Cornell Probability Summer School 2006
|
|
|
- Nora Henderson
- 6 years ago
- Views:
Transcription
1 Colon Cancer Cornell Probability Summer School 2006 Simon Tavaré Lecture 5 Stem Cells Potten and Loeffler (1990): A small population of relatively undifferentiated, proliferative cells that maintain their population size when they divide, while at the same time producing progeny that enter a dividing transit population within which further rounds of division occur together with differentiation events which resulted ultimately in the production of functional cell types required of the tissue Embryonic stem cells (ES) enormous division potential can produce all differentiated cell types needed by organism Adult stem cells restricted range of differentiated products (?) Colon crypts Stem cells are undifferentiated cells residing in a specific location (niche) in a tissue can produce a variety of somatic cell types needed for tissue renewal produce intermediate (Transit Amplifying) cells that can divide rapidly and differentiate into various types of tissue cell must be maintained, as only they can effect continuous renewal Colon lined with 15 million crypts Problem: no way to identify stem cells
2 Colon crypts How do stem cells maintain their numbers? Model A (Deterministic) Small number of stem cells in niche each generates a single stem cell and a single TA cell on division (asymmetric division) each stem cell is immortal How do stem cells maintain their numbers? Model A (Deterministic) Small number of stem cells in niche each generates a single stem cell and a single TA cell on division each stem cell is immortal How do we distinguish between the two? We need a marker that changes rapidly during cell division Mutations in DNA? Model B (Stochastic) Many stem cells in a niche each stem cell produces 0,1 or 2 stem cells (and 2, 1 or 0 TA cells) on division How do we distinguish between the two? We need a marker that changes rapidly during cell division Mutations in DNA? We use CpG methylation patterns... epigenetic changes that survive mitotic division Methylation CpG islands In human genome, CpG dinucleotides are relatively rare CpG pairs undergo a process called methylation that modifies the C nucleotide A methylated C can (with relatively high probability) mutate to a T Promoter regions are CpG rich These regions are not methylated, and thus mutate less often
3 Fragile X Syndrome Methylation... Causes repression of gene expression CpG islands often located around promoters of housekeeping genes these are not usually methylated Inactive genes often methylated Methylation patterns Methylation patterns vary with time Can be detected by bisulfite sequencing Bisulfite treatment changes unmethylated C into U, but leaves methylated C alone. Sequencing identifies methylated sites as C, unmethylated as T. Data We studied methylation patterns in three genes not expressed in colon crypt cells MYOD1(5),CSX(8) BGN(9) X-chromosome locus, 130 bp island 7 male patients 7 9normalcryptsperperson 8 24 molecules studied per crypt Experimental Method Methylation in a Single Individual
4 A stochastic model Start with N stem cells in crypt Assume constant number N of stem cells after every replication Each cell that is not a stem cell is a TA cell TA cells initiate independent branching processes that grow for a fixed number of replications and then die out The branching mechanism reflects the fact that a crypt contains about 2000 cells A Cannings model Start with N stem cells in crypt X 1,...,X N iid with IP(X i =1)=p, IP(X i =0)=IP(X i =2) Assume a constant number of stem cells after every replication The joint distribution of the numbers ν 1,...,ν N of stem cells copied from stem cells 1,..., N is given by L(ν 1,...,ν N )=L(X 1,...,X N ΣX i = N) Describing methylation patterns After g generations crypt contains a number of cells from which we sample a few for bisulfite treatment, PCR amplification, cloning and sequencing Superimpose effect of changes in methylation during mitotic division Aim: infer something about the number of stem cells, given the observed methylation patterns A number of summary statistics: percent methylation number of unique tags ( alleles ) pairwise difference statistics number of segregating sites Which Model? Reminder: ABC Table 2. Observed and expected variance of unique tags per crypt Stem cell model Observed variance 2 immortal, p cell niche, p 0.95 Variance under model, average (CI) 256-cell niche, p 0.89 CSX A ( ) 0.83 ( ) 1.1 ( ) B ( ) 0.84 ( ) 1.0 ( ) C ( ) 1.0 ( ) 1.3 ( ) D ( ) 0.86 ( ) 1.1 ( ) E ( ) 0.83 ( ) 1.1 ( ) F ( ) 0.94 ( ) 1.2 ( ) G ( ) 0.83 ( ) 0.98 ( ) H ( ) 0.81 ( ) 1.0 ( ) I ( ) 0.99 ( ) 1.2 ( ) BGN D (0 0.33) 0.63 ( ) 0.79 ( ) F (0 0.21) 0.80 ( ) 1.0 ( ) H (0 0.14) 0.75 ( ) 0.90 ( ) I (0 0.24) 0.68 ( ) 0.84 ( ) M (0 0.21) 0.67 ( ) 0.90 ( ) Can simulate (forwards) from this model easily, so... Simulate θ from prior π Simulate data D sim from model with parameter θ Accept θ if d(d sim, D) is small Start over The art is in choosing summary statistics
5 ABC Approach Back to crypts... Priors: N U(1, 100),P U(0.9, 1.0),µ U(10 5, ) Priors: N U(1, 100),P U(0.9, 1.0),µ U(10 5, ) Summary statistics: number of unique tags, percent methylation, mean distance, number of segregating sites Back to crypts... Back to crypts... Priors: N U(1, 100),P U(0.9, 1.0),µ U(10 5, ) Summary statistics: number of unique tags, percent methylation, mean distance, number of segregating sites What is close? Small relative error: d = s i,sim s i,obs s i,obs +1 Priors: N U(1, 100),P U(0.9, 1.0),µ U(10 5, ) Summary statistics: number of unique tags, percent methylation, mean distance, number of segregating sites What is close? Small relative error d = s i,sim s i,obs s i,obs +1 Small run 755 points Posterior for N Posterior for P
6 Pierre Nicolas, INRA A Continuous-time Model Say a stem cell dies if it is replaced by two TA cells Life span of a stem cell is Exponential, mean 1/γ When cell dies, another stem cell having two stem cell offspring is copied to replace it The genealogy of stem cells looks like a coalescent Crypt contains N equal-sized subpopulations, each the progeny of a single stem cell A pair of stem cells coalesces at rate 2γ N 1 Modelling Methylation Patterns All islands unmethylated at birth of individual Independent sites model µ =(µ m,µ u ) methylation rates Context-dependent model methylation/demethylation events occur at rate that depends on number of methylated sites ɛ sequencing error rate per site per molecule Genealogy of TA cells Descendants of a Stem Cell TA cells have small, fixed number of divisions (g) Time scale of process expressed in arbitrary units g=5 Rate η of methylation process relative to time scale of TA part stem cell stage 1 stage 2 stage 3 stage 4 cells from a same stem cell progeny stage 5 dead cells removed from the crypt Take η µ Genealogy modeled as a coalescent with expansion Star-like Genealogy of Sample Parameterization and Priors N uniform g=5 λ = γ N 1 λ 1 uniform ν = µ/λ each component is log-normal(0,σ) σ exponential, mean 1 η = αν α exponential, mean 1 g uniform ɛ U(0,1)
7 MCMC algorithm Find posterior of θ =(N,λ,g,σ,ν,α,ɛ) given methylation patterns X And then a miracle occurs! MCMC Non-stationary coalescent Augmented state space: (θ, Λ,Y ) Λ is collection of genealogies of methylation patterns Y denotes methylation patterns in nodes of Λ Updating N is hard embed model in one where N is allowed to vary by ±1 between crypts Simulated Dataset I Moves around Λ via Wilson/Balding (avoids peeling) the values in Y are used to propose changes Many different types of updates are combined in this approach Run for 5,000,000 iterations, and record (θ, Λ,Y ) every 100 steps after first 500,000 No apparent convergence problems PCR: 10 days per run density N= N Simulated Dataset II Predictive assessment of model fitness density N=24 Five within-crypt statistics: number of distinct patterns number of polymorphic sites average distance between patterns number of unmethylated patterns number of singletons N Compare distribution of inter-crypt average and standard deviation with actual values
8 # distinct patterns, polymorphic sites Intercrypt average Intercrypt sd Independent methylation process ave dist, # unmethylated, # singletons Dependent methylation process # distinct patterns, polymorphic sites ave dist, # unmethylated, # singletons Intercrypt average Intercrypt sd
9 Patient X Robustness # distinct patterns, polymorphic sites ave dist, # unmethylated, # singletons Intercrypt average Intercrypt sd Posteriors: shape of genealogy Posteriors Density N Density Density g
10 Posteriors: polymorphism, given genealogy Current work µ 2.0 Nb of methylated sites Density Density Generating much bigger data sets more CpG islands, different lengths experimental issues e.g. PCR errors Getting data from other tissue types have endometrium, small intestine, hair doing blood, brain, heart Develop better markers? Model spatial structure in crypts Inference about crypts: ABC approach References Nicolas P, Shibata D & Tavaré S. Posterior inference on the stem cell population of the human colon crypt through analysis of methylation patterns. In preparation. Shibata D & Tavaré S. Counting divisions in a human somatic cell tree: how, what and why. Cell Cycle, 5, , Kim JY, Tavaré S & Shibata D. Counting human somatic cell replications: Methylation mirrors human endometrial stem cell divisions. Proc Natl Acad Sci USA, 102, , Kim JY, Siegmund KD, Tavaré S&ShibataD. Age-related human small intestine methylation: evidence for stem cell niches. BMC Medicine. 3:10, Calabrese P, Mecklin JP, Järvinen HJ, Aaltonen LA, Tavaré S & Shibata D. Numbers of mutations to different types of colorectal cancer. BMC Cancer, 5:126, Calabrese P, Tavaré S & Shibata D. Pre-tumor progression: clonal evolution of human stem cell populations. Am. J. Pathol., 164, , 2004.
On polyclonality of intestinal tumors
 Michael A. University of Wisconsin Chaos and Complex Systems April 2006 Thanks Linda Clipson W.F. Dove Rich Halberg Stephen Stanhope Ruth Sullivan Andrew Thliveris Outline Bio Three statistical questions
Michael A. University of Wisconsin Chaos and Complex Systems April 2006 Thanks Linda Clipson W.F. Dove Rich Halberg Stephen Stanhope Ruth Sullivan Andrew Thliveris Outline Bio Three statistical questions
CS262 Lecture 12 Notes Single Cell Sequencing Jan. 11, 2016
 CS262 Lecture 12 Notes Single Cell Sequencing Jan. 11, 2016 Background A typical human cell consists of ~6 billion base pairs of DNA and ~600 million bases of mrna. It is time-consuming and expensive to
CS262 Lecture 12 Notes Single Cell Sequencing Jan. 11, 2016 Background A typical human cell consists of ~6 billion base pairs of DNA and ~600 million bases of mrna. It is time-consuming and expensive to
Characterization of Allele-Specific Copy Number in Tumor Genomes
 Characterization of Allele-Specific Copy Number in Tumor Genomes Hao Chen 2 Haipeng Xing 1 Nancy R. Zhang 2 1 Department of Statistics Stonybrook University of New York 2 Department of Statistics Stanford
Characterization of Allele-Specific Copy Number in Tumor Genomes Hao Chen 2 Haipeng Xing 1 Nancy R. Zhang 2 1 Department of Statistics Stonybrook University of New York 2 Department of Statistics Stanford
GATCGTGCACGATCTCGGCAATTCGGGATGCCGGCTCGTCACCGGTCGCT
 Problem. (pts) A. (5pts) Your colleague professor Eugene Mathew Lateed generated a genome-wide DNA methylation map for normal colon cells using MRE-seq and MeDIP-seq. In an intergenic region, he found
Problem. (pts) A. (5pts) Your colleague professor Eugene Mathew Lateed generated a genome-wide DNA methylation map for normal colon cells using MRE-seq and MeDIP-seq. In an intergenic region, he found
12/8/09 Comp 590/Comp Fall
 12/8/09 Comp 590/Comp 790-90 Fall 2009 1 One of the first, and simplest models of population genealogies was introduced by Wright (1931) and Fisher (1930). Model emphasizes transmission of genes from one
12/8/09 Comp 590/Comp 790-90 Fall 2009 1 One of the first, and simplest models of population genealogies was introduced by Wright (1931) and Fisher (1930). Model emphasizes transmission of genes from one
APPLICATION NOTE
 APPLICATION NOTE www.swiftbiosci.com NGS Library Preparation Produces Balanced, Comprehensive Methylome Coverage from Low Input Quantities ABSTRACT Next-generation sequencing (NGS) of bisulfite-converted
APPLICATION NOTE www.swiftbiosci.com NGS Library Preparation Produces Balanced, Comprehensive Methylome Coverage from Low Input Quantities ABSTRACT Next-generation sequencing (NGS) of bisulfite-converted
I See Dead People: Gene Mapping Via Ancestral Inference
 I See Dead People: Gene Mapping Via Ancestral Inference Paul Marjoram, 1 Lada Markovtsova 2 and Simon Tavaré 1,2,3 1 Department of Preventive Medicine, University of Southern California, 1540 Alcazar Street,
I See Dead People: Gene Mapping Via Ancestral Inference Paul Marjoram, 1 Lada Markovtsova 2 and Simon Tavaré 1,2,3 1 Department of Preventive Medicine, University of Southern California, 1540 Alcazar Street,
Identifying CpG islands using hidden Markov models
 Identifying CpG islands using hidden Markov models Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Fall 2016 M. Macauley (Clemson)
Identifying CpG islands using hidden Markov models Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Fall 2016 M. Macauley (Clemson)
Identifying CpG islands using hidden Markov models
 Identifying CpG islands using hidden Markov models Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Spring 2017 M. Macauley (Clemson)
Identifying CpG islands using hidden Markov models Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Spring 2017 M. Macauley (Clemson)
Statistical Methods for Quantitative Trait Loci (QTL) Mapping
 Statistical Methods for Quantitative Trait Loci (QTL) Mapping Lectures 4 Oct 10, 011 CSE 57 Computational Biology, Fall 011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 1:00-1:0 Johnson
Statistical Methods for Quantitative Trait Loci (QTL) Mapping Lectures 4 Oct 10, 011 CSE 57 Computational Biology, Fall 011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 1:00-1:0 Johnson
Lecture 9. Eukaryotic gene regulation: DNA METHYLATION
 Lecture 9 Eukaryotic gene regulation: DNA METHYLATION Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Lecture 9 Eukaryotic gene regulation: DNA METHYLATION Recap.. Eukaryotic RNA polymerases Core promoter elements General transcription factors Enhancers and upstream activation sequences Transcriptional
Human SNP haplotypes. Statistics 246, Spring 2002 Week 15, Lecture 1
 Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Undergraduate research and graduate school so far
 Undergraduate research and graduate school so far Jason Xu, Department of Statistics ACMS Seminar Jan 14, 2016 Why undergraduate research? First of all, it s fun Classroom assignments can feel boring and
Undergraduate research and graduate school so far Jason Xu, Department of Statistics ACMS Seminar Jan 14, 2016 Why undergraduate research? First of all, it s fun Classroom assignments can feel boring and
Quantitative analysis of methylation at multiple CpG sites by Pyrosequencing TM
 Quantitative analysis of methylation at multiple CpG sites by Pyrosequencing TM Robert England and Monica Pettersson Pyrosequencing from Biotage 1 improves virtually every aspect of CpG methylation analysis:
Quantitative analysis of methylation at multiple CpG sites by Pyrosequencing TM Robert England and Monica Pettersson Pyrosequencing from Biotage 1 improves virtually every aspect of CpG methylation analysis:
Genetic Basis of Development & Biotechnologies
 Genetic Basis of Development & Biotechnologies 1. Steps of embryonic development: cell division, morphogenesis, differentiation Totipotency and pluripotency 2. Plant cloning 3. Animal cloning Reproductive
Genetic Basis of Development & Biotechnologies 1. Steps of embryonic development: cell division, morphogenesis, differentiation Totipotency and pluripotency 2. Plant cloning 3. Animal cloning Reproductive
Introduction to Quantitative Genomics / Genetics
 Introduction to Quantitative Genomics / Genetics BTRY 7210: Topics in Quantitative Genomics and Genetics September 10, 2008 Jason G. Mezey Outline History and Intuition. Statistical Framework. Current
Introduction to Quantitative Genomics / Genetics BTRY 7210: Topics in Quantitative Genomics and Genetics September 10, 2008 Jason G. Mezey Outline History and Intuition. Statistical Framework. Current
Measurement of Molecular Genetic Variation. Forces Creating Genetic Variation. Mutation: Nucleotide Substitutions
 Measurement of Molecular Genetic Variation Genetic Variation Is The Necessary Prerequisite For All Evolution And For Studying All The Major Problem Areas In Molecular Evolution. How We Score And Measure
Measurement of Molecular Genetic Variation Genetic Variation Is The Necessary Prerequisite For All Evolution And For Studying All The Major Problem Areas In Molecular Evolution. How We Score And Measure
Chapter 15 Gene Technologies and Human Applications
 Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
Why do we need statistics to study genetics and evolution?
 Why do we need statistics to study genetics and evolution? 1. Mapping traits to the genome [Linkage maps (incl. QTLs), LOD] 2. Quantifying genetic basis of complex traits [Concordance, heritability] 3.
Why do we need statistics to study genetics and evolution? 1. Mapping traits to the genome [Linkage maps (incl. QTLs), LOD] 2. Quantifying genetic basis of complex traits [Concordance, heritability] 3.
Population Genetics II. Bio
 Population Genetics II. Bio5488-2016 Don Conrad dconrad@genetics.wustl.edu Agenda Population Genetic Inference Mutation Selection Recombination The Coalescent Process ACTT T G C G ACGT ACGT ACTT ACTT AGTT
Population Genetics II. Bio5488-2016 Don Conrad dconrad@genetics.wustl.edu Agenda Population Genetic Inference Mutation Selection Recombination The Coalescent Process ACTT T G C G ACGT ACGT ACTT ACTT AGTT
HISTORICAL LINGUISTICS AND MOLECULAR ANTHROPOLOGY
 Third Pavia International Summer School for Indo-European Linguistics, 7-12 September 2015 HISTORICAL LINGUISTICS AND MOLECULAR ANTHROPOLOGY Brigitte Pakendorf, Dynamique du Langage, CNRS & Université
Third Pavia International Summer School for Indo-European Linguistics, 7-12 September 2015 HISTORICAL LINGUISTICS AND MOLECULAR ANTHROPOLOGY Brigitte Pakendorf, Dynamique du Langage, CNRS & Université
Recombination, and haplotype structure
 2 The starting point We have a genome s worth of data on genetic variation Recombination, and haplotype structure Simon Myers, Gil McVean Department of Statistics, Oxford We wish to understand why the
2 The starting point We have a genome s worth of data on genetic variation Recombination, and haplotype structure Simon Myers, Gil McVean Department of Statistics, Oxford We wish to understand why the
DNA METHYLATION RESEARCH TOOLS
 SeqCap Epi Enrichment System Revolutionize your epigenomic research DNA METHYLATION RESEARCH TOOLS Methylated DNA The SeqCap Epi System is a set of target enrichment tools for DNA methylation assessment
SeqCap Epi Enrichment System Revolutionize your epigenomic research DNA METHYLATION RESEARCH TOOLS Methylated DNA The SeqCap Epi System is a set of target enrichment tools for DNA methylation assessment
Analysis of gene function
 Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
BST227 Introduction to Statistical Genetics. Lecture 3: Introduction to population genetics
 BST227 Introduction to Statistical Genetics Lecture 3: Introduction to population genetics!1 Housekeeping HW1 will be posted on course website tonight 1st lab will be on Wednesday TA office hours have
BST227 Introduction to Statistical Genetics Lecture 3: Introduction to population genetics!1 Housekeeping HW1 will be posted on course website tonight 1st lab will be on Wednesday TA office hours have
Model based inference of mutation rates and selection strengths in humans and influenza. Daniel Wegmann University of Fribourg
 Model based inference of mutation rates and selection strengths in humans and influenza Daniel Wegmann University of Fribourg Influenza rapidly evolved resistance against novel drugs Weinstock & Zuccotti
Model based inference of mutation rates and selection strengths in humans and influenza Daniel Wegmann University of Fribourg Influenza rapidly evolved resistance against novel drugs Weinstock & Zuccotti
HST.161 Molecular Biology and Genetics in Modern Medicine Fall 2007
 MIT OpenCourseWare http://ocw.mit.edu HST.161 Molecular Biology and Genetics in Modern Medicine Fall 2007 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
MIT OpenCourseWare http://ocw.mit.edu HST.161 Molecular Biology and Genetics in Modern Medicine Fall 2007 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
Pyrosequencing for quantitative analysis of methylation at multiple CpG sites
 Application Note for Nature Methods Pyrosequencing for quantitative analysis of methylation at multiple CpG sites Robert England and Monica Pettersson Pyrosequencing from Biotage 1 improves virtually every
Application Note for Nature Methods Pyrosequencing for quantitative analysis of methylation at multiple CpG sites Robert England and Monica Pettersson Pyrosequencing from Biotage 1 improves virtually every
Summary. Introduction
 doi: 10.1111/j.1469-1809.2006.00305.x Variation of Estimates of SNP and Haplotype Diversity and Linkage Disequilibrium in Samples from the Same Population Due to Experimental and Evolutionary Sample Size
doi: 10.1111/j.1469-1809.2006.00305.x Variation of Estimates of SNP and Haplotype Diversity and Linkage Disequilibrium in Samples from the Same Population Due to Experimental and Evolutionary Sample Size
B) You can conclude that A 1 is identical by descent. Notice that A2 had to come from the father (and therefore, A1 is maternal in both cases).
 Homework questions. Please provide your answers on a separate sheet. Examine the following pedigree. A 1,2 B 1,2 A 1,3 B 1,3 A 1,2 B 1,2 A 1,2 B 1,3 1. (1 point) The A 1 alleles in the two brothers are
Homework questions. Please provide your answers on a separate sheet. Examine the following pedigree. A 1,2 B 1,2 A 1,3 B 1,3 A 1,2 B 1,2 A 1,2 B 1,3 1. (1 point) The A 1 alleles in the two brothers are
Experimental validation of candidates of tissuespecific and CpG-island-mediated alternative polyadenylation in mouse
 Karin Fleischhanderl; Martina Fondi Experimental validation of candidates of tissuespecific and CpG-island-mediated alternative polyadenylation in mouse 108 - Biotechnologie Abstract --- Keywords: Alternative
Karin Fleischhanderl; Martina Fondi Experimental validation of candidates of tissuespecific and CpG-island-mediated alternative polyadenylation in mouse 108 - Biotechnologie Abstract --- Keywords: Alternative
Nature Genetics: doi: /ng.3254
 Supplementary Figure 1 Comparing the inferred histories of the stairway plot and the PSMC method using simulated samples based on five models. (a) PSMC sim-1 model. (b) PSMC sim-2 model. (c) PSMC sim-3
Supplementary Figure 1 Comparing the inferred histories of the stairway plot and the PSMC method using simulated samples based on five models. (a) PSMC sim-1 model. (b) PSMC sim-2 model. (c) PSMC sim-3
Exome Sequencing Exome sequencing is a technique that is used to examine all of the protein-coding regions of the genome.
 Glossary of Terms Genetics is a term that refers to the study of genes and their role in inheritance the way certain traits are passed down from one generation to another. Genomics is the study of all
Glossary of Terms Genetics is a term that refers to the study of genes and their role in inheritance the way certain traits are passed down from one generation to another. Genomics is the study of all
Computational Models for Cell Reprogramming
 Computational Models for Cell Reprogramming Computational Models for Cell Reprogramming 1 Pluripotent Cells EuroStemCell web page Computational Models for Cell Reprogramming 2 Early Embryonic Development
Computational Models for Cell Reprogramming Computational Models for Cell Reprogramming 1 Pluripotent Cells EuroStemCell web page Computational Models for Cell Reprogramming 2 Early Embryonic Development
Personal and population genomics of human regulatory variation
 Personal and population genomics of human regulatory variation Benjamin Vernot,, and Joshua M. Akey Department of Genome Sciences, University of Washington, Seattle, Washington 98195, USA Ting WANG 1 Brief
Personal and population genomics of human regulatory variation Benjamin Vernot,, and Joshua M. Akey Department of Genome Sciences, University of Washington, Seattle, Washington 98195, USA Ting WANG 1 Brief
c) Assuming he does not run another endurance race, will the steady-state populations be affected one year later? If so, explain how.
 LS1a Fall 06 Problem Set #8 (100 points total) all questions including the (*extra*) one should be turned in 1. (18 points) Erythrocytes, mature red blood cells, are essential for transporting oxygen to
LS1a Fall 06 Problem Set #8 (100 points total) all questions including the (*extra*) one should be turned in 1. (18 points) Erythrocytes, mature red blood cells, are essential for transporting oxygen to
Population Genetics (Learning Objectives)
 Population Genetics (Learning Objectives) Define the terms population, species, allelic and genotypic frequencies, gene pool, and fixed allele, genetic drift, bottle-neck effect, founder effect. Explain
Population Genetics (Learning Objectives) Define the terms population, species, allelic and genotypic frequencies, gene pool, and fixed allele, genetic drift, bottle-neck effect, founder effect. Explain
SLiM: Simulating Evolution with Selection and Linkage
 Genetics: Early Online, published on May 24, 2013 as 10.1534/genetics.113.152181 SLiM: Simulating Evolution with Selection and Linkage Philipp W. Messer Department of Biology, Stanford University, Stanford,
Genetics: Early Online, published on May 24, 2013 as 10.1534/genetics.113.152181 SLiM: Simulating Evolution with Selection and Linkage Philipp W. Messer Department of Biology, Stanford University, Stanford,
BST227 Introduction to Statistical Genetics. Lecture 3: Introduction to population genetics
 BST227 Introduction to Statistical Genetics Lecture 3: Introduction to population genetics 1 Housekeeping HW1 due on Wednesday TA office hours today at 5:20 - FXB G11 What have we studied Background Structure
BST227 Introduction to Statistical Genetics Lecture 3: Introduction to population genetics 1 Housekeeping HW1 due on Wednesday TA office hours today at 5:20 - FXB G11 What have we studied Background Structure
RecQ Helicases and GI Cancers. Mark Derleth, R3 Supervisor: Bill Grady
 RecQ Helicases and GI Cancers Mark Derleth, R3 Supervisor: Bill Grady Learn to perform and interpret several lab assays including PCR, MSP, and Bisulfite sequencing Personal Goal Research Goal To examine
RecQ Helicases and GI Cancers Mark Derleth, R3 Supervisor: Bill Grady Learn to perform and interpret several lab assays including PCR, MSP, and Bisulfite sequencing Personal Goal Research Goal To examine
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Supplementary Figure 1 Workflow for multimodal analysis using sc-gem on a programmable microfluidic device (Fluidigm). 1) Cells are captured and lysed, 2) RNA from lysed single cell is reverse-transcribed
Overview of Human Genetics
 Overview of Human Genetics 1 Structure and function of nucleic acids. 2 Structure and composition of the human genome. 3 Mendelian genetics. Lander et al. (Nature, 2001) MAT 394 (ASU) Human Genetics Spring
Overview of Human Genetics 1 Structure and function of nucleic acids. 2 Structure and composition of the human genome. 3 Mendelian genetics. Lander et al. (Nature, 2001) MAT 394 (ASU) Human Genetics Spring
Supplementary Tables. Primers and probes. Target Primer / probe sequences Chemistry Human HBB F: AACTGTGTTCACTAGCAACCTCAAA
 Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Supplementary Tables Primers and probes Target Primer / probe sequences Chemistry H F: AACTGTGTTCACTAGCAACCTCAAA promoter R: ACAGGGCAGTAACGGCAGACT H Downstream Actb alpha (162) alpha (162.) (Pro) (16)
Great Ideas of Biology
 Great Ideas of Biology Lecture 3 Alan Mortimer PhD Molecular Biology The Great Idea Darwinian Evolution Darwin proposed a theory of evolution based on: Species overproduce There occurs variation in the
Great Ideas of Biology Lecture 3 Alan Mortimer PhD Molecular Biology The Great Idea Darwinian Evolution Darwin proposed a theory of evolution based on: Species overproduce There occurs variation in the
7.03 Final Exam. TA: Alex Bagley Alice Chi Dave Harris Max Juchheim Doug Mills Rishi Puram Bethany Redding Nate Young
 7.03 Final Exam Name: TA: Alex Bagley Alice Chi Dave Harris Max Juchheim Doug Mills Rishi Puram Bethany Redding Nate Young Section time: There are 13 pages including this cover page Please write your name
7.03 Final Exam Name: TA: Alex Bagley Alice Chi Dave Harris Max Juchheim Doug Mills Rishi Puram Bethany Redding Nate Young Section time: There are 13 pages including this cover page Please write your name
Exam 3 4/25/07. Total of 7 questions, 100 points.
 Exam 3 4/25/07 BISC 4A P. Sengupta Total of 7 questions, 100 points. QUESTION 1. Circle the correct answer. Total of 40 points 4 points each. 1. Which of the following is typically attacked by the antibody-mediated
Exam 3 4/25/07 BISC 4A P. Sengupta Total of 7 questions, 100 points. QUESTION 1. Circle the correct answer. Total of 40 points 4 points each. 1. Which of the following is typically attacked by the antibody-mediated
Stem Cells & Neurological Disorders. Said Ismail Faculty of Medicine University of Jordan
 Stem Cells & Neurological Disorders Said Ismail Faculty of Medicine University of Jordan Outline: - Introduction - Types & Potency of Stem Cells - Embryonic Stem Cells - Adult Stem Cells - ipscs -Tissue
Stem Cells & Neurological Disorders Said Ismail Faculty of Medicine University of Jordan Outline: - Introduction - Types & Potency of Stem Cells - Embryonic Stem Cells - Adult Stem Cells - ipscs -Tissue
Bio 311 Learning Objectives
 Bio 311 Learning Objectives This document outlines the learning objectives for Biol 311 (Principles of Genetics). Biol 311 is part of the BioCore within the Department of Biological Sciences; therefore,
Bio 311 Learning Objectives This document outlines the learning objectives for Biol 311 (Principles of Genetics). Biol 311 is part of the BioCore within the Department of Biological Sciences; therefore,
BIOINF/BENG/BIMM/CHEM/CSE 184: Computational Molecular Biology. Lecture 2: Microarray analysis
 BIOINF/BENG/BIMM/CHEM/CSE 184: Computational Molecular Biology Lecture 2: Microarray analysis Genome wide measurement of gene transcription using DNA microarray Bruce Alberts, et al., Molecular Biology
BIOINF/BENG/BIMM/CHEM/CSE 184: Computational Molecular Biology Lecture 2: Microarray analysis Genome wide measurement of gene transcription using DNA microarray Bruce Alberts, et al., Molecular Biology
Simple haplotype analyses in R
 Simple haplotype analyses in R Benjamin French, PhD Department of Biostatistics and Epidemiology University of Pennsylvania bcfrench@upenn.edu user! 2011 University of Warwick 18 August 2011 Goals To integrate
Simple haplotype analyses in R Benjamin French, PhD Department of Biostatistics and Epidemiology University of Pennsylvania bcfrench@upenn.edu user! 2011 University of Warwick 18 August 2011 Goals To integrate
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Sample to Insight. Dr. Bhagyashree S. Birla NGS Field Application Scientist
 Dr. Bhagyashree S. Birla NGS Field Application Scientist bhagyashree.birla@qiagen.com NGS spans a broad range of applications DNA Applications Human ID Liquid biopsy Biomarker discovery Inherited and somatic
Dr. Bhagyashree S. Birla NGS Field Application Scientist bhagyashree.birla@qiagen.com NGS spans a broad range of applications DNA Applications Human ID Liquid biopsy Biomarker discovery Inherited and somatic
NimbleGen Arrays and LightCycler 480 System: A Complete Workflow for DNA Methylation Biomarker Discovery and Validation.
 Cancer Research Application Note No. 9 NimbleGen Arrays and LightCycler 480 System: A Complete Workflow for DNA Methylation Biomarker Discovery and Validation Tomasz Kazimierz Wojdacz, PhD Institute of
Cancer Research Application Note No. 9 NimbleGen Arrays and LightCycler 480 System: A Complete Workflow for DNA Methylation Biomarker Discovery and Validation Tomasz Kazimierz Wojdacz, PhD Institute of
Chapter 14: Genes in Action
 Chapter 14: Genes in Action Section 1: Mutation and Genetic Change Mutation: Nondisjuction: a failure of homologous chromosomes to separate during meiosis I or the failure of sister chromatids to separate
Chapter 14: Genes in Action Section 1: Mutation and Genetic Change Mutation: Nondisjuction: a failure of homologous chromosomes to separate during meiosis I or the failure of sister chromatids to separate
Concepts: What are RFLPs and how do they act like genetic marker loci?
 Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
Complete Sample to Analysis Solutions for DNA Methylation Discovery using Next Generation Sequencing
 Complete Sample to Analysis Solutions for DNA Methylation Discovery using Next Generation Sequencing SureSelect Human/Mouse Methyl-Seq Kyeong Jeong PhD February 5, 2013 CAG EMEAI DGG/GSD/GFO Agilent Restricted
Complete Sample to Analysis Solutions for DNA Methylation Discovery using Next Generation Sequencing SureSelect Human/Mouse Methyl-Seq Kyeong Jeong PhD February 5, 2013 CAG EMEAI DGG/GSD/GFO Agilent Restricted
b. (3 points) The expected frequencies of each blood type in the deme if mating is random with respect to variation at this locus.
 NAME EXAM# 1 1. (15 points) Next to each unnumbered item in the left column place the number from the right column/bottom that best corresponds: 10 additive genetic variance 1) a hermaphroditic adult develops
NAME EXAM# 1 1. (15 points) Next to each unnumbered item in the left column place the number from the right column/bottom that best corresponds: 10 additive genetic variance 1) a hermaphroditic adult develops
Personal Genomics Platform White Paper Last Updated November 15, Executive Summary
 Executive Summary Helix is a personal genomics platform company with a simple but powerful mission: to empower every person to improve their life through DNA. Our platform includes saliva sample collection,
Executive Summary Helix is a personal genomics platform company with a simple but powerful mission: to empower every person to improve their life through DNA. Our platform includes saliva sample collection,
Physical Anthropology 1 Milner-Rose
 Physical Anthropology 1 Milner-Rose Chapter 3 Genetics: Reproducing Life and Producing Variation Our Origins By Clark Spencer Larsen Natural Selection operates on the levels of the 1. living, behaving
Physical Anthropology 1 Milner-Rose Chapter 3 Genetics: Reproducing Life and Producing Variation Our Origins By Clark Spencer Larsen Natural Selection operates on the levels of the 1. living, behaving
Molecular Evolution. COMP Fall 2010 Luay Nakhleh, Rice University
 Molecular Evolution COMP 571 - Fall 2010 Luay Nakhleh, Rice University Outline (1) The neutral theory (2) Measures of divergence and polymorphism (3) DNA sequence divergence and the molecular clock (4)
Molecular Evolution COMP 571 - Fall 2010 Luay Nakhleh, Rice University Outline (1) The neutral theory (2) Measures of divergence and polymorphism (3) DNA sequence divergence and the molecular clock (4)
QTL Mapping, MAS, and Genomic Selection
 QTL Mapping, MAS, and Genomic Selection Dr. Ben Hayes Department of Primary Industries Victoria, Australia A short-course organized by Animal Breeding & Genetics Department of Animal Science Iowa State
QTL Mapping, MAS, and Genomic Selection Dr. Ben Hayes Department of Primary Industries Victoria, Australia A short-course organized by Animal Breeding & Genetics Department of Animal Science Iowa State
CSE /CSE6602E - Soft Computing Winter Lecture 9. Genetic Algorithms & Evolution Strategies. Guest lecturer: Xiangdong An
 CSE3 3./CSE66E - Soft Computing Winter Lecture 9 Genetic Algorithms & Evolution Strategies Guest lecturer: Xiangdong An xan@cs.yorku.ca Genetic algorithms J. Holland, Adaptation in Natural and Artificial
CSE3 3./CSE66E - Soft Computing Winter Lecture 9 Genetic Algorithms & Evolution Strategies Guest lecturer: Xiangdong An xan@cs.yorku.ca Genetic algorithms J. Holland, Adaptation in Natural and Artificial
Supplementary information ATLAS
 Supplementary information ATLAS Vivian Link, Athanasios Kousathanas, Krishna Veeramah, Christian Sell, Amelie Scheu and Daniel Wegmann Section 1: Complete list of functionalities Sequence data processing
Supplementary information ATLAS Vivian Link, Athanasios Kousathanas, Krishna Veeramah, Christian Sell, Amelie Scheu and Daniel Wegmann Section 1: Complete list of functionalities Sequence data processing
Genomic models in bayz
 Genomic models in bayz Luc Janss, Dec 2010 In the new bayz version the genotype data is now restricted to be 2-allelic markers (SNPs), while the modeling option have been made more general. This implements
Genomic models in bayz Luc Janss, Dec 2010 In the new bayz version the genotype data is now restricted to be 2-allelic markers (SNPs), while the modeling option have been made more general. This implements
Mutations during meiosis and germ line division lead to genetic variation between individuals
 Mutations during meiosis and germ line division lead to genetic variation between individuals Types of mutations: point mutations indels (insertion/deletion) copy number variation structural rearrangements
Mutations during meiosis and germ line division lead to genetic variation between individuals Types of mutations: point mutations indels (insertion/deletion) copy number variation structural rearrangements
4.1. Genetics as a Tool in Anthropology
 4.1. Genetics as a Tool in Anthropology Each biological system and every human being is defined by its genetic material. The genetic material is stored in the cells of the body, mainly in the nucleus of
4.1. Genetics as a Tool in Anthropology Each biological system and every human being is defined by its genetic material. The genetic material is stored in the cells of the body, mainly in the nucleus of
Additional Practice Problems for Reading Period
 BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
BS 50 Genetics and Genomics Reading Period Additional Practice Problems for Reading Period Question 1. In patients with a particular type of leukemia, their leukemic B lymphocytes display a translocation
Introducing new DNA into the genome requires cloning the donor sequence, delivery of the cloned DNA into the cell, and integration into the genome.
 Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Stem cells in Development
 ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
Additional levels of regulation
 Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) P. Matthias, May 26th, 2010 Additional levels of regulation Gene organization/localization Allelic exclusion AIRE: a
Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) P. Matthias, May 26th, 2010 Additional levels of regulation Gene organization/localization Allelic exclusion AIRE: a
Genetics Lecture Notes Lectures 6 9
 Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Stem cells in Development
 ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
ANAT 2341 Embryology Lab 10 8 Oct 2009 Therapeutic Use of Stem Cells Practical Hurdles & Ethical Issues Stem cells in Development Blastocyst Cord blood Antonio Lee PhD Neuromuscular & Regenerative Medicine
Lecture 12. Genomics. Mapping. Definition Species sequencing ESTs. Why? Types of mapping Markers p & Types
 Lecture 12 Reading Lecture 12: p. 335-338, 346-353 Lecture 13: p. 358-371 Genomics Definition Species sequencing ESTs Mapping Why? Types of mapping Markers p.335-338 & 346-353 Types 222 omics Interpreting
Lecture 12 Reading Lecture 12: p. 335-338, 346-353 Lecture 13: p. 358-371 Genomics Definition Species sequencing ESTs Mapping Why? Types of mapping Markers p.335-338 & 346-353 Types 222 omics Interpreting
What is Epigenetics? Watch the video
 EPIGENETICS What is Epigenetics? The study of environmental factors on gene expression in DNA. The molecule is called methylation controls when genes are turned on. Methylation turns off genes. Acetylation
EPIGENETICS What is Epigenetics? The study of environmental factors on gene expression in DNA. The molecule is called methylation controls when genes are turned on. Methylation turns off genes. Acetylation
Recombinant DNA recombinant DNA DNA cloning gene cloning
 DNA Technology Recombinant DNA In recombinant DNA, DNA from two different sources, often two species, are combined into the same DNA molecule. DNA cloning permits production of multiple copies of a specific
DNA Technology Recombinant DNA In recombinant DNA, DNA from two different sources, often two species, are combined into the same DNA molecule. DNA cloning permits production of multiple copies of a specific
SUPPLEMENTARY INFORMATION
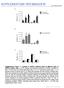 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
UNIT MOLECULAR GENETICS AND BIOTECHNOLOGY
 UNIT MOLECULAR GENETICS AND BIOTECHNOLOGY Standard B-4: The student will demonstrate an understanding of the molecular basis of heredity. B-4.1-4,8,9 Effective June 2008 All Indicators in Standard B-4
UNIT MOLECULAR GENETICS AND BIOTECHNOLOGY Standard B-4: The student will demonstrate an understanding of the molecular basis of heredity. B-4.1-4,8,9 Effective June 2008 All Indicators in Standard B-4
Lesson 7A Specialized Cells, Stem Cells & Cellular Differentiation
 Lesson 7A Specialized Cells, Stem Cells & Cellular Differentiation Learning Goals I can explain the concept of cell differentiation and cell specialization. I can explain how the cell structure relates
Lesson 7A Specialized Cells, Stem Cells & Cellular Differentiation Learning Goals I can explain the concept of cell differentiation and cell specialization. I can explain how the cell structure relates
CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016
 CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016 Topics Genetic variation Population structure Linkage disequilibrium Natural disease variants Genome Wide Association Studies Gene
CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016 Topics Genetic variation Population structure Linkage disequilibrium Natural disease variants Genome Wide Association Studies Gene
Genome annotation & EST
 Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
BIOTECHNOLOGY. Unit 8
 BIOTECHNOLOGY Unit 8 PART 1 BASIC/FUNDAMENTAL SCIENCE VS. APPLIED SCIENCE! Basic/Fundamental Science the development and establishment of information to aid our understanding of the world.! Applied Science
BIOTECHNOLOGY Unit 8 PART 1 BASIC/FUNDAMENTAL SCIENCE VS. APPLIED SCIENCE! Basic/Fundamental Science the development and establishment of information to aid our understanding of the world.! Applied Science
MARKOV CHAIN MONTE CARLO SAMPLING OF GENE GENEALOGIES CONDITIONAL ON OBSERVED GENETIC DATA
 MARKOV CHAIN MONTE CARLO SAMPLING OF GENE GENEALOGIES CONDITIONAL ON OBSERVED GENETIC DATA by Kelly M. Burkett M.Sc., Simon Fraser University, 2003 B.Sc., University of Guelph, 2000, THESIS SUBMITTED IN
MARKOV CHAIN MONTE CARLO SAMPLING OF GENE GENEALOGIES CONDITIONAL ON OBSERVED GENETIC DATA by Kelly M. Burkett M.Sc., Simon Fraser University, 2003 B.Sc., University of Guelph, 2000, THESIS SUBMITTED IN
Algorithms for Genetics: Introduction, and sources of variation
 Algorithms for Genetics: Introduction, and sources of variation Scribe: David Dean Instructor: Vineet Bafna 1 Terms Genotype: the genetic makeup of an individual. For example, we may refer to an individual
Algorithms for Genetics: Introduction, and sources of variation Scribe: David Dean Instructor: Vineet Bafna 1 Terms Genotype: the genetic makeup of an individual. For example, we may refer to an individual
Monitoring genetic change in wild populations of fish &wildlife
 Monitoring genetic change in wild populations of fish &wildlife Fred W. Allendorf University of Montana Michael K. Schwartz U.S. Forest Service GeM: Genetic Monitoring First Working Group jointly funded
Monitoring genetic change in wild populations of fish &wildlife Fred W. Allendorf University of Montana Michael K. Schwartz U.S. Forest Service GeM: Genetic Monitoring First Working Group jointly funded
CSE 427. Markov Models and Hidden Markov Models
 CSE 427 Markov Models and Hidden Markov Models http://upload.wikimedia.org/wikipedia/commons/b/ba/calico_cat 2 Dosage Compensation and X-Inactivation 2 copies (mom/dad) of each chromosome 1-23 Mostly,
CSE 427 Markov Models and Hidden Markov Models http://upload.wikimedia.org/wikipedia/commons/b/ba/calico_cat 2 Dosage Compensation and X-Inactivation 2 copies (mom/dad) of each chromosome 1-23 Mostly,
Admission Exam for the Graduate Course in Bioinformatics. November 17 th, 2017 NAME:
 1 Admission Exam for the Graduate Course in Bioinformatics November 17 th, 2017 NAME: This exam contains 30 (thirty) questions divided in 3 (three) areas (maths/statistics, computer science, biological
1 Admission Exam for the Graduate Course in Bioinformatics November 17 th, 2017 NAME: This exam contains 30 (thirty) questions divided in 3 (three) areas (maths/statistics, computer science, biological
Supplementary Methods
 Supplemental Information for funtoonorm: An improvement of the funnorm normalization method for methylation data from multiple cell or tissue types. Kathleen Oros Klein et al. Supplementary Methods funtoonorm
Supplemental Information for funtoonorm: An improvement of the funnorm normalization method for methylation data from multiple cell or tissue types. Kathleen Oros Klein et al. Supplementary Methods funtoonorm
Neutrality Test. Neutrality tests allow us to: Challenges in neutrality tests. differences. data. - Identify causes of species-specific phenotype
 Neutrality Test First suggested by Kimura (1968) and King and Jukes (1969) Shift to using neutrality as a null hypothesis in positive selection and selection sweep tests Positive selection is when a new
Neutrality Test First suggested by Kimura (1968) and King and Jukes (1969) Shift to using neutrality as a null hypothesis in positive selection and selection sweep tests Positive selection is when a new
Linking Genetic Variation to Important Phenotypes: SNPs, CNVs, GWAS, and eqtls
 Linking Genetic Variation to Important Phenotypes: SNPs, CNVs, GWAS, and eqtls BMI/CS 776 www.biostat.wisc.edu/bmi776/ Colin Dewey cdewey@biostat.wisc.edu Spring 2012 1. Understanding Human Genetic Variation
Linking Genetic Variation to Important Phenotypes: SNPs, CNVs, GWAS, and eqtls BMI/CS 776 www.biostat.wisc.edu/bmi776/ Colin Dewey cdewey@biostat.wisc.edu Spring 2012 1. Understanding Human Genetic Variation
Happy Monday! Have out: 15.1 Notes (due today) Pen or pencil. Upcoming: 15.1 Quiz on block day 15.2 Notes due Friday (2/1)
 Happy Monday! Have out: 15.1 Notes (due today) Pen or pencil Upcoming: 15.1 Quiz on block day 15.2 Notes due Friday (2/1) Plan for today Check 15.1 Notes Go over 15.1 Practice problems 15.1: Human Chromosomes
Happy Monday! Have out: 15.1 Notes (due today) Pen or pencil Upcoming: 15.1 Quiz on block day 15.2 Notes due Friday (2/1) Plan for today Check 15.1 Notes Go over 15.1 Practice problems 15.1: Human Chromosomes
Introduction to human genomics and genome informatics
 Introduction to human genomics and genome informatics Session 1 Prince of Wales Clinical School Dr Jason Wong ARC Future Fellow Head, Bioinformatics & Integrative Genomics Adult Cancer Program, Lowy Cancer
Introduction to human genomics and genome informatics Session 1 Prince of Wales Clinical School Dr Jason Wong ARC Future Fellow Head, Bioinformatics & Integrative Genomics Adult Cancer Program, Lowy Cancer
Molecular Genetics FINAL page 1 of 7 Thursday, Dec. 14, 2006 Your name:
 Molecular Genetics FNAL page 1 of 7 1. (5 points) Here is the sequence of the template strand of a DNA fragment: GAAGTACGACGAGTTCGACCTTCTCGCGAGCGCA Which of the following would be the complementary, nontemplate,
Molecular Genetics FNAL page 1 of 7 1. (5 points) Here is the sequence of the template strand of a DNA fragment: GAAGTACGACGAGTTCGACCTTCTCGCGAGCGCA Which of the following would be the complementary, nontemplate,
Genetic Technologies.notebook March 05, Genetic Technologies
 Genetic Testing Genetic Technologies Tests can be used to diagnose disorders and/or identify those individuals with an increased risk of inheriting a disorder. Prenatal Screening A fetus may be screened
Genetic Testing Genetic Technologies Tests can be used to diagnose disorders and/or identify those individuals with an increased risk of inheriting a disorder. Prenatal Screening A fetus may be screened
FORENSIC GENETICS. DNA in the cell FORENSIC GENETICS PERSONAL IDENTIFICATION KINSHIP ANALYSIS FORENSIC GENETICS. Sources of biological evidence
 FORENSIC GENETICS FORENSIC GENETICS PERSONAL IDENTIFICATION KINSHIP ANALYSIS FORENSIC GENETICS Establishing human corpse identity Crime cases matching suspect with evidence Paternity testing, even after
FORENSIC GENETICS FORENSIC GENETICS PERSONAL IDENTIFICATION KINSHIP ANALYSIS FORENSIC GENETICS Establishing human corpse identity Crime cases matching suspect with evidence Paternity testing, even after
POPULATION GENETICS. Evolution Lectures 4
 POPULATION GENETICS Evolution Lectures 4 POPULATION GENETICS The study of the rules governing the maintenance and transmission of genetic variation in natural populations. Population: A freely interbreeding
POPULATION GENETICS Evolution Lectures 4 POPULATION GENETICS The study of the rules governing the maintenance and transmission of genetic variation in natural populations. Population: A freely interbreeding
DNA METHYLATION NH 2 H 3. Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr
 NH 2 DNA METHYLATION H 3 C Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr NH 2 N N H PAGE 3 Understanding DNA Methylation DNA methylation
NH 2 DNA METHYLATION H 3 C Solutions using bisulfite conversion and immunocapture Ideal for NGS, Sanger sequencing, Pyrosequencing, and qpcr NH 2 N N H PAGE 3 Understanding DNA Methylation DNA methylation
Inference of the Properties of the Recombination Process from Whole Bacterial Genomes
 Genetics: Early Online, published on October 30, 2013 as 10.1534/genetics.113.157172 Inference of the Properties of the Recombination Process from Whole Bacterial Genomes M. Azim Ansari and Xavier Didelot
Genetics: Early Online, published on October 30, 2013 as 10.1534/genetics.113.157172 Inference of the Properties of the Recombination Process from Whole Bacterial Genomes M. Azim Ansari and Xavier Didelot
Introduction to Population Genetics. Spezielle Statistik in der Biomedizin WS 2014/15
 Introduction to Population Genetics Spezielle Statistik in der Biomedizin WS 2014/15 What is population genetics? Describes the genetic structure and variation of populations. Causes Maintenance Changes
Introduction to Population Genetics Spezielle Statistik in der Biomedizin WS 2014/15 What is population genetics? Describes the genetic structure and variation of populations. Causes Maintenance Changes
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Gene Expression. Chapters 11 & 12: Gene Conrtrol and DNA Technology. Cloning. Honors Biology Fig
 Chapters & : Conrtrol and Technology Honors Biology 0 Cloning Produced by asexual reproduction and so it is genetically identical to the parent st large cloned mammal: Dolly the sheep Animals that are
Chapters & : Conrtrol and Technology Honors Biology 0 Cloning Produced by asexual reproduction and so it is genetically identical to the parent st large cloned mammal: Dolly the sheep Animals that are
