SUPPLEMENTAL MATERIAL FOR. Structure of Bacterial Transcription Factor SpoIIID and Evidence for a Novel
|
|
|
- Clinton Pitts
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTAL MATERIAL FOR Structure of Bacterial Transcription Factor SpoIIID and Evidence for a Novel Mode of DNA Binding Bin Chen, Paul Himes, Yu Liu, Yang Zhang, Zhenwei Lu, Aizhuo Liu, Honggao Yan, and Lee Kroos Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI USA 1
2 Table S1. Intermolecular NOEs observed in the SpoIIID DNA complex SpoIIID resonances Possible DNA resonances a DNA resonance chemical shifts b (ppm) Ser35 HN CYT H5 or any ribose H Thr36 HN ADE H6 or CYT H Glu43 HN Ribose H2, H2, H4, H5 or H Arg44 HN Ribose H2, H2, H4, H5 or H5 3.52, 3.26 Lys64 HN Ribose H2, H2, H4, H5 or H Arg67 HN Ribose H2, H2, H4, H5 or H ADE H2, H8 or CYT H6 or THY H6 or GUA 7.56 H8 or ADE H6 or CYT H4 or GUA H2 Gly71 HN Ribose H2, H2, H4, H5 or H5 3.62, 3.96 CYT H5 or any ribose H1 5.91, 6.14 Gly72 HN CYT H5 or any ribose H ADE H2, H8 or CYT H6 or THY H6 or GUA 7.79 H8 or ADE H6 or CYT H4 or GUA H2 Ala74 HN Ribose H2, H2, H4, H5 or H5 3.54, 3.96 ADE H2, H8 or CYT H6 or THY H6 or GUA 7.79 H8 or ADE H6 or CYT H4 or GUA H2 Thr75 HN Ribose H2, H2, H4, H5 or H5 3.96, 4.37 CYT H5 or any ribose H1 6.08, 6.40 Lys76 HN Ribose H2, H2, H4, H5 or H CYT H5 or any ribose H a The ambiguous chemical shift assignments were based on the DNA chemical shift data in the Biological Magnetic Resonance Data Bank ( b All intermolecular NOEs were obtained from the 3D 13 C, 15 N-filtered (F1), 15 N-edited (F3) NOESY spectrum in comparison with the 3D 15 N-edited NOESY spectrum. 2
3 Table S2. Substitutions in SpoIIID and corresponding plasmid and E. coli strain designations SpoIIID substitution pph4 derivative a E. coli strain b K34A pph251 PH251B H38A pph252 PH252B K39A pph253 PH253B E43A pph254 PH254B R44A pph255 PH255B K76A pph258 PH258B K78A pph259 PH259B K80A pph260 PH260B K81A pph261 PH261B a pph4 is a pet-21b (Novagen) derivative in which spoiiid is expressed from a T7 RNA polymerase promoter (1). b Plasmids were transformed into E. coli BL21 (DE3) (Novagen). 3
4 Table 3S. Oligonucleotides used in this study Oligonucleotide a Sequence b LK2331 (K34A) c 5 -ggaatttggtgtttccgcaagtacagtacacaagg-3 LK ccttgtgtactgtacttgcggaaacaccaaattcc-3 LK2333 (H38A) 5 -ggtgtttccaaaagtacagtagccaaggatttaacagagcgtc-3 LK gacgctctgttaaatccttggctactgtacttttggaaacacc-3 LK2335 (K39A) 5 -caaaagtacagtacacgcggatttaacagagcgtc-3 LK gacgctctgttaaatccgcgtgtactgtacttttg-3 LK2337 (E43A) 5 -gtacacaaggatttaacagcgcgtctgcctgaaattaac-3 LK gttaatttcaggcagacgcgctgttaaatccttgtgtac-3 LK2339 (R44A) 5 -cacaaggatttaacagaggcgctgcctgaaattaaccccg-3 LK cggggttaatttcaggcagcgcctctgttaaatccttgtg-3 LK2345 (K76A) 5 -ggaggagaagcgacagcgctcaaatataaaaaag-3 LK cttttttatatttgagcgctgtcgcttctcctcc-3 LK2347 (K78A) 5 -gaggagaagcgacaaagctcgcgtataaaaaagatg-3 LK catcttttttatacgcgagctttgtcgcttctcctc-3 LK2349 (K80A) 5 -gcgacaaagctcaaatatgcgaaagatgaaattctcgaag-3 LK cttcgagaatttcatctttcgcatatttgagctttgtcgc-3 LK2351 (K81A) 5 -gacaaagctcaaatataaagcggatgaaattctcgaagg-3 LK ccttcgagaatttcatccgctttatatttgagctttgtc-3 a Oligonucleotides are shown in pairs that were used for mutagenesis. b Boldface type indicates a mutation. c For each pair of oligonucleotides used for mutagenesis, the substitution is indicated in parentheses. 4
5 Table S4. Ambiguous interaction restraints for modeling the SpoIIID DNA complex Active residues for SpoIIID Passive residues for dsdna Method determined Lys34, His38, Lys39, Lys78, Any DNA nucleotide Mutational analysis Lys80, Lys81 HN of Ser35, Thr36, Glu43, See Supplementary Table S1 Intermolecular NOEs Arg44, Lys64, Arg67, Gly71, Gly72, Ala74, Thr75, Lys76 Active residues for dsdna a Passive residues for SpoIIID CYT4, THY5, THY6, GUA7, THY8, CYT9, CYT10, GUA19, GUA20, ADE21, CYT22, Any SpoIIID residue (1-81) Mutational analysis and/or sequence conservation ADE23, ADE24, GUA25 a See Figure S2 for base numbering. 5
6 mother cell forespore engulfment SpoIIID E polar septum coat K mother cell lysis Figure S1. Morphological changes and transcription factors during B. subtilis sporulation. Nutrient limitation initiates the process, causing a polar septum to form, which divides the cell into mother cell and forespore compartments. Differential transcription in the two cell types is directed by specific sigma subunits of RNA polymerase (2,3). After E RNA polymerase becomes active in the mother cell, it directs transcription of the gene for SpoIIID. The mother cell membrane of the septum engulfs the forespore (orange arrows), and channels (green) form between the two cell types (4-7). Fission of the mother cell membrane near the pole pinches the forespore off as a free protoplast inside the mother cell. In the absence of SpoIIID, some genes under E control are still expressed and engulfment is completed, but then the process of endospore formation stops. Although the blockage is not completely understood, one crucial function of SpoIIID is to activate transcription by E RNA polymerase of the gene for K (8,9). Genes under K control produce proteins that assemble a coat around the forespore, lyse the mother cell, and prepare the spore to germinate when nutrients are sensed (10). 6
7 5 1 GC 28 2 CG 27 3 GC 26 4 CG 25 5 TA 24 6 TA 23 7 GC 22 8 TA 21 9 CG CG TA AT AT TA 15 5 Figure S2. Numbering of bases in the 14-bp DNA duplex bound by SpoIIID. The idealized binding site consensus sequence indicated by the arrow in Figure 5A and in Figure S5A is bases
8 Wt Wt or Position of Ala substitution M Wt SpoIIID Figure S3. Purified SpoIIID proteins. SDS-PAGE followed by Coomassie blue staining of SeeBlue Plus 2 Prestained markers (M) (Invitrogen) with sizes (kda) indicated, a 2-fold dilution series of wild-type (Wt) SpoIIID starting at 0.5 g, unknown amounts of SpoIIID K76A and SpoIIID R44A, and 1 g of wild-type or single-ala-substituted SpoIIID as indicated. Based on the result, it was estimated that 0.5 g and 0.25 g of SpoIIID K76A and SpoIIID R44A had been loaded on the gel. 8
9 9
10 Figure S4. Secondary structural elements and NOEs of SpoIIID. (a) The secondary structural elements of SpoIIID and a summary of the sequential and medium-range NOEs, presence of HN- H 2 O cross-peaks, 13 C chemical shift indices for C and C, and cartoon representation of - helices. Residues that exhibited a cross-peak between HN and H 2 O resonances in the 3D 1 H- 15 N NOESY-HSQC spectrum are marked by *. Triangles indicate proline residues. Chemical shift indices (CSI) for C, C, and C characteristic for -helical regions are represented by ( ). (b) Distribution of NOEs along the sequence of SpoIIID. The number of intra-residue (white), sequential (light gray), medium-range (dark gray), and long-range (black) NOEs at each position is graphed. 10
11 a b Probe 10 CATTAGGACAAGCGCT 18 CATTAGGACAAACGCT 19 CATTAGGACAAGTGCT 20 CATTAGGACAAGCACT 21 CATTAGGACAAGCGTT 22 CATTAGGACGAGCGTT K d Position of Ala substitution SpoIIID: (nm) B Probe: U Figure S5. Binding of SpoIIID to DNA. (a) Sequences of DNA probes and apparent K d for binding of wild-type SpoIIID. Only one strand of each probe is shown. The arrow denotes the idealized binding site consensus sequence in probe 10 and underlined bases in the other probes indicate differences from probe 10. Apparent K d s are the average of a least 3 determinations ± 1 standard deviation. (b) EMSAs of SpoIIID R44A and SpoIIID K39A binding to probes Altered SpoIIID proteins (840 nm) were tested with different probes (indicated below the panel) (0.1 nm). Bound (B) and unbound (U) probe are indicated. 11
12 Figure S6. Schematic summary of hydrogen bonding interactions between SpoIIID and DNA in the top 10 models (a) and in the best model (b). Black lines in (a) indicate that at least one hydrogen bonding interaction is formed in all 10 models between a residue of SpoIIID and a phosphate oxygen, sugar, or base of DNA, while red lines indicate at least one interaction in greater than 8 models, blue lines greater than 5 models, and green lines less than 5 models. In the best model (b), some residues of SpoIIID form 2 or 3 hydrogen bonds with phosphate oxygens of DNA, as indicated by the numbers. 12
13 Helix 1 Helix 2 Helix 3 Helix 4 Helix 5 Figure S7. Alignment of SpoIIID orthologs. The highest scoring sequence from each species was identified using blastp (11, 12) with B. subtilis SpoIIID as the query, and the sequences were aligned using Clustal W (13). The results were visualized using ESPript (14). Numbers refer to residues in B. subtilis SpoIIID, which is marked with a star to the left. White, red, and black letters indicate identical, conserved, and less-conserved residues, respectively. Regions of B. subtilis SpoIIID that are -helical are indicated above the sequences. Adapted from (1).
14 REFERENCES 1. Himes, P., McBryant, S., and Kroos, L. (2010) Two regions of Bacillus subtilis transcription factor SpoIIID allow a monomer to bind DNA. J. Bacteriol. 192, Kroos, L. (2007) The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41, Losick, R., and Stragier, P. (1992) Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature 355, Camp, A. H., and Losick, R. (2008) A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69, Camp, A. H., and Losick, R. (2009) A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23, Doan, T., Morlot, C., Meisner, J., Serrano, M., Henriques, A. O., Moran, C. P., Jr., and Rudner, D. Z. (2009) Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5, e Meisner, J., Wang, X., Serrano, M., Henriques, A. O., and Moran, C. P., Jr. (2008) A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. USA 105, Halberg, R., and Kroos, L. (1994) Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J. Mol. Biol. 243, Kroos, L., Kunkel, B., and Losick, R. (1989) Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science 243, Eichenberger, P., Fujita, M., Jensen, S. T., Conlon, E. M., Rudner, D. Z., Wang, S. T., Ferguson, C., Haga, K., Sato, T., Liu, J. S., and Losick, R. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2, Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman Basic local alignment search tool. J. Mol. Biol. 215: Gish, W., and D. J. States Identification of protein coding regions by database similarity search. Nat. Genet. 3: Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins Clustal W and Clustal X version 2.0. Bioinformatics 23: Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis
 Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Key Area 1.3: Gene Expression
 Key Area 1.3: Gene Expression RNA There is a second type of nucleic acid in the cell, called RNA. RNA plays a vital role in the production of protein from the code in the DNA. What is gene expression?
Key Area 1.3: Gene Expression RNA There is a second type of nucleic acid in the cell, called RNA. RNA plays a vital role in the production of protein from the code in the DNA. What is gene expression?
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplemental Information. Structural Basis for Guide RNA Processing and. Seed-Dependent DNA Targeting by CRISPR-Cas12a
 Molecular Cell, Volume 66 Supplemental Information Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a Daan C. Swarts, John van der Oost, and Martin Jinek Figure
Molecular Cell, Volume 66 Supplemental Information Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a Daan C. Swarts, John van der Oost, and Martin Jinek Figure
-7 ::(teto) ::(teto) 120. GFP membranes merge. overexposed. Supplemental Figure 1 (Marquis et al.)
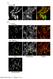 P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words).
 1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
Solutions to Quiz II
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Solutions to 7.014 Quiz II Class Average = 79 Median = 82 Grade Range % A 90-100 27 B 75-89 37 C 59 74 25 D 41 58 7 F 0 40 2 Question 1
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Solutions to 7.014 Quiz II Class Average = 79 Median = 82 Grade Range % A 90-100 27 B 75-89 37 C 59 74 25 D 41 58 7 F 0 40 2 Question 1
Gene regulation II Biochemistry 302. Bob Kelm March 1, 2004
 Gene regulation II Biochemistry 302 Bob Kelm March 1, 2004 Lessons to learn from bacteriophage λ in terms of transcriptional regulation Similarities to E. coli Cis-elements (operator elements) are adjacent
Gene regulation II Biochemistry 302 Bob Kelm March 1, 2004 Lessons to learn from bacteriophage λ in terms of transcriptional regulation Similarities to E. coli Cis-elements (operator elements) are adjacent
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
Ch 10 Molecular Biology of the Gene
 Ch 10 Molecular Biology of the Gene For Next Week Lab -Hand in questions from 4 and 5 by TUES in my mailbox (Biology Office) -Do questions for Lab 6 for next week -Lab practical next week Lecture Read
Ch 10 Molecular Biology of the Gene For Next Week Lab -Hand in questions from 4 and 5 by TUES in my mailbox (Biology Office) -Do questions for Lab 6 for next week -Lab practical next week Lecture Read
Supplementary Figure 1
 Supplementary Figure 1 2 Supplementary Figure 1: Sequence alignment of HsHSD17B8 and HsCBR4 of with KAR orthologs. The secondary structure elements as calculated by DSSP and residue numbers are displayed
Supplementary Figure 1 2 Supplementary Figure 1: Sequence alignment of HsHSD17B8 and HsCBR4 of with KAR orthologs. The secondary structure elements as calculated by DSSP and residue numbers are displayed
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
Supplementary Information for. Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction
 Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
What Are the Chemical Structures and Functions of Nucleic Acids?
 THE NUCLEIC ACIDS What Are the Chemical Structures and Functions of Nucleic Acids? Nucleic acids are polymers specialized for the storage, transmission, and use of genetic information. DNA = deoxyribonucleic
THE NUCLEIC ACIDS What Are the Chemical Structures and Functions of Nucleic Acids? Nucleic acids are polymers specialized for the storage, transmission, and use of genetic information. DNA = deoxyribonucleic
Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs.
 Supplementary Figure 1 Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs. (a) Sequence alignment of the ε-exonuclease homologs from four different
Supplementary Figure 1 Mycobacterium tuberculosis (Mtb) and Mycobacterium smegmatis (Msmeg) contain two ε (dnaq) exonuclease homologs. (a) Sequence alignment of the ε-exonuclease homologs from four different
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Bundle 6 Test Review
 Bundle 6 Test Review DNA vs. RNA DNA Replication Gene Mutations- Protein Synthesis 1. Label the different components and complete the complimentary base pairing. What is this molecule called? Deoxyribonucleic
Bundle 6 Test Review DNA vs. RNA DNA Replication Gene Mutations- Protein Synthesis 1. Label the different components and complete the complimentary base pairing. What is this molecule called? Deoxyribonucleic
Big Idea 3C Basic Review
 Big Idea 3C Basic Review 1. A gene is a. A sequence of DNA that codes for a protein. b. A sequence of amino acids that codes for a protein. c. A sequence of codons that code for nucleic acids. d. The end
Big Idea 3C Basic Review 1. A gene is a. A sequence of DNA that codes for a protein. b. A sequence of amino acids that codes for a protein. c. A sequence of codons that code for nucleic acids. d. The end
Bioinformatics. ONE Introduction to Biology. Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012
 Bioinformatics ONE Introduction to Biology Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012 Biology Review DNA RNA Proteins Central Dogma Transcription Translation
Bioinformatics ONE Introduction to Biology Sami Khuri Department of Computer Science San José State University Biology/CS 123A Fall 2012 Biology Review DNA RNA Proteins Central Dogma Transcription Translation
Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from
 Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Biochemistry 111. Carl Parker x A Braun
 Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
Biochemistry 111 Carl Parker x6368 101A Braun csp@caltech.edu Central Dogma of Molecular Biology DNA-Dependent RNA Polymerase Requires a DNA Template Synthesizes RNA in a 5 to 3 direction Requires ribonucleoside
IB HL Biology Test: Topics 1 and 3
 October 26, 2011 IB HL Biology Test: Topics 1 and 3 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. What conditions must be met for the t-test to be applied?
October 26, 2011 IB HL Biology Test: Topics 1 and 3 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. What conditions must be met for the t-test to be applied?
Videos. Lesson Overview. Fermentation
 Lesson Overview Fermentation Videos Bozeman Transcription and Translation: https://youtu.be/h3b9arupxzg Drawing transcription and translation: https://youtu.be/6yqplgnjr4q Objectives 29a) I can contrast
Lesson Overview Fermentation Videos Bozeman Transcription and Translation: https://youtu.be/h3b9arupxzg Drawing transcription and translation: https://youtu.be/6yqplgnjr4q Objectives 29a) I can contrast
Section 14.1 Structure of ribonucleic acid
 Section 14.1 Structure of ribonucleic acid The genetic code Sections of DNA are transcribed onto a single stranded molecule called RNA There are two types of RNA One type copies the genetic code and transfers
Section 14.1 Structure of ribonucleic acid The genetic code Sections of DNA are transcribed onto a single stranded molecule called RNA There are two types of RNA One type copies the genetic code and transfers
Gene expression. What is gene expression?
 Gene expression What is gene expression? Methods for measuring a single gene. Northern Blots Reporter genes Quantitative RT-PCR Operons, regulons, and stimulons. DNA microarrays. Expression profiling Identifying
Gene expression What is gene expression? Methods for measuring a single gene. Northern Blots Reporter genes Quantitative RT-PCR Operons, regulons, and stimulons. DNA microarrays. Expression profiling Identifying
Gene and DNA structure. Dr Saeb Aliwaini
 Gene and DNA structure Dr Saeb Aliwaini 2016 DNA during cell cycle Cell cycle for different cell types Molecular Biology - "Study of the synthesis, structure, and function of macromolecules (DNA, RNA,
Gene and DNA structure Dr Saeb Aliwaini 2016 DNA during cell cycle Cell cycle for different cell types Molecular Biology - "Study of the synthesis, structure, and function of macromolecules (DNA, RNA,
Student name ID # Second Mid Term Exam, Biology 2020, Spring 2002 Scores Total
 Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
1. DNA replication. (a) Why is DNA replication an essential process?
 ame Section 7.014 Problem Set 3 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68120 by 5:00pm on Friday
ame Section 7.014 Problem Set 3 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68120 by 5:00pm on Friday
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Module 3. Lecture 5. Regulation of Gene Expression in Prokaryotes
 Module 3 Lecture 5 Regulation of Gene Expression in Prokaryotes Recap So far, we have looked at prokaryotic gene regulation using 3 operon models. lac: a catabolic operon which displays induction via negative
Module 3 Lecture 5 Regulation of Gene Expression in Prokaryotes Recap So far, we have looked at prokaryotic gene regulation using 3 operon models. lac: a catabolic operon which displays induction via negative
Supporting Information
 Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
Supporting Information Wiley-VCH 2006 69451 Weinheim, Germany RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch Günter Mayer, Marie-Sophie L. Raddatz, Julia D. Grunwald,
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway
 Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Nucleic acids. How DNA works. DNA RNA Protein. DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology
 Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION A human XRCC4-XLF complex bridges DNA ends. Sara N. Andres 1, Alexandra Vergnes 2, Dejan Ristic 3, Claire Wyman 3, Mauro Modesti 2,4, and Murray Junop 2,4 1 Department of Biochemistry
SUPPLEMENTARY INFORMATION A human XRCC4-XLF complex bridges DNA ends. Sara N. Andres 1, Alexandra Vergnes 2, Dejan Ristic 3, Claire Wyman 3, Mauro Modesti 2,4, and Murray Junop 2,4 1 Department of Biochemistry
Daily Agenda. Warm Up: Review. Translation Notes Protein Synthesis Practice. Redos
 Daily Agenda Warm Up: Review Translation Notes Protein Synthesis Practice Redos 1. What is DNA Replication? 2. Where does DNA Replication take place? 3. Replicate this strand of DNA into complimentary
Daily Agenda Warm Up: Review Translation Notes Protein Synthesis Practice Redos 1. What is DNA Replication? 2. Where does DNA Replication take place? 3. Replicate this strand of DNA into complimentary
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system
 SUPPLEMENTARY DATA Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system Daniel Gleditzsch 1, Hanna Müller-Esparza 1, Patrick Pausch 2,3, Kundan Sharma 4, Srivatsa Dwarakanath 1,
SUPPLEMENTARY DATA Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system Daniel Gleditzsch 1, Hanna Müller-Esparza 1, Patrick Pausch 2,3, Kundan Sharma 4, Srivatsa Dwarakanath 1,
Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with
 Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with transcription buffer with or without a high salt concentration
Supplementary Fig. 1. Characteristics of transcription elongation by YonO. a. YonO forms a saltstable EC. Immobilized ECs were washed with transcription buffer with or without a high salt concentration
BIO 101 : The genetic code and the central dogma
 BIO 101 : The genetic code and the central dogma NAME Objectives The purpose of this exploration is to... 1. design experiments to decipher the genetic code; 2. visualize the process of protein synthesis;
BIO 101 : The genetic code and the central dogma NAME Objectives The purpose of this exploration is to... 1. design experiments to decipher the genetic code; 2. visualize the process of protein synthesis;
LABS 9 AND 10 DNA STRUCTURE AND REPLICATION; RNA AND PROTEIN SYNTHESIS
 LABS 9 AND 10 DNA STRUCTURE AND REPLICATION; RNA AND PROTEIN SYNTHESIS OBJECTIVE 1. OBJECTIVE 2. OBJECTIVE 3. OBJECTIVE 4. Describe the structure of DNA. Explain how DNA replicates. Understand the structure
LABS 9 AND 10 DNA STRUCTURE AND REPLICATION; RNA AND PROTEIN SYNTHESIS OBJECTIVE 1. OBJECTIVE 2. OBJECTIVE 3. OBJECTIVE 4. Describe the structure of DNA. Explain how DNA replicates. Understand the structure
From Gene to Protein
 8.2 Structure of DNA From Gene to Protein deoxyribonucleic acid - (DNA) - the ultimate source of all information in a cell This information is used by the cell to produce the protein molecules which are
8.2 Structure of DNA From Gene to Protein deoxyribonucleic acid - (DNA) - the ultimate source of all information in a cell This information is used by the cell to produce the protein molecules which are
The Structure of Proteins The Structure of Proteins. How Proteins are Made: Genetic Transcription, Translation, and Regulation
 How Proteins are Made: Genetic, Translation, and Regulation PLAY The Structure of Proteins 14.1 The Structure of Proteins Proteins - polymer amino acids - monomers Linked together with peptide bonds A
How Proteins are Made: Genetic, Translation, and Regulation PLAY The Structure of Proteins 14.1 The Structure of Proteins Proteins - polymer amino acids - monomers Linked together with peptide bonds A
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
MBMB451A Section1 Fall 2008 KEY These questions may have more than one correct answer
 MBMB451A Section1 Fall 2008 KEY These questions may have more than one correct answer 1. In a double stranded molecule of DNA, the ratio of purines : pyrimidines is (a) variable (b) determined by the base
MBMB451A Section1 Fall 2008 KEY These questions may have more than one correct answer 1. In a double stranded molecule of DNA, the ratio of purines : pyrimidines is (a) variable (b) determined by the base
Molecular Biology (1)
 Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 49-52, 118-119, 130 Nucleic acids 2 types: Deoxyribonucleic acid
Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 49-52, 118-119, 130 Nucleic acids 2 types: Deoxyribonucleic acid
7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions will be posted on the web.
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
Molecular Biology. IMBB 2017 RAB, Kigali - Rwanda May 02 13, Francesca Stomeo
 Molecular Biology IMBB 2017 RAB, Kigali - Rwanda May 02 13, 2017 Francesca Stomeo Molecular biology is the study of biology at a molecular level, especially DNA and RNA - replication, transcription, translation,
Molecular Biology IMBB 2017 RAB, Kigali - Rwanda May 02 13, 2017 Francesca Stomeo Molecular biology is the study of biology at a molecular level, especially DNA and RNA - replication, transcription, translation,
What is DNA??? DNA = Deoxyribonucleic acid IT is a molecule that contains the code for an organism s growth and function
 Review DNA and RNA 1) DNA and RNA are important organic compounds found in cells, called nucleic acids 2) Both DNA and RNA molecules contain the following chemical elements: carbon, hydrogen, oxygen, nitrogen
Review DNA and RNA 1) DNA and RNA are important organic compounds found in cells, called nucleic acids 2) Both DNA and RNA molecules contain the following chemical elements: carbon, hydrogen, oxygen, nitrogen
Problem Set Unit The base ratios in the DNA and RNA for an onion (Allium cepa) are given below.
 Problem Set Unit 3 Name 1. Which molecule is found in both DNA and RNA? A. Ribose B. Uracil C. Phosphate D. Amino acid 2. Which molecules form the nucleotide marked in the diagram? A. phosphate, deoxyribose
Problem Set Unit 3 Name 1. Which molecule is found in both DNA and RNA? A. Ribose B. Uracil C. Phosphate D. Amino acid 2. Which molecules form the nucleotide marked in the diagram? A. phosphate, deoxyribose
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
Chromatin Structure. a basic discussion of protein-nucleic acid binding
 Chromatin Structure 1 Chromatin DNA packaging g First a basic discussion of protein-nucleic acid binding Questions to answer: How do proteins bind DNA / RNA? How do proteins recognize a specific nucleic
Chromatin Structure 1 Chromatin DNA packaging g First a basic discussion of protein-nucleic acid binding Questions to answer: How do proteins bind DNA / RNA? How do proteins recognize a specific nucleic
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 10 Nucleic Acids
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 10 Nucleic Acids 2 3 DNA vs RNA DNA RNA deoxyribose ribose A, C, G, T A, C, G, U 10 3 10 8 nucleotides 10 2 10 4 nucleotides nucleus cytoplasm double-stranded
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 10 Nucleic Acids 2 3 DNA vs RNA DNA RNA deoxyribose ribose A, C, G, T A, C, G, U 10 3 10 8 nucleotides 10 2 10 4 nucleotides nucleus cytoplasm double-stranded
DNA. ncrna. Non-coding RNA analysis for gene expression regulation using bioinformatics tools
 non-coding RNA 1 2 2 DNA RNA RNA non-coding RNA ncrna ncrna ncrna ncrna ncrna ncrna ncrna ncrna ncrna non-coding RNA Non-coding RNA analysis for gene expression regulation using bioinformatics tools Hayashi
non-coding RNA 1 2 2 DNA RNA RNA non-coding RNA ncrna ncrna ncrna ncrna ncrna ncrna ncrna ncrna ncrna non-coding RNA Non-coding RNA analysis for gene expression regulation using bioinformatics tools Hayashi
DNA, Replication and RNA
 DNA, Replication and RNA The structure of DNA DNA, or Deoxyribonucleic Acid, is the blue prints for building all of life. DNA is a long molecule made up of units called NUCLEOTIDES. Each nucleotide is
DNA, Replication and RNA The structure of DNA DNA, or Deoxyribonucleic Acid, is the blue prints for building all of life. DNA is a long molecule made up of units called NUCLEOTIDES. Each nucleotide is
1/4/18 NUCLEIC ACIDS. Nucleic Acids. Nucleic Acids. ECS129 Instructor: Patrice Koehl
 NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS. ECS129 Instructor: Patrice Koehl
 NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
MOLECULAR GENETICS PROTEIN SYNTHESIS. Molecular Genetics Activity #2 page 1
 AP BIOLOGY MOLECULAR GENETICS ACTIVITY #2 NAME DATE HOUR PROTEIN SYNTHESIS Molecular Genetics Activity #2 page 1 GENETIC CODE PROTEIN SYNTHESIS OVERVIEW Molecular Genetics Activity #2 page 2 PROTEIN SYNTHESIS
AP BIOLOGY MOLECULAR GENETICS ACTIVITY #2 NAME DATE HOUR PROTEIN SYNTHESIS Molecular Genetics Activity #2 page 1 GENETIC CODE PROTEIN SYNTHESIS OVERVIEW Molecular Genetics Activity #2 page 2 PROTEIN SYNTHESIS
7.014 Solution Set 4
 7.014 Solution Set 4 Question 1 Shown below is a fragment of the sequence of a hypothetical bacterial gene. This gene encodes production of HWDWN, protein essential for metabolizing sugar yummose. The
7.014 Solution Set 4 Question 1 Shown below is a fragment of the sequence of a hypothetical bacterial gene. This gene encodes production of HWDWN, protein essential for metabolizing sugar yummose. The
University of Groningen
 University of Groningen Coupling dttp Hydrolysis with DNA Unwinding by the DNA Helicase of Bacteriophage T7 Satapathy, Ajit K.; Kulczyk, Arkadiusz W.; Ghosh, Sharmistha; van Oijen, Antonius; Richardson,
University of Groningen Coupling dttp Hydrolysis with DNA Unwinding by the DNA Helicase of Bacteriophage T7 Satapathy, Ajit K.; Kulczyk, Arkadiusz W.; Ghosh, Sharmistha; van Oijen, Antonius; Richardson,
Nucleic acid and protein Flow of genetic information
 Nucleic acid and protein Flow of genetic information References: Glick, BR and JJ Pasternak, 2003, Molecular Biotechnology: Principles and Applications of Recombinant DNA, ASM Press, Washington DC, pages.
Nucleic acid and protein Flow of genetic information References: Glick, BR and JJ Pasternak, 2003, Molecular Biotechnology: Principles and Applications of Recombinant DNA, ASM Press, Washington DC, pages.
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
When times are good and when times are bad: Stringent response Stationary phase
 When times are good and when times are bad: Stringent response Stationary phase Reading Chapter 13 p,571-572, 573-579, 580-581, 582-584, 554-556,, 598-602 Example of catabolite control Cells shifted from
When times are good and when times are bad: Stringent response Stationary phase Reading Chapter 13 p,571-572, 573-579, 580-581, 582-584, 554-556,, 598-602 Example of catabolite control Cells shifted from
Lecture for Wednesday. Dr. Prince BIOL 1408
 Lecture for Wednesday Dr. Prince BIOL 1408 THE FLOW OF GENETIC INFORMATION FROM DNA TO RNA TO PROTEIN Copyright 2009 Pearson Education, Inc. Genes are expressed as proteins A gene is a segment of DNA that
Lecture for Wednesday Dr. Prince BIOL 1408 THE FLOW OF GENETIC INFORMATION FROM DNA TO RNA TO PROTEIN Copyright 2009 Pearson Education, Inc. Genes are expressed as proteins A gene is a segment of DNA that
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Unit 3c. Microbial Gene0cs
 Unit 3c Microbial Gene0cs Microbial Genetics! Gene0cs: the science of heredity Genome: the gene0c informa0on in the cell Genomics: the sequencing and molecular characteriza0on of genomes Gregor Mendel
Unit 3c Microbial Gene0cs Microbial Genetics! Gene0cs: the science of heredity Genome: the gene0c informa0on in the cell Genomics: the sequencing and molecular characteriza0on of genomes Gregor Mendel
DNA & DNA : Protein Interactions BIBC 100
 DNA & DNA : Protein Interactions BIBC 100 Sequence = Information Alphabet = language L,I,F,E LIFE DNA = DNA code A, T, C, G CAC=Histidine CAG=Glutamine GGG=Glycine Protein = Protein code 20 a.a. LIVE EVIL
DNA & DNA : Protein Interactions BIBC 100 Sequence = Information Alphabet = language L,I,F,E LIFE DNA = DNA code A, T, C, G CAC=Histidine CAG=Glutamine GGG=Glycine Protein = Protein code 20 a.a. LIVE EVIL
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
STRUCTURE, DYNAMICS AND INTERACTIONS OF PROTEINS BY NMR SPECTROSCOPY
 STRUCTURE, DYNAMICS AND INTERACTIONS OF PROTEINS BY NMR SPECTROSCOPY Constantin T. Craescu INSERM & Institut Curie - Recherche Orsay, France A SHORT INTRODUCTION TO PROTEIN STRUCTURE Chemical composition
STRUCTURE, DYNAMICS AND INTERACTIONS OF PROTEINS BY NMR SPECTROSCOPY Constantin T. Craescu INSERM & Institut Curie - Recherche Orsay, France A SHORT INTRODUCTION TO PROTEIN STRUCTURE Chemical composition
Molecular Biology (1)
 Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2017-2018 Resources This lecture Cooper, pp. 49-52, 118-119, 130 What is molecular biology? Central dogma
Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2017-2018 Resources This lecture Cooper, pp. 49-52, 118-119, 130 What is molecular biology? Central dogma
DNA RNA PROTEIN. Professor Andrea Garrison Biology 11 Illustrations 2010 Pearson Education, Inc. unless otherwise noted
 DNA RNA PROTEIN Professor Andrea Garrison Biology 11 Illustrations 2010 Pearson Education, Inc. unless otherwise noted DNA Molecule of heredity Contains all the genetic info our cells inherit Determines
DNA RNA PROTEIN Professor Andrea Garrison Biology 11 Illustrations 2010 Pearson Education, Inc. unless otherwise noted DNA Molecule of heredity Contains all the genetic info our cells inherit Determines
Zool 3200: Cell Biology Exam 3 3/6/15
 Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Site directed mutagenesis, Insertional and Deletion Mutagenesis. Mitesh Shrestha
 Site directed mutagenesis, Insertional and Deletion Mutagenesis Mitesh Shrestha Mutagenesis Mutagenesis (the creation or formation of a mutation) can be used as a powerful genetic tool. By inducing mutations
Site directed mutagenesis, Insertional and Deletion Mutagenesis Mitesh Shrestha Mutagenesis Mutagenesis (the creation or formation of a mutation) can be used as a powerful genetic tool. By inducing mutations
Introduction to Cellular Biology and Bioinformatics. Farzaneh Salari
 Introduction to Cellular Biology and Bioinformatics Farzaneh Salari Outline Bioinformatics Cellular Biology A Bioinformatics Problem What is bioinformatics? Computer Science Statistics Bioinformatics Mathematics...
Introduction to Cellular Biology and Bioinformatics Farzaneh Salari Outline Bioinformatics Cellular Biology A Bioinformatics Problem What is bioinformatics? Computer Science Statistics Bioinformatics Mathematics...
Videos. Bozeman Transcription and Translation: Drawing transcription and translation:
 Videos Bozeman Transcription and Translation: https://youtu.be/h3b9arupxzg Drawing transcription and translation: https://youtu.be/6yqplgnjr4q Objectives 29a) I can contrast RNA and DNA. 29b) I can explain
Videos Bozeman Transcription and Translation: https://youtu.be/h3b9arupxzg Drawing transcription and translation: https://youtu.be/6yqplgnjr4q Objectives 29a) I can contrast RNA and DNA. 29b) I can explain
Gene Expression - Transcription
 DNA Gene Expression - Transcription Genes are expressed as encoded proteins in a 2 step process: transcription + translation Central dogma of biology: DNA RNA protein Transcription: copy DNA strand making
DNA Gene Expression - Transcription Genes are expressed as encoded proteins in a 2 step process: transcription + translation Central dogma of biology: DNA RNA protein Transcription: copy DNA strand making
The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR
 The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR S. Thomas, C. S. Fernando, J. Roach, U. DeSilva and C. A. Mireles DeWitt The objective
The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR S. Thomas, C. S. Fernando, J. Roach, U. DeSilva and C. A. Mireles DeWitt The objective
Nucleic Acid Structure:
 Genetic Information In Microbes: The genetic material of bacteria and plasmids is DNA. Bacterial viruses (bacteriophages or phages) have DNA or RNA as genetic material. The two essential functions of genetic
Genetic Information In Microbes: The genetic material of bacteria and plasmids is DNA. Bacterial viruses (bacteriophages or phages) have DNA or RNA as genetic material. The two essential functions of genetic
MOLECULAR STRUCTURE OF DNA
 MOLECULAR STRUCTURE OF DNA Characteristics of the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell (generation to generation) 2. Information Storage Biologically
MOLECULAR STRUCTURE OF DNA Characteristics of the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell (generation to generation) 2. Information Storage Biologically
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Problem Set #2
 20.320 Problem Set #2 Due on September 30rd, 2011 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all of your assumptions, and provide step-by-step solutions
20.320 Problem Set #2 Due on September 30rd, 2011 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all of your assumptions, and provide step-by-step solutions
1 Fig. 5.1 shows part of a DNA strand. Fig (a) (i) Name the base represented by the letter T. ... [1]
![1 Fig. 5.1 shows part of a DNA strand. Fig (a) (i) Name the base represented by the letter T. ... [1] 1 Fig. 5.1 shows part of a DNA strand. Fig (a) (i) Name the base represented by the letter T. ... [1]](/thumbs/81/84079258.jpg) 1 Fig. 5.1 shows part of a DNA strand. Fig. 5.1 (a) (i) Name the base represented by the letter T.... [1] On Fig. 5.1, draw a section of the mrna strand that is complementary to the section of the DNA
1 Fig. 5.1 shows part of a DNA strand. Fig. 5.1 (a) (i) Name the base represented by the letter T.... [1] On Fig. 5.1, draw a section of the mrna strand that is complementary to the section of the DNA
Transcription and Translation. DANILO V. ROGAYAN JR. Faculty, Department of Natural Sciences
 Transcription and Translation DANILO V. ROGAYAN JR. Faculty, Department of Natural Sciences Protein Structure Made up of amino acids Polypeptide- string of amino acids 20 amino acids are arranged in different
Transcription and Translation DANILO V. ROGAYAN JR. Faculty, Department of Natural Sciences Protein Structure Made up of amino acids Polypeptide- string of amino acids 20 amino acids are arranged in different
Genetics: Analysis of Genes and Genomes, Ninth Edition. Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION
 CHAPTER 1 STUDENT NOT STUDY FOR SALE GUIDE OR DISTRIBUTION TEST BANK Daniel Jones L. Hartl & Bartlett and Bruce Learning, J. Cochrane LLC Multiple Choice 1. NOT What FOR happens SALE if organisms OR DISTRIBUTION
CHAPTER 1 STUDENT NOT STUDY FOR SALE GUIDE OR DISTRIBUTION TEST BANK Daniel Jones L. Hartl & Bartlett and Bruce Learning, J. Cochrane LLC Multiple Choice 1. NOT What FOR happens SALE if organisms OR DISTRIBUTION
BIO303, Genetics Study Guide II for Spring 2007 Semester
 BIO303, Genetics Study Guide II for Spring 2007 Semester 1 Questions from F05 1. Tryptophan (Trp) is encoded by the codon UGG. Suppose that a cell was treated with high levels of 5- Bromouracil such that
BIO303, Genetics Study Guide II for Spring 2007 Semester 1 Questions from F05 1. Tryptophan (Trp) is encoded by the codon UGG. Suppose that a cell was treated with high levels of 5- Bromouracil such that
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Can have defects in any of the steps in the synthesis of arginine. With arginine in the medium, all arg mutants can grow on minimal medium.
 Molecular Biology I Biochemistry studying a single component in an organism Genetics studying an organism without that component Biochemical Genetics Look at the Arginine biosynthetic pathway: A B C Arginine
Molecular Biology I Biochemistry studying a single component in an organism Genetics studying an organism without that component Biochemical Genetics Look at the Arginine biosynthetic pathway: A B C Arginine
Synthetic Biology for
 Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
A Discovery Laboratory Investigating Bacterial Gene Regulation
 Chapter 8 A Discovery Laboratory Investigating Bacterial Gene Regulation Robert Moss Wofford College 429 N. Church Street Spartanburg, SC 29307 mosssre@wofford.edu Bob Moss is an Associate Professor of
Chapter 8 A Discovery Laboratory Investigating Bacterial Gene Regulation Robert Moss Wofford College 429 N. Church Street Spartanburg, SC 29307 mosssre@wofford.edu Bob Moss is an Associate Professor of
Feedback D. Incorrect! No, although this is a correct characteristic of RNA, this is not the best response to the questions.
 Biochemistry - Problem Drill 23: RNA No. 1 of 10 1. Which of the following statements best describes the structural highlights of RNA? (A) RNA can be single or double stranded. (B) G-C pairs have 3 hydrogen
Biochemistry - Problem Drill 23: RNA No. 1 of 10 1. Which of the following statements best describes the structural highlights of RNA? (A) RNA can be single or double stranded. (B) G-C pairs have 3 hydrogen
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2013-2014 MOLECULAR BIOLOGY BIO-2B02 Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question
A. Incorrect! This feature does help with it suitability as genetic material.
 College Biology - Problem Drill 08: Gene Structures and Functions No. 1 of 10 1. Which of the statements below is NOT true in explaining why DNA is a suitable genetic material? #01 (A) Its double helix
College Biology - Problem Drill 08: Gene Structures and Functions No. 1 of 10 1. Which of the statements below is NOT true in explaining why DNA is a suitable genetic material? #01 (A) Its double helix
Lac Operon contains three structural genes and is controlled by the lac repressor: (1) LacY protein transports lactose into the cell.
 Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
Supplementary Figure 1 Sequence alignment of representative CbbQ sequences.
 Walker A Pore loop 1 Walker B Supplementary Figure 1 Sequence alignment of representative CbbQ sequences. Amino acid sequences of CbbQ1 and CbbQ2 proteins from selected chemoautotrophic bacteria were aligned
Walker A Pore loop 1 Walker B Supplementary Figure 1 Sequence alignment of representative CbbQ sequences. Amino acid sequences of CbbQ1 and CbbQ2 proteins from selected chemoautotrophic bacteria were aligned
