Supplementary Figure 1. Isolation of GFPHigh cells.
|
|
|
- Gillian Richardson
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1. Isolation of GFP High cells. (A) Schematic diagram of cell isolation based on Wnt signaling activity. Colorectal cancer (CRC) cell lines were stably transduced with lentivirus encoding ionatenin reporter (7TGP). After selection, cells were processed for fluorescenceactivated cell sorting (FACS), based on GFP expression. Parental HCT116 cells (left panel) served as negative controls for cell sorting. Scale bars = 100 μm. (B and C) Isolation of GFP High cells from CRC cells. After FACS, HCT116-7TGP cells were further cultured. GFP High (B) and GFP Low cells (C). Scale bars = 50 μm. (D) Increased β-catenin expression in GFP High cells. Immunofluorescent (IF) staining of SW620-7TGP cells. Scale bars = 20 μm. Arrows: cells highly expressing β-catenin. (E and F) Upregulation of β- catenintarget genes in GFP High cells. IF staining of SW620-7TGP cells for β- catenin (D), CD44 (E), and CD133 (F). Scale bars = 20 μm. Arrows: cells highly expressing CD44 or CD133. (G and H) Increased sphere formation of GFP High cells. GFP High and GFP Low HCT116-7TGP cells were grown in low attachment condition for 14 days. Images (G) and quantification of the number and size of spheres (H). Scale bars = 200 μm; Student s t- test; N = 3. Error bars = SEM. 1
2 Supplementary Figure 2. No difference in cell proliferation between GFP High and GFP Low cells HCT116-7TGP cells were sorted based on GFP expression using FACS. 5,000 cells were plated and counted at day 1, 3, and 5, using Biorad cell counter (TC10). Student s t-test; N = 3. Error bars = SEM. 2
3 Supplementary Figure 3. No effects of icrt14 on CRC cell proliferation. HCT116 cells were treated with icrt14 (0~75 μm) and analyzed for cell counting at day 1, 3, and 5. 5,000 cells were plated initially. Student s t-test; N = 3. Error bars = SEM. 3
4 Supplementary Figure 4. Radiosensitization of GFP High CRC cells by icrt14. SW620-GFP High and HCT116-GFP High cells were analyzed for clonogenic assays under IR (0, 2, 4, and 8 Gy). Cells were continuously treated with icrt14 (50 μm). Student s t- test; N = 3. Error bars = SEM. 4
5 Supplementary Figure 5. Validation of TCE cassette Prior to generating TERTTCE knock-in mescs and mouse model, we characterized tdtomato-creert2 (TCE) cassette. (A) Illustration of TCE fusion protein. pa: poly (A) signal. (B and C) Nuclear translocation of TCE by 4-hydroxy-tamoxigen (4OHT) treatment. HeLa cells were transiently transfected with TCE-pcDNA3.1 plasmid. 24 hours after transfection, cells were treated with 100 μm 4OHT (in ethanol) and analyzed for IF staining of TCE (tdtomato red fluorescent signal). 24 hours after 4OHT treatment, TCE was completely localized in the nucleus. At 48 hours after 4OHT treatment, the cytosolic TCE was also observed (B). However, tdtomato (a negative control) displayed subcellular localization both in the nucleus and in the cytosol, regardless of 4OHT treatment (C). 5
6 Supplementary Figure 6. Establishment of TERT TCE genetically engineered mouse model (A) Gene targeting (knock-in) strategy. We constructed targeting vector containing 5 homologous arm, tdtomato-creert2-pa, Neo cassette (for positive selection) flanked by FRT sites, 3 homologous arm, and diphtheria toxin A (DTA) (for negative selection) cassette. Of note, TCE cassette was inserted into TERT allele in-frame. Linearized targeting plasmids were electroporated into G4 mescs to generate TERT TCE-Neo mescs. mescs were injected into the blastocysts to generate chimeric mice. Mice confirmed for germ line transmission were further bred with FLPeR deletor strain to remove Neo selection cassette (TERT TCE strain). (B) Genotyping of TERT TCE strain. For genotyping, insertion of 5 and 3 homologous arms were confirmed by PCR of genomic DNA of TERT TCE-Neo mescs. Clones #1, 8, and 10 were selected for blastocyst injection. 6
7 Supplementary Figure 7. Change in cell adhesion components by radiation (A) Localization change of p120-catenin by radiation. Mice were treated with WBI (10 Gy). 24 hours later, mouse small intestine samples were collected for immunostaining for p120-catenin (Cell Signaling). Of note, in normal intestine, p120-catenin is specifically associated with E-cadherin. However, WBI-treated small intestine displays the cytosolic and the cell adhesion-associated p120-catenin. (B and C) The decrease of E-cadherin by WBI. Untreated and WBI (10 Gy)-treated mouse small intestine samples were subjected to immunostaining of E-cadherin (B). Using two different antibodies for E-cadherin detection similarly exhibited the significant decrease of E-cadherin by WBI. Consistently, immunoblotting of intestinal epithelial cells isolated from mouse small intestine (control and WBI) also showed the decreased level of E-cadherin (C). Representative images were shown. All images were captured under the same exposure time for comparison. Scale bars = 20 μm. 7
8 Supplementary Figure 8. Analysis of lentiviral reporter plasmid integration (A) Quantification of lentiviral DNA integration. 12 GFP High (H1~H12) and 11 GFP Low (L1~L11) clones were clonally picked and cultured for genomic DNA (gdna) extraction ug of gdna was used for PCR. GFP PCR amplification is similar between 12 GFP High and 11 GFP Low clones at 28 and 34 PCR cycles. AXIN2 promoter DNA was also amplified as internal controls. (B) Expression of GFP in the sorted cells. SW620-7TGP cells were sorted by GFP expression (GFP high and GFP low ). Then, clonally selected cells were grown from a single cell for 10 days, and analyzed by GFP expression (500 msec exposure). Scale bars = 20 μm 8
9 Supplementary Figure 9. Uncropped immunoblot images of Figure 5C. 9
10 Supplementary Table 1. Primer sequences for qrt-pcr and ChIP-PCR qrt-pcr from 5' to 3' PRKDC-F AGGACTGGCGTGTGAAACTT PRKDC-R ACAGCAGCATGTCATGGAAG RAD21-F TGGCTGGCTATGAAAACAGA RAD21-R CTGCTCGGAAGCTACACCTC RAD50-F TCCACGATAGGTACTTCGCC RAD50-R TGAGGACAACAGAACTTGTGAAC RAD51-F GGTCTGGTGGTCTGTGTTGA RAD51-R GGTGAAGGAAAGGCCATGTA RAD51C-F GTGTGACTCAGATGTACCAGCA RAD51C-R CTAGAGGTGAAACCCTCCGA RAD51B-F GCTCCACTCAGATGGGTTGT RAD51B-R GATGCACAACTTCAAGGCAA RAD51D-F CCACATTTGCTGCCATACAG RAD51D-R TACTGCCATCCTGTCCACTG RAD52-F CTGGCACTGTCCAAAGCATA RAD52-R TAGATCGAGCTCCCTGTGTG RAD54L-F AGAGCCCAGAGGACCTTGAT RAD54L-R TCCTTTTCGGAAACCTTTGA XRCC2-F TCTACCTTCAAGTCGGGCAA XRCC2-R TAGAGTCTGCGCAGTTGGTG XRCC3-F CGTCTTCCGTGCAGATGTAG XRCC3-R CATCACTGAGCTGGCCG XRCC4-F TTTCAGCTGAGATGTGCTCC XRCC4-R AGGAGACAGCGAATGCAAAG XRCC5-F GAAGGCTCGGATGCAGTCTA XRCC5-R CCTGCTGAAAACTTCCGTGT XRCC6-F TGGTTCATTTGTTTCCCGAT XRCC6-R AGACCAGGAAGCGAGCACT NHEJ1-F TGCAGATTCATGACAAAGGG NHEJ1-R ACTACCAGGAGAGTGGGGCT ChIP-promoter scanning from 5' to 3' LIG4ChIP-1F AGTGATGGCCATGCTTCTCT LIG4ChIP-1R GGGAGCCTGCAGTGATATTC LIG4ChIP-2F AATGGGGTCCACAAGATCAG LIG4ChIP-2R TTCCCTCATTTACCGTGAGC LIG4ChIP-3F AACCCAGGAGTCGAGGTTG LIG4ChIP-3R CATTTGCTGGTGGCAGAAT LIG4ChIP-4F GCACAGCTGAAATGGAAACA LIG4ChIP-4R GATGCTGTCTGAACCATCCA 10
11 LIG4ChIP-5F LIG4ChIP-5R LIG4ChIP-6F LIG4ChIP-6R LIG4ChIP-7F LIG4ChIP-7R LIG4ChIP-8F LIG4ChIP-8R CCCAGGGCAATTTGGTTAG CACACTGCCTCCCACTGAC CACCAGGACTCCCTCCAGT TGCGACACCCTTAACAACC TCCCTTGGGCTTTCCTTATT AGAGGCTGAGGCGAGAGAAT GGTCAAAGCTGGGTTCTTGA TTCAAAGCCCAGTCTCCATT 11
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
 Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Mouse Engineering Technology. Musculoskeletal Research Center 2016 Summer Educational Series David M. Ornitz Department of Developmental Biology
 Mouse Engineering Technology Musculoskeletal Research Center 2016 Summer Educational Series David M. Ornitz Department of Developmental Biology Core service and new technologies Mouse ES core Discussions
Mouse Engineering Technology Musculoskeletal Research Center 2016 Summer Educational Series David M. Ornitz Department of Developmental Biology Core service and new technologies Mouse ES core Discussions
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary Figure 1 Characterization of OKSM-iNSCs
 Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
Supplementary Figure 1 Characterization of OKSM-iNSCs (A) Gene expression levels of Sox1 and Sox2 in the indicated cell types by microarray gene expression analysis. (B) Dendrogram cluster analysis of
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Pei et al. Supplementary Figure S1
 Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Analysis of gene function
 Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy
 Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Supplementary Figure 1. IFN-γ induces TRC dormancy. a, IFN-γ induced dormancy of various tumor type TRCs, including H22 (murine hepatocarcinoma) and CT26 (murine colon cancer). Bar, 50 µm. b, B16 cells
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
SUPPLEMENTARY FIGURE LEGENDS Supplementary Fig. 1. (A) Working model. The pluripotency transcription factor OCT4 directly up-regulates the expression of NIPP1 and CCNF that together inhibit protein phosphatase
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
To assess the localization of Citrine fusion proteins, we performed antibody staining to
 Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Trinh et al 1 SUPPLEMENTAL MATERIAL FlipTraps recapitulate endogenous protein localization To assess the localization of Citrine fusion proteins, we performed antibody staining to compare the expression
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
Supplementary Figure 1 Generation of the AARE-Gene system construct. (a) Position and sequence alignment of AAREs extracted from human Trb3, Chop or Atf3 promoters. AARE core sequences are boxed in grey.
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Primers used for PCR of conductin, SGK1 and GAPDH have been described in (Dehner et al,
 Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary METHODS Flow Cytometry (FACS) For FACS analysis, trypsinized cells were fixed in ethanol, rehydrated in PBS and treated with 40μg/ml propidium iodide and 10μ/ml RNase for 30 min at room temperature.
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected
 Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Supplementary Figure 1. (a) The qrt-pcr for lnc-2, lnc-6 and lnc-7 RNA level in DU145, 22Rv1, wild type HCT116 and HCT116 Dicer ex5 cells transfected with the sirna against lnc-2, lnc-6, lnc-7, and the
Percent survival. Supplementary fig. S3 A.
 Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Generating stable cell lines expressing various Cas9 proteins from AAVS1 locus
 IGI Protocol prepared by Chong Park 1 Generating stable cell lines expressing various Cas9 proteins from AAVS1 locus This is modified from original protocol from the Conklin laboratory (Mandegar et al.
IGI Protocol prepared by Chong Park 1 Generating stable cell lines expressing various Cas9 proteins from AAVS1 locus This is modified from original protocol from the Conklin laboratory (Mandegar et al.
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Analyses of ECTRs by C-circle and T-circle assays.
 Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Supplementary Figure 1 Analyses of ECTRs by C-circle and T-circle assays. (a) C-circle and (b) T-circle amplification reactions using genomic DNA from different cell lines in the presence (+) or absence
Student Learning Outcomes (SLOS) - Advanced Cell Biology
 Course objectives The main objective is to develop the ability to critically analyse and interpret the results of the scientific literature and to be able to apply this knowledge to afford new scientific
Course objectives The main objective is to develop the ability to critically analyse and interpret the results of the scientific literature and to be able to apply this knowledge to afford new scientific
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
Figure S2. Response of mouse ES cells to GSK3 inhibition. Mentioned in discussion
 Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
Stem Cell Reports, Volume 1 Supplemental Information Robust Self-Renewal of Rat Embryonic Stem Cells Requires Fine-Tuning of Glycogen Synthase Kinase-3 Inhibition Yaoyao Chen, Kathryn Blair, and Austin
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 Giovanna Gambarotta- Only for teaching purposes. TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random
Giovanna Gambarotta- Only for teaching purposes. TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Supplementary Material. Mutation of the RAD51C gene in a Fanconi anemia-like disorder
 Supplementary Material Mutation of the RAD51C gene in a Fanconi anemia-like disorder Fiona Vaz, Helmut Hanenberg, Beatrice Schuster, Karen Barker, Constanze Wiek, Verena Erven, Kornelia Neveling, Daniela
Supplementary Material Mutation of the RAD51C gene in a Fanconi anemia-like disorder Fiona Vaz, Helmut Hanenberg, Beatrice Schuster, Karen Barker, Constanze Wiek, Verena Erven, Kornelia Neveling, Daniela
Adenoviral Expression Systems. Lentivirus is not the only choice for gene delivery. Adeno-X
 Adenoviral Expression Systems Lentivirus is not the only choice for gene delivery 3 Adeno-X Why choose adenoviral gene delivery? Table I: Adenoviral vs. Lentiviral Gene Delivery Lentivirus Adenovirus Infects
Adenoviral Expression Systems Lentivirus is not the only choice for gene delivery 3 Adeno-X Why choose adenoviral gene delivery? Table I: Adenoviral vs. Lentiviral Gene Delivery Lentivirus Adenovirus Infects
Supplemental Data. Borg et al. Plant Cell (2014) /tpc
 Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1 - Alignment of selected angiosperm DAZ1 and DAZ2 homologs Multiple sequence alignment of selected DAZ1 and DAZ2 homologs. A consensus sequence built using default parameters is shown
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Introducing new DNA into the genome requires cloning the donor sequence, delivery of the cloned DNA into the cell, and integration into the genome.
 Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
A Low salt diet. C Low salt diet + mf4-31c1 3. D High salt diet + mf4-31c1 3. B High salt diet
 A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
SUPPLEMENTARY NOTE 2. Supplememtary Note 2, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 2 A recombinase reporter system for permanent reporter activation We made use of the Cre-loxP recombinase system for a maximal amplification and complete kinetic uncoupling within single
SUPPLEMENTARY NOTE 2 A recombinase reporter system for permanent reporter activation We made use of the Cre-loxP recombinase system for a maximal amplification and complete kinetic uncoupling within single
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably
 Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplementary Information Supplementary Figure S1. The tetracycline-inducible CRISPR system. A) Hela cells stably expressing shrna sequences against TRF2 were examined by western blotting. shcon, shrna
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera
 SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
SUPPLEMENTARY INFORMATION Roles of human POLD1 and POLD3 in genome stability Emanuela Tumini, Sonia Barroso, Carmen Pérez Calero and Andrés Aguilera SUPPLEMENTARY METHODS Cell proliferation After sirna
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3
 Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Supplementary Figure S1 Evi/Wls and Wnt3 are overexpressed in epithelial cells in colon cancers. (a) Validation of the specificity of the Wnt3 antibody. HCT116 cells were reverse transfected with sicontrol
Supplementary Materials for
 www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
www.advances.sciencemag.org/cgi/content/full/1/7/e1500454/dc1 Supplementary Materials for CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation Xin Han, Zongbin Liu, Myeong chan Jo,
0.9 5 H M L E R -C tr l in T w is t1 C M
 a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
a. b. c. d. e. f. g. h. 2.5 C elltiter-g lo A ssay 1.1 5 M T S a s s a y Lum inescence (A.U.) 2.0 1.5 1.0 0.5 n s H M L E R -C tr l in C tr l C M H M L E R -C tr l in S n a il1 C M A bsorbance (@ 490nm
CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression
 Supplementary information for: CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression Qian Liang, Lili Li, Jianchao Zhang, Yang Lei, Liping Wang, Dong-Xu Liu,
Supplementary information for: CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression Qian Liang, Lili Li, Jianchao Zhang, Yang Lei, Liping Wang, Dong-Xu Liu,
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell
 Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Supplementary Information Alternative splicing of CD44 mrna by ESRP1 enhances lung colonization of metastatic cancer cell Supplementary Figures S1-S3 Supplementary Methods Supplementary Figure S1. Identification
Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced
 Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Supplementary Data Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nucleaseinduced mutations Suresh Ramakrishna 1, Seung Woo Cho 2, Sojung Kim 2, Myungjae Song 1, Ramu Gopalappa
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Nature Medicine: doi: /nm.4169
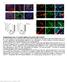 Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Fig.1. EC-specific deletion of Ccm3 by Cdh5-CreERT2. a. mt/mg reporter mice were bred with Cdh5CreERT2 deleter mice followed by tamoxifen feeding from P1 to P3. mg expression was specifically
Supplementary Data. Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter
 Supplementary Data Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter We first introduced an expression unit composed of a tetresponse
Supplementary Data Generation and Characterization of Mixl1- Inducible Embryonic Stem Cells Under the Control of Oct3/4 Promoter or CAG Promoter We first introduced an expression unit composed of a tetresponse
Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More. Ed Davis, Ph.D.
 TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
TECHNICAL NOTE Genome Editing: Cas9 Stable Cell Lines for CRISPR sgrna Validation, Library Screening, and More Introduction Ed Davis, Ph.D. The CRISPR-Cas9 system has become greatly popular for genome
supplementary information
 DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
DOI: 10.1038/ncb2156 Figure S1 Depletion of p114rhogef with different sirnas. Caco-2 (a) and HCE (b) cells were transfected with individual sirnas, pools of the two sirnas or the On Target (OnT) sirna
Supplementary Figure 1. Mouse genetic models used for identification of Axin2-
 Supplementary Figure 1. Mouse genetic models used for identification of Axin2- expressing cells and their derivatives. Schematic representations illustrate genetic cell labeling strategies to identify
Supplementary Figure 1. Mouse genetic models used for identification of Axin2- expressing cells and their derivatives. Schematic representations illustrate genetic cell labeling strategies to identify
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Quantitative analysis of recombination in YFP and CFP gene of FRET biosensor induced by lentiviral or retroviral gene transfer.
 Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
DOI: 10.1038/ncb2774 Figure S1 TRF2 dosage modulates the tumorigenicity of mouse and human tumor cells. (a) Left: immunoblotting with antibodies directed against the Myc tag of the transduced TRF2 forms
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction)
 Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Table S1. Primers used in RT-PCR studies (all in 5 to 3 direction) Epo Fw CTGTATCATGGACCACCTCGG Epo Rw TGAAGCACAGAAGCTCTTCGG Jak2 Fw ATCTGACCTTTCCATCTGGGG Jak2 Rw TGGTTGGGTGGATACCAGATC Stat5A Fw TTACTGAAGATCAAGCTGGGG
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
TRANSGENIC ANIMALS. transient. stable. - Two methods to produce transgenic animals:
 Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
SUPPLEMENTARY INFORMATION
 A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
A diphtheria toxin resistance marker for in vitro and in vivo selection of stably transduced human cells Gabriele Picco, Consalvo Petti, Livio Trusolino, Andrea Bertotti and Enzo Medico SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
TRIM31 is recruited to mitochondria after infection with SeV.
 Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
Supplementary Figure 1 TRIM31 is recruited to mitochondria after infection with SeV. (a) Confocal microscopy of TRIM31-GFP transfected into HEK293T cells for 24 h followed with SeV infection for 6 h. MitoTracker
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
DOI: 10.1038/ncb3363 Supplementary Figure 1 Several WNTs bind to the extracellular domains of PKD1. (a) HEK293T cells were co-transfected with indicated plasmids. Flag-tagged proteins were immunoprecipiated
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
Supplementary Figure 1
 Supplementary Figure 1 Cell cycle distribution (%) 1 8 6 4 2 Cell Cycle G1 S G2 Viability (%) 5 4 3 2 1 Viability Supplementary Figure 1. Cell cycle distribution and viability during DSB repair measurements.
Supplementary Figure 1 Cell cycle distribution (%) 1 8 6 4 2 Cell Cycle G1 S G2 Viability (%) 5 4 3 2 1 Viability Supplementary Figure 1. Cell cycle distribution and viability during DSB repair measurements.
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion
 Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion Supplementary Figure 1. The presence of Plp::EGFP + /O4 + cells 21 days after transgene induction.
Supplementary Material for Yang et al., Generation of oligodendroglial cells by direct lineage conversion Supplementary Figure 1. The presence of Plp::EGFP + /O4 + cells 21 days after transgene induction.
Return to Web Version
 Page 1 of 7 Page 1 of 7 Return to Web Version ZFN Technology Biowire Volume 10 Article 1 Have your genomic work cut out for you The genomes of several organisms, including humans, have been sequenced,
Page 1 of 7 Page 1 of 7 Return to Web Version ZFN Technology Biowire Volume 10 Article 1 Have your genomic work cut out for you The genomes of several organisms, including humans, have been sequenced,
Generation of Hprt-disrupted rat through mouse rat ES chimeras
 Supplementary Information Generation of Hprt-disrupted rat through mouse rat ES chimeras Ayako Isotani 1,2, *, Kazuo Yamagata 2,3, Masaru Okabe 2 and Masahito Ikawa 1,2 1. Immunology Frontier Research
Supplementary Information Generation of Hprt-disrupted rat through mouse rat ES chimeras Ayako Isotani 1,2, *, Kazuo Yamagata 2,3, Masaru Okabe 2 and Masahito Ikawa 1,2 1. Immunology Frontier Research
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Supplemental Information. Quiescence Exit of Tert + Stem Cells. by Wnt/b-Catenin Is Indispensable. for Intestinal Regeneration
 Cell Reports, Volume 21 Supplemental Information Quiescence Exit of Tert + Stem Cells by Wnt/b-Catenin Is Indispensable for Intestinal Regeneration Han Na Suh, Moon Jong Kim, Youn-Sang Jung, Esther M.
Cell Reports, Volume 21 Supplemental Information Quiescence Exit of Tert + Stem Cells by Wnt/b-Catenin Is Indispensable for Intestinal Regeneration Han Na Suh, Moon Jong Kim, Youn-Sang Jung, Esther M.
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells
 OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
OmicsLink shrna Clones guaranteed knockdown even in difficult-to-transfect cells OmicsLink shrna clone collections consist of lentiviral, and other mammalian expression vector based small hairpin RNA (shrna)
The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells
 Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
Supplementary Information The non-muscle-myosin-ii heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells Bing Zhao 1,3, Zhen Qi 1,3, Yehua Li 1,3, Chongkai Wang 2,
CRISPR/Cas9 Gene Editing Tools
 CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
CRISPR/Cas9 Gene Editing Tools - Guide-it Products for Successful CRISPR/Cas9 Gene Editing - Why choose Guide-it products? Optimized methods designed for speed and ease of use Complete kits that don t
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
DOI: 10.1038/ncb3209 Supplementary Figure 1 IR induces the association of FH with chromatin. a, U2OS cells synchronized by thymidine double block (2 mm) underwent no release (G1 phase) or release for 2
Supplementary Figure 1.
 Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
Supplementary Figure 1. (A) UCSC Genome Browser view of region immediately downstream of the NEUROG3 start codon. All candidate grna target sites which meet the G(N 19 )NGG constraint are aligned to illustrate
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
Comparative assessment of vaccine vectors encoding ten. malaria antigens identifies two protective liver-stage
 Supplementary Information Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates Rhea J. Longley 1,,,*, Ahmed M. Salman 1,2,, Matthew G.
Supplementary Information Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates Rhea J. Longley 1,,,*, Ahmed M. Salman 1,2,, Matthew G.
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
