Prof. dr. ir. Jan Steyaert
|
|
|
- Ellen Wells
- 6 years ago
- Views:
Transcription
1 Prof. dr. ir. Jan Steyaert Structural Biology Brussels Vrije Univestiteit Brussel & Structural Biology Research Center Vlaams Instituut Biotechnologie Pleinlaan Brussel Belgium Phone Fax Mobile Jan.Steyaert@vub.ac.be PERSONAL INFORMATION Full name STEYAERT JAN, Stefaan, Peter, Jozef Place of birth Ukkel (Belgium) Date of birth 21 April 1964 Nationality Belgian Languages Dutch (Native), English (Fluent), French (Fluent) URL for web site CURRENT POSITIONS Full Professor & Francqui Research Professor, Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium. Director, Department of Structural Biology Brussels, Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium Deputy-Director, Structural Biology Research Center, Vlaams Instituut Biotechnologie (VIB), Brussels, Belgium. EDUCATION Curriculum vitae of Jan Steyaert November 6,
2 PhD in Bioengineering Sciences, Plant Genetic Systems and Vrije Universiteit Brussel (VUB), Brussels, Belgium. Maxima cum laude MSc in Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium. Maxima cum laude BSc in Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium. Cum laude POSITIONS AND EMPLOYEMENT Francqui Research Professor, Francqui foundation, Sabbatical leave, visiting the labs of Brian Kobilka (Stanford, USA), Robert Lefkowitz (Duke, USA) and Gebhard Shertler (PSI, Switzerland) 2011-current Deputy-Director, Structural Biology Research Center, Vlaams Instituut Biotechnologie (VIB), Brussels, Belgium Director at interim, Structural Biology Research Center, Vlaams Instituut Biotechnologie (VIB), Brussels, Belgium 2008-current 2006-current Director, Structural Biology Brussels, Vrije Universiteit Brussel (VUB), Brussels, Belgium Full professor (Gewoon Hoogleraar), Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium Full professor (Hoogleraar), Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium Associate professor (Hoofddocent), Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel, Brussels, Belgium 1998-current Groupleader, Structural Biology Research Center, Vlaams Instituut Biotechnologie (VIB), Brussels, Belgium Assistant professor (Docent), Faculty of Sciences and Bioengineering Sciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium Postdoctoral fellow, International Laboratory for Research on Animal Diseases (ILRI), Nairobi, Kenya Postdoctoral fellow (FWO), Institute for molecular Biology & Biotechnology, Department of Ultrastructure, Vrije Universiteit Brussel (VUB), Sint-Genesius-Rode, Belgium. PUBLICATION LIST For actual h-index and citations: Google scolar: 1. Ehrnstorfer, I.A., Geertsma, E.R., Pardon, E., Steyaert, J. & Dutzler, R. (2014) The structural basis for transition metal ion selectivity in transporters of the SLC11/NRAMP family. Nature Struct Mol Biol 21, (ISI IF2013 = ) 2
3 2. Somme, J., Van Laer, B., Roovers, M., Steyaert, J., Versées, W., & Droogmans, L. (2014) Characterisation of two homologous 2 -O-methyltransferases showing different specificities for their trna substrates, RNA 20 (8), (ISI IF2012 = 5.088) 3. Oyen, D., Steyaert, J., & Barlow, J. (2014) Inhibition of ligand exchange kinetics via active-site trapping with an antibody fragment. Biochemistry 53, (ISI IF2012 = 3.377) 4. Fislage, M., Brosens, E., Deyaert, Egon., Spilotros, A., Pardon, E., Loris, R., Steyaert, J., Garcia-Pino, A., & Versees, W. (2014) SAXS analysis of the trna-modifying enzyme complex MnmE/MnmG reveals a novel interaction mode and GTP-induced oligomerization, Nucleic acids res, in press. (ISI IF2012 = 8.278) 5. Pardon, E., Laeremans, T., Triest, S., Rasmussen, S. G. F., Wohlkönig, A., Ruf, A., Muyldermans, S., Hol, W.J.G., Kobilka, B.K. & Steyaert, J. (2014) A general protocol for the generation of Nanobodies for structural biology. Nature protocols 9, Featured protocol (ISI IF2012 = 7.960) 6. Pathare, G.R., Nagy, I., Śledź, P., Anderson, D.J., Zhou, H.-J., Pardon, E., Steyaert, J., Förster, F., Bracher, A. & Baumeister, W. (2014) Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci U S A 111, (ISI IF2012 = 9.737) 7. Abskharon, R.N.N., Giachin, G., Wohlkonig, A., Soror, S.H., Pardon, E., Legname, G. & Steyaert, J. (2014) Probing the N-terminal β-sheet conversion in the crystal structure of the full-length human prion protein bound to a Nanobody, J Am Chem Soc 136, (ISI IF2012 = ) 8. Staus, D.P., Wingler, L. M., Strachan, R. T., Rasmussen, S. G. F., Pardon, E., Ahn, S., Steyaert, J., Kobilka, B.K., & Lefkowitz, R. J. (2014) Regulation of Beta-2-Adrenergic Receptor Function by Conformationally Selective Singledomain Intrabodies. Mol Pharm 85, (ISI IF2012 = 4.411) 9. Kruse, A.C., Ring, A.M., Manglik, A., Hu, J., Hu, K., Eitel, K., Hübner, H., Pardon, E., Valant, V., Sexton, P.M., Christopoulos, A., Felder, C., Gmeiner, P., Steyaert, J., Weis, W.I., K., Garcia, K., Wess, J., & Kobilka, B.K. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, (ISI IF2012 = ) 10. Oyen, D., Wechselberger, R., Srinivasan, V., Steyaert, J., & Barlow, J. N. (2013). Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihydrofolate reductase of Escherichia coli. Biochim Biophys Acta 1834, (ISI IF2012 = 3.733) 11. Löw, C., Yau, Y.H., Pardon, E., Jegerschöld, C., Wåhlin, L., Quistgaard, E.M., Moberg, P., Geifman-Shochat, S., Steyaert, J. & Nordlund, P. (2013). Nanobody Mediated Crystallization of an Archeal Mechanosensitive Channel. PloS one 8, e (ISI IF2012 = 3.730) 12. Yuan, J., Zhan, Y.-A., Abskharon, R., Xiao, X., Martinez, M.C., Zhou, X., Kneale, G., Mikol, J., Lehmann, S., Surewicz, W.K., Castilla, J., Steyaert, J., Zhang, Z., Kong, Q., Petersen, R.B., Wohlkonig, A. & Zou, W.-Q. (2013) Recombinant Human Prion Protein Inhibits Prion Propagation in vitro. Nature Sci Rep 3, (ISI IF2012 = 2.927) 13. Saskia Vanderhaegen, S., Fislage, M., Domanska, K., Versées, W., Pardon, E., Bellotti, V., & Steyaert, J. (2013) Structure of an early native-like intermediate of β2-microglobulin amyloidogenesis. Protein Sci 22, (ISI IF2012 = 2.735) 14. Ward, A., Szewczyk, P., Grimar, V., Lee, C.-W., Matrinez, L., Cay, A., Villaluz, M., Pardon, E., Cregger, C., Falsone, P., Urbatsch, I., Govaerts, C., Steyaert, J., & Chang, G. (2013) Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain, Proc Natl Acad Sci U S A 110, (ISI IF2012 = 9.737) 15. Khamrui, S., Turley, S., Pardon, E., Steyaert, J., Fan, E., Verlinde, C., Bergman, L.W. & Hol, W. (2013) The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a Nanobody, Mol Biochem Parasitol 190, (ISI IF2012 = 2.734) 16. Sohier, J.S., Laurent C., Chevigne A., Pardon E., Srinivasan V., Wernery U., Steyaert J., Galleni M. (2013) Allosteric inhibition of VIM metallo-beta-lactamases by a camelid nanobody. Biochem J 15, (ISI IF2012 = 4.654) 17. Banner, D. W., Gsell, B., Benz, J., Bertschinger, J., Burger, D., Brack, S., Cuppuleri, S., Debulpaep, M., Gast, A., Grabulovski, D., Hennig, M., Hilpert, H., Huber, W., Kuglstatter, A., Kusznir, E., Laeremans, T., Matile, H., Miscenic, C., Rufer, A. C., Schlatter, D., Steyaert, J., Stihle, M., Thoma, R., Weber, M. & Ruf, A. (2013) Mapping 3
4 the conformational space accessible to BACE2 using surface mutants and cocrystals with Fab fragments, Fynomers and Xaperones. Acta Crystallographica Section D 69, (ISI IF2012 = ) 18. Vuchelen, A., Pardon, E., Steyaert, J., Gamain, B., Loris, R., van Nuland, N.A. J., Ramboarina, S. (2013) Production, crystallization and X-ray diffraction analysis of two nanobodies against the Duffy binding-like (DBL) domain DBL6 -FCR3 of the Plasmodium falciparum VAR2CSA protein. Acta Crystallogr F Struct Biol Cryst Commun 69, ISI IF12 = Vercruysse,T., Boons, E., Venken, T., Vanstreels, E., Voet, A., Steyaert, J., De Maeyer, M., Daelemans, D. (2013) Mapping the Binding Interface between an HIV-1 Inhibiting Intrabody and the Viral Protein Rev. PLoS one 8, e ISI IF12 = Guilliams, T., El-Turk, F., Buell, A.K., O'Day, E.M. Aprile, F.A., Esbjörner, E.K., Vendruscolo, M., Cremades, N., Pardon, E., Wyns, L., Welland, M.E., Steyaert, J., Christodoulou, J., Dobson, C.M. & De Genst, E. (2013) Nanobodies raised against monomeric α-synuclein distinguish between fibrils at different maturation stages. J Mol Biol 245, ISI IF12 = a. JMB commentary: The Mysterious C-Terminal Tail of Alpha-Synuclein: Nanobody's Guess. J Mol Biol 245, Rivera-Calzada, A., Fronzes, R., Savva C. G., Chandran, V., Lian, P. W., Laeremans, T., Pardon, E., Steyaert, J., Remaut, H., Waksman, G., & Orlova E. V. (2013) Structure of a bacterial type IV secretion core complex at subnanometer resolution. EMBO J. 32, ISI IF11 = Irannejad, R., Tomshine, J.C., Tomshine, J.R., Steyaert, J., Rasmussen, S., Sunahara, R., Chevalier, M., El-Samad, H., Huang, B., & von Zastrow, M. (2013) Conformational biosensors reveal adrenoceptor signaling from endosomes. Nature 495, ISI IF11 = a. News & views: Lohse, M.J. & Calebiro, D. (2013) Cell biology: Receptor signals come in waves. Nature 495, b. Methods to watch: Pastrana, E. (2014) Intracellular mini-binders. Nature Methods 11, Park, Y.-J., Budiarto, T., Wu, M., Pardon, E., Steyaert, J., & Hol, W.G.J. (2012) The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA editing ligase L2, Nucleic acids res 40, ISI IF10 = Baranova, E., Fronzes, R., Garcia-Pino, A., Van Gerven, N, Papapostolou, D., Péhau-Arnaudet, Pardon, E., Steyaert, J., Howorka, S. & Remaut, H. (2012) SbsB structure and lattice reconstruction unveil Ca 2+ triggered S- layer assembly, Nature 487, ISI IF12 = Park, Y.J., Pardon, E., Wu, M., Steyaert, J. & Hol, W.G.J (2012) Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody. Nucleic acids res 40, ISI IF10 = Abskharon, R.N.N., Ramboarina, S., El Hassan, H., Gad, W., Apostol, M.I., Giachin, G., Legname, G., Steyaert, J., Messens, J., Soror, S.H., & Wohlkonig, A. (2012) A novel expression system for production of soluble prion proteins in E. coli. Microb Cell Fact 11, 6. ISI IF10 = Abskharon, R.N.N., Soror, S.H., Pardon, E, El Hassan, H., Legname, G., Steyaert, J., Wohlkonig, A. (2011) Combining in-situ proteolysis and microseed matrix screening to promote crystallization of PrP(c)-nanobody complexes. Protein Eng Des Sel 24, ISI IF11 = Westfield, G., Rasmussen, S.G.F, Su M., Dutta, S, DeVree, B.T., Chung, K.Y., Calinski, D., Velez-Ruiz, G., Oleskie, A.N., Pardon, E., Chae, P.S., Liu, T., Li, S., Woods Jr., V.L., Steyaert, J., Kobilka, B.K., Sunahara, R.K. & Skiniotis, G. (2011) Structural flexibility of the Gαs α-helical domain in the β2-adrenoceptor Gs Complex. Proc Natl Acad Sci U S A 108, ISI IF11= Korotkov K.V., Johnson T.L., Jobling M.G., Pruneda J., Pardon E., Héroux, A., Turley, S., Steyaert, J., Holmes, R.K., Sandkvist, M., & Hol, W.G.J. (2011) Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System. PLoS Pathog 7(9): e ISI IF10 =
5 30. Rasmussen, S.G.F, DeVree, B.T., Zou, Y, Kruse, A.C., Chung, K.Y.,Thian, T.S., Thian, F.S., Chae, P.S., Pardon, E., Calinski, D., Mathiesen, J.M., Shah, S.T.A., Lyons, J.A., Caffrey, M., Gellman, S.H., Steyaert, J., Skiniotis, G., Weis, W., Sunahara, R.K., Kobilka, B.K. (2011) Crystal Structure of the 2 Adrenergic Receptor-Gs protein complex. Nature 477, (with cover) ISI IF11 = & F1000 factor = 30 a. News in Focus: Buchen L (2011) Cell signaling caught in the act. Nature 475, b. New & Views: Schwartz, T.W. & Sakmar, T.P. (2011) Structural biology: Snapshot of a signalling complex. Nature 477, c. 365 days: Interactive timeline: Van Noorden R. (2011) A clickable calendar. Nature doi: /nature d. Nature news: Van Noorden R. (2012) Cell-signalling work nets chemistry Nobel. Nature doi: /nature Steyaert, J. & Kobilka, B.K. (2011) Nanobody stabilization of G protein coupled receptor conformational states. Curr Opin Struct Biol 21, ISI IF11 = Vercruysse, T., Pawar, S., De Borggraeve, W., Pardon, E., Pavlakis, G.N., Pannecouque, C., Steyaert, J., Balzarini, J. & Daelemans, D. (2011) Measuring cooperative Rev protein-protein interactions on Rev Responsive RNA by fluorescence resonance energy transfer. RNA Biol 8, ISI IF10 = Domanska, K., Vanderhaegen, S, Srinivasan, V., Pardon, E., Dupeux, F., Marquez,J.A., Gorgetti, S., Stoppini, M., Wyns, L., Bellotti, V. & Steyaert, J. (2011) Atomic structure of a nanobody trapped intermediate of β2m amyloidogenesis, Proc Natl Acad Sci USA 108, ISI IF11= Oyen, D., Srinivasan, V., Steyaert, J., & Barlow, JN. (2011) Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition. J Mol Biol 407, ISI IF10 = Rasmussen, S.G.F., Choi, H.-J., Fung, J.-J., Pardon, E., Casarosa, P., Seok Chae, P., DeVree, B.T., Rosenbaum, D.M., Thian, F.S., Kobilka, T.S., Schnapp, A., Konetzki, I., Sunahara, R.K., Gellman, S.H., Pautsch, A., Steyaert, J., Weis, W.I., & Kobilka, B.K. (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, ISI IF10 = a. News and Views: Sprang, S.R. (2011) Cell signalling: Binding the receptor at both ends. Nature 469, Wu, M., Park, Y.-U., Pardon, E., Guo, X., Turley, S., Stuart, K. S., Hayhurst, A., Deng, J., Steyaert, J. and Hol, W.G.J. (2011) Structures of a key interaction protein from the trypanosoma brucei editosome in complex with single domain antibodies, J Struct Biol 174, ISI IF10 = Abskharon R.N., Soror S.H., Pardon E., El Hassan H., Legname G., Steyaert J. & Wohlkonig A. (2010) Crystallization and preliminary X-ray diffraction analysis of a specific VHH domain against mouse prion protein. Acta Crystallogr F Struct Biol Cryst Commun 66, ISI IF10 = De Genst, E., Guilliams, T., Wellens, J., O' Day, E., Waudby, C., Meehan, C., Dumoulin, M., Hsu, S.T-D., Cremades, N., Verschueren, K., Pardon, E., Wyns, L., Steyaert, J., Christodoulou, J. & Dobson, C. (2010) Structure and properties of a complex of α-synuclein and a single domain camelid antibody, J Mol Biol 402, ISI IF10 = Vandemeulebroucke, A., Minici, C., Bruno, I., Muzzolini, L., Tornaghi, P., Parkin, D., Schramm, V.L., Versées, W., Steyaert, J., & Degano, M. (2010) Structure, function, and inhibition of the principal nucleotide catabolic enzyme in the purine-auxotrophic Trypanosoma brucei brucei. Biochemistry 49, ISI IF10 = Berg, M., Kohl, L., Van der Veken, P., Joossens, J., Al-Salabi, MI., Castagna, V., Giannese, F., Cos, P., Versées, W., Steyaert, J., Grellier, P., Haemers, A., Degano, M., Maes, L., de Koning, H.P. & Augustyns, K. (2010) Evaluation of nucleoside hydrolase inhibitors for treatment of African trypanosomiasis. Antimicrob agents chemother 54, ISI IF10 =
6 41. Vercruysse, T., Pardon, E., Vanstreels, E., Steyaert, J., & Daelemans, D. (2010). An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 rev prevents viral production. J Biol Chem 285, ISI IF10 = Van den Abbeele, A., De Clercq, S., De Ganck, A., De Corte, V, Van Loo, B., Soror, S. H., Srinivasan, V., Steyaert, J., Vandekerckhove, J., and Gettemans, J. (2010) A llama-derived gelsolin single domain antibody blocks gelsolin G-actin interaction. Cell Mol Life Sci 67, ISI IF10 = Ryckaert, S., Pardon, E., Steyaert, J., & Callewaert, N. (2010) Isolation of antigen-binding camelid heavy chain antibody fragments (nanobodies) from an immune library displayed on the surface of Pichia pastoris. J Biotech 145, ISI IF10 = De Vocht, C., Ranquin, A., Willaert, R., Van Ginderachter, J., Vanhaecke, T., Rogiers, V., Versées, W., Van Gelder, P. & Steyaert, J. (2009) Assessment of stability, toxicity and immunogenicity of new polymeric nanoreactors for use in enzyme replacement therapy of MNGIE. J Control Release 137, ISI IF09 = Barlow, J., Conrath, K., & Steyaert, J. (2009) Substrate-dependent modulation of enzyme activity by allosteric effector antibodies. Biochim Biophys Acta 1794, ISI IF09 = Versées, W., Goeminne, A., Berg, M., Vandemeulebroucke, A., Haemers, A., Augustyns, K., & Steyaert, J. (2009) Crystal structures of T. vivax nucleoside hydrolase in complex with new potent and specific inhibitors Biochim Biophys Acta 1794, ISI IF09 = Berg, M., Bal, G., Goeminne, A., Van Der Veken, P., Versées, W., Steyaert, J., Haemers, A., Augustyns, K. (2009) Synthesis of bicyclic N-arylmethyl-substituted iminoribitol derivatives as selective nucleoside hydrolase inhibitors. Chemmedchem 4, ISI IF09 = Korotkov K. V., Pardon E., Steyaert J. & Hol W. (2009) Crystal Structure of the N-Terminal Domain of the Secretin GspD from ETEC Determined with the Assistance of a Nanobody. Structure 17, ISI IF09 = Lam, A. Y., Pardon, E., Korotkov K. V., Hol, W.G.J & Steyaert, J. (2009) Crystal structure of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus in complex with a nanobody, J Struct Biol 166, ISI IF09 = Vandemeulebroucke, A., De Vos, S., Van Holsbeke, E., Steyaert, J. & Versées, W. (2008) A flexible loop as a functional element in the catalytic mechanism of Nucleoside hydrolase from Trypanosoma vivax. J Biol Chem 283, ISI IF08 = Goeminne, A., Berg, M., Mcnaughton, M., Bal, G., Surpateanu, G., Van Der Veken, P., De Prol, S., Versées, W., Steyaert, J., Haemers, A. & Augustyns, K. (2008) N-Arylmethyl substituted iminoribitol derivatives as inhibitors of a purine specific nucleoside hydrolase Bioorg Med Chem 16, ISI IF08 = Goeminne, A., Mcnaughton, M., Bal, G., Surpateanu, G., Van Der Veken, P., De Prol, S., Versees, W., Steyaert, J., Haemers, A., Augustyns, K. (2008) Synthesis and biochemical evaluation of guanidino-alkyl-ribitol derivatives as nucleoside hydrolase inhibitors. Eur J Med Chem 43, ISI IF08 = Barlow, J. N. & Steyaert, J (2007) Examination of the mechanism and energetic contribution of leaving group activation in the purine-specific nucleoside hydrolase from Trypanosoma vivax. Biochim Biophys Acta 1774, ISI IF07 = Spaepen, S., Versées, W., Gocke, D., Pohl, M., Steyaert, J., & Vanderleyden, J (2007) Characterization of Phenylpyruvate Decarboxylase, Involved in Auxin Production of Azospirillum brasilense. J Bacteriol 189, ISI IF07 = Versees, W., Spaepen, S., Wood, M., Leeper, F., Vanderleyden, J., & Steyaert, J. (2007) Molecular Mechanism of allosteric substrate activation in a thiamine diphosphate-dependent decarboxylase, J Biol Chem 282, ISI IF07 = Goeminne, A., McNaughton, M., Bal, G., Surpateanu, G., Van Der Veken, P., De Prol, S., Versées, W., Steyaert, J., Haemers, A., Augustyns, K. (2007) 1,2,3-Triazolylalkylribitol derivatives as nucleoside hydrolase inhibitors. Bioorg Med Chem Lett 17, ISI IF07 = Versees, W., Spaepen, S., Vanderleyden, J., Steyaert J. (2007) The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. FEBS J 274, ISI IF07 =
7 58. Vandemeulebroucke, A., Versées, W., Steyaert, J. & Barlow, J. (2006) Multiple transients in the pre-steadystate of nucleoside hydrolase reveal complex substrate binding, product base release, and two apparent rates of chemistry. Biochemistry 45, ISI IF06 = Versees, W., Barlow, J., & Steyaert, J. (2006) Transition-state complex of the purine-specific nucleoside hydrolase of T. vivax: enzyme conformational changes and implications for catalysis, J Mol Biol 359, ISI IF06 = Muzzolini, L., Versees, W., Tornaghi, P., Van Holsbeke, E., Steyaert, J. & Degano, M. (2006) New insights into the mechanism of nucleoside hydrolases from the crystal structure of the Escherichia coli YbeK protein bound to the reaction product. Biochemistry 24, ISI IF06 = Loverix, S., Versees, W., Steyaert, J. & Geerlings, P. (2006) Quantum Chemical Study of Leaving Group Activation in T. vivax Nucleoside Hydrolase. Int J Quantum Chem 106, ISI IF06 = Ranquin, A., Wim Versées, W., Meier, W., Steyaert, J, and Van Gelder, P. (2005) Therapeutic Nanoreactors: Combining Chemistry and Biology in a Novel Triblock Copolymer Drug Delivery System. Nanoletters 5, ISI IF06 = Huysmans G., Ranquin A., Wyns L., Steyaert J. & Van Gelder P. (2005) Encapsulation of therapeutic nucleoside hydrolase in functionalised nanocapsules. J Controled Release 102, Mignon, P., Loverix, S., Steyaert, J. & Geerlings, P. (2005) Influence of the p p interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res 33, Loverix, S., Geerlings, P., McNaughton, P., Augustyns, K., Vandemeulebroucke, A., Steyaert, J. & Versées, W. (2005) Substrate-assisted Leaving Group Activation in Enzyme-catalyzed N-Glycosidic Bond Cleavage, J Biol Chem 280, Versées, W, Loverix, S., Vandemeulebroucke, A., Geerlings, P., & Steyaert, J. (2004) Leaving group activation by aromatic stacking: an alternative to general acid catalysis. J Mol Biol 338, Mignon, P., Loverix, S., Steyaert, J. & Geerlings, P. (2004) Functional assesment of in vivo and in silico mutations in the Guanie Binding Site of RNase T1: A DFT Study, Int J Quantum Chem 99, Vandemeulebroucke, A., Versées, W., De Vos, S., Van Holsbeke, E., & Steyaert, J. (2003) Pre-Steady State Analysis of the Nucleoside Hydrolase of Trypanosoma vivax. Evidence for half-of- the-sites reactivity and rate limiting product release. Biochemistry 44, Versées, W. & Steyaert, J. (2003) Catalysis by inosine-adenosine-guanosine-preferring nucleoside hydrolases. Curr opin struct biol 13, Versées, W., Van Holsbeke, E., De Vos, S., Decanniere, K., Zegers, I. & Steyaert, J. (2003) Cloning, preliminary characterization, and crystallization of nucleoside hydrolases from Caenorhabditis elegans and Campylobacter jejuni. Acta Crystallogr D Biol Crystallogr D59, Loverix, S. & Steyaert, J. (2003) Ribonucleases: from prototypes to therapeutic targets? Curr Med Chem 10, Mignon, P., Steyaert, J., Loris, R., Geerlings, P. & Loverix, S. (2002) A nucleophyle activation dyad in ribonucleases: a combined X-ray crystallographic/ ab initio quantum chemical study. J Biol Chem 277, Versées, W., Decanniere, K., Van Holsbeke E., Devroede, N. & Steyaert, J. (2002) Enzyme-Substrate Interactions in the Purine-Specific Nucleoside Hydrolase from Trypanosoma vivax. J Biol Chem 277, De Vos, S., Backmann, J., Steyaert, J., & Loris, R. (2001) Hydrophobic core manipulations in RNase T1, Biochemistry 40, Deswarte, J., De Vos, S., Langhorst, U., Steyaert, J., & Loris, R. (2001) The contribution of metal ions to the conformational stability of RNase T1: crystal vs. solution. Eur J Biochem 268, Versées, W., Decanniere, K., Pellé, R., Depoorter, J., Parkin, D.W., & Steyaert, J. (2001) Structure and Function of a novel Purine-Specific Nucleoside Hydrolase from Trypanosoma vivax. J Mol Biol 307, Loverix, S., & Steyaert, J. (2001) Mechanistic analyses of RNase T1. Methods Enzymol 341, Loverix, S., Winquist, A., Strömberg, R., & Steyaert, J. (2000) Mechanism of RNase T1: concerted triester-like phosphoryl transfer via a catalytic three-centered hydrogen bond. Chem Biol 7,
8 79. Langhorst, U., Backmann, J., Loris, R., & Steyaert, J. (2000) Water mediated interactions in proteins: thermodynamic analysis of a hydration site in RNase T1, Biochemistry 39, Huyghues-Despointes, B.M., Langhorst, U., Steyaert, J., Pace, C.N., & Scholtz, J.M. (1999) Hydrogen-exchange stabilities of RNase T1 and variants with buried and solvent-exposed Ala>Gly mutations in the helix. Biochemistry 38, Loris, R., De Vos, S., Langhorst, U., Decanniere, K., Bouckaert, J., Maes, D., Transue, T.R., & Steyaert, J. (1999) Conserved Water Molecules in a Large Family of Microbial Ribonucleases, Proteins 36, Langhorst, U., Loris, R., Denisov, V.P., Doumen, J., Roose, P., Maes, D., Halle, B., & Steyaert, J. (1999) Dissection of the structural and functional role of a conserved hydration site in RNase T1, Protein sci 8, Loverix, S., Laus, G., Martins, J., Wyns, L., & Steyaert, J. (1998) Reconsidering the energetics of Ribonuclease Catalyzed RNA hydrolysis. Eur J Bioch 257, Dao-Thi, T.R. Transue, Pellé, R., Murphy, N.B., Poortmans, F., & Steyaert, J. (1998) Expression, Purification, Crystallization and preliminary X-ray analysis of cyclophylin of the bovine parasite Trypanosoma bucei brucei. Acta Crystallogr D Biol Crystallogr D54, Loverix, S., Winquist, A., Strömberg, R., & Steyaert, J. (1998) An engineered Ribonuclease specific for thiophosphate RNA. Nat Struct Biol 5, Zegers,I., Loris, R., Dehollander,G., Haikal, A., F., Poortmans, F., Steyaert, J., & Wyns, L. (1998) The hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analysed by time-resolved crystallography. Nat Struct Biol 5, De Vos, S., Doumen, J., Langhorst, U., & Steyaert, J. (1998) Dissecting Histidine Interactions of Ribonuclease T1 with Asparagine and Glutamine Replacements: Analysis of Double Mutant Cycles at one Position. J Mol Biol 275, Steyaert, J. (1997) Review. A decade of protein engineering on Ribonuclease T1. Eur J Biochem 247, Loverix, S., Doumen, J., & Steyaert, J. (1997) Additivity of Protein-Guanine Interactions in Ribonucleases T1. J Biol Chem 272, Doumen, J., Gonciarz, M., Zegers, I., Loris, R. Wyns, L., & Steyaert, J. (1996) A catalytic function for the structurally conserved residue Phe100 of ribonuclease T1. Protein Sci 5, Steyaert, J., & Engelborghs, Y. (1995) A two binding site kinetic model for the ribonuclease catalysed depolymerisation of dinucleoside phosphate substrates. Eur J Biochem 233, Fish, W., R., Nkhungulu, Z., M., Muriuki, C., W., Ndegwa, D., M., Londsdale-Eccles, J., D., & Steyaert, J. (1995) Primary structure and partial characterization of a life-cycle-regulated cysteine protease from Trypanosoma (Nannomonas) congolense. Gene 161, Steyaert, J., Haikal, A. F., and Wyns, L. (1994) Investigation of the functional interplay between the primary site and the subsite of RNase T1 : Kinetic analysis of single and double mutants for modified substrates Proteins 18, Pletinckx J., Steyaert J., Hui-Woog Choe, Heinemann, U., & Wyns, L. (1994) Crystallographic study of Glu58 RNase T1*2'-guanosine monophosphate at 1.9 A resolution. Biochemistry 33, Loris, R., Steyaert, J., Maes, D., Lisgarten J., Pickersgill, R., & Wyns, L. (1993) Crystal structure determination and refinement at 2.3 A resolution of the lentil lectin. Biochemistry 32, Bernard, P., Kezdi, K., E., Van Melderen, L., Steyaert, J., Wyns, L., Pato, M. L., Higgins, P., N., and Couturier, M. (1993) The F plasmid ccdb protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol 234, Steyaert, J., Van Melderen, L., Bernard, P., Dao Thi, M. H., Loris, R., Wyns, L., & Couturier, M. (1993) Purification, Circular dichroism analysis, crystallization and primary X-ray diffraction analysis of the F plasmid CcdB killer protein. J Mol Biol 203, Steyaert, J., & Wyns, L. (1993) Functional interactions among the His40, Glu58 and His92 catalysts of Ribonuclease T 1 as studied by double and triple mutants. J Mol Biol 229, Zegers, I., Verhelst, P., Choe, H.-W., Steyaert, J., Heinemann, U., Saenger, W., & Wyns, L. (1992) Role of Histidine-40 in Ribonuclease T 1 Catalysis: Three-Dimensional Structures of the Partially Active His40Lys Mutant. Biochemistry 31,
9 99. Steyaert, J., Haikal, A. F., Stanssens, P., & Wyns, L. (1992) Dissection of the Ribonuclease T 1 subsite: the transesterification kinetics of Asn36Ala and Asn98Ala RNase T 1 for minimal dinucleoside phosphates. Eur J Biochem 203, Steyaert, J., Haikel A. F., Wyns, L., & Stanssens, P. (1991) Subsite Interactions of Ribonuclease T 1 : Asn36 and Asn98 Accelerate GpN Transesterification through Interactions with the Leaving Nucleoside N. Biochemistry 30, Steyaert, J., Wyns, L., & Stanssens, P. (1991) Subsite Interations of Ribonuclease T 1 : Viscosity Effects Indicate That the Rate-Limiting Step of GpN Transesterification Depends on the Nature of N. Biochemistry 30, Steyaert, J., Opsomer, C., Wyns, L., & Stanssens, P. (1991) Quantitative Analysis of the Contribution of Glu46 and Asn98 to the Guanosine Specificity of Ribonuclease T 1. Biochemistry 30, Steyaert, J., Hallenga, K., Wyns, L., & Stanssens, P. (1990) Histidine-40 of Ribonuclease T1 Acts as Base Catalyst When the True Catalytic Base, Glutamic Acid-58, Is Replaced by Alanine. Biochemistry 29, Shirley, B., A., Stanssens, P., Steyaert, J., & Pace, N. (1989) Conformational Stability and Activity of Ribonuclease T 1 and Mutants. J Biol Chem 264, INVITED LECTURES OR SEMINARS Nanobodies to study GPCR transmembrane signaling: from structures to function to drugs (2014) Keynote lecture at the New Strategies For Macromolecular Complexes analysis symposium, Strasbourg, France, November Nanobodies to study GPCR transmembrane signaling: from structures to function to drugs (2014) To be presented at MedChem 2014 Annual one-day meeting on Medicinal chemistry of SRC & KVCV, Braine-L alleud, Belgium, November 21. Innovative Methods for the Generation of Nanobodies against Membrane Proteins (2014) Presented at the 1th annual Discovery on Target meeting: Antibody generation, selection and screening, Boston, CA, October Nanobody-enabled fragment screening on active-state constrained GPCRs (2014) Presented at the 1th annual Discovery on Target meeting: GPCR-Based Drug Discovery, Boston, CA, October 8-9. Nanobodies to study GPCR transmembrane signaling: from structures to function to drugs (2014) Keynote lecture presented at the annual GLISTEN meeting, Budapest, Hungary, October 2-4. Nanobodies to study GPCR transmembrane signaling: from structure to function to drugs. (2014) Presented at the MipTec Conference, Basel, Switzerland, September GPCR Structural Biology: New Therapeutic Opportunities (2014) Presented at the 23 th EFMC-ISMC International Symposium on Medicinal Chemistry 2014, Lisbon, Portugal, September 7-11 (Session Chair) Nanobodies for the investigation of GPCR transmembrane signaling: from structure to function to drugs (2014) Seminar at the Research Centre for Natural Sciences of the Hungarian Academy of Sciences, Budapest, Hungary, June 25 Nanobody-enabled fragment screening on active-state constrained GPCRs (2014) Presented at the GTC Drug Discovery summit: GPCR structure, function and drug discovery, Cambridge, MA, May
10 Nanobody-enabled fragment screening on active-state constrained GPCRs (2014) Presented at the Cambridge Healttech Institutes s 9th Annual Fragment-Based Drug Discovery meeting, San Diego, CA, April Nanobodies as tools for the structural and functional investigation of GPCR transmembrane signaling (2014) Structural Biology Symposium, Leuven, Belgium, March 17 Nanobodies for the investigation of GPCR transmembrane signaling: from structure to function to drugs (2014) Seminar at the Centre de Genetic Fonctionelle, CNRS, Montpellier, France, January 10 Nanobodies for the structural and functional investigation of GPCR transmembrane signaling (2013) Seminar at Ablynx, Gent, Belgium, December 20 Nanobodies for the structural and functional investigation of GPCR transmembrane signaling (2013) Presented at the FWO Anniversary meeting: Kennismakers, al 85 jaar Zuurstof voor Onderzoek en Ontwikkeling, Gent, Belgium, December 17 Nanobodies for the structural investigation of GPCR transmembrane signaling (2013) Seminar at Novartis, Basel, October 25 Nanobodies for the structural and functional investigation of GPCR transmembrane signaling (2013) Presented at the 23rd Solvay Conference on Chemistry: New Chemistry and New Opportunities from the Expanding Protein Universe, Brussels, Belgium, October Nanobodies to study GPCR transmembrane signaling: from structure to function to drugs (2013) Presented at the 2nd Annual meeting of the GDR-3545: RCPG-Physio-Med, Strasbourg, France, October Nanobodies to study GPCR transmembrane signaling from structure to function to drugs (2013) Presented at Discovery on Target: Antibodies against Membrane Protein Targets. Boston, USA, September Nanobodies to study GPCR transmembrane signaling from structure to function to drugs (2013) Presented at the Benzon Symposium No 59 Membrane proteins: Structure, Function and Dynamics, Copenhagen, Denmark, August Nanobodies to study GPCR transmembrane signaling from structure to function to drugs (2013) Presented at GPCR Medicinal Chemistry Roots, fruits and fertilizers, VU University Amsterdam, Amsterdam, June Nanobodies as tools for the structural and functional investigation of GPCR transmembrane signaling (2013) Presented at the Instruct Biennial Structural Biology Conference, EMBL Advanced Training Centre, Heidelberg, Germany, May Nanobodies as tools for the structural and functional investigation of GPCR transmembrane signaling (2013) Presented at the Cold Spring Harbor Asia Conferences: Membrane Protein Structure and Function, Dushu Lake Conference Center, Suzhou, China, May Nanobodies as tools for the structural and functional investigation of GPCR transmembrane signaling (2013) Presented at the Gordon Conference on Molecular Pharmacology, Renaissance Tuscany Il Ciocco Resort, Lucca (Barga), Italy, April 28-May 3 Nanobodies for the Structural and Functional Investigation of GPCR Transmembrane Signaling (2013) Presented at the Drug Discovery Chemistry Conference, Hilton San Diego Resort & Spa, San Diego, CA, April Nanobodies for the structural analysis of transmembrane signaling (2012) Presented at the NIH roadmap to membrane protein structures and complexes. The 4th Membrane Protein Technologies Meeting, San Francisco, CA, USA, November
11 Keynote lecture: Nanobodies for the structural analysis of transmembrane signaling (2012) Presented at PEGS Europe, Protein & Antibody Engineering summit, Vienna, Austria, November 6-8 Nanobodies for the structural investigation of transmembrane signaling (2012) Seminar at Pfizer, Groton, CT, October 4. Nanobodies for the structural analysis of transmembrane signaling (2012) Presented at Discovery on Target, Boston, USA, October 1-3 Structural investigation of GPCR transmembrane signaling by use of nanobodies (2012) Presented at G-protein-coupled-receptors: from structural insights to functional mechanisms. Monash University Prato Centre, Prato, Italy, September Nanobodies for the structural investigation of transmembrane signaling (2012) Presented at the Gordon Research Conferences: Phosphorylation & G-Protein Mediated Signaling Networks, University of New England, Biddeford, ME, June Why are Nanobodies the key to crystallize membrane-bound proteins? (2012) Presented at the International Workshop: New Approaches in Drug Design & Discovery, Schloss Rauischholzhausen, Marburg, March Nanobodies for the structural analysis of transmembrane signaling (2012) Seminar at the Biocenter Oulu and Department of Biochemistry, University of Oulu, Oulu, Finland, March 13 Structural Investigation of GPCR Transmembrane Signaling by Use of Nanobodies (2012) Presented at the Molecular Med TRI-CON 2012: Phage and yeast display of difficult targets, Moscone North Convention Center, San Francisco, CA, February Nanobodies for the structural analysis of transmembrane signaling (2011) Presented at the GPCR Workshop 2011, Hyatt Regency, Maui, Hawaii, December 4-8 Structural investigation of GPCR transmembrane signaling by use of Nanobodies (2011) Presented at the 19th PSDI meeting: Protein Structure Determination in Industry Goteburg, November. Structural investigation of GPCR transmembrane signaling by use of Nanobodies (2011) Seminar at UCB, Braine-L alleud, Belgium, October 25. Nanobodies for the structural analysis of GPCR transmembrane signaling (2011) Presented at the RAMMC 2011 meeting: Recent Advances in Macromolecular Crystallization, Strasbourg, September Conformation selective Xaperones for GPCR research (2011) Presented at Select Biosciences: Advances in Protein crystallography, Hamburg, Germany, June 30 July 1 Conformation selective nanobodies for GPCR research (2011) Presented at the International conference on Structural Genomics, Toronto, Canada, May Xaperone assisted X-ray crystallography: conformation selective nanobodies to solve active state GPCR Structures (2011) DRA mini-symposium on Receptor Structure and Function, Copenhagen, Denmark, May 4 Nanobodies with G protein like properties stabilize a G protein-coupled receptor active state (2011) Presented at the 9th annual Informa congress on G Protein-Coupled Receptors in Drug Discovery, Berlin, Germany, March Nanobodies with G protein like properties stabilize a G protein-coupled receptor active state (2011) Seminar at the LMB, Cambridge, UK, February 22 Single chain camelid antibodies in GPCR research (2011) Seminar at the Paul Scherrer Institut, Villigen, Switzerland, February 11 11
12 Single chain camelid antibodies in GPCR research (2010) GPCR Workshop 2010, Honolulu, Hawaii, December 7-10 Nanobodies with G protein like properties stabilize a G Protein-Coupled Receptor Active State (2010) Presented at the Structural Biology and Drug Discovery Zing Conference, Cancun, Mexico, December 4-7 Nanobodies with G protein like properties stabilize a G Protein-Coupled Receptor Active State (2010) Seminar at Pfizer, Sandwich, UK, November 18 Nanobodies with G protein like properties stabilize a G Protein-Coupled Receptor Active State (2010) Seminar at EVOTEC, Milton Park, Abingdon, UK, November 17 Nanobodies with G protein like properties stabilize a G Protein-Coupled Receptor Active State (2010) Presented at the PSDI 2010 meeting on Protein Structure Determination in Industry, Oxford, UK, November Nanobodies with G protein like properties stabilize a G Protein-Coupled Receptor Active State (2010) Presented at the G protein-coupled receptors, from structure to Diseases conference, Barcelona, Spain, November 5-6 Nanobodies with G protein like properties stabilize a G Protein-Coupled Receptor Active State (2010) 1st Symphosium on single domain antibodies, Gent, Belgium, October Nanotools for Biomedical Targets Nanobodies Stabilize Active State of G-Protein Coupled Receptors (2010) 21st EFMC-ISMC International Symposium on Medicinal Chemistry, Brussels, Belgium, September 5-9 New catalytic strategies in the nucleoside hydrolase family (2009) Pesented at the symphosium in honour pf Professor Jean-Marie Frère: Penicillin-recognizing enzymes: from enzyme kinetics to protein folding, University of Liège, July 1-3. New catalytic strategies for leaving group activation in nucleoside hydrolases (2009) Presented at the 13th International Symposium on Purine and Pyrimidine Metabolism in Man (PP09), Skogshem- Wijk, Stockholm, Sweden, June New catalytic strategies in the nucleoside hydrolase family (2005) Presented at the Frontiers in Chemical Biology: Mechanistic Enzymology and Biocatalysis meeting, University of Exeter, UK, September A decade of Protein engineering, RNase T1 undressed (2002) Presented at the 6th International meeting on ribonucelases, Bath, UK, June. From protein structure to function: RNase T1 as an example (1999) Presented at the Proteins from Structure to Function meeting on the occasion of the conferment of the title of Doctor Honoris Causa to Prof. A. Fersht, VUB, Brussels, November 19. Protein engineering of RNase T1 to alter substrate specificity (1999) Presented at the fifth International Meeting on Ribonucleases, Warrenton, Virginia, USA, May Mutant enzymes and modified substrates to analyse the structure-function relationship of Ribonuclease T1 (1997) Presented at the 3rd International Engelhardt conference on molecular Biology, Moscow, Russia, June 9-14 The use of mutant enzymes and modified substrates to analyse Ribonuclease T1 function (1996) Presented at the EMBO Workshop 1996, Nucleotidyl and phosphoryl Transfer in the Protein and RNA world, Xanten, Germany, September 29-October 3 12
13 The use of mutant enzymes and modified substrates to analyse Ribonuclease T1 function (1996) Presented at the 4th international meeting on Ribonucleases: Chemistry, Biology, Biotechnology, Groningen, the Netherlands, July
The dynamic process of GPCR activation: Insights from the human β 2 AR
 The structural basis of G protein coupled receptor signaling The dynamic process of GPCR activation: Insights from the human β 2 AR Brian Kobilka Department of Molecular and Cellular Physiology Brian Kobilka
The structural basis of G protein coupled receptor signaling The dynamic process of GPCR activation: Insights from the human β 2 AR Brian Kobilka Department of Molecular and Cellular Physiology Brian Kobilka
CURRICULUM VITAE FREDERICK S. GIMBLE ASSOCIATE PROFESSOR OF BIOCHEMISTRY
 CURRICULUM VITAE FREDERICK S. GIMBLE ASSOCIATE PROFESSOR OF BIOCHEMISTRY ADDRESS: Department of Biochemistry Purdue University 175 S. University St. West Lafayette, IN 47907-1153 Phone: (765) 494-1653
CURRICULUM VITAE FREDERICK S. GIMBLE ASSOCIATE PROFESSOR OF BIOCHEMISTRY ADDRESS: Department of Biochemistry Purdue University 175 S. University St. West Lafayette, IN 47907-1153 Phone: (765) 494-1653
The Confo body technology, a new platform to enable fragment screening on GPCRs
 The Confo body technology, a new platform to enable fragment screening on GPCRs Christel Menet, PhD CSO Miptec, 216 Confo Therapeutics Incorporated in June 215 Located in Brussels, Belgium Confo Therapeutics
The Confo body technology, a new platform to enable fragment screening on GPCRs Christel Menet, PhD CSO Miptec, 216 Confo Therapeutics Incorporated in June 215 Located in Brussels, Belgium Confo Therapeutics
a) JOURNAL OF BIOLOGICAL CHEMISTRY b) PNAS c) NATURE
 a) JOURNAL OF BIOLOGICAL CHEMISTRY b) c) d) ........................ JOURNAL OF BIOLOGICAL CHEMISTRY MOLECULAR PHARMACOLOGY TRENDS IN PHARMACOLOGICAL S AMERICAN JOURNAL OF PHYSIOLOGY-HEART AND CIRCULATORY
a) JOURNAL OF BIOLOGICAL CHEMISTRY b) c) d) ........................ JOURNAL OF BIOLOGICAL CHEMISTRY MOLECULAR PHARMACOLOGY TRENDS IN PHARMACOLOGICAL S AMERICAN JOURNAL OF PHYSIOLOGY-HEART AND CIRCULATORY
CURRICULUM VITAE. Lecturer (Pharmaceutical Chemistry Department, Faculty of Pharmacy, Al- Azhar University, Cairo, Egypt)
 CURRICULUM VITAE Farag F. S. Selim, PhD CONTACT DETAILS Name: Farag Farouk Sherbiny Selim Nationality: Egyptian Position: Lecturer (Pharmaceutical Chemistry Department, Faculty of Pharmacy, Al- Azhar University,
CURRICULUM VITAE Farag F. S. Selim, PhD CONTACT DETAILS Name: Farag Farouk Sherbiny Selim Nationality: Egyptian Position: Lecturer (Pharmaceutical Chemistry Department, Faculty of Pharmacy, Al- Azhar University,
Current Status and Future Prospects p. 46 Acknowledgements p. 46 References p. 46 Hammerhead Ribozyme Crystal Structures and Catalysis
 Ribozymes and RNA Catalysis: Introduction and Primer What are Ribozymes? p. 1 What is the Role of Ribozymes in Cells? p. 1 Ribozymes Bring about Significant Rate Enhancements p. 4 Why Study Ribozymes?
Ribozymes and RNA Catalysis: Introduction and Primer What are Ribozymes? p. 1 What is the Role of Ribozymes in Cells? p. 1 Ribozymes Bring about Significant Rate Enhancements p. 4 Why Study Ribozymes?
CURRICULUM VITAE. Beena Krishnan
 CURRICULUM VITAE Beena Krishnan Dept. of Biochemistry & Molecular Biology LGRT Room No. 1209 University of Massachusetts-Amherst Amherst, Massachusetts 01003 beena@biochem.umass.edu beenakrish@gmail.com
CURRICULUM VITAE Beena Krishnan Dept. of Biochemistry & Molecular Biology LGRT Room No. 1209 University of Massachusetts-Amherst Amherst, Massachusetts 01003 beena@biochem.umass.edu beenakrish@gmail.com
Supporting Information
 Supporting Information Drugs Modulate Interactions Between the First Nucleotide-Binding Domain and the Fourth Cytoplasmic Loop of Human P-glycoprotein Tip W. Loo and David M. Clarke Department of Medicine
Supporting Information Drugs Modulate Interactions Between the First Nucleotide-Binding Domain and the Fourth Cytoplasmic Loop of Human P-glycoprotein Tip W. Loo and David M. Clarke Department of Medicine
Genomics Research Center: Current Status & Future Development
 Genomics Research Center: Current Status & Future Development Introduction Research in the life sciences has entered a new era after completion of the human genome project and the sequencing of the genomes
Genomics Research Center: Current Status & Future Development Introduction Research in the life sciences has entered a new era after completion of the human genome project and the sequencing of the genomes
CRISTINA IFTODE. Current position: Associate Professor, Rowan University, Dept. Biological Sciences. Research Interests. Education.
 CRISTINA IFTODE Work address Rowan University Dept. Biological Sciences 201 Mullica Hill Rd. Glassboro, NJ 08028 Phone: (856) 256-4500 ext.3586 E-mail: iftode@rowan.edu URL: http://www.rowan.edu/colleges/las/departments/biologicalsci/facultystaff/cristinaiftode.htm
CRISTINA IFTODE Work address Rowan University Dept. Biological Sciences 201 Mullica Hill Rd. Glassboro, NJ 08028 Phone: (856) 256-4500 ext.3586 E-mail: iftode@rowan.edu URL: http://www.rowan.edu/colleges/las/departments/biologicalsci/facultystaff/cristinaiftode.htm
Protein Structure/Function Relationships
 Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
DNA. Branden & Tooze, Ch. 7 Deoxyribose nucleic acids are made of three parts
 DNA Branden & Tooze, Ch. 7 Deoxyribose nucleic acids are made of three parts base: adenine, cytosine, guanine, thymine sugar: deoxyribose phosphate: will form the phosphate backbone wide narrow DNA binding
DNA Branden & Tooze, Ch. 7 Deoxyribose nucleic acids are made of three parts base: adenine, cytosine, guanine, thymine sugar: deoxyribose phosphate: will form the phosphate backbone wide narrow DNA binding
Module Overview. Lecture
 Module verview Day 1 2 3 4 5 6 7 8 Lecture Introduction SELEX I: Building a Library SELEX II: Selecting RNA with target functionality SELEX III: Technical advances & problem-solving Characterizing aptamers
Module verview Day 1 2 3 4 5 6 7 8 Lecture Introduction SELEX I: Building a Library SELEX II: Selecting RNA with target functionality SELEX III: Technical advances & problem-solving Characterizing aptamers
Bi 8 Lecture 7. Ellen Rothenberg 26 January Reading: Ch. 3, pp ; panel 3-1
 Bi 8 Lecture 7 PROTEIN STRUCTURE, Functional analysis, and evolution Ellen Rothenberg 26 January 2016 Reading: Ch. 3, pp. 109-134; panel 3-1 (end with free amine) aromatic, hydrophobic small, hydrophilic
Bi 8 Lecture 7 PROTEIN STRUCTURE, Functional analysis, and evolution Ellen Rothenberg 26 January 2016 Reading: Ch. 3, pp. 109-134; panel 3-1 (end with free amine) aromatic, hydrophobic small, hydrophilic
DNA and RNA: Structure and Function. 阮雪芬 May 14, 2004
 DNA and RNA: Structure and Function 阮雪芬 May 14, 2004 Two Fundamental types of nucleic acids participate as genetic molecules DNA: deoxyribonucleic acid Found in the chromosome form in the cell s nucleus
DNA and RNA: Structure and Function 阮雪芬 May 14, 2004 Two Fundamental types of nucleic acids participate as genetic molecules DNA: deoxyribonucleic acid Found in the chromosome form in the cell s nucleus
DNA Structures. Biochemistry 201 Molecular Biology January 5, 2000 Doug Brutlag. The Structural Conformations of DNA
 DNA Structures Biochemistry 201 Molecular Biology January 5, 2000 Doug Brutlag The Structural Conformations of DNA 1. The principle message of this lecture is that the structure of DNA is much more flexible
DNA Structures Biochemistry 201 Molecular Biology January 5, 2000 Doug Brutlag The Structural Conformations of DNA 1. The principle message of this lecture is that the structure of DNA is much more flexible
Antibody-Antigen recognition. Structural Biology Weekend Seminar Annegret Kramer
 Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Chapter 3 Nucleic Acids, Proteins, and Enzymes
 3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
BC 367, Exam 2 November 13, Part I. Multiple Choice (3 pts each)- Please circle the single best answer.
 Name BC 367, Exam 2 November 13, 2008 Part I. Multiple Choice (3 pts each)- Please circle the single best answer. 1. The enzyme pyruvate dehydrogenase catalyzes the following reaction. What kind of enzyme
Name BC 367, Exam 2 November 13, 2008 Part I. Multiple Choice (3 pts each)- Please circle the single best answer. 1. The enzyme pyruvate dehydrogenase catalyzes the following reaction. What kind of enzyme
DEPARTMENT OF BIOCHEMISTRY AND MOLECULAR BIOLOGY
 Department of Biochemistry and Molecular Biology 1 DEPARTMENT OF BIOCHEMISTRY AND MOLECULAR BIOLOGY Office in Molecular and Radiological Biosciences Building, Room 111 (970) 491-5602 bmb.colostate.edu
Department of Biochemistry and Molecular Biology 1 DEPARTMENT OF BIOCHEMISTRY AND MOLECULAR BIOLOGY Office in Molecular and Radiological Biosciences Building, Room 111 (970) 491-5602 bmb.colostate.edu
Protein-Protein Interactions II
 Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Nucleic Acids, Proteins, and Enzymes
 3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
DNA and RNA are both made of nucleotides. Proteins are made of amino acids. Transcription can be reversed but translation cannot.
 INFORMATION TRANSFER Information in cells Properties of information Information must be able to be stored, accessed, retrieved, transferred, read and used. Information is about order, it is basically the
INFORMATION TRANSFER Information in cells Properties of information Information must be able to be stored, accessed, retrieved, transferred, read and used. Information is about order, it is basically the
Module 2 overview SPRING BREAK
 1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
BASIC MOLECULAR GENETIC MECHANISMS Introduction:
 BASIC MOLECULAR GENETIC MECHANISMS Introduction: nucleic acids. (1) contain the information for determining the amino acid sequence & the structure and function of proteins (1) part of the cellular structures:
BASIC MOLECULAR GENETIC MECHANISMS Introduction: nucleic acids. (1) contain the information for determining the amino acid sequence & the structure and function of proteins (1) part of the cellular structures:
Optimizing Synthetic DNA for Metabolic Engineering Applications. Howard Salis Penn State University
 Optimizing Synthetic DNA for Metabolic Engineering Applications Howard Salis Penn State University Synthetic Biology Specify a function Build a genetic system (a DNA molecule) Genetic Pseudocode call producequorumsignal(luxi
Optimizing Synthetic DNA for Metabolic Engineering Applications Howard Salis Penn State University Synthetic Biology Specify a function Build a genetic system (a DNA molecule) Genetic Pseudocode call producequorumsignal(luxi
B. Information for Spring Semester TIMES: Lecture 1 M,W,F 12:00-12:50 Room Science A109 Lab 1 T 11:00-13:50 Science D118
 SYLLABUS for CHEMISTRY 260 ELEMENTARY BIOCHEMISTRY Spring 2006 INSTRUCTOR Dr. Thomas M. Zamis I. Course Description A. Biochemistry (Prerequisites: Chem 220; or 326 and 328) Elementary Biochemistry is
SYLLABUS for CHEMISTRY 260 ELEMENTARY BIOCHEMISTRY Spring 2006 INSTRUCTOR Dr. Thomas M. Zamis I. Course Description A. Biochemistry (Prerequisites: Chem 220; or 326 and 328) Elementary Biochemistry is
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015
 Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
Chemistry 1050 Exam 4 Study Guide
 Chapter 19 Chemistry 1050 Exam 4 Study Guide 19.1 and 19.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges
Chapter 19 Chemistry 1050 Exam 4 Study Guide 19.1 and 19.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges
Chemistry 1120 Exam 3 Study Guide
 Chemistry 1120 Exam 3 Study Guide Chapter 9 9.1 and 9.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges of
Chemistry 1120 Exam 3 Study Guide Chapter 9 9.1 and 9.2 Know there are 20 common amino acids that can polymerize into proteins. Know why amino acids are called alpha amino acids. Identify the charges of
High Profile Publishing in Molecular Biology. Hélène Hodak, Marina Ostankovitch,
 High Profile Publishing in Molecular Biology Hélène Hodak, h.hodak@elsevier.com Marina Ostankovitch, m.ostankovitch@elsevier.com The field of Molecular Biology, inception to current trends Preparing a
High Profile Publishing in Molecular Biology Hélène Hodak, h.hodak@elsevier.com Marina Ostankovitch, m.ostankovitch@elsevier.com The field of Molecular Biology, inception to current trends Preparing a
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
The Two-Hybrid System
 Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
BIOLOGY 200 Molecular Biology Students registered for the 9:30AM lecture should NOT attend the 4:30PM lecture.
 BIOLOGY 200 Molecular Biology Students registered for the 9:30AM lecture should NOT attend the 4:30PM lecture. Midterm date change! The midterm will be held on October 19th (likely 6-8PM). Contact Kathy
BIOLOGY 200 Molecular Biology Students registered for the 9:30AM lecture should NOT attend the 4:30PM lecture. Midterm date change! The midterm will be held on October 19th (likely 6-8PM). Contact Kathy
BIOCHEM + MOLE BIOLOGY- BC (BC)
 Biochem + Mole Biology-BC (BC) 1 BIOCHEM + MOLE BIOLOGY- BC (BC) Courses BC 192 Biochemistry Freshman Seminar Credits: 2 (1-0-1) Introduction to curriculum and career options for biochemistry majors. Registration
Biochem + Mole Biology-BC (BC) 1 BIOCHEM + MOLE BIOLOGY- BC (BC) Courses BC 192 Biochemistry Freshman Seminar Credits: 2 (1-0-1) Introduction to curriculum and career options for biochemistry majors. Registration
Biocatalysis. Amanda Garner September 14, Trypsin Aminotransferase. RNA Ligase
 Biocatalysis TIFF (Uncompressed) decompressortiff (Uncompressed) decompressor TIFF (Uncompressed) decompressor RNA Ligase Trypsin Aminotransferase Amanda Garner September 14, 2007 All enzyme structures
Biocatalysis TIFF (Uncompressed) decompressortiff (Uncompressed) decompressor TIFF (Uncompressed) decompressor RNA Ligase Trypsin Aminotransferase Amanda Garner September 14, 2007 All enzyme structures
Gary Ketner, PhD Johns Hopkins University. Treatment of Infectious Disease: Drugs and Drug Resistance
 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this material constitutes acceptance of that license and the conditions of use of materials on this
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this material constitutes acceptance of that license and the conditions of use of materials on this
Andrew D. Hollenbach, PhD Louisiana State University Health Sciences Center Department of Genetics 533 Bolivar St. New Orleans, LA 70112
 Andrew D. Hollenbach, PhD Louisiana State University Health Sciences Center Department of Genetics 533 Bolivar St. New Orleans, LA 70112 Phone: (504)568-2431 e-mail: aholle@lsuhsc.edu EDUCATION Research
Andrew D. Hollenbach, PhD Louisiana State University Health Sciences Center Department of Genetics 533 Bolivar St. New Orleans, LA 70112 Phone: (504)568-2431 e-mail: aholle@lsuhsc.edu EDUCATION Research
CHEMISTRY 450: Protein Structure and Function
 CHEMISTRY 450: Protein Structure and Function CHEM 450: Fall 2017, Jayasinghe, page 1 of 8 Term: Fall, 2017 Prerequisites: CHEM 341/351 (or equivalent with a minimum grade of C (grade point 2.0). Class
CHEMISTRY 450: Protein Structure and Function CHEM 450: Fall 2017, Jayasinghe, page 1 of 8 Term: Fall, 2017 Prerequisites: CHEM 341/351 (or equivalent with a minimum grade of C (grade point 2.0). Class
Drug DNA interaction. Modeling DNA ligand interaction of intercalating ligands
 Drug DNA interaction DNA as carrier of genetic information is a major target for drug interaction because of the ability to interfere with transcription (gene expression and protein synthesis) and DNA
Drug DNA interaction DNA as carrier of genetic information is a major target for drug interaction because of the ability to interfere with transcription (gene expression and protein synthesis) and DNA
Biochemistry 302, February 11, 2004 Exam 1 (100 points) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed DNA
 1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Ph.D Chemical Engineering, The University of Texas at Austin. B.S Chemical Engineering, University of Minnesota, Minneapolis-St.
 Patrick S. Daugherty, Ph.D. Department of Chemical Engineering University of California, Santa Barbara, CA, 93106 Ph: 805-893-2610, Email: psd@engineering.ucsb.edu Education Ph.D. 1999 Chemical Engineering,
Patrick S. Daugherty, Ph.D. Department of Chemical Engineering University of California, Santa Barbara, CA, 93106 Ph: 805-893-2610, Email: psd@engineering.ucsb.edu Education Ph.D. 1999 Chemical Engineering,
Fig. 1. A schematic illustration of pipeline from gene to drug : integration of virtual and real experiments.
 1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015
 Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
Examination I PHRM 836 Biochemistry for Pharmaceutical Sciences II September 29, 2015 PHRM 836 Exam I - 1 Name: Instructions 1. Check your exam to make certain that it has 9 pages including this cover
BEH.462/3.962J Molecular Principles of Biomaterials Spring 2003
 Lecture 17: Drug targeting Last time: Today: Intracellular drug delivery Drug targeting Reading: T.J. Wickham, Ligand-directed targeting of genes to the site of disease, Nat. Med. 9(1) 135-139 (2003) Drug
Lecture 17: Drug targeting Last time: Today: Intracellular drug delivery Drug targeting Reading: T.J. Wickham, Ligand-directed targeting of genes to the site of disease, Nat. Med. 9(1) 135-139 (2003) Drug
Microbiology, Molecular Biology and Biochemistry
 Microbiology, Molecular Biology and Biochemistry Bruce L. Miller, Interim Dept. Head, Dept. of Microbiology, Molecular Biology and Biochemistry (142 Life Sc. Bldg. 83844-3052; phone 208/885-7966; mmbb@uidaho.edu;
Microbiology, Molecular Biology and Biochemistry Bruce L. Miller, Interim Dept. Head, Dept. of Microbiology, Molecular Biology and Biochemistry (142 Life Sc. Bldg. 83844-3052; phone 208/885-7966; mmbb@uidaho.edu;
BIO 311C Spring Lecture 16 Monday 1 March
 BIO 311C Spring 2010 Lecture 16 Monday 1 March Review Primary Structure of a portion of a polypeptide chain backbone of Polypeptide chain R-groups of amino acids Native conformation of a dimeric protein,
BIO 311C Spring 2010 Lecture 16 Monday 1 March Review Primary Structure of a portion of a polypeptide chain backbone of Polypeptide chain R-groups of amino acids Native conformation of a dimeric protein,
Outline. Background. Background. Background and past work Introduction to present work ResultsandDiscussion Conclusions Perspectives BIOINFORMATICS
 Galactosemia Foundation 2012 Conference Breakout Session G July 20th, 2012, 14:15 15:15 Computational Biology Strategy for the Development of Ligands of GALK Enzyme as Potential Drugs for People with Classic
Galactosemia Foundation 2012 Conference Breakout Session G July 20th, 2012, 14:15 15:15 Computational Biology Strategy for the Development of Ligands of GALK Enzyme as Potential Drugs for People with Classic
Expressive. Bacterial surface display for the identification of active ingredients for the pharmaceutical and cosmetics industry
 Active Ingredient Research Expressive Bacterial surface display for the identification of active ingredients for the pharmaceutical and cosmetics industry Prof. Dr. Joachim Jose, Institute of Pharmaceutical
Active Ingredient Research Expressive Bacterial surface display for the identification of active ingredients for the pharmaceutical and cosmetics industry Prof. Dr. Joachim Jose, Institute of Pharmaceutical
Lecture 13. Motor Proteins I
 Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Lecture 13 Motor Proteins I Introduction: The study of motor proteins has become a major focus in cell and molecular biology. Motor proteins are very interesting because they do what no man-made engines
Introduction to Proteins
 Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Polypharmacology. Giulio Rastelli Molecular Modelling and Drug Design Lab
 Polypharmacology Giulio Rastelli Molecular Modelling and Drug Design Lab www.mmddlab.unimore.it Dipartimento di Scienze della Vita Università di Modena e Reggio Emilia giulio.rastelli@unimore.it Magic
Polypharmacology Giulio Rastelli Molecular Modelling and Drug Design Lab www.mmddlab.unimore.it Dipartimento di Scienze della Vita Università di Modena e Reggio Emilia giulio.rastelli@unimore.it Magic
Molekulare Mechanismen der Signaltransduktion
 Molekulare Mechanismen der Signaltransduktion 12 Mechanism of auxin perception slides: http://tinyurl.com/modul-mms doi: 10.1038/nature05731 previous model http://www.plantcell.org/content/17/9/2425/f1.large.jpg
Molekulare Mechanismen der Signaltransduktion 12 Mechanism of auxin perception slides: http://tinyurl.com/modul-mms doi: 10.1038/nature05731 previous model http://www.plantcell.org/content/17/9/2425/f1.large.jpg
Protein Folding in vivo. Biochemistry 530 David Baker
 Protein Folding in vivo Biochemistry 530 David Baker I. Protein folding in vivo: Molecular Chaperones II. Protein unfolding in vivo: ATP driven unfolding in mitochondrial import and the proteasome. III.
Protein Folding in vivo Biochemistry 530 David Baker I. Protein folding in vivo: Molecular Chaperones II. Protein unfolding in vivo: ATP driven unfolding in mitochondrial import and the proteasome. III.
5/15/13. Agenda. Agenda. Introduction practical assignment: Identification of VHH against ErbB1/ EGFR. Structure of heavy chain antibodies
 Introduction practical assignment: Identification of VHH against ErbB1/ EGFR Erik Hofman, Alex Klarenbeek, Rachid El Khoulathi 15-05-2013 Structure of heavy chain antibodies C H 1 C L V H V L V HH C H
Introduction practical assignment: Identification of VHH against ErbB1/ EGFR Erik Hofman, Alex Klarenbeek, Rachid El Khoulathi 15-05-2013 Structure of heavy chain antibodies C H 1 C L V H V L V HH C H
Structures of the human! 2 Adrenoceptor ! 2 AR-T4L. ! 2 AR-Fab5. Expression Purification of functional receptor Stabilization!Protein engineering!!
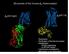 Structures of the human! 2 Adrenoceptor! 2 AR-Fab5! 2 AR-T4L Expression Purification of functional receptor Stabilization!Protein engineering!! 2 AR-Fab complex Lipid-based crystallization Microbeam X-ray
Structures of the human! 2 Adrenoceptor! 2 AR-Fab5! 2 AR-T4L Expression Purification of functional receptor Stabilization!Protein engineering!! 2 AR-Fab complex Lipid-based crystallization Microbeam X-ray
Paper : 03 Structure and Function of Biomolecules II Module: 02 Nucleosides, Nucleotides and type of Nucleic Acids
 Paper : 03 Structure and Function of Biomolecules II Module: 02 Nucleosides, Nucleotides and type of Nucleic Acids Principal Investigator Paper Coordinators Prof. Sunil Kumar Khare, Professor, Department
Paper : 03 Structure and Function of Biomolecules II Module: 02 Nucleosides, Nucleotides and type of Nucleic Acids Principal Investigator Paper Coordinators Prof. Sunil Kumar Khare, Professor, Department
DNA delivery and DNA Vaccines
 DNA delivery and DNA Vaccines Last Time: Today: Reading: intracellular drug delivery: enhancing cross priming for vaccines DNA vaccination D.W. Pack, A.S. Hoffman, S. Pun, and P.S. Stayton, Design and
DNA delivery and DNA Vaccines Last Time: Today: Reading: intracellular drug delivery: enhancing cross priming for vaccines DNA vaccination D.W. Pack, A.S. Hoffman, S. Pun, and P.S. Stayton, Design and
Molecular biology WID Masters of Science in Tropical and Infectious Diseases-Transcription Lecture Series RNA I. Introduction and Background:
 Molecular biology WID 602 - Masters of Science in Tropical and Infectious Diseases-Transcription Lecture Series RNA I. Introduction and Background: DNA and RNA each consists of only four different nucleotides.
Molecular biology WID 602 - Masters of Science in Tropical and Infectious Diseases-Transcription Lecture Series RNA I. Introduction and Background: DNA and RNA each consists of only four different nucleotides.
Biophysical characterization of proteinprotein
 Biophysical characterization of proteinprotein interactions Rob Meijers EMBL Hamburg EMBO Global Exchange Lecture Course Hyderabad 2012 Bottom up look at protein-protein interactions Role of hydrogen bonds
Biophysical characterization of proteinprotein interactions Rob Meijers EMBL Hamburg EMBO Global Exchange Lecture Course Hyderabad 2012 Bottom up look at protein-protein interactions Role of hydrogen bonds
Enzymes Part III: regulation I. Dr. Mamoun Ahram Summer, 2017
 Enzymes Part III: regulation I Dr. Mamoun Ahram Summer, 2017 Mechanisms of regulation Expression of isoenzymes Regulation of enzymatic activity Inhibitors Conformational changes Allostery Modulators Reversible
Enzymes Part III: regulation I Dr. Mamoun Ahram Summer, 2017 Mechanisms of regulation Expression of isoenzymes Regulation of enzymatic activity Inhibitors Conformational changes Allostery Modulators Reversible
From mechanism to medicne
 From mechanism to medicne a look at proteins and drug design Chem 342 δ δ δ+ M 2009 δ+ δ+ δ M Drug Design - an Iterative Approach @ DSU Structural Analysis of Receptor Structural Analysis of Ligand-Receptor
From mechanism to medicne a look at proteins and drug design Chem 342 δ δ δ+ M 2009 δ+ δ+ δ M Drug Design - an Iterative Approach @ DSU Structural Analysis of Receptor Structural Analysis of Ligand-Receptor
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
Four levels of protein Structure
 Proteins (polypeptides) Four levels of protein Structure Primary Structure (1 structure): Secondary Structure (2 structure): Tertiary Structure (3 structure): Quaternary Structure (4 structure): Proteins
Proteins (polypeptides) Four levels of protein Structure Primary Structure (1 structure): Secondary Structure (2 structure): Tertiary Structure (3 structure): Quaternary Structure (4 structure): Proteins
Science June 3, 1988 v240 n4857 p1310(7) Page 1
 Science June 3, 1988 v240 n4857 p1310(7) Page 1 by Brian K. Kobilka, Tong Sun Kobilka, Kiefer Daniel, John W. Regan, Marc G. Caron and Robert J. Lefkowitz COPYRIGHT 1988 American Association for the Advancement
Science June 3, 1988 v240 n4857 p1310(7) Page 1 by Brian K. Kobilka, Tong Sun Kobilka, Kiefer Daniel, John W. Regan, Marc G. Caron and Robert J. Lefkowitz COPYRIGHT 1988 American Association for the Advancement
DNA Cloning with Cloning Vectors
 Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Chemistry 171: Biological Synthesis
 Chemistry 171: Biological Synthesis Harvard University Spring, 2012 Tuesday and Thursday, 10 11:30 AM Location: Pfizer Lecture Hall, Mallinckrodt Lab 12 Oxford St., Cambridge Nature is a remarkable synthetic
Chemistry 171: Biological Synthesis Harvard University Spring, 2012 Tuesday and Thursday, 10 11:30 AM Location: Pfizer Lecture Hall, Mallinckrodt Lab 12 Oxford St., Cambridge Nature is a remarkable synthetic
NANYANG TECHNOLOGICAL UNIVERSITY SEMESTER I EXAMINATION CBC922 Medicinal Chemistry. NOVEMBER TIME ALLOWED: 120 min
 AYAG TECLGICAL UIVERSITY SEMESTER I EXAMIATI 2006-2007 CBC922 Medicinal Chemistry VEMBER 2006 - TIME ALLWED 120 min ISTRUCTIS T CADIDATES 1. This examination paper contains TW (2) parts and comprises SIX
AYAG TECLGICAL UIVERSITY SEMESTER I EXAMIATI 2006-2007 CBC922 Medicinal Chemistry VEMBER 2006 - TIME ALLWED 120 min ISTRUCTIS T CADIDATES 1. This examination paper contains TW (2) parts and comprises SIX
Bacteriophages get a foothold on their prey
 Bacteriophages get a foothold on their prey Long and thin, the receptor-binding needle of bacteriophage T4 Bacterial viruses, bacteriophages or phages, have served as a tool to decipher principles of molecular
Bacteriophages get a foothold on their prey Long and thin, the receptor-binding needle of bacteriophage T4 Bacterial viruses, bacteriophages or phages, have served as a tool to decipher principles of molecular
A New Cellular and Molecular Engineering Curriculum at Rice University
 Session A New Cellular and Molecular Engineering Curriculum at Rice University Ka-Yiu San, Larry V. McIntire, Ann Saterbak Department of Bioengineering, Rice University Houston, Texas 77005 Abstract The
Session A New Cellular and Molecular Engineering Curriculum at Rice University Ka-Yiu San, Larry V. McIntire, Ann Saterbak Department of Bioengineering, Rice University Houston, Texas 77005 Abstract The
Nucleic acid and protein Flow of genetic information
 Nucleic acid and protein Flow of genetic information References: Glick, BR and JJ Pasternak, 2003, Molecular Biotechnology: Principles and Applications of Recombinant DNA, ASM Press, Washington DC, pages.
Nucleic acid and protein Flow of genetic information References: Glick, BR and JJ Pasternak, 2003, Molecular Biotechnology: Principles and Applications of Recombinant DNA, ASM Press, Washington DC, pages.
CHEM 4420 Exam I Spring 2013 Page 1 of 6
 CHEM 4420 Exam I Spring 2013 Page 1 of 6 Name Use complete sentences when requested. There are 100 possible points on this exam. The multiple choice questions are worth 2 points each. All other questions
CHEM 4420 Exam I Spring 2013 Page 1 of 6 Name Use complete sentences when requested. There are 100 possible points on this exam. The multiple choice questions are worth 2 points each. All other questions
Chemistry-Biology Interface Training Program
 ChemistryBiology Interface Training Program University of Massachusetts Amherst Biochemistry & Molecular Biology Chemistry Polymer cience & Engineering Chemical Engineering Veterinary & Animal ciences
ChemistryBiology Interface Training Program University of Massachusetts Amherst Biochemistry & Molecular Biology Chemistry Polymer cience & Engineering Chemical Engineering Veterinary & Animal ciences
Parasitological and serological methods. New tools for improved diagnosis. Dr. Magdalena Radwanska FIND 27JUN07
 Parasitological and serological methods New tools for improved diagnosis Dr. Magdalena Radwanska FIND FIND HAT diagnostic projects Parasite detection Improved maect Trapping the parasite on the filter
Parasitological and serological methods New tools for improved diagnosis Dr. Magdalena Radwanska FIND FIND HAT diagnostic projects Parasite detection Improved maect Trapping the parasite on the filter
Lecture Series 10 The Genetics of Viruses and Prokaryotes
 Lecture Series 10 The Genetics of Viruses and Prokaryotes The Genetics of Viruses and Prokaryotes A. Using Prokaryotes and Viruses for Genetic Experiments B. Viruses: Reproduction and Recombination C.
Lecture Series 10 The Genetics of Viruses and Prokaryotes The Genetics of Viruses and Prokaryotes A. Using Prokaryotes and Viruses for Genetic Experiments B. Viruses: Reproduction and Recombination C.
Unit 2 Review: DNA, Protein Synthesis & Enzymes
 1. One of the functions of DNA is to A. secrete vacuoles.. make copies of itself.. join amino acids to each other. D. carry genetic information out of the nucleus. 2. Two sugars found in nucleic acids
1. One of the functions of DNA is to A. secrete vacuoles.. make copies of itself.. join amino acids to each other. D. carry genetic information out of the nucleus. 2. Two sugars found in nucleic acids
Nucleotides & Nucleic Acids. Central Dogma of Biology
 Roles: Energy currency (ATP, GTP) Chemical links in response of cells to hormones (camp) Involved in cofactors (NAD, FAD, CoA) Metabolic intermediates (acetyl CoA) Constituents of nucleic acids, DNA and
Roles: Energy currency (ATP, GTP) Chemical links in response of cells to hormones (camp) Involved in cofactors (NAD, FAD, CoA) Metabolic intermediates (acetyl CoA) Constituents of nucleic acids, DNA and
Synthetic Biology. Sustainable Energy. Therapeutics Industrial Enzymes. Agriculture. Accelerating Discoveries, Expanding Possibilities. Design.
 Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
Synthetic Biology Accelerating Discoveries, Expanding Possibilities Sustainable Energy Therapeutics Industrial Enzymes Agriculture Design Build Generate Solutions to Advance Synthetic Biology Research
The 5' cap (red) is added before synthesis of the primary transcript is complete. A non coding sequence following the last exon is shown in orange.
 RNA PROCESSING The 5' cap (red) is added before synthesis of the primary transcript is complete. A non coding sequence following the last exon is shown in orange. Splicing can occur either before or after
RNA PROCESSING The 5' cap (red) is added before synthesis of the primary transcript is complete. A non coding sequence following the last exon is shown in orange. Splicing can occur either before or after
Discovery on Target. Short Course Preview Deck
 Discovery on Target Short Course Preview Deck Targeting of GPCRs with Monoclonal Antibodies Barbara Swanson, Ph.D. Sorrento Therapeutics, Inc. October 2014 GPCR Short Course Overview Introductions Brief
Discovery on Target Short Course Preview Deck Targeting of GPCRs with Monoclonal Antibodies Barbara Swanson, Ph.D. Sorrento Therapeutics, Inc. October 2014 GPCR Short Course Overview Introductions Brief
AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA
 AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA Encodes hereditary information Used to specify the amino
AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA Encodes hereditary information Used to specify the amino
Feature Article. Development of an Intracellular ph Measurement Method using DNA as a Sensing Material
 Feature Article The Winner's Article of the First Dr. Masao Horiba's Award Development of an Intracellular ph Measurement Method using DNA as a Sensing Material Naoki Sugimoto, Tatsuo Ohmichi While carrying
Feature Article The Winner's Article of the First Dr. Masao Horiba's Award Development of an Intracellular ph Measurement Method using DNA as a Sensing Material Naoki Sugimoto, Tatsuo Ohmichi While carrying
RNA synthesis/transcription I Biochemistry 302. February 6, 2004 Bob Kelm
 RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
BIOCHEMISTRY Nucleic Acids
 BIOCHEMISTRY Nucleic Acids BIOB111 CHEMISTRY & BIOCHEMISTRY Session 17 Session Plan Types of Nucleic Acids Nucleosides Nucleotides Primary Structure of Nucleic Acids DNA Double Helix DNA Replication Types
BIOCHEMISTRY Nucleic Acids BIOB111 CHEMISTRY & BIOCHEMISTRY Session 17 Session Plan Types of Nucleic Acids Nucleosides Nucleotides Primary Structure of Nucleic Acids DNA Double Helix DNA Replication Types
RESEARCH AREA NEUROSCIENCE
 RESEARCH AREA NEUROSCIENCE coordinator MATHEW E. DIAMOND area committee Alessandro Treves Raffaella Rumiati Giuseppe Legname Andrea Nistri Dario Olivieri A variety of research groups are active in Neuroscience.
RESEARCH AREA NEUROSCIENCE coordinator MATHEW E. DIAMOND area committee Alessandro Treves Raffaella Rumiati Giuseppe Legname Andrea Nistri Dario Olivieri A variety of research groups are active in Neuroscience.
Conformational properties of enzymes
 Conformational properties of enzymes; Physics of enzyme substrate interactions; Electronic conformational interactions, cooperative properties of enzymes Mitesh Shrestha Conformational properties of enzymes
Conformational properties of enzymes; Physics of enzyme substrate interactions; Electronic conformational interactions, cooperative properties of enzymes Mitesh Shrestha Conformational properties of enzymes
By Dr. Abo Bakr Eltayeb
 Assiut University Workshop Protein electrophoresis and immunoblot (Western blot) By Dr. Abo Bakr Eltayeb March 27-28 2011 Introduction of protein structure and isolation Protein Function Enzymes - proteases,
Assiut University Workshop Protein electrophoresis and immunoblot (Western blot) By Dr. Abo Bakr Eltayeb March 27-28 2011 Introduction of protein structure and isolation Protein Function Enzymes - proteases,
ELSEVIER ACADEMIC PRESS INC, 525 B STREET, SUITE 1900, SAN DIEGO, USA, CA,
 SCIENCE CITATION INDEX - GENETICS & HEREDITY - JOURNAL LIST Total journals: 95 1. ADVANCES IN GENETICS Irregular ISSN: 0065-2660 ELSEVIER ACADEMIC PRESS INC, 525 B STREET, SUITE 1900, SAN DIEGO, USA, CA,
SCIENCE CITATION INDEX - GENETICS & HEREDITY - JOURNAL LIST Total journals: 95 1. ADVANCES IN GENETICS Irregular ISSN: 0065-2660 ELSEVIER ACADEMIC PRESS INC, 525 B STREET, SUITE 1900, SAN DIEGO, USA, CA,
Chem Lecture 5 Catalytic Strategies
 Chem 452 - Lecture 5 Catalytic Strategies 111026 Enzymes have evolved an array of different strategies or enhancing the power and specificity of the reactions they catalyze. For numerous enzymes the details
Chem 452 - Lecture 5 Catalytic Strategies 111026 Enzymes have evolved an array of different strategies or enhancing the power and specificity of the reactions they catalyze. For numerous enzymes the details
ProteoGenix. Life Sciences Services and Products. From gene to biotherapeutics Target Validation to Lead optimisation
 ProteoGenix Life Sciences Services and Products From gene to biotherapeutics Target Validation to Lead optimisation ProteoGenix Philippe FUNFROCK, founder and CEO French company located in Strasbourg,
ProteoGenix Life Sciences Services and Products From gene to biotherapeutics Target Validation to Lead optimisation ProteoGenix Philippe FUNFROCK, founder and CEO French company located in Strasbourg,
Astronomy picture of the day (4/21/08)
 Biol 205 Spring 2008 Astronomy picture of the day (4/21/08) http://antwrp.gsfc.nasa.gov/apod/ap080421.html Are viruses alive? http://serc.carleton.edu/microbelife/yellowstone/viruslive.html 1 Week 3 Lecture
Biol 205 Spring 2008 Astronomy picture of the day (4/21/08) http://antwrp.gsfc.nasa.gov/apod/ap080421.html Are viruses alive? http://serc.carleton.edu/microbelife/yellowstone/viruslive.html 1 Week 3 Lecture
Introduction practical assignment: Identification of VHH/nanobodies against ErbB1/EGFR
 Introduction practical assignment: Identification of VHH/nanobodies against ErbB1/EGFR Sofia Doulkeridou, Rachid El Khoulati, Paul van Bergen en Henegouwen 12-05-2014 Agenda Introduction to Nanobodies
Introduction practical assignment: Identification of VHH/nanobodies against ErbB1/EGFR Sofia Doulkeridou, Rachid El Khoulati, Paul van Bergen en Henegouwen 12-05-2014 Agenda Introduction to Nanobodies
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA
 RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
Custom Antibody Services. Antibodies Designed Just for You. HuCAL Recombinant Monoclonal Antibody Generation Service
 Custom Antibody Services Antibodies Designed Just for You HuCAL Recombinant Monoclonal Antibody Generation Service Antibodies Designed Just for You What if there was a technology that would allow you to
Custom Antibody Services Antibodies Designed Just for You HuCAL Recombinant Monoclonal Antibody Generation Service Antibodies Designed Just for You What if there was a technology that would allow you to
fibrils, however, oligomeric structures and amorphous protein aggregates were
 Supplementary Figure 1: Effect of equimolar EGCG on αs and Aβ aggregate formation. A D B E αs C F Figure S1: (A-C) Analysis of EGCG treated αs (1 µm) aggregation reactions by EM. A 1:1 molar ratio of EGCG
Supplementary Figure 1: Effect of equimolar EGCG on αs and Aβ aggregate formation. A D B E αs C F Figure S1: (A-C) Analysis of EGCG treated αs (1 µm) aggregation reactions by EM. A 1:1 molar ratio of EGCG
Chun Wu Research Assistant Professor University of California Santa Barbara
 Accurate bio-molecule model(force field) + China super computer: upgrade China s bio-research, pharmaceutical & bio-tech industries using molecular dynamics simulation Chun Wu Research Assistant Professor
Accurate bio-molecule model(force field) + China super computer: upgrade China s bio-research, pharmaceutical & bio-tech industries using molecular dynamics simulation Chun Wu Research Assistant Professor
ProCode TM. Life Science, Inc. A Rapid Flexible MAb-Like Discovery Platform for Creating Diagnostic Antibodies.
 ProCode TM A Rapid Flexible MAb-Like Discovery Platform for Creating Diagnostic Antibodies Life Science, Inc. www.meridianlifescience.com Custom Monoclonal Development Meridian Life Science, Inc. (MLS)
ProCode TM A Rapid Flexible MAb-Like Discovery Platform for Creating Diagnostic Antibodies Life Science, Inc. www.meridianlifescience.com Custom Monoclonal Development Meridian Life Science, Inc. (MLS)
COURSES IN BIOLOGICAL SCIENCE. Undergraduate Courses Postgraduate Courses
 COURSES IN BIOLOGICAL SCIENCE Undergraduate Courses Postgraduate Courses Undergraduate Courses: BISC 001 Appreciation of Biological Sciences [3-0-0:3] Diversity of life forms; origin of life; chemical
COURSES IN BIOLOGICAL SCIENCE Undergraduate Courses Postgraduate Courses Undergraduate Courses: BISC 001 Appreciation of Biological Sciences [3-0-0:3] Diversity of life forms; origin of life; chemical
