% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
|
|
|
- Tyrone Turner
- 6 years ago
- Views:
Transcription
1 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy) UV (2 J/m 2 ) 75mM HU.25µg/ml 4-NQO 5µg/ml CPT Drug concentration nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Supplementary Figure 1. Sensitivity of Isw2 and Ino8 complex mutants to inhibitors of replication. (a) MMS sensitivity of nhp1 and ino8 9 mutants. Wild type (W1588-4c), (YTT18), nhp1 (YTT2), nhp1 (YTT219), ino8 9 (YTT4179), and ino8 9 (YTT427) stationary phase cells were spotted across YPD plates containing gradients of MMS. The maximum concentration of the drug is indicated at the left. See Supplementary Methods for the protocol to make drug gradient plates. (b) Survival of nhp1 mutants during MMS treatment in culture. Strains as in (a) were grown to early log phase, treated with the indicated concentrations of MMS for one hour, then plated. Percent survival (+/- the standard deviation) of wild type, nhp1, and mag1 (YTT277, an MMS sensitive control strain) was determined by plating known numbers of cells before and after drug treatment and counting colonies. (c) Spot tests of UV and gamma irradiated cells. 1-fold serial dilutions of saturated cultures (a) were spotted to YPD plates then exposed to the doses of radiation indicated. Note that the number of colonies at each spot is comparable in each strain. (d) Sensitivity of nhp1 mutants to other DNA replication inhibitors. Stationary phase cells were spotted across YPD plates containing gradients of Hydroxyurea (HU), 4-nitroquinoline 1-oxide (4-NQO), or Camptothecin (CPT) as in (a).
2 Supplementary Figure 2. Replication profiles of MMS treated cells. (a) Schematic diagram of the dense isotope transfer experiments conducted in the presence of.15% MMS, and the generation of replication profiles from isotope labelled DNA collected throughout S phase. (b) FACS analyses of cell collection for generation of replication profiles. Wild type (YTT1831) and nhp1 (YTT336) cells were treated as outlined in (a) and collected for FACS at the indicated times after release from α-factor arrest. Samples used for generation of replication profiles shown in Figure 2 and in (c) are indicated by asterisks. Gray profiles are from cells collected prior to G1 arrest. (c) Replication profiles were generated from the samples indicated in (b). The collection of nhp1 mutants began at minutes post-release because of a slight delay in cell cycle initiation following alpha-factor arrest characteristic of the double mutant (b). Profiles from corresponding collection times are the same color. The total percentage of replication for the whole genome at each time-point was determined by slot blot of both replicated and unreplicated fractions used for microarray analysis, followed by hybridization with radioactively labeled and fragmented genomic DNA. Positions of confirmed and likely ARSs, defined at are indicated at the bottom of each graph. Filled triangles correspond to origins identified in two studies of replication timing 1,2 as being amongst the earliest 25% of origins replicated in a normal S phase. Open triangles represent origins that are amongst the earliest 25% in only one of the two studies. The filled circles correspond to remaining origins. Origins that are active in wild type cells treated with 2mM Hydroxyurea 3 are indicated in red. Positions of centromeres are indicated by the black circle on the x-axis. Replication profiles from all 16 chromosomes are displayed to facilitate comparison between genotypes
3 Supplementary Figure 2 a b 1) Grow in dense ( 13 C 15 N) media α-factor 15 min 2) Transfer to light ( 12 C 14 N) media α-factor 75 min MMS min Time 3 nhp1 3) Release in light media + MMS 4) Collect cells throughout S phase 5) Prepare and digest genomic DNA 6) Separate replicated DNA on CsCl gradient 7) Label replicated & unreplicated DNA replicated unreplicated ) Compete replicated & unreplicated DNA on microarray 9) Generate replication profiles Chromosome Coordinate late-s mid-s early-s
4 c 1 8 Chr I 1 8 Supplementary Figure 2 Chr II nhp nhp Chr III Chr V time % total genomic replication nhp nhp nhp % 66% % 35% 32% 27% 2% % 14% % 1 8 Chr IV nhp
5 c Supplementary Figure Chr VI 1 8 Chr VII nhp Chr VIII 1 8 Chr IX nhp nhp Chr X time % total genomic replication nhp nhp % 66% % 35% 32% 27% 2% 4 31% 14% %
6 c Supplementary Figure Chr XI nhp Chr XII nhp Chr XIII time % total genomic replication nhp nhp % 66% % 31% 35% 32% 27% 2% 14% %
7 c Supplementary Figure Chr XIV nhp Chr XV nhp Chr XVI nhp time % total genomic replication 81% 66% % 31% 12% nhp1 35% 32% 27% 2% 14%
8 a Time post G1 release (+MMS): Bubble arc (active origin) ARS7 (early) nhp1 Y arc d a 1n ARS1 (early-mid) nhp1 b S-phase (-MMS) ARS152 (late) nhp1 nhp1 ARS152 (late) ARS3 (late) nhp1 nhp1 ARS3 (late) Supplementary Figure 3. Patterns of origin firing in nhp1 mutants. (a) Origin firing during MMS treatment. The bubble arc is detected when an origin of replication is active in the probed restriction fragment. The ascending portion of the Y arcs ( a ; diagrammed on the right) arises when the probed region is replicated from forks originating outside of the restriction fragment, while the descending portion ( d ) can result from both passive replication and large replication bubbles from an active origin within the fragment. Genomic DNA for 2-D electrophoresis was isolated from (YTT1831) and nhp1 (YTT336) cells treated as in Supplementary Figure 2a. ARS7 is an early origin, ARS1 is an early-middle origin, while ARS152 and ARS3 initiate later in S phase. The grey arrows indicate faint bubble arcs at ARS3 and ARS152. Equivalent amounts of DNA were loaded as indicated by the intensity of 1N signals. See Supplementary Methods for the protocol. (b), 2-D gel analysis of late origin firing in the absence of MMS treatment. Genomic DNA for 2-D electrophoresis was isolated from strains undergoing S phase in the absence of MMS.
9 a ARS7 DPB4 % budded Time b Time 15 3 nhp nhp1 ARS7 DPB4 % budded Time c 3 C nhp d Time Time 16 C nhp Time Time 24 3 Supplementary Figure 4. Fork progression analyses. (a) Comparison of replication kinetics of early and late-replicating loci during MMS treatment. Percent replication at an early-replicating locus, ARS7 (open circles), and a locus in a late-replicating region of Chromosome IV, DPB4 (Chromosome coordinate 694 kb, filled circles), was determined for each sample isolated in (b) as described in Methods. The percentage of budded cells is indicated by the dashed line. (b) FACS analysis of ARS8/ARS9 double deletion strains collected for fork progression-rate determination. Wild type (YTT3528) and nhp1 (YTT3531) cells were treated as outlined in Supplementary Figure 2a and collected for FACS analysis and isolation of genomic DNA at the indicated times after release from α-factor arrest. (c) Replication kinetics of the right arm of Chromosome VI at 3 C. Wild type and nhp1 cells, as in (b), were treated as in Supplemental Figure 2c except that MMS was omitted. Percent replication of regions 1-5 on chromosome VI, designated by blue lines of increasing intensity (as diagrammed in Figure 3a), was determined as described in Methods. The percentage of budded cells is indicated by the dashed line. The horizontal dotted line corresponds to the percent replication value designated as one-half of the maximum obtained for that experiment. The point of intersection of the horizontal dotted line with the fitted curve is designated as the T rep for that region. (d) Replication kinetics of the right arm of Chromosome VI at 16 C. As described in (c).
10 a 1) Grow cells to early log-phase 2) Split culture b vs. nhp1 nhp1 vs. nhp1 Untreated 2 hours 3 C.2% MMS Genes w/ 1.5 -fold change nhp1 specific changes Genes w/ 1.5 -fold change 3a) Harvest 3b) Harvest c log expression level nhp log expression level log expression level nhp log expression level nhp1 v nhp1 (MMS) (control) Supplementary Figure 5. Summary of gene-expression analysis of MMS-treated nhp1 mutants e4 s: JAV18 : MMS v MMS nhp1 (3 replicates) (Default Interpretation), v is... s: JAV18 : MMS v MMS nhp1 (3 replicates) (Default Interpretation), v is... Colored by: JAV18 : MMS v MMS nhp1 (3 replicates) (Default Interpretation) Gene List: all genes (68), 195 genes selected (a) MMS treatment and culture conditions. (YTT18), nhp1 (YTT2), and nhp1 (YTT219) strains were treated as shown before isolation of RNA. (b) Definition of nhp1 specific changes in gene-expression. Transcript levels in an nhp1 mutant were compared to transcript levels from either (blue) or nhp1 (orange) by co-hybridizing labeled mrna samples from the single and double mutant on the same microarray (indicated at the top of the diagram). Changes in transcript levels specific to the double mutant were defined as those common to both co-hybridization experiments. (c) Scatterplot of gene expression levels for MMS treated or nhp1 single mutants versus the expression levels of MMS treated nhp1 double mutants. The black data points correspond to 26 genes involved in replication, repair, or the DNA damage checkpoint. Blue lines mark 1.5-fold changes in gene expression. This gene list was compiled based on descriptions provided at the Saccharomyces Genome Database.
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
SUPPLEMENTARY INFORMATION
 Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1 Origin use and efficiency are similar among WT, rrm3, pif1-m2, and pif1-m2; rrm3 strains. A. Analysis of fork progression around confirmed and likely origins (from cerevisiae.oridb.org).
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
DNA replication in eukaryotic cells is a complex
 Copyright Ó 2008 by the Genetics Society of America DOI: 10.1534/genetics.108.094359 The Temporal Program of Chromosome Replication: Genomewide Replication in clb5d Saccharomyces cerevisiae Heather J.
Copyright Ó 2008 by the Genetics Society of America DOI: 10.1534/genetics.108.094359 The Temporal Program of Chromosome Replication: Genomewide Replication in clb5d Saccharomyces cerevisiae Heather J.
Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
 Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes Hannah G. Blitzblau 1,2, Clara S. Chan 1, Andreas Hochwagen 2,3, Stephen P. Bell 1 * 1 Department of Biology, Howard
Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes Hannah G. Blitzblau 1,2, Clara S. Chan 1, Andreas Hochwagen 2,3, Stephen P. Bell 1 * 1 Department of Biology, Howard
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
PIE1 ARP6 SWC6 KU70 ARP6 PIE1. HSA SNF2_N HELICc SANT. pie1-3 A1,A2 K1,K2 K1,K3 K3,LB2 A3, A4 A3,LB1 A1,A2 K1,K2 K1,K3. swc6-1 A3,A4.
 A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
A B N-terminal SWC2 H2A.Z SWC6 ARP6 PIE1 HSA SNF2_N HELICc SANT C pie1-3 D PIE1 ARP6 5 Kb A1 200 bp A3 A2 LB1 arp6-3 A4 E A1,A2 A3, A4 A3,LB1 K1,K2 K1,K3 K3,LB2 SWC6 swc6-1 A1,A2 A3,A4 K1,K2 K1,K3 100
SUPPLEMENTARY INFORMATION
 doi:.8/nature979 Financial support: CS is a Royal Swedish Academy of Sciences Research Fellow supported by Knut and Alice Wallenbergs Foundation. This work was financed European research council (ERC starting
doi:.8/nature979 Financial support: CS is a Royal Swedish Academy of Sciences Research Fellow supported by Knut and Alice Wallenbergs Foundation. This work was financed European research council (ERC starting
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric
 Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Protocol CRISPR Genome Editing In Cell Lines
 Protocol CRISPR Genome Editing In Cell Lines Protocol 2: HDR donor plasmid applications (gene knockout, gene mutagenesis, gene tagging, Safe Harbor ORF knock-in) Notes: 1. sgrna validation: GeneCopoeia
Protocol CRISPR Genome Editing In Cell Lines Protocol 2: HDR donor plasmid applications (gene knockout, gene mutagenesis, gene tagging, Safe Harbor ORF knock-in) Notes: 1. sgrna validation: GeneCopoeia
supplementary information
 DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
7.06 Problem Set Four, 2006
 7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Contents. 1 Basic Molecular Microbiology of Bacteria... 1 Exp. 1.1 Isolation of Genomic DNA Introduction Principle...
 Contents 1 Basic Molecular Microbiology of Bacteria... 1 Exp. 1.1 Isolation of Genomic DNA... 1 Introduction... 1 Principle... 1 Reagents Required and Their Role... 2 Procedure... 3 Observation... 4 Result
Contents 1 Basic Molecular Microbiology of Bacteria... 1 Exp. 1.1 Isolation of Genomic DNA... 1 Introduction... 1 Principle... 1 Reagents Required and Their Role... 2 Procedure... 3 Observation... 4 Result
Supplemental Figure 1
 Supplemental Figure 1 Supplemental Figure 1 (previous page): Heavy chain gene content of ARS/Igh66 transgenic lines. A. Tail DNA from the indicated transgenic lines was amplified for the indicated gene
Supplemental Figure 1 Supplemental Figure 1 (previous page): Heavy chain gene content of ARS/Igh66 transgenic lines. A. Tail DNA from the indicated transgenic lines was amplified for the indicated gene
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
ATP-dependent chromatin remodeling shapes the DNA replication landscape
 1 ATP-dependent chromatin remodeling shapes the DNA replication landscape Jack A. Vincent, Tracey J. Kwong, and Toshio Tsukiyama Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100
1 ATP-dependent chromatin remodeling shapes the DNA replication landscape Jack A. Vincent, Tracey J. Kwong, and Toshio Tsukiyama Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Gene Expression Technology
 Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Gene Expression Technology Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Gene expression Gene expression is the process by which information from a gene
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
C24. ros1-1. ros1-1 rdm18-1. ros1-1 rdm18-2. ros1-1 nrpe1
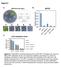 Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
Figure S1 Methylation level 1 0.8 0.6 0.4 0.2 0 p35s methylation levels 24 ros1-1 ros1-1 rdm18-1 ros1-1 rdm18-2 ros1-1 nrpe1 mg mhg mhh Figure S1. RM18/PKL promotes silencing at the p35s-npt II transgene
of NOBOX. Supplemental Figure S1F shows the staining profile of LHX8 antibody (Abcam, ab41519), which is a
 SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
SUPPLEMENTAL MATERIALS AND METHODS LHX8 Staining During the course of this work, it came to our attention that the NOBOX antibody used in this study had been discontinued. There are a number of other nuclear
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
MID-TERM EXAMINATION
 PLNT3140 INTRODUCTORY CYTOGENETICS MID-TERM EXAMINATION 1 p.m. to 2:15 p.m. Thursday, October 18, 2012 Answer any combination of questions totalling to exactly 100 points. If you answer questions totalling
PLNT3140 INTRODUCTORY CYTOGENETICS MID-TERM EXAMINATION 1 p.m. to 2:15 p.m. Thursday, October 18, 2012 Answer any combination of questions totalling to exactly 100 points. If you answer questions totalling
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination.
 Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figure 1 qrt-pcr expression analysis of NLP8 with and without KNO 3 during germination. Seeds of Col-0 were harvested from plants grown at 16 C, stored for 2 months, imbibed for indicated
Supplementary Figures Montero et al._supplementary Figure 1
 Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Montero et al_suppl. Info 1 Supplementary Figures Montero et al._supplementary Figure 1 Montero et al_suppl. Info 2 Supplementary Figure 1. Transcripts arising from the structurally conserved subtelomeres
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C
 CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
CORRECTION NOTICE Nat. Genet. 47, 598 606 (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C Borbala Mifsud, Filipe Tavares-Cadete, Alice N Young, Robert Sugar,
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
New Statistical Algorithms for Monitoring Gene Expression on GeneChip Probe Arrays
 GENE EXPRESSION MONITORING TECHNICAL NOTE New Statistical Algorithms for Monitoring Gene Expression on GeneChip Probe Arrays Introduction Affymetrix has designed new algorithms for monitoring GeneChip
GENE EXPRESSION MONITORING TECHNICAL NOTE New Statistical Algorithms for Monitoring Gene Expression on GeneChip Probe Arrays Introduction Affymetrix has designed new algorithms for monitoring GeneChip
American Society of Cytopathology Core Curriculum in Molecular Biology
 American Society of Cytopathology Core Curriculum in Molecular Biology American Society of Cytopathology Core Curriculum in Molecular Biology Chapter 3 Molecular Techniques Separation and Detection, Part
American Society of Cytopathology Core Curriculum in Molecular Biology American Society of Cytopathology Core Curriculum in Molecular Biology Chapter 3 Molecular Techniques Separation and Detection, Part
Name Genotype Reference
 Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Supplemental Data Supplemental Table 1 S. cerevisiae strains. Name Genotype Reference RMY200 UKY403 W303-1a MATa ade2-101 (och) his3 200 lys2-801 (amp) trp1 901 ura3-52 hht1,hhf1::leu2 hht2,hhf2::his3
Nature Methods: doi: /nmeth Supplementary Figure 1. Pilot CrY2H-seq experiments to confirm strain and plasmid functionality.
 Supplementary Figure 1 Pilot CrY2H-seq experiments to confirm strain and plasmid functionality. (a) RT-PCR on HIS3 positive diploid cell lysate containing known interaction partners AT3G62420 (bzip53)
Supplementary Figure 1 Pilot CrY2H-seq experiments to confirm strain and plasmid functionality. (a) RT-PCR on HIS3 positive diploid cell lysate containing known interaction partners AT3G62420 (bzip53)
plasmid shuffle strain
 Histone-Histone Interactions and Centromere Function LYNN GLOWCZEWSKI, PEIRONG YANG, TATYANA KALASHNIKOVA, MARIA SOLEDAD SANTISTEBAN, AND M. MITCHELL SMITH* Department of Microbiology and Cancer Center,
Histone-Histone Interactions and Centromere Function LYNN GLOWCZEWSKI, PEIRONG YANG, TATYANA KALASHNIKOVA, MARIA SOLEDAD SANTISTEBAN, AND M. MITCHELL SMITH* Department of Microbiology and Cancer Center,
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Intracellular receptors specify complex patterns of gene expression that are cell and gene
 SUPPLEMENTAL RESULTS AND DISCUSSION Some HPr-1AR ARE-containing Genes Are Unresponsive to Androgen Intracellular receptors specify complex patterns of gene expression that are cell and gene specific. For
SUPPLEMENTAL RESULTS AND DISCUSSION Some HPr-1AR ARE-containing Genes Are Unresponsive to Androgen Intracellular receptors specify complex patterns of gene expression that are cell and gene specific. For
Supplementary Information
 Supplementary Information Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae Junhyun Jeon, Jaeyoung
Supplementary Information Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae Junhyun Jeon, Jaeyoung
Marathon TM cdna Amplification Kit Protocol-at-a-Glance
 (PT1115-2) Marathon cdna amplification is a fairly complex, multiday procedure. Please read the User Manual before using this abbreviated protocol, and refer to it often for interpretation of results during
(PT1115-2) Marathon cdna amplification is a fairly complex, multiday procedure. Please read the User Manual before using this abbreviated protocol, and refer to it often for interpretation of results during
reverse transcription! RT 1! RT 2! RT 3!
 Supplementary Figure! Entire workflow repeated 3 times! mirna! stock! -fold dilution series! to yield - copies! per µl in PCR reaction! reverse transcription! RT! pre-pcr! mastermix!!!! 3!!! 3! ddpcr!
Supplementary Figure! Entire workflow repeated 3 times! mirna! stock! -fold dilution series! to yield - copies! per µl in PCR reaction! reverse transcription! RT! pre-pcr! mastermix!!!! 3!!! 3! ddpcr!
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum. Blood-stage Development
 Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Supplementary Information to: Genome-wide Real-time in vivo Transcriptional Dynamics During Plasmodium falciparum Blood-stage Development Painter et al. 1 of 8 Supplementary Figure 1: Supplementary Figure
Expressed genes profiling (Microarrays) Overview Of Gene Expression Control Profiling Of Expressed Genes
 Expressed genes profiling (Microarrays) Overview Of Gene Expression Control Profiling Of Expressed Genes Genes can be regulated at many levels Usually, gene regulation, are referring to transcriptional
Expressed genes profiling (Microarrays) Overview Of Gene Expression Control Profiling Of Expressed Genes Genes can be regulated at many levels Usually, gene regulation, are referring to transcriptional
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
7.03, 2005, Lecture 20 EUKARYOTIC GENES AND GENOMES I
 7.03, 2005, Lecture 20 EUKARYOTIC GENES AND GENOMES I For the last several lectures we have been looking at how one can manipulate prokaryotic genomes and how prokaryotic genes are regulated. In the next
7.03, 2005, Lecture 20 EUKARYOTIC GENES AND GENOMES I For the last several lectures we have been looking at how one can manipulate prokaryotic genomes and how prokaryotic genes are regulated. In the next
Supplementary Information
 Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Supplementary Information Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing Won-Ki Cho 1, Namrata Jayanth 1, Susan Mullen
Erhard et al. (2013). Plant Cell /tpc
 Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Supplemental Figure 1. c1-hbr allele structure. Diagram of the c1-hbr allele found in stocks segregating 1:1 for rpd1-1 and rpd1-2 homozygous mutants showing the presence of a 363 base pair (bp) Heartbreaker
Table 1.C.1 Example of Hazardous Materials Identification System. Name of chemical. Personal protection equipment. Table 1.C.2 Example NFPA table.
 Table 1.C.1 Example of Hazardous Materials Identification System. Name of chemical Health Flammability Reactivity Personal protection equipment Table 1.C.2 Example NFPA table. Fire Health Reactivity Specific
Table 1.C.1 Example of Hazardous Materials Identification System. Name of chemical Health Flammability Reactivity Personal protection equipment Table 1.C.2 Example NFPA table. Fire Health Reactivity Specific
NAME Gene 603 Exam III November 30, 2,001
 NAME Gene 603 Exam III November 30, 2,001 I.. Suppose that a single nucleotide polymorphism exists in the centromere of chromosome 5 in Neurospora crassa. Use as many of the "ordered asci" as needed in
NAME Gene 603 Exam III November 30, 2,001 I.. Suppose that a single nucleotide polymorphism exists in the centromere of chromosome 5 in Neurospora crassa. Use as many of the "ordered asci" as needed in
d. reading a DNA strand and making a complementary messenger RNA
 Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Wu et al., Determination of genetic identity in therapeutic chimeric states. We used two approaches for identifying potentially suitable deletion loci
 SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in
 Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1: Overexpression of EBV-encoded proteins Western blot analysis of the expression levels of EBV-encoded latency III proteins in BL2 cells. The Ponceau S staining of the membranes or
Supplementary Figure 1. Additional RNAi screen data
 Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Supplementary Figure 1. Additional RNAi screen data A. Cisplatin induced ATR autophosphorylation. Western blot illustrating ATR and phospho-atr (T1989) in cells exposed to 1 µm cisplatin for 24 hours prior
Zhang et al., RepID facilitates replication Initiation. Supplemental Information:
 Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Supplemental Information: a b 1 Supplementary Figure 1 (a) DNA sequence of all the oligonucleotides used in this study. Only one strand is shown. The unshaded nucleotide sequences show changes from the
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supporting Information
 Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Supplemental Figure 1 A
 Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Supplemental Figure A prebleach postbleach 2 min 6 min 3 min mh2a.-gfp mh2a.2-gfp mh2a2-gfp GFP-H2A..9 Relative Intensity.8.7.6.5 mh2a. GFP n=8.4 mh2a.2 GFP n=4.3 mh2a2 GFP n=2.2 GFP H2A n=24. GFP n=7.
Figure S1. Replication initiation sites at efficient ORIs do not coincide with nucleosomedepleted
 Figure S1. Replication initiation sites at efficient ORIs do not coincide with nucleosomedepleted regions in the mouse genome. Typical examples of SNS profiles (Cayrou et al., 11) and nucleosome occupancies
Figure S1. Replication initiation sites at efficient ORIs do not coincide with nucleosomedepleted regions in the mouse genome. Typical examples of SNS profiles (Cayrou et al., 11) and nucleosome occupancies
sicker; i.e. to test whether we can reverse the mutant phenotype of mec1-100 when the suppressor gene is ectopically over-expressed in mec1-100.
 Internship Report ~FMI (Basel, Switzerland)~ Kyoto University Faculty of Medicine 1 st grade Yuiko Hirata This summer I went to the FMI (Friedrich Miescher Institute for Biomedical Research) in Basel.
Internship Report ~FMI (Basel, Switzerland)~ Kyoto University Faculty of Medicine 1 st grade Yuiko Hirata This summer I went to the FMI (Friedrich Miescher Institute for Biomedical Research) in Basel.
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
AGATCCACTAGTTCTAGAGCTCAACTTGTTGGTTTGTCTTT TGTCCAAAAACGGTTTCCAG TGCCTTTCTGTTCGATTGAT
 1 Table S1. PCR primers Primer Use Sequence (5 3 ) JC17 SAT1 flipper GGCCCCCCCTCGAGGAAGTT JC18 SAT1 flipper GCTCTAGAACTAGTGGATCT JC57 5 NCR of CNA1 AATGATGGAGTTTATGGGAA JC58 5 NCR of CNA1 AACTTCCTCGAGGGGGGGCCGAAAAAAAAAAAGGGGGGGG
1 Table S1. PCR primers Primer Use Sequence (5 3 ) JC17 SAT1 flipper GGCCCCCCCTCGAGGAAGTT JC18 SAT1 flipper GCTCTAGAACTAGTGGATCT JC57 5 NCR of CNA1 AATGATGGAGTTTATGGGAA JC58 5 NCR of CNA1 AACTTCCTCGAGGGGGGGCCGAAAAAAAAAAAGGGGGGGG
Supplementary Material
 Supplementary Material The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection Eriston Vieira Gomes 1, Mariana
Supplementary Material The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection Eriston Vieira Gomes 1, Mariana
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Biology 201 (Genetics) Exam #3 120 points 20 November Read the question carefully before answering. Think before you write.
 Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Supplementary Information
 Supplementary Information Supplement to Genome-wide allele- and strand-specific expression profiling, Julien Gagneur, Himanshu Sinha, Fabiana Perocchi, Richard Bourgon, Wolfgang Huber and Lars M. Steinmetz
Supplementary Information Supplement to Genome-wide allele- and strand-specific expression profiling, Julien Gagneur, Himanshu Sinha, Fabiana Perocchi, Richard Bourgon, Wolfgang Huber and Lars M. Steinmetz
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days,
 Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
Supplementary Figure 1. Expressions of stem cell markers decreased in TRCs on 2D plastic. TRCs were cultured on plastic for 1, 3, 5, or 7 days, respectively, and their mrnas were quantified by real time
AP Biology. Chapter 20. Biotechnology: DNA Technology & Genomics. Biotechnology. The BIG Questions. Evolution & breeding of food plants
 What do you notice about these phrases? radar racecar Madam I m Adam Able was I ere I saw Elba a man, a plan, a canal, Panama Was it a bar or a bat I saw? Chapter 20. Biotechnology: DNA Technology & enomics
What do you notice about these phrases? radar racecar Madam I m Adam Able was I ere I saw Elba a man, a plan, a canal, Panama Was it a bar or a bat I saw? Chapter 20. Biotechnology: DNA Technology & enomics
4000/ Sal I. Eco RI and Sma I/Probe 1 Bam HI and Sma I/Probe 2. Increasing Shape. Increasing Shape
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 7.28 Spring 2005 Name Exam One Question 1 (28 Points). Your lab is studying a novel thermophilic eukaryote called S. mokin that has an optimum
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 7.28 Spring 2005 Name Exam One Question 1 (28 Points). Your lab is studying a novel thermophilic eukaryote called S. mokin that has an optimum
(Hadlock, Journal of Virology, 2000), (Keck, Journal of Virology, 2008) CBH-5 Domain B 412, 416, 417, 418, 420, 421, 422, 423, 483, 484, 485, 488,
 Supplementary Table. mabs analyzed mab Antigenic Critical Binding Residues by Citations Domains Alanine Scanning 2 CBH-4B Domain A NA (Hadlock, Journal of Virology, 2), (Keck, Journal of Virology, 24)
Supplementary Table. mabs analyzed mab Antigenic Critical Binding Residues by Citations Domains Alanine Scanning 2 CBH-4B Domain A NA (Hadlock, Journal of Virology, 2), (Keck, Journal of Virology, 24)
Nuclear Mitochondrial DNA Activates Replication in Saccharomyces cerevisiae
 Nuclear Mitochondrial DNA Activates Replication in Saccharomyces cerevisiae Laurent Chatre 1,2, Miria Ricchetti 1,2 * 1 Departement d Immunologie, Institut Pasteur, Paris, France, 2 Unité de Génétique
Nuclear Mitochondrial DNA Activates Replication in Saccharomyces cerevisiae Laurent Chatre 1,2, Miria Ricchetti 1,2 * 1 Departement d Immunologie, Institut Pasteur, Paris, France, 2 Unité de Génétique
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
Supplementary Figure 1 Endogenous gene tagging to study subcellular localization and chromatin binding. a, b, Schematic of experimental set-up to endogenously tag RNAi factors using the CRISPR Cas9 technology,
BIO 202 Midterm Exam Winter 2007
 BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
Supplementary Data Supplementary Figure 1. Knockdown of VentX with a different sirna sequence reduces terminal monocyte to macrophage
 Supplementary Data Supplementary Figure 1. Knockdown of VentX with a different sirna sequence reduces terminal monocyte to macrophage differentiation. Monocytes were transfected with either a scrambled
Supplementary Data Supplementary Figure 1. Knockdown of VentX with a different sirna sequence reduces terminal monocyte to macrophage differentiation. Monocytes were transfected with either a scrambled
Technical Review. Real time PCR
 Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Technical Review Real time PCR Normal PCR: Analyze with agarose gel Normal PCR vs Real time PCR Real-time PCR, also known as quantitative PCR (qpcr) or kinetic PCR Key feature: Used to amplify and simultaneously
Biotechnology: DNA Technology & Genomics
 Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 1 The BIG Questions! How can we use our knowledge of DNA to: " diagnose disease or defect? " cure disease or defect? " change/improve organisms?!
Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 1 The BIG Questions! How can we use our knowledge of DNA to: " diagnose disease or defect? " cure disease or defect? " change/improve organisms?!
Computational Biology I
 Computational Biology I Microarray data acquisition Gene clustering Practical Microarray Data Acquisition H. Yang From Sample to Target cdna Sample Centrifugation (Buffer) Cell pellets lyse cells (TRIzol)
Computational Biology I Microarray data acquisition Gene clustering Practical Microarray Data Acquisition H. Yang From Sample to Target cdna Sample Centrifugation (Buffer) Cell pellets lyse cells (TRIzol)
Chapter 20 DNA Technology & Genomics. If we can, should we?
 Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
DNA Methylation. (B-Value) 3BKO. (B-Value) Supplementary Figure BKO HCT116 DKO8
 Supplementary Figure 1 a 1 DNA Methylation 0 3BKO DKO8 b DNA Methylation 0.5 3BKO DKO8 Supplementary Fig. 1. 3BKO and DKO8 cells compared to the parental cell line. (a) Heatmap showing the 10% (n=39,605)
Supplementary Figure 1 a 1 DNA Methylation 0 3BKO DKO8 b DNA Methylation 0.5 3BKO DKO8 Supplementary Fig. 1. 3BKO and DKO8 cells compared to the parental cell line. (a) Heatmap showing the 10% (n=39,605)
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
XXII DNA cloning and sequencing. Outline
 XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
DESIGNER GENES SAMPLE TOURNAMENT
 DESIGNER GENES SAMPLE TOURNAMENT PART ONE- GENETICS PROBLEMS In dogs, the inheritance of hair color involves a gene (B) for black hair and a gene (b) for brown hair. A dominant (C) is also involved. It
DESIGNER GENES SAMPLE TOURNAMENT PART ONE- GENETICS PROBLEMS In dogs, the inheritance of hair color involves a gene (B) for black hair and a gene (b) for brown hair. A dominant (C) is also involved. It
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Genetics. I Name the following: 1. It is the mechanism of inheritance of characters from the parental generation to the offspring.
 Grade Subject Topic : HS : Science : Genetics Genetics I Name the following: 1. It is the mechanism of inheritance of characters from the parental generation to the offspring. ---------------------------------------------------------
Grade Subject Topic : HS : Science : Genetics Genetics I Name the following: 1. It is the mechanism of inheritance of characters from the parental generation to the offspring. ---------------------------------------------------------
Figure S1- Targeting strategy for generating SIK1-T182A, SIK2-T175A and SIK3-T163A KI mice.
 Figure S1- Targeting strategy for generating SIK1-T12A, SIK2-T175A and SIK3-T163A mice. Left panel represents a depiction of the genomic loci of each gene, the targeting strategy and the final recombined
Figure S1- Targeting strategy for generating SIK1-T12A, SIK2-T175A and SIK3-T163A mice. Left panel represents a depiction of the genomic loci of each gene, the targeting strategy and the final recombined
