Supplementary Figure 1
|
|
|
- Timothy George
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use TRMPV- Neo (Genbank HQ456315), which is selectable with G418. TRMPV-ns (no selection, Genbank HQ456316) is a simplified backbone that lacks a drug-selectable marker. TRMPV-Hygro (Genbank HQ456314) is Hygromycin-selectable. TtTMPV-Neo (Genbank HQ456319) features a TurboRFP reporter (Evrogen) and an optimized TRE promoter (TREtight) 1,2, which reduces leakiness in the absence of dox resulting from basal TRE activity by ~40-fold 3, while retaining strong expression levels comparable to conventional TRE in the induced state. This vector variant is particularly recommended whenever leaky shrna expression presents a concern, or when cells are transduced at high MOI - a setting that is generally associated with increased leaky expression levels (data not shown). TRMPVIR (Genbank HQ456317) enforces rtta expression in a single vector as described in Fig. 1e-g, and TtRMPVIR (Genbank HQ456318) is the TREtight version of TRMPVIR. All vectors are constructed in the pqcxix self-inactivating (SIN) retroviral backbone. Unique restriction sites for shrna cloning and reporter exchange are indicated.
2 Supplementary Figure 2 Supplementary Figure 2: Quantification of Renilla Luciferase and Rpa3 shrna potency; testing of reversibility and re-inducibility of TRMPV-based shrna expression. (a) Luciferase activity in NIH3T3 cells that stably express Renilla Luciferase (parental) and different shrnas targeting Renilla or a
3 control shrna (Trp ). Stable NIH3T3-Renilla luciferase cells were transduced with LMP retroviruses containing each shrna, selected for one week and luminescence measured in whole cell protein extract. (b) Relative luciferase activity in shren.713 transgenic embryonic stem (ES) cells. ES cell clones containing dox-inducible shren.713 were transduced with a retrovirus that expresses Renilla luciferase and treated with or without dox for 4 days. Dox-dependent shrna induction caused a 10-fold reduction in Renilla luciferase activity. Error bars represent the s.e.m., n=5. (c) Quantification of Rpa3 immunoblot (see Fig. 1b). Rpa3 band intensities were quantified using ImageJ software according to method 1 outlined at (gray bars), and normalized to β-actin bands from the same gel (black bars). (d) Uncropped images of immunoblots shown in Fig. 1b. (e) Representative flow cytometry plots of immortalized Rosa26-rtTA- M2 MEFs transduced with TRMPV. Cells were maintained under selection, and cultured in dox for 3 days, then in the absence of dox for 6 days, then again in dox for 3 days. shren cultures show that dsred expression can be quickly reversed and subsequently re-induced when a neutral shrna is expressed. shrpa3 cultures show reversibility in a subset of cells, though a rapid selection for cells that fail to induce the TRE (Venus + dsred - ) or both reporters (Venus - dsred - ) also occurs.
4 Supplementary Figure 3 Supplementary Figure 3: Characterization of Tet-On MLL-AF9;Nras G12D induced leukemia. (a) Schematic of retroviral vectors and experimental strategies used to generate a Tet-On competent, bioluminescent mouse model of AML. (b) Bioluminescence imaging of recipient mice 15 days after transplantation of HSPCs transduced with MSCV-Luciferase-IRES-Nras G12D alone or in combination with MSCV-rtTA3-IRES-MLL-AF9. (c) Kaplan-Meier survival curve of syngeneic recipient mice of HSPC transduced with MSCV-rtTA3-IRES-MLL-AF9 and/or MSCV-Luciferase-IRES-Nras G12D. (d) May- Grünwald-Giemsa-stained peripheral blood smears showing typical morphology of MLL-AF9;Nras G12D AML at an advanced stage. Original magnification 100x (top) and 500x (bottom). (e) Representative immunophenotyping of MLL-AF9;Nras G12D AML. Overall, disease generated with rtta-linked vectors showed no difference in disease penetrance, latency or phenotype compared to similar AML models driven by vectors lacking rtta3 4, suggesting that expression of rtta3 does not influence disease biology.
5 Supplementary Figure 4 Supplementary Figure 4: Competitive proliferation assays in Tet-On MLL-AF9;Nras G12D AML. (a) Quantification of shrna-expressing (Venus + dsred + ) cells in competitive proliferation assays of Tet-On competent MLL-AF9;Nras G12D AML. Leukemia cells were transduced with indicated TRMPV shrnas, drug selected, mixed with untranduced cells, and passaged in media containing dox for 12 days. Venus + dsred + cells were quantified each day starting the first day after dox treatment. Each series of bars represents day 1 to day 12 from left to right. Rpa3 shrna-expressing AML cells are depleted over time. Both the kinetics and the level of depletion correlate with Rpa3 shrna potency (see also Fig. 1b, Supplementary Fig. 2c). (b) Representative flow cytometry plots of competitive proliferation assays in (a) 10 days after dox-induction of indicated shrnas. Note the outgrowth of Venus + dsred - cells that evade expression of Rpa3 shrnas.
6 Supplementary Figure 5 Supplementary Figure 5: TRMPV clones exhibit dramatic differences in TRE induction in response to dox. (a) Representative flow cytometry histograms showing distribution of Venus fluorescence in bulk and clonal MLL-AF9;Nras G12D AML cells transduced with TRMPV.shRpa Clonal populations were derived through expansion after limiting dilution and show homogeneous Venus levels, likely due to expression from the same proviral genomic integration site. (b) Flow cytometry plots of bulk and clonal populations in competitive proliferation assays. The cell populations in (a) were mixed 1:1 with untransduced cells and passaged in the presence/absence of dox for 3 days. On dox (lower panel), the bulk transduced population shows heterogeneous TRE induction and depletion relative to untransduced (double-negative) cells. Clone 1 shows strong homogeneous TRE induction on dox, and rapid depletion, while clone 4 does not respond to dox treatment. (c) Spleen histology of untreated and dox-treated mice 4 days after start of treatment. Mice were transplanted with clonal MLL-AF9;Nras G12D AML harboring TRMPV.shRpa3.455 and treated with/without dox at disease onset. Scale bar: 50 μm.
7 Supplementary Figure 6 Supplementary Figure 6: Estimation of average retroviral integration numbers in MLL-AF9; Nras G12D AML cells. Cells were co-transduced with a green (MSCV-GFP) and a red (MSCV-dsRed) fluorescence-tagged retrovirus to estimate multiplicity of infection (MOI) at different overall infection efficiencies. (a) Representative flow cytometry plots of transduced AML cells. Left panel (high MOI): In conditions where 44% of cells receive at least one retrovirus, a large proportion is GFP + dsred + indicating that they harbor >1 retroviral integration. Right panel (low MOI): In conditions where 5% of cells are transduced, a small proportion is GFP + dsred + double positive. Given percentages indicate frequencies of GFP +, dsred + and GFP + dsred + cells in all transduced cells. (b) Estimation of cells harboring a single retrovirus relative to overall transduction efficiency. We assumed that at least one third of cells harboring multiple integration events are GFP + dsred +, since two integrations of MSCV-GFP or two integrations of MSCV-dsRed can also occur. Therefore, the frequency of cells with a single integration among all infected cells was calculated as 100% - 3*(% GFP + dsred + ).
8 Supplementary Figure 7 Supplementary Figure 7: Pooled negative selection RNAi screening in vivo - library representation in individual mice and comparison of untreated and dox-treated mice. (a) Scatter plots of normalized reads per shrna in T0 (prior to transplantation) compared to individual untreated mice (off dox mouse 1-
9 3), or to the average of untreated mice (off dox mean). Blue dots represent examples of shrnas detected as positively selected in the offdox mean. Typically, such outliers are enriched in only one of the individual mice, suggesting that they represent stochastic clonal events during disease engraftment and progression. Red and green dots represent Rpa3 and Ren control shrnas. r, nonparametric (Spearman) correlation coefficient. (b) Scatter plots of normalized reads per shrna in T0 (prior to transplantation) compared to individual dox-treated mice (ondox mouse 1-3), or the average of dox-treated mice (ondox mean). Red and green dots represent Rpa3 and Ren control shrnas. r, nonparametric (Spearman) correlation coefficient. (c) Relative abundance of all 824 shrnas in Venus + dsred + -sorted leukemia cells from dox-treated mice compared to untreated mice. The means of normalized reads in dox-treated mice (n=3) was divided by the mean in untreated mice (n=3); shrnas are plotted according to the resulting ratios in ascending order. All three shrnas targeting Rpa3 are among the 25 most depleted shrnas, while neutral shren.713 is not altered.
10 Supplementary Figure 8 Supplementary Figure 8: FACS purification of shrna expressing cells in TRMPV-based pooled shrna screening. Representative flow cytometry profiles of bone marrow and spleen cells (1:1 mix) from moribund untreated (off dox) and dox-treated (on dox) recipient mice of MLL-AF9;Nras G12D AML transduced with a complex TRMPV-shRNA library (824 shrnas). Gates for quantifying Venus + dsred - and Venus + dsred + cells are shown; the Venus + dsred + gate was used for sorting. Both axes are on a biexponential log scale.
11 Supplementary Table 2 Rpa3.53 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCCGCCAGCATGTTACCACAGTATAGTGAAGCCACAGATGTA TACTGTGGTAACATGCTGGCGTTGCCTACTGCCTCGGAATTC Rpa3.276 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCAAGGAAGATACTAATCGCTTTTAGTGAAGCCACAGATGTAA AAGCGATTAGTATCTTCCTTATGCCTACTGCCTCGGAATTC Rpa3.429 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCAAGGAAGACTCCTGCAGTTTATAGTGAAGCCACAGATGTAT AAACTGCAGGAGTCTTCCTTATGCCTACTGCCTCGGAATTC Rpa3.455 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCGCGACTCCTATAATTTCTAATTAGTGAAGCCACAGATGTAA TTAGAAATTATAGGAGTCGCTTGCCTACTGCCTCGGAATTC Rpa3.561 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCAAAAGTGATACTTCAATATATTAGTGAAGCCACAGATGTAA TATATTGAAGTATCACTTTTATGCCTACTGCCTCGGAATTC Ren.27 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGAAACAAAGGAAACGGATGATAATAGTGAAGCCACAGATGTA TTATCATCCGTTTCCTTTGTTCTGCCTACTGCCTCGGAATTC Ren.660 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGACTCGTGAAATCCCGTTAGTAATAGTGAAGCCACAGATGTAT TACTAACGGGATTTCACGAGGTGCCTACTGCCTCGGAATTC Ren.702 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGATACAAATTGTTAGGAATTATATAGTGAAGCCACAGATGTAT ATAATTCCTAACAATTTGTACTGCCTACTGCCTCGGAATTC Ren.713 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTAT AGATAAGCATTATAATTCCTATGCCTACTGCCTCGGAATTC Ren.826 CTCGAGAAGGTATATTGCTGTTGACAGTGAGCGCTACTGAATTTGTCAAAGTAAATAGTGAAGCCACAGATGTAT TTACTTTGACAAATTCAGTATTGCCTACTGCCTCGGAATTC Supplementary Table 2: Sequences of shrnas. 116 nt XhoI/EcoRI mir30-shrna fragments are provided. shrnas were designated by the number of the first nucleotide of the 22-nt target site in the mrna transcript at the time of design.
12 Supplementary references 1. Sipo, I. et al. An improved Tet-On regulatable FasL-adenovirus vector system for lung cancer therapy. J Mol Med 84, (2006). 2. Agha-Mohammadi, S. et al. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J Gene Med 6, (2004). 3. Clontech Gene Expression Systems. Clontechniques XVIII, 1 (2003). 4. Zuber, J. et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev 23, (2009).
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
Supplementary Figure 1 Negative selection analysis of sgrnas targeting all Brd4 exons comparing day 2 to day 10 time points. Systematic evaluation of 64 Brd4 sgrnas in negative selection experiments, targeting
Supporting Information
 Supporting Information McJunkin et al. 1.173/pnas.1149718 SI Text shrn Library Production. puromycin-selectable retrovirus (plm-puro) was cloned by insertion of an MluI site in the multiple cloning site
Supporting Information McJunkin et al. 1.173/pnas.1149718 SI Text shrn Library Production. puromycin-selectable retrovirus (plm-puro) was cloned by insertion of an MluI site in the multiple cloning site
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
sherwood - UltramiR shrna Collections
 sherwood - UltramiR shrna Collections Incorporating advances in shrna design and processing for superior potency and specificity sherwood - UltramiR shrna Collections Enabling Discovery Across the Genome
sherwood - UltramiR shrna Collections Incorporating advances in shrna design and processing for superior potency and specificity sherwood - UltramiR shrna Collections Enabling Discovery Across the Genome
initial single-cell analysis, with a pragmatic focus on surface markers with the highest potential for
 Supplementary Figure 1: Summary of the exclusionary approach to surface marker selection for initial single-cell analysis, with a pragmatic focus on surface markers with the highest potential for protein
Supplementary Figure 1: Summary of the exclusionary approach to surface marker selection for initial single-cell analysis, with a pragmatic focus on surface markers with the highest potential for protein
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplemental Table S1. RT-PCR primers used in this study
 Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Activation capacity of tettale-ad compared to tet trans-activator (ttas) using different teto variants. (A) HeLa cells were co-transfected with activation
Supplementary Figure 1 Supplementary Figure 1. Activation capacity of tettale-ad compared to tet trans-activator (ttas) using different teto variants. (A) HeLa cells were co-transfected with activation
SUPPLEMENTARY INFORMATION
 Elastase levels and activity are increased in dystrophic muscle and impair myoblast cell survival, proliferation and differentiation N. Arecco 1,2,3, C.J. Clarke 1,2, F.K. Jones 1,2, D. Simpson 1,4, D.
Elastase levels and activity are increased in dystrophic muscle and impair myoblast cell survival, proliferation and differentiation N. Arecco 1,2,3, C.J. Clarke 1,2, F.K. Jones 1,2, D. Simpson 1,4, D.
Supplementary Figure 1: MYCER protein expressed from the transgene can enhance
 Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Relative luciferase activity Relative luciferase activity MYC is a critical target FBXW7 MYC Supplementary is a critical Figures target 1-7. FBXW7 Supplementary Material A E-box sequences 1 2 3 4 5 6 HSV-TK
Supplementary
 Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary information Supplementary Material and Methods Plasmid construction The transposable element vectors for inducible expression of RFP-FUS wt and EGFP-FUS R521C and EGFP-FUS P525L were derived
Supplementary Material & Methods
 Supplementary Material & Methods Affymetrix micro-array Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, USA). RNA concentration and quality was determined using the ND-1000 spectrophotometer
Supplementary Material & Methods Affymetrix micro-array Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, USA). RNA concentration and quality was determined using the ND-1000 spectrophotometer
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1. Reconstitution of human-acquired lymphoid system in
 Supplementary Figure 1. Reconstitution of human-acquired lymphoid system in mouse NOD/SCID/Jak3 null mice were transplanted with human CD34 + hematopoietic stem cells. (Top) Four weeks after the transplantation
Supplementary Figure 1. Reconstitution of human-acquired lymphoid system in mouse NOD/SCID/Jak3 null mice were transplanted with human CD34 + hematopoietic stem cells. (Top) Four weeks after the transplantation
SUPPLEMENTARY INFORMATION
 A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION
 doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
doi:.38/nature899 Supplementary Figure Suzuki et al. a c p7 -/- / WT ratio (+)/(-) p7 -/- / WT ratio Log X 3. Fold change by treatment ( (+)/(-)) Log X.5 3-3. -. b Fold change by treatment ( (+)/(-)) 8
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Nature Biotechnology: doi: /nbt.4086
 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Supplementary Figure 1. Isolation of GFPHigh cells.
 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding
Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan).
 1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
1 2 3 4 5 6 7 8 Supplemental Materials and Methods Cell proliferation assay Cell proliferation was measured with Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). GCs were plated at 96-well
Supplementary Data. Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter
 Supplementary Data Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter We first introduced an expression unit composed of a tetresponse
Supplementary Data Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter We first introduced an expression unit composed of a tetresponse
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Reintroduction of HDAC1 in HDAC1-/- ES cells reverts the phenotype of HDAC1-/- teratomas. 3x10 6 HDAC1 reintroduced (HDAC1-/-re) and empty vector infected
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 CSRP2 PFKP ADFP ADM C10orf10 GPI LOX PLEKHA2 WIPF1
 Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplemental Table 1 Gene Symbol FDR corrected p-value PLOD1 4.52E-18 PDK1 6.77E-18 CSRP2 4.42E-17 PFKP 1.23E-14 MSH2 3.79E-13 NARF_A 5.56E-13 ADFP 5.56E-13 FAM13A1 1.56E-12 FAM29A_A 1.22E-11 CA9 1.54E-11
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Supplementary Information for: CRISPR/Cas9-based target validation. for p53-reactivating model compounds
 for: CRISPR/Cas9-based target validation for p53-reactivating model compounds Michael Wanzel 1,5, Jonas B. Vischedyk 1, Miriam P. Gittler 1, Niklas Gremke 1, Julia R. Seiz 1, Mirjam Hefter 1, Magdalena
for: CRISPR/Cas9-based target validation for p53-reactivating model compounds Michael Wanzel 1,5, Jonas B. Vischedyk 1, Miriam P. Gittler 1, Niklas Gremke 1, Julia R. Seiz 1, Mirjam Hefter 1, Magdalena
Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression
 Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression LVP336 LVP336-PBS LVP339 LVP339-PBS LVP297 LVP297-PBS LVP013 LVP013-PBS LVP338 LVP338-PBS LVP027 LVP027-PBS LVP337 LVP337-PBS
Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression LVP336 LVP336-PBS LVP339 LVP339-PBS LVP297 LVP297-PBS LVP013 LVP013-PBS LVP338 LVP338-PBS LVP027 LVP027-PBS LVP337 LVP337-PBS
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1
 Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
Hyper sensitive protein detection by Tandem-HTRF reveals Cyclin D1 dynamics in adult mouse Alexandre Zampieri, Julien Champagne, Baptiste Auzemery, Ivanna Fuentes, Benjamin Maurel and Frédéric Bienvenu
SUPPLEMENTARY INFORMATION
 Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Figure S1 Colony-forming efficiencies using transposition of inducible mouse factors. (a) Morphologically distinct growth foci appeared from rtta-mefs 6-8 days following PB-TET-mFx cocktail
Supplementary Information
 Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
Supplementary Information Supplementary Figures Supplementary Figure 1. MLK1-4 phosphorylate MEK in the presence of RAF inhibitors. (a) H157 cells were transiently transfected with Flag- or HA-tagged MLK1-4
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State
 Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Electromagnetic Fields Mediate Efficient Cell Reprogramming Into a Pluripotent State Soonbong Baek, 1 Xiaoyuan Quan, 1 Soochan Kim, 2 Christopher Lengner, 3 Jung- Keug Park, 4 and Jongpil Kim 1* 1 Lab
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Information Supplementary Figure 1
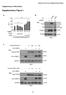 Bararia, Kwok et al, Supplementary Page 1 Supplementary Information Supplementary Figure 1 S1 Bararia, Kwok et al, Supplementary Page 2 Supplementary Figure 1 (cont.) S2 Bararia, Kwok et al, Supplementary
Bararia, Kwok et al, Supplementary Page 1 Supplementary Information Supplementary Figure 1 S1 Bararia, Kwok et al, Supplementary Page 2 Supplementary Figure 1 (cont.) S2 Bararia, Kwok et al, Supplementary
* * IgL locus (V - J ) λ λ
 Supplementary Figures and Figure Legend Supplementary Figure S1 A C F Vav-P-BCL2 doner mice 8 6 4 2 Lymphoma free (%)1 Tumor Pathology Score Retroviral transduction Vav-P-BCL2 HPCs HPC + shrna VavP-Bcl2
Supplementary Figures and Figure Legend Supplementary Figure S1 A C F Vav-P-BCL2 doner mice 8 6 4 2 Lymphoma free (%)1 Tumor Pathology Score Retroviral transduction Vav-P-BCL2 HPCs HPC + shrna VavP-Bcl2
Supplemental data and methods Molecular transformation from committed progenitor to leukemia stem cell initiated by MLL-AF9
 Supplemental data and methods Molecular transformation from committed progenitor to leukemia stem cell initiated by MLL-AF9 Andrei V. Krivtsov, David Twomey, Zhaohui Feng, Matthew C. Stubbs, Yingzi Wang,
Supplemental data and methods Molecular transformation from committed progenitor to leukemia stem cell initiated by MLL-AF9 Andrei V. Krivtsov, David Twomey, Zhaohui Feng, Matthew C. Stubbs, Yingzi Wang,
Supplementary Figure 1. Mouse genetic models used for identification of Axin2-
 Supplementary Figure 1. Mouse genetic models used for identification of Axin2- expressing cells and their derivatives. Schematic representations illustrate genetic cell labeling strategies to identify
Supplementary Figure 1. Mouse genetic models used for identification of Axin2- expressing cells and their derivatives. Schematic representations illustrate genetic cell labeling strategies to identify
A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo.
 Gus Frangou, Stephanie Palmer & Mark Groudine Fred Hutchinson Cancer Research Center, Seattle- USA A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo. During mammalian development and differentiation
Gus Frangou, Stephanie Palmer & Mark Groudine Fred Hutchinson Cancer Research Center, Seattle- USA A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo. During mammalian development and differentiation
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna
 Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Supplementary Figure 1: sgrna library generation and the length of sgrnas for the functional screen. (a) A diagram of the retroviral vector for sgrna expression. It contains a U6-promoter-driven sgrna
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Premade Lentiviral Particles for human ips Stem Factors
 Premade Lentiviral Particles for human ips Stem Factors For generating induced pluripotent stem (ips) cells or other applications RESEARCH USE ONLY, not for diagnostics or therapeutics Cat# Product Name
Premade Lentiviral Particles for human ips Stem Factors For generating induced pluripotent stem (ips) cells or other applications RESEARCH USE ONLY, not for diagnostics or therapeutics Cat# Product Name
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM
 Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Pre-made Reporter Lentivirus for p53 Signal Pathway
 Pre-made Reporter for p53 Signal Pathway Cat# Product Name Amounts LVP977-P or: LVP977-P-PBS P53-GFP (Puro) LVP978-P or: LVP978-P-PBS P53-RFP (Puro) LVP979-P or: LVP979-P-PBS P53-Luc (Puro) LVP980-P or:
Pre-made Reporter for p53 Signal Pathway Cat# Product Name Amounts LVP977-P or: LVP977-P-PBS P53-GFP (Puro) LVP978-P or: LVP978-P-PBS P53-RFP (Puro) LVP979-P or: LVP979-P-PBS P53-Luc (Puro) LVP980-P or:
Supplementary Figure 1
 Supplementary Figure 1 CITE-seq library preparation. (a) Illustration of the DNA-barcoded antibodies used in CITE-seq. (b) Antibody-oligonucleotide complexes appear as a high-molecular-weight smear when
Supplementary Figure 1 CITE-seq library preparation. (a) Illustration of the DNA-barcoded antibodies used in CITE-seq. (b) Antibody-oligonucleotide complexes appear as a high-molecular-weight smear when
Partial list of differentially expressed genes from cdna microarray, comparing MUC18-
 Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Supplemental Figure legends Table-1 Partial list of differentially expressed genes from cdna microarray, comparing MUC18- silenced and NT-transduced A375SM cells. Supplemental Figure 1 Effect of MUC-18
Protocol Using a Dox-Inducible Polycistronic m4f2a Lentivirus to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the transduction and reprogramming of one well of Oct4-GFP mouse embryonic fibroblasts (MEF) using the Dox Inducible Reprogramming Polycistronic
STEMGENT Page 1 OVERVIEW The following protocol describes the transduction and reprogramming of one well of Oct4-GFP mouse embryonic fibroblasts (MEF) using the Dox Inducible Reprogramming Polycistronic
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot
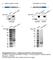 Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1
 number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
number of cells, normalized number of cells, normalized number of cells, normalized Supplementary Figure CD CD53 Cd3e fluorescence intensity fluorescence intensity fluorescence intensity Supplementary
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1.
 Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
Supplementary Figure 1. Characterization and high expression of Lnc-β-Catm in liver CSCs. (a) Heatmap of differently expressed lncrnas in Liver CSCs (CD13 + CD133 + ) and non-cscs (CD13 - CD133 - ) according
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
MLN8237 induces proliferation arrest, expression of differentiation markers and
 Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides
 Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Figure 1. Confirmation of sirna in PC3 and H1299 cells PC3 (a) and H1299 (b) cells were transfected with sirna oligonucleotides targeting RCP (SMARTPool (RCP) or two individual oligos (RCP#1
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Protocol Using the Reprogramming Ecotropic Retrovirus Set: Mouse OSKM to Reprogram MEFs into ips Cells
 STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
STEMGENT Page 1 OVERVIEW The following protocol describes the reprogramming of one well of mouse embryonic fibroblast (MEF) cells into induced pluripotent stem (ips) cells in a 6-well format. Transduction
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo
 Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Gaussia Luciferase-a Novel Bioluminescent Reporter for Tracking Stem Cells Survival, Proliferation and Differentiation in Vivo Rampyari Raja Walia and Bakhos A. Tannous 1 2 1 Pluristem Innovations, 1453
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/8/347/347ra94/d Supplementary Materials for n mirn-mediated therapy for S6 blocks IRES-driven translation of the N second cistron Yu Miyazaki, Xiaofei
www.sciencetranslationalmedicine.org/cgi/content/full/8/347/347ra94/d Supplementary Materials for n mirn-mediated therapy for S6 blocks IRES-driven translation of the N second cistron Yu Miyazaki, Xiaofei
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix
 Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix Authors Kerim B. Kaylan, 1,2 Viktoriya Ermilova, 1,2 Ravi Chandra Yada,
Pre-made Lentiviral Particles for nuclear permeant CRE recombinase expression
 Pre-made Lentiviral Particles for nuclear permeant CRE recombinase expression Cat# Product Name Amounts* LVP336 NLS-CRE (Bsd) LVP336-PBS NLS-CRE (Bsd), in vivo ready LVP339 NLS-CRE (Puro) LVP339-PBS NLS-CRE
Pre-made Lentiviral Particles for nuclear permeant CRE recombinase expression Cat# Product Name Amounts* LVP336 NLS-CRE (Bsd) LVP336-PBS NLS-CRE (Bsd), in vivo ready LVP339 NLS-CRE (Puro) LVP339-PBS NLS-CRE
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
a Award Number: W81XWH TITLE:
 AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
AD Award Number: W81XWH-04-1-0138 TITLE: Imaging Metastatic Prostate Cancer After Genetic Manipulation of Transcriptional Memory Regulators EZH2 and EED PRINCIPAL INVESTIGATOR: Lily Wu, M.D., Ph.D. CONTRACTING
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles
 Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Rejuvenation of the muscle stem cell population restores strength to injured aged muscles Benjamin D Cosgrove, Penney M Gilbert, Ermelinda Porpiglia, Foteini Mourkioti, Steven P Lee, Stephane Y Corbel,
Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity
 Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity Sebastian Virreira Winter, Arturo Zychlinsky and Bart W. Bardoel Department of Cellular
Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity Sebastian Virreira Winter, Arturo Zychlinsky and Bart W. Bardoel Department of Cellular
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Supplementary Information. Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications
 Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation
 LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
Premade Lentiviral Particles for ips Stem Factors: Single vector system. For generation of induced pluripotent stem (ips) cells and other applications
 Premade Lentiviral Particles for ips Stem Factors: Single vector system For generation of induced pluripotent stem (ips) cells and other applications RESEARCH USE ONLY, not for diagnostics or therapeutics
Premade Lentiviral Particles for ips Stem Factors: Single vector system For generation of induced pluripotent stem (ips) cells and other applications RESEARCH USE ONLY, not for diagnostics or therapeutics
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Table S1
 Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
Primers used in RT-qPCR, ChIP and Bisulphite-Sequencing. Quantitative real-time RT-PCR primers Supplementary Table S1 gene Forward primer sequence Reverse primer sequence Product TRAIL CAACTCCGTCAGCTCGTTAGAAAG
TITLE: Identification of Prostate Cancer Metastasis-Suppressor Genes Using Genomic shrna Libraries
 AD Award Number: W81XWH-07-1-0184 TITLE: Identification of Prostate Cancer Metastasis-Suppressor Genes Using Genomic shrna Libraries PRINCIPAL INVESTIGATOR: Irwin H. Gelman, Ph.D. CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-07-1-0184 TITLE: Identification of Prostate Cancer Metastasis-Suppressor Genes Using Genomic shrna Libraries PRINCIPAL INVESTIGATOR: Irwin H. Gelman, Ph.D. CONTRACTING ORGANIZATION:
Nature Biotechnology doi: /nbt Supplementary Figure 1. Substrate-linked protein evolution components.
 Supplementary Figure 1 Substrate-linked protein evolution components. (a) loxbtr subsites used for the combinational directed-evolution process to generate Brec1. Sequence differences (compared to the
Supplementary Figure 1 Substrate-linked protein evolution components. (a) loxbtr subsites used for the combinational directed-evolution process to generate Brec1. Sequence differences (compared to the
Antibody used for FC Figure S1. Multimodal characterization of NIR dyes in vitro Figure S2. Ex vivo analysis of HL60 cells homing
 Antibody used for FC The following antibodies were used following manufacturer s instructions: anti-human CD4 (clone HI3, IgG1, k - Becton Dickinson), anti-human CD33 (clone WM3, IgG1, k- Becton Dickinson),
Antibody used for FC The following antibodies were used following manufacturer s instructions: anti-human CD4 (clone HI3, IgG1, k - Becton Dickinson), anti-human CD33 (clone WM3, IgG1, k- Becton Dickinson),
SUPPLEMENTARY NOTE 2. Supplememtary Note 2, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 2 A recombinase reporter system for permanent reporter activation We made use of the Cre-loxP recombinase system for a maximal amplification and complete kinetic uncoupling within single
SUPPLEMENTARY NOTE 2 A recombinase reporter system for permanent reporter activation We made use of the Cre-loxP recombinase system for a maximal amplification and complete kinetic uncoupling within single
Quality Control Assays
 QUALITY CONTROL An integral part of the Penn Vector Core is its robust quality control program which is carried out by a separate quality control group. Quality control assays have been developed and optimized
QUALITY CONTROL An integral part of the Penn Vector Core is its robust quality control program which is carried out by a separate quality control group. Quality control assays have been developed and optimized
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
SUPPLEMENTARY INFORMATION
 Supplementary Figure a 3 factors 4 factors Nanog-GFP phase b c.5 Endogenous Oct3/4 2 Total Oct3/4.5 Relative expression (ES = ).5.5 6 3 Endogenous Sox2 Nanog Relative expression (ES = ) 5 25 2 Total Sox2
Supplementary Figure a 3 factors 4 factors Nanog-GFP phase b c.5 Endogenous Oct3/4 2 Total Oct3/4.5 Relative expression (ES = ).5.5 6 3 Endogenous Sox2 Nanog Relative expression (ES = ) 5 25 2 Total Sox2
Medicinal Chemistry of Modern Antibiotics
 Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2012 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2012 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Medicinal Chemistry of Modern Antibiotics
 Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2008 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2008 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
Adenoviral Expression Systems. Lentivirus is not the only choice for gene delivery. Adeno-X
 Adenoviral Expression Systems Lentivirus is not the only choice for gene delivery 3 Adeno-X Why choose adenoviral gene delivery? Table I: Adenoviral vs. Lentiviral Gene Delivery Lentivirus Adenovirus Infects
Adenoviral Expression Systems Lentivirus is not the only choice for gene delivery 3 Adeno-X Why choose adenoviral gene delivery? Table I: Adenoviral vs. Lentiviral Gene Delivery Lentivirus Adenovirus Infects
Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95
 Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
Supplementary Information Regulation of transcription by the complex and MLL complex-associated Hao Jiang, Xiangdong Lu, Miho Shimada, Yali Dou, Zhanyun Tang, and Robert G. Roeder Input HeLa NE IP lot:
gacgacgaggagaccaccgctttg aggcacattgaaggtctcaaacatg
 Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
Supplementary information Supplementary table 1: primers for cloning and sequencing cloning for E- Ras ggg aat tcc ctt gag ctg ctg ggg aat ggc ttt gcc ggt cta gag tat aaa gga agc ttt gaa tcc Tpbp Oct3/4
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More. Ed Davis, Ph.D.
 TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
