Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD
|
|
|
- Angela McDaniel
- 6 years ago
- Views:
Transcription
1 Data collection Table S1. Crystallographic Data and Refinement Statistics, Related to Experimental Procedures hsddb1-drddb2 CPD #1 hsddb1-drddb2 CPD #2 Values in parentheses are for the highest resolution shell hsddb1-drddb2 CPD hsddb1-drddb2 CPD #3 #4 CRL4A DDB2 SeMet-CRL4A DDB2 #522 SeMet-CRL4A DDB2 #566 SeMet-CRL4A DDB2 #523 CAND1-CUL4B CRL4B DDB2 DDB1-CSA Beamline SLS X10SA SLS X10SA SLS X06DA SLS X10SA SLS X10SA SLS X06DA SLS X06DA SLS X06DA SLS X10SA SLS X06DA SLS X06DA/X10SA Wavelength (Å) Space Group P P C222 1 P C2 C2 C2 C2 P2 1 P2 1 P3 221 Cell parameters a, b, c (Å) , , , , 154, , , , , , 78.02, , 77.40, , 77.70, , 77.20, , , , , , , α, β, γ (deg) 90, 90, 90 90, 90, 90 90, 90, 90 90, 90, 90 90, 108.5, 90 90, 108, 90 90, 108.7, 90 90, 108.1, 90 90, 89.4, 90 90, 94.2, 90 90, 90, 120 Resolution (Å) ( ) ( ) ( ) ( ) ( ) ( ) ( ) ( ) ( ) ( ) ( ) Completeness (%) 99.4 (100.0) 99.8 (99.6) 99.4 (99.2) 99.7 (99.7) 98.5 (99.9) 73.2 (35.0) 81.5 (39.1) 85.1 (39.2) 99.7 (99.8) 97.5 (99.1) 92.4 (78.4) Unique reflections (3465) (3471) (1834) (2758) (823) 8396 (392) 5674 (264) 7329 (326) (4435) (1013) (2352) Redundancy 6.3 (6.3) 7.3 (7.6) 5.3 (5.4) 7.4 (7.6) 4.9 (5.3) 3.0 (2.0) 6.5 (4.8) 6.8 (3.2) 3.8 (3.8) 3.7 (3.7) 2.2 (1.9) R sym (%) 11.6 (58.7) 12.9 (57.7) 23.3 (57.2) 14.3 (62.1) 13.9 (55.3) 8.3 (36.9) 13.3 (41.9) 7.3 (41.0) 9.2 (51.1) 9.0 (62.2) 10.4 (43.1) I/σI 14.2 (4.1) 16.0 (4.5) 8.7 (3.4) 11.9 (3.2) 8.07 (2.41) 19.9 (2.0) 20 (2.5) 24.4 (2.1) 10.2 (2.8) 10.2 (2.3) 7.0 (2.0) Refinement PDB code 4A08 4A09 4A0A 4A0B 4A0K 4A0C 4A0L 4A11 R work/r free 23.4/ / / / / / / /23.3 Reflections (working set) Reflections (test set) Number of Atoms Protein DNA Water Ligand R.m.s. deviations Bond lengths (Å) * * Bond Angles (deg) * * Ramachandran favoured * 88.5 * 95.9 disallowed * 1.5 * 0 Avg. B-factor (Å 2 ) Crystallization Reservoir 100 mm MES, 25 mm NaOH, 18% PEG NaOH, 21% PEG mm MES, 28 mm NaOH, 16% PEG NaOH, 18% PEG 8.3, 33% PEG , 35% PEG , 29% PEG 200 a ; b R free is the same as R work, but calculated on the reflections set aside from refinement. * Subunits from high resolution search models (2HYE, 3EI2 and 4A0C) have been placed by molecular replacement and were rigid body refined. 8.5, 31% PEG mm MES ph6.3, 30% PEG200, 2% PEG mm MES ph 6.2, 3.1% PEG 6000, 4% ethyleneglycol M NaKPO 4, 0.1 M NaMalonate, M Li 2SO 4 DDB1/DDB2/ DDB1/DDB2/ DDB1/DDB2/ DDB1/DDB2/ CAND1/CUL4B/ DDB1/DDB2/ DDB1-E194A/DDB2 DDB1-E194A/DDDB2 DDB1-E224K/DDB2 DDB1-E224S/DDB2 DDB1/CSA Protein CUL4A/RBX1 CUL4A/RBX1 CUL4A/RBX1 CUL4A/RBX1 RBX1 CUL4B/RBX1 Concentration (mg/ml) Cryoprotectant 100 mm MES, 35 mm NaOH, 35% PEG, 20mM CaAcetate NaOH, 35% PEG 200, 20mM CaAcetate Paratone N 100 mm MES, 35 mm NaOH, 35% PEG, 20mM CaAcetate Reservoir Reservoir Reservoir Reservoir Reservoir 100 mm MES ph 6.5, 5% PEG 6000, 30% ethyleneglycol 2M NaMalonate, 0.1M Tris-HCl ph 8.0
2 Table S2. Minor/Major Groove Width for CPD#1, Related to Figure 1 Major groove Minor groove Base (D U+m; m=4) Width (Å) Base (D U+m; m=-3) Width (Å) C(D-5) T(U+1) G(D-4) C(U-1) C(D-3) C(U-2) G(D-2) C(U-3) A(D-1) G(U-4) CPD(D+1) C(U-5) CPD(D+2) G(U-6) G(D+3) G(U-7) C(D+4) G(U-8) G(D-2) G(U+5) A(D-1) C(U+4) CPD(D+1) G(U+3) CPD(D+2) C(U+2) G(D+3) T(U+1) C(D+4) C(U-1) G(D+5) C(U-2) C(D+6) C(U-3) Table S3. Phosphate-Phosphate Distance for CPD#1, Related to Figure 1 Damaged Strand PP-distance (Å) PP-distance (Å) Undamaged strand D-5 D U+5 U+4 D-4 D U+4 U+3 D-3 D D-2 D-1 U+3 U+2 D-1 D U+2 U+1 D+1 D U+1 U-1 D+2 D K U-1 U-2 D+3 D U-2 U-3 D+4 D K U-3 U-4 D+5 D U-4 U U-5 U-6 U-6 U-7 U-7 U-8
3 Table S4. Oligonucleotides Used for Crystallization, Related to Experimental Procedures DDB1/DDB2-CPD #1 damaged strand 5'-ACGCGA(CPD)GCGCCC-3 5'-TGGGCGCCCTCGCG-3' DDB1/DDB2-CPD #2 damaged strand 5'-GGTGAAA(CPD)AGCAGG -3' 5'-CCTGCTCCTTTCACCC-3' DDB1/DDB2-CPD #3 damaged strand 5'-GGGTGAAT(CPD)AGCAGG-3' DDB1/DDB2-CPD #4 damaged strand 5'- GGGTGAAT(CPD)AGCAGG -3 CRL4A DDB2 -THF damaged strand: 5'-GCTACT(THF)ACGCA-3 : 5'-TGCGTAAGTAGC-3' CRL4A DDB2 (1x C3) linked strand 5 -TGCGTAAGTAGCT(C3)CGATCT(THF)ACGGAA-3' complementary 1 complementary 2 5 -TCCGTAAGATCG-3' 5 -GCTACT(THF)ACGCAA-3' CRL4A DDB2 (2x C3) linked strand 5 -TGCGTAAGTAGCT(C3)(C3)CGATCT(THF)ACGGAA-3' complementary 1 complementary 2 5 -TCCGTAAGATCG-3' 5 -GCTACT(THF)ACGCAA-3'
4 Table S5. Oligonucleotides Used for EMSA and Biochemical Assays, Related to Figure 5 15bp DNA THF -Cy5 damaged strand 5'-AAATGAAT(THF)AAGCAGG-3 5'-(Cy5)CCTGCTTTATTCATTT-3' 15bp DNA THF damaged strand 5'-AAATGAAT(THF)AAGCAGG-3 5'-CCTGCTTTATTCATTT-3' 15bp DNA 5'-AAATGAATAAAGCAGG-3 5'-CCTGCTTTATTCATTT-3' 24bp DNA THF damaged strand 5'-GTCCTGAATGAAT(THF)ACGCAA -3' 5'-TTGCGTAATTCATTCAGGAC -3' 21bp DNA 6-4PP damaged strand 5'-TTTCCTAGACT(6-4PP)GCCCAATTA -3 5'-ATAATTGGGCAAAGTCTAGGAA -3' 16bp DNA CPD damaged strand 5'- GGGTGAAT(CPD)AGCAGG -3
5 Table S6. Reported CSA Mutations in Cockayne Syndrome Patients, Related to Figure 6 Mutations Patient CS class Location Expected Effect Q106P CS2SE CS I Blade 1 hydrophobic core, pointing into the central cavity of the WD40 propeller A160T CS852VI CS III Blade 3 hydrophobic core of the blade Minor effect on the fold affecting DDB1 binding and possibly A160V CS3BE CS I Blade 3 hydrophobic core of the blade Minor effect on the fold affecting DDB1 binding and possibly W194C CS655VI CS I Blade 4 hydrophobic cavity that appears no longer filled by the W > S mutations L202S 08STR3 CS I Blade 4 hydrophobic cavity, in which a more polar residues is positioned T204K 4_1, 4_2 CS I Blade 4 hydrogen bonds with Trp214, His185 and Lys212. The T > K mutation would be expected to result in significant steric clashes A205P AGO7075 CS I Blade 4 hydrophobic core of the propeller. The A > P mutations likely interferes with the β-sheet conformation, due to disallowed Ramachandran space integrity of the fold: impaired DDB1 &. Lack of DDB1 binding has been demonstrated D266G CS861VI, CS I Blade 5 tight turn (Asp is the canonical Likely interfering with on 08STR2, WD40 residues in this position). The the narrow side of the WD40 propeller 08STR2_2, change to Gly is conservative, however, and 3_1, 3_2 expected to have mostly local effects Deletion A207-S209 CS794VI CS I Blade: truncates the loop between strands B and C. Effect is expected to be local Likely interfering with on the narrow side of the WD40 propeller W361C UV S SVI UV S S Blade 7: Cys mutations results in the hydrophobic cavity not being properly filled. Conservative mutation, however, expected to cause only limited damage Mildly interfering with the integrity of the WD40 fold: impaired DDB1 & substrate binding Mutations of CSA were identified in 41 CS patients: 8 different point mutations, 8 frameshift mutations, 4 deletions and 3 stop-codon insertions have been described (Laugel et al., 2010). CS has been classified into three different sub-groups according to the severity and onset of the disease: the most severe form (class 1) has an onset in utero (COFS) or during the first years of life; (class 2) manifests itself in the early teens, while class 3 becomes apparent after the second or third decade of life. The least severe phenotype is observed in UV sensitivity syndrome (UV S S), which is also associated with mutations in CSA (Laugel et al., 2010) and comes with no obvious developmental phenotype, unlike classes I, II and III. On the basis of the structure, there are three conceivable ways to impair CSA function: (i) point mutations in the structured HLH-WD40 domain abolishing DDB1 binding, which has been directly observed in the A205P mutation (Jin et al., 2006); (ii) mutations preventing on the narrow face of the WD40 propeller;
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
SUPPLEMENTARY INFORMATION
 Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport
 Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Dr. R. Sankar, BSE 631 (2018)
 Pauling, Corey and Branson Diffraction of DNA http://www.nature.com/scitable/topicpage/dna-is-a-structure-that-encodes-biological-6493050 In short, stereochemistry is important in determining which helices
Pauling, Corey and Branson Diffraction of DNA http://www.nature.com/scitable/topicpage/dna-is-a-structure-that-encodes-biological-6493050 In short, stereochemistry is important in determining which helices
Supporting Online Material. Av1 and Av2 were isolated and purified under anaerobic conditions according to
 Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
SUPPLEMENTARY INFORMATION Figures. Supplementary Figure 1 a. Page 1 of 30. Nature Chemical Biology: doi: /nchembio.2528
 SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
Supplementary Information For. A genetically encoded tool for manipulation of NADP + /NADPH in living cells
 Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
SUPPLEMENTARY INFORMATION
 Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
BETA STRAND Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna
 Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna E-mail: a.hochkoeppler@unibo.it C-ter NH and CO groups: right, left, right (plane of the slide)
Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna E-mail: a.hochkoeppler@unibo.it C-ter NH and CO groups: right, left, right (plane of the slide)
Acceleration of protein folding by four orders of magnitude through a single amino acid substitution
 Acceleration of protein folding by four orders of magnitude through a single amino acid substitution Daniel J. A. Roderer 1, Martin A. Schärer 1, Marina Rubini 2 * and Rudi Glockshuber 1 AUTHOR ADDRESS
Acceleration of protein folding by four orders of magnitude through a single amino acid substitution Daniel J. A. Roderer 1, Martin A. Schärer 1, Marina Rubini 2 * and Rudi Glockshuber 1 AUTHOR ADDRESS
Supplemental Material Liu et al.
 Supplemental Material Liu et al. Supplemental Methods Protein expression, purification Recombinant etud11 (a.a 2344-2515) and Tud7-11 (a.a. 1617-2515) proteins (Tud7-11) were expressed in the Rosetta strain
Supplemental Material Liu et al. Supplemental Methods Protein expression, purification Recombinant etud11 (a.a 2344-2515) and Tud7-11 (a.a. 1617-2515) proteins (Tud7-11) were expressed in the Rosetta strain
1/4/18 NUCLEIC ACIDS. Nucleic Acids. Nucleic Acids. ECS129 Instructor: Patrice Koehl
 NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS. ECS129 Instructor: Patrice Koehl
 NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
NUCLEIC ACIDS ECS129 Instructor: Patrice Koehl Nucleic Acids Nucleotides DNA Structure RNA Synthesis Function Secondary structure Tertiary interactions Wobble hypothesis DNA RNA Replication Transcription
Comparative Modeling Part 1. Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center
 Comparative Modeling Part 1 Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center Function is the most important feature of a protein Function is related to structure Structure is
Comparative Modeling Part 1 Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center Function is the most important feature of a protein Function is related to structure Structure is
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
SUPPLEMENTARY INFORMATION
 This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
Nature Structural & Molecular Biology: doi: /nsmb.3428
 Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
SUPPLEMENTARY INFORMATION. Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly
 SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
Supplemental Material
 Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it Life The main actors in the chemistry of life are molecules called proteins nucleic acids Proteins: many different
Structure/function relationship in DNA-binding proteins
 PHRM 836 September 22, 2015 Structure/function relationship in DNA-binding proteins Devlin Chapter 8.8-9 u General description of transcription factors (TFs) u Sequence-specific interactions between DNA
PHRM 836 September 22, 2015 Structure/function relationship in DNA-binding proteins Devlin Chapter 8.8-9 u General description of transcription factors (TFs) u Sequence-specific interactions between DNA
Structure formation and association of biomolecules. Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München
 Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Structural bioinformatics
 Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Gene Regulation & Mutation 8.6,8.7
 Gene Regulation & Mutation 8.6,8.7 Eukaryotic Gene Regulation Transcription factors: ensure proteins are made at right time and in right amounts. One type forms complexes that guide & stabilize binding
Gene Regulation & Mutation 8.6,8.7 Eukaryotic Gene Regulation Transcription factors: ensure proteins are made at right time and in right amounts. One type forms complexes that guide & stabilize binding
Antibody-Antigen recognition. Structural Biology Weekend Seminar Annegret Kramer
 Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I
 BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
Gene and DNA structure. Dr Saeb Aliwaini
 Gene and DNA structure Dr Saeb Aliwaini 2016 DNA during cell cycle Cell cycle for different cell types Molecular Biology - "Study of the synthesis, structure, and function of macromolecules (DNA, RNA,
Gene and DNA structure Dr Saeb Aliwaini 2016 DNA during cell cycle Cell cycle for different cell types Molecular Biology - "Study of the synthesis, structure, and function of macromolecules (DNA, RNA,
Combine Cross-Diffusion Microbatch Method with Seeding Technique to Enhance Protein Crystallization Based on a Common Dispersing Agent
 Electronic Supplementary Material (ESI) for CrystEngComm. This journal is The Royal Society of Chemistry 2017 Supplementary Material Combine Cross-Diffusion Microbatch Method with Seeding Technique to
Electronic Supplementary Material (ESI) for CrystEngComm. This journal is The Royal Society of Chemistry 2017 Supplementary Material Combine Cross-Diffusion Microbatch Method with Seeding Technique to
SUPPLEMENTARY INFORMATION
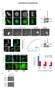 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
Τάσος Οικονόµου ιαλεξη 8. Kινηση, λειτουργια, ελεγχος.
 Τάσος Οικονόµου ιαλεξη 8 Kινηση, λειτουργια, ελεγχος http://ecoserver.imbb.forth.gr/bio321.htm εν ξεχνω. Cell The peptide bond Polypeptides are stabilized by: 1. Covalent bonds= amide bond 2. Noncovalent,
Τάσος Οικονόµου ιαλεξη 8 Kινηση, λειτουργια, ελεγχος http://ecoserver.imbb.forth.gr/bio321.htm εν ξεχνω. Cell The peptide bond Polypeptides are stabilized by: 1. Covalent bonds= amide bond 2. Noncovalent,
Cell, Volume 135 Supplemental Data Structural Basis of UV DNA-Damage Recognition by the DDB1-DDB2 Complex
 Cell, Volume 135 Supplemental Data Structural Basis of UV DNA-Damage Recognition by the DDB1-DDB2 Complex Andrea Scrima, Renata Koníčková, Bryan K. Czyzewski, Yusuke Kawasaki, Philip D. Jeffrey, Regina
Cell, Volume 135 Supplemental Data Structural Basis of UV DNA-Damage Recognition by the DDB1-DDB2 Complex Andrea Scrima, Renata Koníčková, Bryan K. Czyzewski, Yusuke Kawasaki, Philip D. Jeffrey, Regina
Nucleic acids. How DNA works. DNA RNA Protein. DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology
 Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
Nucleic acid chemistry and basic molecular theory Nucleic acids DNA (deoxyribonucleic acid) RNA (ribonucleic acid) Central Dogma of Molecular Biology Cell cycle DNA RNA Protein Transcription Translation
produces an RNA copy of the coding region of a gene
 1. Transcription Gene Expression The expression of a gene into a protein occurs by: 1) Transcription of a gene into RNA produces an RNA copy of the coding region of a gene the RNA transcript may be the
1. Transcription Gene Expression The expression of a gene into a protein occurs by: 1) Transcription of a gene into RNA produces an RNA copy of the coding region of a gene the RNA transcript may be the
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Chapter 8. Microbial Genetics. Lectures prepared by Christine L. Case. Copyright 2010 Pearson Education, Inc.
 Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Biology Day 67. Tuesday, February 24 Wednesday, February 25, 2015
 Biology Day 67 Tuesday, February 24 Wednesday, February 25, 2015 Do#Now:' Brainstorm+8.5 + 1. Write today s FLT! 2. Identify 3 specific differences between DNA and RNA.! 3. is the process of making RNA
Biology Day 67 Tuesday, February 24 Wednesday, February 25, 2015 Do#Now:' Brainstorm+8.5 + 1. Write today s FLT! 2. Identify 3 specific differences between DNA and RNA.! 3. is the process of making RNA
If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial:
 Name (s): Swiss PDB viewer assignment chapter 4. If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial: http://spdbv.vital-it.ch/themolecularlevel/spvtut/index.html
Name (s): Swiss PDB viewer assignment chapter 4. If you wish to have extra practice with swiss pdb viewer or to familiarize yourself with how to use the program here is a tutorial: http://spdbv.vital-it.ch/themolecularlevel/spvtut/index.html
A Brief Introduction to Structural Biology and Protein Crystallography
 A Brief Introduction to Structural Biology and Protein Crystallography structural biology of H2O http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/water-structure.jpg Protein polymers fold up into
A Brief Introduction to Structural Biology and Protein Crystallography structural biology of H2O http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/water-structure.jpg Protein polymers fold up into
Protein Structure. Protein Structure Tertiary & Quaternary
 Lecture 4 Protein Structure Protein Structure Tertiary & Quaternary Dr. Sameh Sarray Hlaoui Primary structure: The linear sequence of amino acids held together by peptide bonds. Secondary structure: The
Lecture 4 Protein Structure Protein Structure Tertiary & Quaternary Dr. Sameh Sarray Hlaoui Primary structure: The linear sequence of amino acids held together by peptide bonds. Secondary structure: The
Programme Good morning and summary of last week Levels of Protein Structure - I Levels of Protein Structure - II
 Programme 8.00-8.10 Good morning and summary of last week 8.10-8.30 Levels of Protein Structure - I 8.30-9.00 Levels of Protein Structure - II 9.00-9.15 Break 9.15-11.15 Exercise: Building a protein model
Programme 8.00-8.10 Good morning and summary of last week 8.10-8.30 Levels of Protein Structure - I 8.30-9.00 Levels of Protein Structure - II 9.00-9.15 Break 9.15-11.15 Exercise: Building a protein model
Specificity: Induced Fit
 Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
The Stringent Response
 The Stringent Response When amino acids are limiting a response is triggered to shut down a wide range of biosynthetic processes This process is called the Stringent Response It results in the synthesis
The Stringent Response When amino acids are limiting a response is triggered to shut down a wide range of biosynthetic processes This process is called the Stringent Response It results in the synthesis
Fundamentals of Biochemistry
 Donald Voet Judith G. Voet Charlotte W. Pratt Fundamentals of Biochemistry Second Edition Chapter 6: Proteins: Three-Dimensional Structure Copyright 2006 by John Wiley & Sons, Inc. 1958, John Kendrew Any
Donald Voet Judith G. Voet Charlotte W. Pratt Fundamentals of Biochemistry Second Edition Chapter 6: Proteins: Three-Dimensional Structure Copyright 2006 by John Wiley & Sons, Inc. 1958, John Kendrew Any
Bi 8 Lecture 7. Ellen Rothenberg 26 January Reading: Ch. 3, pp ; panel 3-1
 Bi 8 Lecture 7 PROTEIN STRUCTURE, Functional analysis, and evolution Ellen Rothenberg 26 January 2016 Reading: Ch. 3, pp. 109-134; panel 3-1 (end with free amine) aromatic, hydrophobic small, hydrophilic
Bi 8 Lecture 7 PROTEIN STRUCTURE, Functional analysis, and evolution Ellen Rothenberg 26 January 2016 Reading: Ch. 3, pp. 109-134; panel 3-1 (end with free amine) aromatic, hydrophobic small, hydrophilic
Basic concepts of molecular biology
 Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
Basic concepts of molecular biology Gabriella Trucco Email: gabriella.trucco@unimi.it What is life made of? 1665: Robert Hooke discovered that organisms are composed of individual compartments called cells
MCB 110:Biochemistry of the Central Dogma of MB. MCB 110:Biochemistry of the Central Dogma of MB
 MCB 110:Biochemistry of the Central Dogma of MB Part 1. DNA replication, repair and genomics (Prof. Alber) Part 2. RNA & protein synthesis. Prof. Zhou Part 3. Membranes, protein secretion, trafficking
MCB 110:Biochemistry of the Central Dogma of MB Part 1. DNA replication, repair and genomics (Prof. Alber) Part 2. RNA & protein synthesis. Prof. Zhou Part 3. Membranes, protein secretion, trafficking
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
has only one nucleotide, U20, between G19 and A21, while trna Glu CUC has two
 SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
Structure of the C-cadherin ectodomain and implications for the mechanism of cell adhesion
 Structure of the C-cadherin ectodomain and implications for the mechanism of cell adhesion Titus J. Boggon, John Murray, Sophie Chappuis-Flament, Ellen Wong, Barry M. Gumbiner, and Lawrence Shapiro Ref.
Structure of the C-cadherin ectodomain and implications for the mechanism of cell adhesion Titus J. Boggon, John Murray, Sophie Chappuis-Flament, Ellen Wong, Barry M. Gumbiner, and Lawrence Shapiro Ref.
Multiple choice questions (numbers in brackets indicate the number of correct answers)
 1 February 15, 2013 Multiple choice questions (numbers in brackets indicate the number of correct answers) 1. Which of the following statements are not true Transcriptomes consist of mrnas Proteomes consist
1 February 15, 2013 Multiple choice questions (numbers in brackets indicate the number of correct answers) 1. Which of the following statements are not true Transcriptomes consist of mrnas Proteomes consist
Supporting Information
 Supporting Information Xiao et al. 10.1073/pnas.1309211110 SI Methods Protein Expression and Purification. Bacmid was generated and recombinant baculovirus amplified using standard procedures (Bac-to-Bac;
Supporting Information Xiao et al. 10.1073/pnas.1309211110 SI Methods Protein Expression and Purification. Bacmid was generated and recombinant baculovirus amplified using standard procedures (Bac-to-Bac;
doi: /nature09408 Figure S1
 doi:10.1038/nature09408 A Figure S1 www.nature.com/nature 1 RESEARCH SUPPLEMENTARY INFORMATION Figure S1 Primary sequence, and structure of NorM VC. A. Amino acid sequence alignment of NorM VC with selected
doi:10.1038/nature09408 A Figure S1 www.nature.com/nature 1 RESEARCH SUPPLEMENTARY INFORMATION Figure S1 Primary sequence, and structure of NorM VC. A. Amino acid sequence alignment of NorM VC with selected
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
1.1 Chemical structure and conformational flexibility of single-stranded DNA
 1 DNA structures 1.1 Chemical structure and conformational flexibility of single-stranded DNA Single-stranded DNA (ssdna) is the building base for the double helix and other DNA structures. All these structures
1 DNA structures 1.1 Chemical structure and conformational flexibility of single-stranded DNA Single-stranded DNA (ssdna) is the building base for the double helix and other DNA structures. All these structures
Homology Modelling. Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen
 Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
7.014 Solution Set 4
 7.014 Solution Set 4 Question 1 Shown below is a fragment of the sequence of a hypothetical bacterial gene. This gene encodes production of HWDWN, protein essential for metabolizing sugar yummose. The
7.014 Solution Set 4 Question 1 Shown below is a fragment of the sequence of a hypothetical bacterial gene. This gene encodes production of HWDWN, protein essential for metabolizing sugar yummose. The
Proteins Higher Order Structures
 Proteins Higher Order Structures Dr. Mohammad Alsenaidy Department of Pharmaceutics College of Pharmacy King Saud University Office: AA 101 msenaidy@ksu.edu.sa Previously on PHT 426!! Protein Structures
Proteins Higher Order Structures Dr. Mohammad Alsenaidy Department of Pharmaceutics College of Pharmacy King Saud University Office: AA 101 msenaidy@ksu.edu.sa Previously on PHT 426!! Protein Structures
KEY CONCEPT DNA was identified as the genetic material through a series of experiments. Found live S with R bacteria and injected
 Section 1: Identifying DNA as the Genetic Material KEY CONCEPT DNA was identified as the genetic material through a series of experiments. VOCABULARY bacteriophage MAIN IDEA: Griffith finds a transforming
Section 1: Identifying DNA as the Genetic Material KEY CONCEPT DNA was identified as the genetic material through a series of experiments. VOCABULARY bacteriophage MAIN IDEA: Griffith finds a transforming
EMSA Mutant SOX10 proteins interfere with the DNA binding of wild type SOX10
 18-05-2010 SOX10 IL FENOTIPO MENO GRAVE È DOVUTO A DOWNREGULATION DA NMD Intron Minigene approach: * * Mutazione introdotta come controllo Northern blotting * mrna levels Inoue et al., Nature Genetics
18-05-2010 SOX10 IL FENOTIPO MENO GRAVE È DOVUTO A DOWNREGULATION DA NMD Intron Minigene approach: * * Mutazione introdotta come controllo Northern blotting * mrna levels Inoue et al., Nature Genetics
Protein Folding Problem I400: Introduction to Bioinformatics
 Protein Folding Problem I400: Introduction to Bioinformatics November 29, 2004 Protein biomolecule, macromolecule more than 50% of the dry weight of cells is proteins polymer of amino acids connected into
Protein Folding Problem I400: Introduction to Bioinformatics November 29, 2004 Protein biomolecule, macromolecule more than 50% of the dry weight of cells is proteins polymer of amino acids connected into
Bi Lecture 3 Loss-of-function (Ch. 4A) Monday, April 8, 13
 Bi190-2013 Lecture 3 Loss-of-function (Ch. 4A) Infer Gene activity from type of allele Loss-of-Function alleles are Gold Standard If organism deficient in gene A fails to accomplish process B, then gene
Bi190-2013 Lecture 3 Loss-of-function (Ch. 4A) Infer Gene activity from type of allele Loss-of-Function alleles are Gold Standard If organism deficient in gene A fails to accomplish process B, then gene
6-Foot Mini Toober Activity
 Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
Supplemental Information. Structural Basis for the Versatile. and Methylation-Dependent Binding of CTCF to DNA
 Molecular ell, Volume 66 Supplemental Information Structural Basis for the Versatile and Methylation-Dependent Binding of TF to DNA Hideharu Hashimoto, Dongxue Wang, John R. Horton, Xing Zhang, Victor.
Molecular ell, Volume 66 Supplemental Information Structural Basis for the Versatile and Methylation-Dependent Binding of TF to DNA Hideharu Hashimoto, Dongxue Wang, John R. Horton, Xing Zhang, Victor.
Chapter 8 DNA Recognition in Prokaryotes by Helix-Turn-Helix Motifs
 Chapter 8 DNA Recognition in Prokaryotes by Helix-Turn-Helix Motifs 1. Helix-turn-helix proteins 2. Zinc finger proteins 3. Leucine zipper proteins 4. Beta-scaffold factors 5. Others λ-repressor AND CRO
Chapter 8 DNA Recognition in Prokaryotes by Helix-Turn-Helix Motifs 1. Helix-turn-helix proteins 2. Zinc finger proteins 3. Leucine zipper proteins 4. Beta-scaffold factors 5. Others λ-repressor AND CRO
SUPPORTING INFORMATION.
 SUPPORTING INFORMATION. Structural and dynamic changes associated with beneficial engineered single amino acid deletion mutations in enhanced Green Fluorescent Protein James A. J. Arpino 1, Pierre J. Rizkallah
SUPPORTING INFORMATION. Structural and dynamic changes associated with beneficial engineered single amino acid deletion mutations in enhanced Green Fluorescent Protein James A. J. Arpino 1, Pierre J. Rizkallah
7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions will be posted on the web.
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
BS 50 Genetics and Genomics Week of Oct 24
 BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
BS 50 Genetics and Genomics Week of Oct 24 Additional Practice Problems for Section Question 1: The following table contains a list of statements that apply to replication, transcription, both, or neither.
Non-standard base pairs Non-standard base pairs play critical roles in the varied structures observed in DNA and RNA.
 DNA ORIENTATION Non-standard base pairs Non-standard base pairs play critical roles in the varied structures observed in DNA and RNA. Non-standard base pairs Wobble and mismatched base pairs still use
DNA ORIENTATION Non-standard base pairs Non-standard base pairs play critical roles in the varied structures observed in DNA and RNA. Non-standard base pairs Wobble and mismatched base pairs still use
Fundamentals of Protein Structure
 Outline Fundamentals of Protein Structure Yu (Julie) Chen and Thomas Funkhouser Princeton University CS597A, Fall 2005 Protein structure Primary Secondary Tertiary Quaternary Forces and factors Levels
Outline Fundamentals of Protein Structure Yu (Julie) Chen and Thomas Funkhouser Princeton University CS597A, Fall 2005 Protein structure Primary Secondary Tertiary Quaternary Forces and factors Levels
Activation of a Floral Homeotic Gene in Arabidopsis
 Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Activation of a Floral Homeotic Gene in Arabidopsis By Maximiliam A. Busch, Kirsten Bomblies, and Detlef Weigel Presentation by Lis Garrett and Andrea Stevenson http://ucsdnews.ucsd.edu/archive/graphics/images/image5.jpg
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
DNA Structures. Biochemistry 201 Molecular Biology January 5, 2000 Doug Brutlag. The Structural Conformations of DNA
 DNA Structures Biochemistry 201 Molecular Biology January 5, 2000 Doug Brutlag The Structural Conformations of DNA 1. The principle message of this lecture is that the structure of DNA is much more flexible
DNA Structures Biochemistry 201 Molecular Biology January 5, 2000 Doug Brutlag The Structural Conformations of DNA 1. The principle message of this lecture is that the structure of DNA is much more flexible
Protein Structure/Function Relationships
 Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
Protein Structure/Function Relationships W. M. Grogan, Ph.D. OBJECTIVES 1. Describe and cite examples of fibrous and globular proteins. 2. Describe typical tertiary structural motifs found in proteins.
From Gene to Protein. How Genes Work
 From Gene to Protein How Genes Work 2007-2008 The Central Dogma Flow of genetic information in a cell How do we move information from DNA to proteins? DNA RNA protein replication phenotype You! Step 1:
From Gene to Protein How Genes Work 2007-2008 The Central Dogma Flow of genetic information in a cell How do we move information from DNA to proteins? DNA RNA protein replication phenotype You! Step 1:
4/10/2011. Rosetta software package. Rosetta.. Conformational sampling and scoring of models in Rosetta.
 Rosetta.. Ph.D. Thomas M. Frimurer Novo Nordisk Foundation Center for Potein Reseach Center for Basic Metabilic Research Breif introduction to Rosetta Rosetta docking example Rosetta software package Breif
Rosetta.. Ph.D. Thomas M. Frimurer Novo Nordisk Foundation Center for Potein Reseach Center for Basic Metabilic Research Breif introduction to Rosetta Rosetta docking example Rosetta software package Breif
Protein Synthesis Notes
 Protein Synthesis Notes Protein Synthesis: Overview Transcription: synthesis of mrna under the direction of DNA. Translation: actual synthesis of a polypeptide under the direction of mrna. Transcription
Protein Synthesis Notes Protein Synthesis: Overview Transcription: synthesis of mrna under the direction of DNA. Translation: actual synthesis of a polypeptide under the direction of mrna. Transcription
Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein
 Paper Presentation PLoS ONE 2011 Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein Debanu Das, Mireille Herve, Julie Feuerhelm, etc. and Dominique
Paper Presentation PLoS ONE 2011 Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein Debanu Das, Mireille Herve, Julie Feuerhelm, etc. and Dominique
Conformational properties of enzymes
 Conformational properties of enzymes; Physics of enzyme substrate interactions; Electronic conformational interactions, cooperative properties of enzymes Mitesh Shrestha Conformational properties of enzymes
Conformational properties of enzymes; Physics of enzyme substrate interactions; Electronic conformational interactions, cooperative properties of enzymes Mitesh Shrestha Conformational properties of enzymes
Assembly and Characteristics of Nucleic Acid Double Helices
 Assembly and Characteristics of Nucleic Acid Double Helices Patterns of base-base hydrogen bonds-characteristics of the base pairs Interactions between like and unlike bases have been observed in crystal
Assembly and Characteristics of Nucleic Acid Double Helices Patterns of base-base hydrogen bonds-characteristics of the base pairs Interactions between like and unlike bases have been observed in crystal
Virtual bond representation
 Today s subjects: Virtual bond representation Coordination number Contact maps Sidechain packing: is it an instrumental way of selecting and consolidating a fold? ASA of proteins Interatomic distances
Today s subjects: Virtual bond representation Coordination number Contact maps Sidechain packing: is it an instrumental way of selecting and consolidating a fold? ASA of proteins Interatomic distances
8.1. DNA was identified as the genetic material through a series of experiments. Injected mice with R bacteria. Injected mice with S bacteria
 SECTION 8.1 IDENTIFYING DNA AS THE GENETIC MATERIAL Study Guide KEY CONCEPT DNA was identified as the genetic material through a series of experiments. VOCABULARY bacteriophage Griffith finds a transforming
SECTION 8.1 IDENTIFYING DNA AS THE GENETIC MATERIAL Study Guide KEY CONCEPT DNA was identified as the genetic material through a series of experiments. VOCABULARY bacteriophage Griffith finds a transforming
Genes and Proteins in Health. and Disease
 Genes and Health and I can describe the structure of proteins All proteins contain the chemical elements Carbon, Hydrogen, Oxygen and Nitrogen. Some also contain sulphur. Proteins are built from subunits
Genes and Health and I can describe the structure of proteins All proteins contain the chemical elements Carbon, Hydrogen, Oxygen and Nitrogen. Some also contain sulphur. Proteins are built from subunits
Station 1: DNA Structure Use the figure above to answer each of the following questions. 1.This is the subunit that DNA is composed of. 2.
 1. Station 1: DNA Structure Use the figure above to answer each of the following questions. 1.This is the subunit that DNA is composed of. 2.This subunit is composed of what 3 parts? 3.What molecules make
1. Station 1: DNA Structure Use the figure above to answer each of the following questions. 1.This is the subunit that DNA is composed of. 2.This subunit is composed of what 3 parts? 3.What molecules make
human Cdc45 Figure 1c. (c)
 1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
Molekulare Mechanismen der Signaltransduktion
 Molekulare Mechanismen der Signaltransduktion 12 Mechanism of auxin perception slides: http://tinyurl.com/modul-mms doi: 10.1038/nature05731 previous model http://www.plantcell.org/content/17/9/2425/f1.large.jpg
Molekulare Mechanismen der Signaltransduktion 12 Mechanism of auxin perception slides: http://tinyurl.com/modul-mms doi: 10.1038/nature05731 previous model http://www.plantcell.org/content/17/9/2425/f1.large.jpg
SUPPLEMENTARY INFORMATION. Design and Characterization of Bivalent BET Inhibitors
 SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
Mutations in unstructured region at N-terminus
 Mutations in unstructured region at N-terminus Nucleotide / Protein c.1a>t p.met1 Tanaka # 3 Implications on protein folding and function Start codon mutated, second ATG starts protein in position Met104.
Mutations in unstructured region at N-terminus Nucleotide / Protein c.1a>t p.met1 Tanaka # 3 Implications on protein folding and function Start codon mutated, second ATG starts protein in position Met104.
Lecture for Wednesday. Dr. Prince BIOL 1408
 Lecture for Wednesday Dr. Prince BIOL 1408 THE FLOW OF GENETIC INFORMATION FROM DNA TO RNA TO PROTEIN Copyright 2009 Pearson Education, Inc. Genes are expressed as proteins A gene is a segment of DNA that
Lecture for Wednesday Dr. Prince BIOL 1408 THE FLOW OF GENETIC INFORMATION FROM DNA TO RNA TO PROTEIN Copyright 2009 Pearson Education, Inc. Genes are expressed as proteins A gene is a segment of DNA that
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Molecular Biology (1)
 Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2017-2018 Resources This lecture Cooper, pp. 49-52, 118-119, 130 What is molecular biology? Central dogma
Molecular Biology (1) DNA structure and basic applications Mamoun Ahram, PhD Second semester, 2017-2018 Resources This lecture Cooper, pp. 49-52, 118-119, 130 What is molecular biology? Central dogma
Homology Modelling. Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen
 Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
