391 Index A Albumins, seed protein extraction, 20, 21, 23 Antibody screening, see Protein microarray Arabidopsis thaliana, see Chloroplast; Plasma membrane B Blue-native polyacrylamide gel (BN-PAGE),
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore, India
 Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1144622/dc1 Supporting Online Material for Genome Transplantation in Bacteria: Changing One Species to Another Carole Lartigue, John I. Glass, * Nina Alperovich, Rembert
www.sciencemag.org/cgi/content/full/1144622/dc1 Supporting Online Material for Genome Transplantation in Bacteria: Changing One Species to Another Carole Lartigue, John I. Glass, * Nina Alperovich, Rembert
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Thursday December 20 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Thursday December 20 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Peptide biomarkers as a way to determine meat authenticity
 Peptide biomarkers as a way to determine meat authenticity Miguel A. Sentandreu Laboratory of Meat Science IATA (CSIC), SPAIN The problem of meat authentication Clear and reliable information about food
Peptide biomarkers as a way to determine meat authenticity Miguel A. Sentandreu Laboratory of Meat Science IATA (CSIC), SPAIN The problem of meat authentication Clear and reliable information about food
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
27041, Week 02. Review of Week 01
 27041, Week 02 Review of Week 01 The human genome sequencing project (HGP) 2 CBS, Department of Systems Biology Systems Biology and emergent properties 3 CBS, Department of Systems Biology Different model
27041, Week 02 Review of Week 01 The human genome sequencing project (HGP) 2 CBS, Department of Systems Biology Systems Biology and emergent properties 3 CBS, Department of Systems Biology Different model
Just launched. Operated by. Karel Harant
 Just launched Operated by Karel Harant 1 Rapid development of instrumentation Decline of MALDI-based instruments 2D protein electrophoresis abandoned 80% of proteomic data acquired in last two years (Somebody,
Just launched Operated by Karel Harant 1 Rapid development of instrumentation Decline of MALDI-based instruments 2D protein electrophoresis abandoned 80% of proteomic data acquired in last two years (Somebody,
Purification: Step 1. Lecture 11 Protein and Peptide Chemistry. Cells: Break them open! Crude Extract
 Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1. Protein and Peptide Chemistry. Lecture 11. Big Problem: Crude extract is not the natural environment. Cells: Break them open!
 Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
Genomics, Transcriptomics and Proteomics
 Genomics, Transcriptomics and Proteomics GENES ARNm PROTEINS Genomics Transcriptomics Proteomics (10 6 protein forms?) Maturation PTMs Partners Localisation EDyP Lab : «Exploring the Dynamics of Proteomes»
Genomics, Transcriptomics and Proteomics GENES ARNm PROTEINS Genomics Transcriptomics Proteomics (10 6 protein forms?) Maturation PTMs Partners Localisation EDyP Lab : «Exploring the Dynamics of Proteomes»
N-Glycan Profiling Analysis of a Monoclonal Antibody Using UHPLC/FLD/Q-TOF
 N-Glycan Profiling Analysis of a Monoclonal Antibody Using UHPLC/FLD/Q-TOF Application Note Authors Xianming Liu, Wei Zhang, Yi Du, Sheng Yin, Hong Que, and Weichang Zhou WuXi AppTec iopharmaceuticals
N-Glycan Profiling Analysis of a Monoclonal Antibody Using UHPLC/FLD/Q-TOF Application Note Authors Xianming Liu, Wei Zhang, Yi Du, Sheng Yin, Hong Que, and Weichang Zhou WuXi AppTec iopharmaceuticals
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins
 Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
