SUPPLEMENTARY INFORMATION. Transcriptional output transiently spikes upon mitotic exit
|
|
|
- Monica Wells
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION Transcriptional output transiently spikes upon mitotic exit Viola Vaňková Hausnerová 1, 2, Christian Lanctôt 1* 1 BIOCEV and Department of Cell Biology, Faculty of Science, Charles University, Prague, Czech Republic, 2 First Faculty of Medicine, Charles University, Prague, Czech Republic 1
2 Supplementary Methods Preparation of metaphase spreads Growing cells (HepG2 and HT-1080) were incubated in medium containing 0.1 g/ml colchicine for 20 minutes. Cells were then trypsinized, incubated in an hypotonic solution (0.56M KCl) for 12 minutes at 37ºC, and fixed with methanol:acetic acid (3:1) for 20 minutes at -20ºC. After several washes, fixed cells were dropped on microscope slides in a humidified chamber in a water bath at 55ºC and left to dry for 2 minutes. Metaphase spreads were aged at room temperature for at least 5 days. FISH on metaphase spreads with BAC probes Bacterial artificial chromosomes (BAC) comprising the human POLR2A gene (RP11-104H15, CHORI BACPAC Resource Center) or sequences 4 Mb centromeric to the human TFRC gene (3q28, RP11-183L23, a kind gift of Dr. Marion Cremer, Ludwig-Maximilians Universität München) were extracted from 1 ml of bacterial cultures and linearly amplified using the GenomiPhi V2 kit according to the manufacturer s instructions (GE Healthcare Life Sciences). Labeling of BAC DNA and FISH on metaphase spreads were performed as previously described 1. Briefly, DNA was labeled either with DIG (POLR2A) or biotin (3q28). Before hybridization, samples were treated with pepsin at 100 g/ml in 0.01N for 8-9 minutes at room temperature, dehydrated and air-dried. Probes (at final concentration of 5 ng/ l) and target DNA were denatured simultaneously by placing the slides on a hot block at 76ºC for 2 minutes. Hybridization was carried out at 37ºC for 2 days. After washes, hybridized probes were detected with antibodies against DIG coupled to Cy5 (POLR2A) or streptavidin coupled to Cy3 (3q28/TFRC). Samples were counterstained with DAPI and imaged on a Nikon TiE widefield fluorescence microscope. 2
3 Analysis of the mean DAPI intensity at transcriptional spots We have quantified the DAPI signal intensity in the regions of transcriptional spots in the telophase/early G1 nuclei in the following way. 1) the z-section containing the focus of a given transcription spot was determined manually; 2) DAPI spot, the pixel value at the spot maxima was determined in the corresponding DAPI channel; 3) the DAPI pixel intensity values were determined in a region-of-interest of ~3 m x 3 m (50 pixels by 50 pixels) drawn around the transcriptional spot and distributed into 256 bins of equal size; 4) the bin comprising the DAPI spot value was noted. The above steps were repeated for 15 spots for each gene. Results are plotted separately for individual spots. Immuno-RNA FISH to detect tubulin along with TFRC and POLR2A RNAs HepG2 cells were grown on uncoated 18 mm x 18 mm #1.5 coverslips, fixed with 4% formaldehyde in 1X PBS for 10 minutes, washed and stored overnight in 70% EtOH at 4 C. After rehydration in PBS, cells were blocked for 7 minutes in PBS containing 1 mg/ml bovine serum albumin and 10mM ribonucleoside vanadyl complex (New England Biolabs), which was also included in all antibody solutions during the subsequent immunostaining procedure. Cells were then incubated in a 1:20 dilution (in PBS) of a mouse monoclonal antibody against -tubulin (clone E7, Developmental Studies Hybridoma Bank at the University of Iowa) for 30 minutes at room temperature. After brief washes with PBS (2 times, 2 minutes each), cells were incubated in a 1:200 dilution (in PBS, final concentration of 6.5 g/ml) of a secondary biotinylated goat anti-mouse antibody (Jackson Labs) for 30 minutes at room temperature. After brief washes with PBS (3 times, 2 minutes each), the immunocomplexes were fixed for 10 minutes with 2% (v/v) formaldehyde in PBS. After successive washes with PBS (1 minute) and 2X SSC (2 minutes), followed by equilibration in 2XSSC/10% formamide (5 minutes), smrna FISH was performed as described in Materials and Methods. After the last 3
4 wash with 2X SSC, cells were further stained for 20 minutes at room temperature.with DAPI (1 g/ml) and a 1:400 dilution of avidin-alexa 488 (final concentration of 2.5 g/ml, ThermoFisher/Life Technologies). Finally, samples were washed twice with 2X SSC and mounted in Prolong Gold. References for supplementary methods 1 Cremer, M. et al. Multicolor 3D Fluorescence In Situ Hybridization for Imaging Interphase Chromosomes. Methods Mol Biol 463, (2008). 4
5 Figure S1. Segregation of mrna molecules during cell division. (A) Frequency distributions of the ratios of mrna counts in pairs of HepG2 daughter cells, in bins of 10%, for TFRC (red) and POLR2A (green). The combined results from 3 independent experiments are plotted (total of 50 pairs of daughter cells). Allowing for a smrna FISH experimental error on the order of 10%, a clear majority of related daughter cells contain similar numbers of mrna molecules (ratios of 0.9-1). However, mrna counts can differ by up to 40% in a significant fraction of daughter cell pairs (ratios ). 5
6 Figure S2. Quantitative smrna FISH results for HT-1080 cells. (A-C) Representative smrna FISH images of HT-1080 cells are shown on the left for each of the target stages (A, interphase; B, metaphase; C, telophase/early G1). The images are projections of consecutive optical sections totaling 2 m in thickness. Green dots, POLR2A RNA molecules; red dots, TFRC RNA molecules. DAPI counterstain in gray. Scale bar, 5 µm. Frequency distributions 6
7 of mrna counts in the population of cells that were analyzed are shown on the right (27 or 26 bins of equal size for each gene). (D-E) mrna counts for TFRC (D, red) and POLR2A (E, green) at each target stage (n = 2 experiments: interphase, open circles, total of 76 cells; metaphase, triangles, total of 42 cells; telophase/early G1, filled circles, total of 98 cells). Each symbol represents the mrna count in an individual cell. Mean values (thick lines) ± standard deviation. ****, p <
8 Figure S3. Determination of gene copy number. (A, C) Representative FISH results on metaphase spreads from cell lines that were used in this study. Red: signals from a probe against a region near the TFRC gene (3q28, ~4 Mb away). Green: signals from a probe comprising the POLR2A gene. DAPI counterstain. (A) HepG2. (C) HT (B, D) Frequency distribution of the number of doublet signals per metaphase spread for a region near TFRC (red) and POLR2A (green). 15 metaphase spreads were analyzed for each marker. Cells are aneuploid and the majority have 4 copies of each target gene. (B) HepG2. (D) HT
9 Figure S4. Increased transcription upon mitotic exit in HT-1080 cells. (A-C) Frequency distribution of the number of active alleles per HT-1080 cell for TFRC (red) and POLR2A (green), at interphase (A, total of 76 cells), metaphase (B, total of 42 cells) or telophase/early G1 (C, total of 98 cells). n = 3 experiments. (D-G) Representative images of smrna FISH signals in a pair of daughter cells shortly after mitotic exit (D-E, POLR2A, green) or in individual nuclei (F, POLR2A, green; G, TFRC, red). Shown are xy (D, F and G) or xz (E) projections of optical sections (thickness of 2 m for panels D and E, 0.5 m for F and G). DAPI counterstain in gray. Scale bar, 5 µm. Arrows point to intense nuclear dots which mark putative nascent transcription sites. Notice that the nuclear dots are often found in regions that weakly stain with DAPI. (H) Number of nascent RNA molecules per active allele in interphase cells (open circles) or in telophase/early G1 (filled circles). TFRC (red): interphase, total of 64 alleles; telophase/early G1, total of 147 alleles. POLR2A (green): interphase, total 9
10 of 46 alleles; telophase/early G1, total of 118 alleles. n = 2 experiments. Mean values (thick lines) ± standard deviation. ns, not significant. 10
11 Figure S5. See legend below. 11
12 Figure S5. The transcriptional spots that are detected in telophase/early G1 cells are preferentially localized in regions of low DAPI intensity. Each plot shows the distributions of DAPI intensities in a ~3x3 µm nuclear region containing a given transcriptional spot. The pixel values were distributed in 256 bins of equal size. For 12
13 each plot, bin 1 corresponds to the lowest DAPI intensity and bin 256, to the highest. Results are shown for 15 POLR2A spots (green) and 15 TFRC spots (red). Arrows point to the bin that comprises the DAPI intensity at the transcriptional spot. Note that these clearly point to low DAPI intensity in 12/15 cases for POLR2A and in 15/15 cases for TFRC. 13
14 Figure S6. Intense nuclear smrna FISH signals disappear upon transcriptional inhibition. Cells were treated with an inhibitor of transcriptional elongation (flavopiridol, 1 M) or with a DNA intercalating compound (actinomycin D, 1.5 g/ml) for 1 hour before being processed for smrna FISH. (A) Frequency distribution of the number of intense nuclear dots per HepG2 cell in interphase cells for TFRC (red) and POLR2A (green) in control cells (CTL, filled bars, 54 cells), actinomycin D-treated cells (ACT, empty bars, 51 cells) and flavopiridol-treated cells (FLA, dashed bars, 50 cells), n = 1 experiment. (B) Frequency distribution of the number of intense nuclear dots per HepG2 cell in telophase/early G1 cells for TFRC (red) and POLR2A (green) in control cells (CTL, filled bars, 32 cells), actinomycin D-treated cells (ACT, empty bars, 38 cells) and flavopiridoltreated cells (FLA, dashed bars, 32 cells), n = 1. 14
15 Figure S7. Transcriptional spikes occur early in the cell cycle. Representative images of HepG2 cells co-stained for -tubulin (left panels), TFRC RNA (middle panels) and POLR2A RNA (right panels). Arrowheads point to abscission midbodies. Arrows point to intense nascent transcriptional spots. The contours of the nuclei are marked by dotted lines. The smrna FISH signals are shown according to the fire color scheme, i.e. strong signals in yellow/white and weak ones in blue. Shown are pseudo-3d view for -tubulin and maximum intensity projections for the smrna FISH signals. (A) Early telophase/g1 cells show intense transcriptional spots for both genes. (B) Daugther cells that are still joined by a midbody but show no signs of nascent transcription of either genes. Scale bar, 5 m. 15
16 SEQUENCE Probe name Probe number Probe pos Percent GC agttgggaggaaaaagccgg hpolr2a_ ,00% tcactacaaaaagcctgcgc hpolr2a_ ,00% gactcaggactccgaactgg hpolr2a_ ,00% agacattcgcttcagttcat hpolr2a_ ,00% tcgtctctgggtatttgatg hpolr2a_ ,00% cagttcaatgtggccaaagt hpolr2a_ ,00% gggttgttagagtccacaag hpolr2a_ ,00% aggtcgtagacatgtgtgag hpolr2a_ ,00% ctgagagtcctcattaacgt hpolr2a_ ,00% ttgatcttcacgatgtcagc hpolr2a_ ,00% tctgtcaatgttgaaggggg hpolr2a_ ,00% agtcaatgcgatcaccattg hpolr2a_ ,00% tgtacggagttgtcacacta hpolr2a_ ,00% ggaacatcaggaggttcatc hpolr2a_ ,00% gagggagaagatttgcttgc hpolr2a_ ,00% agaagaggcgagtgatgtca hpolr2a_ ,00% gatgaggagccagttgttaa hpolr2a_ ,00% tagtgttctgaatgtcctgg hpolr2a_ ,00% tcgatgacctctattacgtc hpolr2a_ ,00% gcatcgttaagaatgcggtt hpolr2a_ ,00% agacagggatttctgagcag hpolr2a_ ,00% gacctgggagatgttaatct hpolr2a_ ,00% cggtgcttgaagccaaatgg hpolr2a_ ,00% aggtaggagttctccacaaa hpolr2a_ ,00% cttgttggaaggcttaagcg hpolr2a_ ,00% caactcgttctggatgtgtg hpolr2a_ ,00% ttcttgctcaattccttgac hpolr2a_ ,00% caggtggatgttgaagagca hpolr2a_ ,00% ggaaatgttgatgagctcct hpolr2a_ ,00% taagcgaaggagtctttggc hpolr2a_ ,00% atcttttcagcaatctgctc hpolr2a_ ,00% catatctgtcagcatgttgg hpolr2a_ ,00% ccgtgatgatgatcttcttc hpolr2a_ ,00% tctccacaatgtcattggac hpolr2a_ ,00% ggagccatcaaaggagatga hpolr2a_ ,00% cacacaagagagccaagtgt hpolr2a_ ,00% ctggcccagcatgatattct hpolr2a_ ,00% aataagagggactctggggt hpolr2a_ ,00% ctatagttgggactggttgg hpolr2a_ ,00% gaatagctgggtgatgttgg hpolr2a_ ,00% ggaaggggagtaacttggtg hpolr2a_ ,00% tataggttggagactgtggt hpolr2a_ ,00% 16
17 gctgggactgtaagaaggac hpolr2a_ ,00% ggtgggtgaatatttgggac hpolr2a_ ,00% ggagatgttggggagtattt hpolr2a_ ,00% ctagtaggtgagtacttggg hpolr2a_ ,00% aacgggatccagaagttcac hpolr2a_ ,00% acccctaagttaaaataccc hpolr2a_ ,00% Table S1. Oligonucleotide sequences of the smrna FISH probe against human POLR2A. Shown in this table are the sequences of individual labeled oligonucleotides (20-mer, from 5 to 3 ), the oligonucleotide identification and number, its position on the mature mrna and its GC contents. The Ensembl Gene ID for human POLR2A is ENSG
18 SEQUENCE (5'->3') Probe name Probe no. Probe pos in intron 1 Percent GC cataagcagcgagaaagcgc hpolr2a_i1_ % ctgctcaactctttgcaaaa hpolr2a_i1_ % catctcggacaaagcgctac hpolr2a_i1_ % ctggagtgtgaaatcagtca hpolr2a_i1_ % atcgcctttaacgtcggtaa hpolr2a_i1_ % gtaaagactccctaggattc hpolr2a_i1_ % ggtttggcttcttaaccaaa hpolr2a_i1_ % cagaagaggaggaagctacc hpolr2a_i1_ % cttagcgccacaagggaaaa hpolr2a_i1_ % cttaactccattctttccag hpolr2a_i1_ % ggtctctatccacaaacagc hpolr2a_i1_ % tgaactttctcccagcaaaa hpolr2a_i1_ % acgttctgactcctgacata hpolr2a_i1_ % ctgttctagctgttcttaca hpolr2a_i1_ % ttcactctgcatacttctga hpolr2a_i1_ % cgtaaatacgttcccacttt hpolr2a_i1_ % tttgtcaacagtgtcccata hpolr2a_i1_ % ccagagatgactctggtaga hpolr2a_i1_ % atcgaagcccaacaatccta hpolr2a_i1_ % aactgcaagacatgcagagc hpolr2a_i1_ % ggtccctcaaaatacagaca hpolr2a_i1_ % ctacccaaaattgaaccgtc hpolr2a_i1_ % caacaagaggcaccttactc hpolr2a_i1_ % atacactctgacccctaaga hpolr2a_i1_ % aattcaaagacccctccgta hpolr2a_i1_ % cttctaacagcaaggacaac hpolr2a_i1_ % caaagctctagtaccaaccc hpolr2a_i1_ % aaattttgtgcacacgctgg hpolr2a_i1_ % gaaatccagcaccctctttc hpolr2a_i1_ % aataggggtcaactactgcg hpolr2a_i1_ % gcattccatgatagacagtt hpolr2a_i1_ % caatgtagcccacactctac hpolr2a_i1_ % caaggatcatcttctcactc hpolr2a_i1_ % tccctcacaagattataggt hpolr2a_i1_ % ctcccacatttatgttctaa hpolr2a_i1_ % acagctttatttggtaccac hpolr2a_i1_ % gcaaagctaacaaggtcctt hpolr2a_i1_ % ggagctgtcattgaatctca hpolr2a_i1_ % cctgaggccacaataacaaa hpolr2a_i1_ % tgggttctctttgggttctg hpolr2a_i1_ % ttaaacacttgagggggtgg hpolr2a_i1_ % ctcactgcctacaggataaa hpolr2a_i1_ % 18
19 cattttaaattagcccccaa hpolr2a_i1_ % tccatcaccaagttcacgaa hpolr2a_i1_ % tgtcctagacagcagacacg hpolr2a_i1_ % tgctttcaggctttctgaga hpolr2a_i1_ % ccacccttctaatgactaat hpolr2a_i1_ % Table S2. Oligonucleotide sequences of the smrna FISH probe against the first intron of human POLR2A. Shown in this table are the sequences of individual labeled oligonucleotides (20-mer, from 5 to 3 ), the oligonucleotide identification and number, its position relative to the first nucleotide of the intron and its GC contents. The Ensembl Gene ID for human POLR2A is ENSG
Replication-independent chromatin loading of Dnmt1 during G2 and M phases
 Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
TR DISTRIBUTION STATEMENT A: Approved for public release; distribution is unlimited.
 Chapter 14. Intracellular detection of viral transcription and replication using RNA FISH i. Summary/Abstract Many hemorrhagic fever viruses require BSL-3 or 4 laboratory containment for study. The necessary
Chapter 14. Intracellular detection of viral transcription and replication using RNA FISH i. Summary/Abstract Many hemorrhagic fever viruses require BSL-3 or 4 laboratory containment for study. The necessary
Spectral-karyotyping- SKY
 Spectral-karyotyping- SKY Content: 1) Introduction 2) Technical tips: 3) Frequently Asked questions 4) Spectral-karytoyping protocol 1) Introduction SKY is a powerful technique that improves karyotype
Spectral-karyotyping- SKY Content: 1) Introduction 2) Technical tips: 3) Frequently Asked questions 4) Spectral-karytoyping protocol 1) Introduction SKY is a powerful technique that improves karyotype
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Figure 1
 Supplementary Figure 1 (A) Schematic of sequential hybridization and barcoding. (B) Schematic of the FISH images of the cell. In each round of hybridization, the same spots are detected, but the dye associated
Supplementary Figure 1 (A) Schematic of sequential hybridization and barcoding. (B) Schematic of the FISH images of the cell. In each round of hybridization, the same spots are detected, but the dye associated
Nature Methods: doi: /nmeth Supplementary Figure 1. Retention of RNA with LabelX.
 Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figures
 Supplementary Figures Wossidlo et al. S1 Supplementary Figure S1 5hmC (clone 63.3) immunostaining on mouse zygotes. a, Representative images of indirect immunostainings using antibodies against 5hmC (clone
Supplementary Figures Wossidlo et al. S1 Supplementary Figure S1 5hmC (clone 63.3) immunostaining on mouse zygotes. a, Representative images of indirect immunostainings using antibodies against 5hmC (clone
Online Supplement ALVEOLAR CELL SENESCENCE IN PATIENTS WITH PULMONARY EMPHYSEMA. Takao Tsuji, Kazutetsu Aoshiba, and Atsushi Nagai
 Online Supplement ALVEOLAR CELL SENESCENCE IN PATIENTS WITH PULMONARY EMPHYSEMA Takao Tsuji, Kazutetsu Aoshiba, and Atsushi Nagai MATERIALS AND METHODS Immunohistochemistry Deparaffinized tissue sections
Online Supplement ALVEOLAR CELL SENESCENCE IN PATIENTS WITH PULMONARY EMPHYSEMA Takao Tsuji, Kazutetsu Aoshiba, and Atsushi Nagai MATERIALS AND METHODS Immunohistochemistry Deparaffinized tissue sections
DNA SPECTRAL KARYOTYPING HYBRIDIZATION & DETECTION PROTOCOL
 2012 1 WI-KIT-005-DNA-Protocol-ROW_2 REV.C DNA SPECTRAL KARYOTYPING HYBRIDIZATION & DETECTION PROTOCOL For Research Use Only - Not for use in Clinical Diagnosis The DNA Spectral Karyotyping Reagents are
2012 1 WI-KIT-005-DNA-Protocol-ROW_2 REV.C DNA SPECTRAL KARYOTYPING HYBRIDIZATION & DETECTION PROTOCOL For Research Use Only - Not for use in Clinical Diagnosis The DNA Spectral Karyotyping Reagents are
ab BrdU Immunohistochemistry Kit
 ab125306 - BrdU Immunohistochemistry Kit Instructions for Use For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells. This product
ab125306 - BrdU Immunohistochemistry Kit Instructions for Use For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells. This product
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex
 Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Materials and Methods Materials Required for Fixing, Embedding and Sectioning. OCT embedding matrix (Thermo Scientific, LAMB/OCT)
 Page 1 Introduction Tissue freezing and sectioning is a rapid method of generating tissue samples (cryosections) for histological analysis, and obviates the need for wax embedding. The method is popular
Page 1 Introduction Tissue freezing and sectioning is a rapid method of generating tissue samples (cryosections) for histological analysis, and obviates the need for wax embedding. The method is popular
Stellaris RNA FISH Protocol for Simultaneous IF + FISH in Adherent Cells
 Stellaris RNA FISH Protocol for Simultaneous IF + FISH in Adherent Cells General Protocol & Storage Product Description A set of Stellaris RNA FISH Probes is comprised of up to 48 singly labeled oligonucleotides
Stellaris RNA FISH Protocol for Simultaneous IF + FISH in Adherent Cells General Protocol & Storage Product Description A set of Stellaris RNA FISH Probes is comprised of up to 48 singly labeled oligonucleotides
Specific DNA fluorescently tagged PROTOCOLO DE UTILIZACIÓN
 Specific DNA fluorescently tagged PROTOCOLO DE UTILIZACIÓN Product: 1. DNA probe fluorescently labeled. 2. Hybridization Buffer 3. Reagents to prepare Mounting Solution (DAPI + antifade) These probes are
Specific DNA fluorescently tagged PROTOCOLO DE UTILIZACIÓN Product: 1. DNA probe fluorescently labeled. 2. Hybridization Buffer 3. Reagents to prepare Mounting Solution (DAPI + antifade) These probes are
HistoMark Double Staining Procedures. Where Better Science Begins.
 HistoMark Double Staining Procedures Where Better Science Begins www.kpl.com HistoMark Double Staining Procedures Researchers often need the ability to visualize multiple proteins in one tissue sample.
HistoMark Double Staining Procedures Where Better Science Begins www.kpl.com HistoMark Double Staining Procedures Researchers often need the ability to visualize multiple proteins in one tissue sample.
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
Stellaris FISH Probes Protocols and Storage
 Stellaris FISH Probes Protocols and Storage Catalog No. SMF-2035-1 Product Name Stellaris FISH Probes, Human MALAT1 with Quasar 570 Dye Product Description Product consists of Quasar 570-labeled oligos
Stellaris FISH Probes Protocols and Storage Catalog No. SMF-2035-1 Product Name Stellaris FISH Probes, Human MALAT1 with Quasar 570 Dye Product Description Product consists of Quasar 570-labeled oligos
Combined Digoxigenin-labeled in situ hybridization/ Immunohistochemistry protocol (for fixed frozen cryostat sections)
 Combined Digoxigenin-labeled in situ hybridization/ Immunohistochemistry protocol (for fixed frozen cryostat sections) A. Digoxigenin-UTP labeling of crna antisense probe Refer to laboratory protocol and
Combined Digoxigenin-labeled in situ hybridization/ Immunohistochemistry protocol (for fixed frozen cryostat sections) A. Digoxigenin-UTP labeling of crna antisense probe Refer to laboratory protocol and
Introduction to Fluorescent In Situ Hybridization (FISH)
 Robert Driscoll, M.F.S. Heather Cunningham, M.S. Introduction to Fluorescent In Situ Hybridization (FISH) Fluorescent In Situ Hybridization (FISH) FISH is a cytogenetic technique used to detect the presence
Robert Driscoll, M.F.S. Heather Cunningham, M.S. Introduction to Fluorescent In Situ Hybridization (FISH) Fluorescent In Situ Hybridization (FISH) FISH is a cytogenetic technique used to detect the presence
Methods of Biomaterials Testing Lesson 3-5. Biochemical Methods - Molecular Biology -
 Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Fluorescent in-situ Hybridization
 Fluorescent in-situ Hybridization Presented for: Presented by: Date: 2 Definition In situ hybridization is the method of localizing/ detecting specific nucleotide sequences in morphologically preserved
Fluorescent in-situ Hybridization Presented for: Presented by: Date: 2 Definition In situ hybridization is the method of localizing/ detecting specific nucleotide sequences in morphologically preserved
DIG-label In Situ Hybridizations (Jessell lab), from Scharen-Wiemers and Gerfin-Moser Histochemistry 100:
 DIG-label In Situ Hybridizations (Jessell lab), 2004 from Scharen-Wiemers and Gerfin-Moser Histochemistry 100:431-440. I. Tissue section: 1 Fix embryo overnight at 4 C in 4% paraformaldehyde, 0.1 M PB.
DIG-label In Situ Hybridizations (Jessell lab), 2004 from Scharen-Wiemers and Gerfin-Moser Histochemistry 100:431-440. I. Tissue section: 1 Fix embryo overnight at 4 C in 4% paraformaldehyde, 0.1 M PB.
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
ZytoLight FISH-Cytology. Implementation Kit. For fluorescence in situ hybridization (FISH) on cytology specimens using any ZytoLight FISH probe
 ZytoLight FISH-Cytology Implementation Kit Z-2099-20 20 For fluorescence in situ hybridization (FISH) on cytology specimens using any ZytoLight FISH probe.... In vitro diagnostic medical device according
ZytoLight FISH-Cytology Implementation Kit Z-2099-20 20 For fluorescence in situ hybridization (FISH) on cytology specimens using any ZytoLight FISH probe.... In vitro diagnostic medical device according
1. Paraffin section slides can be stored at room temperature for a long time.
 Immunohistochemistry (IHC) Protocols Immunohistochemistry (IHC) Protocol of Paraffin Section 1. Fix dissected tissues with 10% formalin for no less than 48 hours at room temperature. Inadequately fixation
Immunohistochemistry (IHC) Protocols Immunohistochemistry (IHC) Protocol of Paraffin Section 1. Fix dissected tissues with 10% formalin for no less than 48 hours at room temperature. Inadequately fixation
Stellaris RNA FISH Protocol for D. Melanogaster Wing Imaginal Discs
 Stellaris RNA FISH Protocol for D. Melanogaster Wing Imaginal Discs General Protocol & Storage Product Description A set of Stellaris RNA FISH Probes is comprised of up to 48 singly labeled oligonucleotides
Stellaris RNA FISH Protocol for D. Melanogaster Wing Imaginal Discs General Protocol & Storage Product Description A set of Stellaris RNA FISH Probes is comprised of up to 48 singly labeled oligonucleotides
Stellaris RNA FISH Protocol for Adherent Cells in 96 Well Glass Bottom Plates
 Stellaris RNA FISH Protocol for Adherent Cells in 96 Well Glass Bottom Plates This protocol is specifically designed for high throughput applications of Stellaris in 96 well glass bottom plates. General
Stellaris RNA FISH Protocol for Adherent Cells in 96 Well Glass Bottom Plates This protocol is specifically designed for high throughput applications of Stellaris in 96 well glass bottom plates. General
Immunohistochemistry guide
 Immunohistochemistry guide overview immunohistochemistry Overview Immunohistochemistry is a laboratory technique utilized for the visual detection of antigens in tissue. When working with cells this technique
Immunohistochemistry guide overview immunohistochemistry Overview Immunohistochemistry is a laboratory technique utilized for the visual detection of antigens in tissue. When working with cells this technique
Propidium Iodide. Catalog Number: Structure: Molecular Formula: C 27H 34I 2N 4. Molecular Weight: CAS #
 Catalog Number: 195458 Propidium Iodide Structure: Molecular Formula: C 27H 34I 2N 4 Molecular Weight: 668.45 CAS # 25535-16-4 Physical Description: Dark red crystals Description: Reagent used for the
Catalog Number: 195458 Propidium Iodide Structure: Molecular Formula: C 27H 34I 2N 4 Molecular Weight: 668.45 CAS # 25535-16-4 Physical Description: Dark red crystals Description: Reagent used for the
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry of cytochrome c and mitochondria.
 PROTOCOL ApoTrack Cytochrome c Apoptosis ICC Antibody Kit 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSA07 Rev.1 DESCRIPTION ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry
PROTOCOL ApoTrack Cytochrome c Apoptosis ICC Antibody Kit 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 MSA07 Rev.1 DESCRIPTION ApoTrack Cytochrome c Apoptosis ICC Antibody Kit: 2 color immunocytochemistry
mrna IN SITU HYBRIDIZATION For Sectioned Zebrafish
 DAYS 1 2: Harvesting fish, tissue fixation, hybridization 1. Harvest and fix embryos in 4% paraformaldehyde overnight at 4C *Fix should be made fresh on the day it will be used. Do not store it for long
DAYS 1 2: Harvesting fish, tissue fixation, hybridization 1. Harvest and fix embryos in 4% paraformaldehyde overnight at 4C *Fix should be made fresh on the day it will be used. Do not store it for long
A Supersandwich Fluorescence in Situ Hybridization (SFISH) Strategy. for Highly Sensitive and Selective mrna Imaging in Tumor Cells
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) A Supersandwich Fluorescence in Situ Hybridization (SFISH)
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) A Supersandwich Fluorescence in Situ Hybridization (SFISH)
Methodology for Immunohistochemistry. Learning Objectives:
 Proteomics Methodology for Immunohistochemistry Methodology for Immunohistochemistry A staining process for identifying the proteins location in cells, tissues by using antigen-antibody property. Immuno
Proteomics Methodology for Immunohistochemistry Methodology for Immunohistochemistry A staining process for identifying the proteins location in cells, tissues by using antigen-antibody property. Immuno
Left: Linearized vector (~4kbp); Right: Final product of RNA probe (~300bp) FISH Protocol AE, adapted from Ressler Lab, edited by YG 6/18/12-1-
 Insitu 1. Linearize vector. Use > 4000 ng vector in a single 50uL RXN. 2. Run on gel (single lane) and band-purify the linearized vector, elute with 8Ul Rnase free water. Take 2 Ul to quantify the DNA
Insitu 1. Linearize vector. Use > 4000 ng vector in a single 50uL RXN. 2. Run on gel (single lane) and band-purify the linearized vector, elute with 8Ul Rnase free water. Take 2 Ul to quantify the DNA
In Situ Hybridization
 In Situ Hybridization A Practical Approach Second Edition Edited by D. G. WILKINSON MMfl, London Oxford New York Tokyo OXFORD UNIVERSITY PRESS 1998 List of con trib u tors Abbreviations 1. The theory and
In Situ Hybridization A Practical Approach Second Edition Edited by D. G. WILKINSON MMfl, London Oxford New York Tokyo OXFORD UNIVERSITY PRESS 1998 List of con trib u tors Abbreviations 1. The theory and
DNA Microarray Technology
 2 DNA Microarray Technology 2.1 Overview DNA microarrays are assays for quantifying the types and amounts of mrna transcripts present in a collection of cells. The number of mrna molecules derived from
2 DNA Microarray Technology 2.1 Overview DNA microarrays are assays for quantifying the types and amounts of mrna transcripts present in a collection of cells. The number of mrna molecules derived from
TUNEL Universal Apoptosis Detection Kit ( Biotin-labeled POD )
 TUNEL Universal Apoptosis Detection Kit ( Biotin-labeled POD ) Cat. No. L00290 Technical Manual No. 0267 Version 03112011 I Description. 1 II Key Features.... 1 III Kit Contents.. 1 IV Storage.. 2 V Procedure...
TUNEL Universal Apoptosis Detection Kit ( Biotin-labeled POD ) Cat. No. L00290 Technical Manual No. 0267 Version 03112011 I Description. 1 II Key Features.... 1 III Kit Contents.. 1 IV Storage.. 2 V Procedure...
BrdU Immunohistochemistry Kit Instruction Manual
 BrdU Immunohistochemistry Kit Instruction Manual Features Easy to use system Reagents titered for success Proven protocol Ordering Information Catalog Number X1545K Size 50 Slides Format Immunohistochemistry
BrdU Immunohistochemistry Kit Instruction Manual Features Easy to use system Reagents titered for success Proven protocol Ordering Information Catalog Number X1545K Size 50 Slides Format Immunohistochemistry
Fluorescent In Situ Hybridization (FISH) Assay
 Fluorescent In Situ Hybridization (FISH) Assay 1 What is FISH 2 Probes 3 FISH Procedure 4 Application Definition, Principle and Sample Types The core of FISH technology A quick and simple FISH protocol
Fluorescent In Situ Hybridization (FISH) Assay 1 What is FISH 2 Probes 3 FISH Procedure 4 Application Definition, Principle and Sample Types The core of FISH technology A quick and simple FISH protocol
IHC staining protocol. Paraffin, frozen and free-floating sections
 IHC staining protocol Paraffin, frozen and free-floating sections IHC staining protocol Contents Paraffin and frozen sections Immunostaining free-floating sections Signal amplification Paraffin and frozen
IHC staining protocol Paraffin, frozen and free-floating sections IHC staining protocol Contents Paraffin and frozen sections Immunostaining free-floating sections Signal amplification Paraffin and frozen
Neural Stem Cell Characterization Kit
 Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Neural Stem Cell Characterization Kit Catalog No. SCR019 FOR RESEARCH USE ONLY Not for use in diagnostic procedures USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23 8026 2233
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Diffusion of MCF-7 and 3T3 cells. (a) High-resolution image (40x): DAPI and Cy5-CellTracker TM stained MCF-7 cells. (b) Corresponding high-resolution (40x)
Supplementary Figures Supplementary Figure 1. Diffusion of MCF-7 and 3T3 cells. (a) High-resolution image (40x): DAPI and Cy5-CellTracker TM stained MCF-7 cells. (b) Corresponding high-resolution (40x)
Product Name Quantity* Product No.
 Page 1 of 10 Lit.# ML017 10/04/12 Label IT Fluorescence In Situ Hybridization (FISH) Kits Product Name Quantity* Product No. Label IT FISH Cy 3 Labeling Kit Label IT FISH TM-Rhodamine Kit Label IT FISH
Page 1 of 10 Lit.# ML017 10/04/12 Label IT Fluorescence In Situ Hybridization (FISH) Kits Product Name Quantity* Product No. Label IT FISH Cy 3 Labeling Kit Label IT FISH TM-Rhodamine Kit Label IT FISH
Department of Microbiology, Lab 016 instructions
 Protocol for standard FISH and DOPE-FISH for prokaryotes (slightly modified from Amann, 1995, note, for other modifications or other microorganisms like eukaryotes, consult special literature or check
Protocol for standard FISH and DOPE-FISH for prokaryotes (slightly modified from Amann, 1995, note, for other modifications or other microorganisms like eukaryotes, consult special literature or check
LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic
 information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
ab BrdU Immunohistochemistry Kit
 ab125306 - BrdU Immunohistochemistry Kit Instructions for Use For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells. This product
ab125306 - BrdU Immunohistochemistry Kit Instructions for Use For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells. This product
rrna subtraction protocol for metatranscriptomics
 rrna subtraction protocol for metatranscriptomics Recommended materials/kits Price Vendor Stock Number MEGAscript transcription kit $249 Ambion AM1334 MEGAclear TM kit $89 Ambion AM1908 RNeasy MinElute
rrna subtraction protocol for metatranscriptomics Recommended materials/kits Price Vendor Stock Number MEGAscript transcription kit $249 Ambion AM1334 MEGAclear TM kit $89 Ambion AM1908 RNeasy MinElute
DAPI mrna intron. Chromosome 19 UBA2 EIF3K
 Supplementary Figure DAPI mrna intron a b c d p-arm Chromosome 9 q-arm.79 3.98.2.76 2.79 7.97 8.68 23. 3.92 39. 39.9.33.3 2.36.39.7 7.28 2.69.7 6.6 e intron mrna Chr. 9 Supplementary Fig.. Targeting 2
Supplementary Figure DAPI mrna intron a b c d p-arm Chromosome 9 q-arm.79 3.98.2.76 2.79 7.97 8.68 23. 3.92 39. 39.9.33.3 2.36.39.7 7.28 2.69.7 6.6 e intron mrna Chr. 9 Supplementary Fig.. Targeting 2
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
!! PLEASE READ BEFORE USE!!
 In situ Proximity Ligation Assay protocols!! PLEASE READ BEFORE USE!! The test protocol is a guideline, user need to determine their optimal experimental condition for best performance. The following protocol
In situ Proximity Ligation Assay protocols!! PLEASE READ BEFORE USE!! The test protocol is a guideline, user need to determine their optimal experimental condition for best performance. The following protocol
ebioscience BrdU Kit for IHC/ICC Colorimetric Catalog Number: RUO: For Research Use Only. Not for use in diagnostic procedures.
 Page 1 of 1 ebioscience BrdU Kit for IHC/ICC Colorimetric Catalog Number: 8800-6599 RUO: For Research Use Only. Not for use in diagnostic procedures. Product Information Contents: ebioscience BrdU Kit
Page 1 of 1 ebioscience BrdU Kit for IHC/ICC Colorimetric Catalog Number: 8800-6599 RUO: For Research Use Only. Not for use in diagnostic procedures. Product Information Contents: ebioscience BrdU Kit
BrdU IHC Kit. For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells
 K-ASSAY BrdU IHC Kit For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells Cat. No. KT-077 For Research Use Only. Not for Use in
K-ASSAY BrdU IHC Kit For the detection and localization of bromodeoxyuridine incorporated into newly synthesized DNA of actively proliferating cells Cat. No. KT-077 For Research Use Only. Not for Use in
Formaldehyde-based Whole-Mount In Situ Hybridization Methodology for the Planarian Schmidtea mediterranea Sánchez Alvarado Lab, November 2008
 Formaldehyde-based Whole-Mount In Situ Hybridization Methodology for the Planarian Schmidtea mediterranea Sánchez Alvarado Lab, November 2008 Animal Preparation Unless noted at particular steps, intact
Formaldehyde-based Whole-Mount In Situ Hybridization Methodology for the Planarian Schmidtea mediterranea Sánchez Alvarado Lab, November 2008 Animal Preparation Unless noted at particular steps, intact
ApoTrack Cytochrome c Apoptosis ICC Antibody
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and nonapoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and nonapoptotic
Single Molecule FISH on Mouse Tissue Sections
 Single Molecule FISH on Mouse Tissue Sections Shalev Itzkovitz, December 2012 Tissue preparation Solutions and s 10X PBS (Ambion #AM9625) 4% formaldehyde or paraformaldehyde in PBS Cryoprotecting solution:
Single Molecule FISH on Mouse Tissue Sections Shalev Itzkovitz, December 2012 Tissue preparation Solutions and s 10X PBS (Ambion #AM9625) 4% formaldehyde or paraformaldehyde in PBS Cryoprotecting solution:
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 215 Supporting Information Quantitative Description of Thermodynamic and Kinetic Properties of the
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 215 Supporting Information Quantitative Description of Thermodynamic and Kinetic Properties of the
ApoTrack Cytochrome c Apoptosis ICC Antibody Kit
 ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
ab110417 ApoTrack Cytochrome c Apoptosis ICC Antibody Kit Instructions for Use For the Immunocytochemistry analysis of cytochrome c and a mitochondrial marker (Complex Vα) in apoptotic cells and non-apoptotic
Tyramide Signal Amplification Kits
 Tyramide Signal Amplification Kits Table 1 Contents and storage Material* Amount Storage Stability Labeled tyramide (Component A) 1 vial Dimethylsulfoxide (DMSO) (Component B) 200 ml HRP-conjugated secondary
Tyramide Signal Amplification Kits Table 1 Contents and storage Material* Amount Storage Stability Labeled tyramide (Component A) 1 vial Dimethylsulfoxide (DMSO) (Component B) 200 ml HRP-conjugated secondary
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
IQFISH on Dako Omnis. Panel for Lung Cancer. Dako FAST RESULTS. ALK, ROS1, RET and MET IQFISH. Dako Omnis. Agilent Pathology Solutions
 PRODUCT INFORMATION Dako Omnis ALK, ROS1, RET and MET IQFISH Dako Agilent Pathology Solutions IQFISH on Dako Omnis Panel for Lung Cancer FAST RESULTS Fast, high-quality FISH Integrated into your IHC workflow
PRODUCT INFORMATION Dako Omnis ALK, ROS1, RET and MET IQFISH Dako Agilent Pathology Solutions IQFISH on Dako Omnis Panel for Lung Cancer FAST RESULTS Fast, high-quality FISH Integrated into your IHC workflow
Figure S6. Detection of anti-gfp antibodies in anti-dna and normal plasma without competition DNA--9
 Supplementary Information Ultrasensitive antibody detection by agglutination-pcr (ADAP) Cheng-ting Tsai 1 *, Peter V. Robinson 1 *, Carole A. Spencer 2 and Carolyn R. Bertozzi 3,4ǂ Department of 1 Chemistry,
Supplementary Information Ultrasensitive antibody detection by agglutination-pcr (ADAP) Cheng-ting Tsai 1 *, Peter V. Robinson 1 *, Carole A. Spencer 2 and Carolyn R. Bertozzi 3,4ǂ Department of 1 Chemistry,
SUPPLEMENTARY INFORMATION FILE
 SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
SUPPLEMENTARY INFORMATION FILE Existence of a microrna pathway in anucleate platelets Patricia Landry, Isabelle Plante, Dominique L. Ouellet, Marjorie P. Perron, Guy Rousseau & Patrick Provost 1. SUPPLEMENTARY
Stellaris RNA FISH Protocol for C. elegans
 Stellaris RNA FISH Protocol for C. elegans General Protocol & Storage Product Description A set of Stellaris RNA FISH Probes is comprised of up to 48 singly labeled oligonucleotides designed to selectively
Stellaris RNA FISH Protocol for C. elegans General Protocol & Storage Product Description A set of Stellaris RNA FISH Probes is comprised of up to 48 singly labeled oligonucleotides designed to selectively
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Histone Acetylation Controls the Inactive X Chromosome. Replication Dynamics
 Histone Acetylation Controls the Inactive X Chromosome Replication Dynamics Corella S. Casas-Delucchi 1,2,, Alessandro Brero 2,, Hans-Peter Rahn 2, Irina Solovei 3, Anton Wutz 4, Thomas Cremer 3, Heinrich
Histone Acetylation Controls the Inactive X Chromosome Replication Dynamics Corella S. Casas-Delucchi 1,2,, Alessandro Brero 2,, Hans-Peter Rahn 2, Irina Solovei 3, Anton Wutz 4, Thomas Cremer 3, Heinrich
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Histology FISH Accessory Kit Step-by-Step Procedure
 PROCEDURE Histology FISH Accessory Kit Step-by-Step Procedure Code K5799 For probes diluted in Formamide based hybridization buffer Reagent Preparation Equilibration Deparaffinization/Rehydration Prepare
PROCEDURE Histology FISH Accessory Kit Step-by-Step Procedure Code K5799 For probes diluted in Formamide based hybridization buffer Reagent Preparation Equilibration Deparaffinization/Rehydration Prepare
Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence
 Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
Supplementary Information Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Yoshinori Kousoku, Aya Ohkuni,
EnzMet HRP Detection Kit for IHC / ISH
 EnzMet HRP Detection Kit for IHC / ISH 95 Horseblock Road, Unit 1, Yaphank NY 11980-9710 Tel: (877) 447-6266 (Toll-Free in US) or (631) 205-9490 Fax: (631) 205-9493 Tech Support: (631) 205-9492 tech@nanoprobes.com
EnzMet HRP Detection Kit for IHC / ISH 95 Horseblock Road, Unit 1, Yaphank NY 11980-9710 Tel: (877) 447-6266 (Toll-Free in US) or (631) 205-9490 Fax: (631) 205-9493 Tech Support: (631) 205-9492 tech@nanoprobes.com
XCyte lab manual MetaSystems
 XCyte lab manual MetaSystems GmbH, Robert Bosch Strasse 6, D-68804 Altlussheim, Germany Tel: +49-6205-39610, FAX: +49-6205-32270 e-mail: probes@metasystems.de web: http://www.metasystems.de 1 Fluorescence
XCyte lab manual MetaSystems GmbH, Robert Bosch Strasse 6, D-68804 Altlussheim, Germany Tel: +49-6205-39610, FAX: +49-6205-32270 e-mail: probes@metasystems.de web: http://www.metasystems.de 1 Fluorescence
TintoDetector Operating Manual
 1 TintoDetector Operating Manual Capillary Gap System for ICC, IHC, CISH and FISH 2 TintoDetector Manual Table of Contents Overview 3 TintoDetector Components 4 TintoDetector Specifications 5 Guidelines
1 TintoDetector Operating Manual Capillary Gap System for ICC, IHC, CISH and FISH 2 TintoDetector Manual Table of Contents Overview 3 TintoDetector Components 4 TintoDetector Specifications 5 Guidelines
Supplementary Figure 1 A green: cytokeratin 8
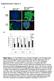 Supplementary Figure 1 A green: cytokeratin 8 green: α-sma red: α-sma blue: DAPI blue: DAPI Panc-1 Panc-1 Panc-1+hPSC Panc-1+hPSC monoculture coculture B Suppl. Figure 1: A, Immunofluorescence staining
Supplementary Figure 1 A green: cytokeratin 8 green: α-sma red: α-sma blue: DAPI blue: DAPI Panc-1 Panc-1 Panc-1+hPSC Panc-1+hPSC monoculture coculture B Suppl. Figure 1: A, Immunofluorescence staining
SUPPLEMENTARY NOTE 3. Supplememtary Note 3, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Monteiro et al., http://www.jcb.org/cgi/content/full/jcb.201306162/dc1 Figure S1. 3D deconvolution microscopy analysis of WASH and exocyst
Supplementary Figures and legend. Heparin affinity purification of extracellular vesicles.
 Supplementary Figures and legend Heparin affinity purification of extracellular vesicles. Leonora Balaj 1, Nadia A. Atai 1,2, Weilin Chen 1, Dakai Mu 1, Bakhos A. Tannous 1, Xandra O. Breakefield 1, Johan
Supplementary Figures and legend Heparin affinity purification of extracellular vesicles. Leonora Balaj 1, Nadia A. Atai 1,2, Weilin Chen 1, Dakai Mu 1, Bakhos A. Tannous 1, Xandra O. Breakefield 1, Johan
Supplementary Protocol. sirna transfection methodology and performance
 Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Protocol sirna transfection methodology and performance sirna oligonucleotides, DNA construct and cell line. Chemically synthesized 21 nt RNA duplexes were obtained from Ambion Europe, Ltd.
Supplementary Methods
 Supplementary Methods Antibodies For immunocytochemistry, the following antibodies were used: mouse anti-γ-h2ax (Upstate), rabbit anti-γ-h2ax (Abcam), rabbit anti-53bp1 (Novus), mouse anti-atm-phosphoserine1981
Supplementary Methods Antibodies For immunocytochemistry, the following antibodies were used: mouse anti-γ-h2ax (Upstate), rabbit anti-γ-h2ax (Abcam), rabbit anti-53bp1 (Novus), mouse anti-atm-phosphoserine1981
The measurement of telomere length was performed by the same method as in the previous study (11).
 1 SUPPLEMENTAL DATA 2 3 METHODS 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 Telomere length measurement by quantitative real-time PCR (q-pcr) The measurement of telomere length was performed
1 SUPPLEMENTAL DATA 2 3 METHODS 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 Telomere length measurement by quantitative real-time PCR (q-pcr) The measurement of telomere length was performed
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
ZytoLight FISH-Tissue Implementation Kit
 ZytoLight FISH-Tissue Implementation Kit Z-2028-20 20 Z-2028-5 5 For fluorescence in situ hybridization (FISH) using any ZytoLight FISH probe.... In vitro diagnostic medical device according to EU directive
ZytoLight FISH-Tissue Implementation Kit Z-2028-20 20 Z-2028-5 5 For fluorescence in situ hybridization (FISH) using any ZytoLight FISH probe.... In vitro diagnostic medical device according to EU directive
Supplemental Figure 1 (Figure S1), related to Figure 1 Figure S1 provides evidence to demonstrate Nfatc1Cre is a mouse line that directed gene
 Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Developmental Cell, Volume 25 Supplemental Information Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair Yiqin Xiong, Wei Li, Ching Shang, Richard M. Chen,
Protein patterning on hydrogels by direct microcontact. printing: application to cardiac differentiation SUPPLEMTARY INFORMATION
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Protein patterning on hydrogels by direct microcontact printing: application to cardiac differentiation
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Protein patterning on hydrogels by direct microcontact printing: application to cardiac differentiation
Propidium Iodide Solution
 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Propidium Iodide Solution (1mg/ml in deionized water) (Cat. # 786 1272, 786 1273) think proteins!
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Propidium Iodide Solution (1mg/ml in deionized water) (Cat. # 786 1272, 786 1273) think proteins!
ViewRNA Cell Plus Assay Catalog Number: RUO: For Research Use Only. Not for use in diagnostic procedures.
 Page 1 of 2 ViewRNA Cell Plus Assay Catalog Number: 88-19000 RUO: For Research Use Only. Not for use in diagnostic procedures. Product Information Contents: ViewRNA Cell Plus Assay Catalog Number: 88-19000
Page 1 of 2 ViewRNA Cell Plus Assay Catalog Number: 88-19000 RUO: For Research Use Only. Not for use in diagnostic procedures. Product Information Contents: ViewRNA Cell Plus Assay Catalog Number: 88-19000
TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells
 Supplementary Information for TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells Barun Mahata 1, Avisek
Supplementary Information for TALEN mediated targeted editing of GM2/GD2-synthase gene modulates anchorage independent growth by reducing anoikis resistance in mouse tumor cells Barun Mahata 1, Avisek
In Situ Hybridization
 In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
Supplementary Figure 1: (a) 3D illustration of the mobile phone microscopy attachment, containing a 3D movable sample holder (red piece for z
 Supplementary Figure 1: (a) 3D illustration of the mobile phone microscopy attachment, containing a 3D movable sample holder (red piece for z movement and blue piece for x-y movement). Illumination sources
Supplementary Figure 1: (a) 3D illustration of the mobile phone microscopy attachment, containing a 3D movable sample holder (red piece for z movement and blue piece for x-y movement). Illumination sources
Table 1.C.1 Example of Hazardous Materials Identification System. Name of chemical. Personal protection equipment. Table 1.C.2 Example NFPA table.
 Table 1.C.1 Example of Hazardous Materials Identification System. Name of chemical Health Flammability Reactivity Personal protection equipment Table 1.C.2 Example NFPA table. Fire Health Reactivity Specific
Table 1.C.1 Example of Hazardous Materials Identification System. Name of chemical Health Flammability Reactivity Personal protection equipment Table 1.C.2 Example NFPA table. Fire Health Reactivity Specific
Chapter 14. FISH Analysis of Human Pluripotent Stem Cells. Suzanne E. Peterson, Jerold Chun, and Jeanne Loring. Abstract. 1.
 Chapter 14 FISH Analysis of Human Pluripotent Stem Cells Suzanne E. Peterson, Jerold Chun, and Jeanne Loring Abstract Human pluripotent stem cells (PSCs) hold promise for treating a multitude of diseases.
Chapter 14 FISH Analysis of Human Pluripotent Stem Cells Suzanne E. Peterson, Jerold Chun, and Jeanne Loring Abstract Human pluripotent stem cells (PSCs) hold promise for treating a multitude of diseases.
Immunofluorescence Staining Protocol for 3 Well Chamber, removable
 Immunofluorescence Staining Protocol for 3 Well Chamber, removable This Application Note presents a simple protocol for the cultivation, fixation, and staining of cells using the 3 Well Chamber, removable.
Immunofluorescence Staining Protocol for 3 Well Chamber, removable This Application Note presents a simple protocol for the cultivation, fixation, and staining of cells using the 3 Well Chamber, removable.
In general, 8- to 10-week-old adult females were ovariectomized and rested for 10 days
 Animal injections and tissue processing In general, 8- to 10-week-old adult females were ovariectomized and rested for 10 days before they received any injections. Mice were injected with sesame seed oil
Animal injections and tissue processing In general, 8- to 10-week-old adult females were ovariectomized and rested for 10 days before they received any injections. Mice were injected with sesame seed oil
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
TECHNICAL BULLETIN. Goat ExtrAvidin Peroxidase Staining Kit. Product Number EXTRA-1 Storage Temperature 2-8 C
 Goat ExtrAvidin Peroxidase Staining Kit Product Number EXTRA-1 Storage Temperature 2-8 C TECHNICAL BULLETIN Product Description The unique avidin reagent, ExtrAvidin, combines the high specific activity
Goat ExtrAvidin Peroxidase Staining Kit Product Number EXTRA-1 Storage Temperature 2-8 C TECHNICAL BULLETIN Product Description The unique avidin reagent, ExtrAvidin, combines the high specific activity
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections
 Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Detection of Histone Modifications at Specific Gene Loci in Single Cells in Histological Sections Delphine Gomez; Laura L Shankman; Anh T Nguyen; Gary K Owens. Supplementary Figures 1-13 Supplementary
Materials and Methods Materials Required for Fixing, Embedding and Sectioning
 Page 1 Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigenspecific antibodies coupled to fluorophores. Depending on the antibody or dye used, proteins,
Page 1 Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigenspecific antibodies coupled to fluorophores. Depending on the antibody or dye used, proteins,
Survivin (BIRC5) Immunohistochemistry Kit
 Survivin (BIRC5) Immunohistochemistry Kit For Immunohistochemical Staining of Survivin (BIRC5) in human FFPE Tissue RUK-KBI01-20 For Research Use Only Riverside Biosciences Inc. 2327 S 5th Ave, North Riverside,
Survivin (BIRC5) Immunohistochemistry Kit For Immunohistochemical Staining of Survivin (BIRC5) in human FFPE Tissue RUK-KBI01-20 For Research Use Only Riverside Biosciences Inc. 2327 S 5th Ave, North Riverside,
Deoxyribonucleic Acid DNA
 Introduction to BioMEMS & Medical Microdevices DNA Microarrays and Lab-on-a-Chip Methods Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
Introduction to BioMEMS & Medical Microdevices DNA Microarrays and Lab-on-a-Chip Methods Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
