Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and
|
|
|
- Adele Davidson
- 6 years ago
- Views:
Transcription
1 Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting Residues in Chain B Chain A GLN A 347 LYS B 360 TYR A 349 SER B 354 ASP B 356 GLU B 357 LYS B 360 THR A 350 SER B 354 ARG B 355 LEU A 351 LEU B 351 SER B 354 THR B 366 SER A 354 TYR B 349 THR B 350 LEU B 351 ARG A 355 THR B 350 ASP A 356 TYR B 349 LYS B 439 GLU A 357 TYR B 349 LYS B 370 LYS A 360 GLN B 347 TYR B 349 SER A 364 LEU B 368 LYS B 370 THR A 366 LEU B 351 TYR B 407 LEU A 368 SER B 364 LYS B 409 LYS A 370 GLU B 357 SER B 364 ASN A 390 SER B 400 LYS A 392 ASP B 399 SER B 400 PHE B 405 THR A 394 THR B 394 VAL B 397 PHE B 405 TYR B 407 PRO A 395 VAL B 397 VAL A 397 THR B 394 PRO B 395 ASP A 399 LYS B 392 LYS B 409 SER A 400 ASN B 390 LYS B 392 PHE A 405 LYS B 392 THR B 394 LYS B 409 TYR A 407 THR B 366 THR B 394 TYR B 407 LYS B 409 LYS A 409 LEU B 368 ASP B 399 PHE B 405 TYR B 407 LYS A 439 ASP B 356 a Positions involving interaction between oppositely charged residues are indicated in bold. Due to the 2-fold symmetry present in the CH3-CH3 domain interaction, each pairwise interaction is represented twice in the structure (for example, Asp A Lys B 439 & Lys A Asp B 356 ; Figure 1). Eu numbering scheme is used here to identify residue positions.
2 Supplementary Table 2: Quantification of percentage of homodimer and heterodimer yields for the SDS-PAGE shown in Figure 2c a scfv-fc WT D399 K D399 K D399 K;E356 K D399 K;E357 K D399 K;E356 K D399 K;E357 K T366 W (Hole) Fc WT K409D;K360D K409D; K392D K409D;K392D K409D;K392D K409D;K439D K409D;K370D T366S;L368A; Y407V (Knob) scfv-fc / scfv-fc ND ND ND ND 13.3 scfv-fc / Fc Fc / Fc ND ND ND a ND stands for Not Detectable in the density based analysis.
3 Supplementary Table 3: CH3-CH3 domain binding free energy for various mutants designed to enhance heterodimer formation, calculated using the EGAD program (1) a Protein Description G (in kcal/mol) G mut (in kcal/mol) Melting Temp. T m (in ºC) b WT Wild Type T366W/Y407 A Knob-Hole T366W/T366 S:L368 A:Y407 V Knob-Hole K409D:K392D/D399 K:D356 K Charge- Charge ND c a Not all possible charge-charge pairs were considered for the binding free energy calculation. Wild type is listed for comparison. G is defined as energy difference between the complex and free states. The binding free energy of a mutant ( G mut ) is defined as difference between the mutant ( G mut ) and wild type ( G WT ) free energies. b CH3 domain melting temperatures (T m ) for the knob-hole mutants and WT are as reported in the literature (2). c Melting temperature for the charge pair mutant was not measured using only the CH3 domain. The DSC profiles shown in the Supplementary Figure 4 includes CH3, CH2, and scfv domains.
4 Supplementary Figure Legends Supplementary Figure 1. Flow chart of the scheme used to analyze the Fc crystal structures and sequences in order to arrive at the strategy (shown in Figure 1d) for engineering heterodimeric Fc. Supplementary Figure 2. Structural conservation of the CH3 domain interface residues is shown here by superimposing the identified Fc crystal structures from the PDB. Only the side chain atoms of the interface residues are shown for clarity. The buried residues (%ASA 10) are colored based on atom type and the exposed residues (%ASA > 10) are colored brown. The structurally not conserved residues are colored in green and their conformations in other structures are not shown for clarity. Supplementary Figure 3. Multiple sequence alignment of (a) human and (b) mouse IgG subclasses CH3 domain sequences. The star (*) indicates residue positions involved in the CH3-CH3 domain interaction identified based on the IgG1 human Fc crystal structure (1L6X). The CH3 domain interface charged residues marked with rectangles are highly conserved among the IgGs. Supplementary Figure 4. Production of scfv-fc/fc heterodimers for Differential Scanning Calorimetric (DSC) analysis. (a) SDS-PAGE and western blot analysis of purified scfv-fc/fc heterodimer created using the charge pair strategy (K409D:K392D / D399 K:E356 K, lane 1), WT (lane 2), and the knobs-into-holes strategy
5 (T366W/T366 S:L368 A:Y407 V, lane 3) under reduced and non-reduced condition. Culture supernatants contain scfv-fc/fc heterodimers were first purified by Protein A affinity chromatography. To separate scfv-fc/fc heterodimer from contaminating homodimers for WT and knob-into-hole constructs, a 6xHis tag was placed at the C- terminal of the scfv-fc construct. scfv-fc/scfv-fc homodimer and scfv-fc/fc heterodimer were separated from Fc/Fc homodimer by a nickel His-trap column and the scfv-fc/fc heterodimer was further purified by Size exclusion chromatography (SEC). Left panel: Western blot of scfv-fc/fc heterodimers preps after Protein A purification using anti-his antibody. Both WT and knob-into-hole constructs still contain scfv- Fc/scFv-Fc homodimer species. Right panel: SDS-PAGE analysis of purified scfv- Fc/Fc heterodimer after additional purification steps. (b) Differential Scanning Calorimetric (DSC) profiles for the three purified scfv-fc/fc constructs described in (a). DSC measurements were carried out for all samples at a concentration of 0.5mg/mL in ph 7.0 PBS buffer. Each sample was scanned at a rate of 0.25 C/min from 30 to 90 C. Reference scan using the PBS buffer was conducted and subtracted from the sample scans. Supplementary Figure 5. Schematic diagram showing the potential products resulting from the co-expression of heavy chain and light chain-fc fusion (a) without any mutations in the CH3 domain and (b) with charge pair mutations at the CH3 domain interface.
6 Supplementary Figure 6. Ribbon representation of the modeled heterodimeric CH3 domain structure showing the charge complementarity with (a) D399K mutation in the first chain and K392 D:K409 D mutations in the second chain and (b) D399K:E356K mutations in the first chain and K392 D:K409 D mutations in the second chain. Although both (a) and (b) design promote heterodimer formation, in the case of (a) the homodimer involving D399K mutation is not completely suppressed. The addition of E356K mutation away from the D399K mutation in the CH3 domain interface structural space, as shown in (b), leads to complete suppression of homodimers. However, the design (a) has higher thermal stability compared to the design (b). Supplementary References 1. Pokala, N., and Handel, T. M. (2005) J Mol Biol 347(1), Atwell, S., Ridgway, J. B., Wells, J. A., and Carter, P. (1997) J Mol Biol 270(1), 26-35
7 Computational Analyses Scheme Identify Fc Crystal Structures from PDB and Select Highest Resolution Human IgG1 Crystal Structure Identify Interface Residues through (1) Contact and (2) Solvent Accessibility Analyses Examine Structural Conservation of Side Chain Conformations Identify Buried Interface Residues and Compare with Structurally Conserved Positions Analyze Amino Acid Conservation at the CH3 Domain Interface Through Sequence Comparisons Examine Charge Complementarity at the Interface Through Vdw / Electrostatic Surface and Contact Analysis Binding Free Energy Estimation and Hotspots Analysis Using Available Computational Methods (e.g., EGAD) Supplementary Figure 1
8 S400 V397 D399 P395 F405 K370 Q347 T394 L368 Y349 K392 Y407 L351 T350 K439 K409 T366 S364 K360 S354 E357 R355 K360 D356 Supplementary Figure 2
9 Supplementary Figure 3
10 (a) Initial material after protein A column Non reduced Reduced Non reduced Final material Reduced kda 97.4 kda 66.5kDa 55.4kDa 36.5kDa 31.0kDa 21.5kDa 14.4kDa 6.0kDa kda 97.4 kda 66.5kDa 55.4kDa 36.5kDa 31.0kDa 21.5kDa 14.4kDa 6.0kDa (b) Cp (kcal/mole/ oc) Charge Pair Mutant WT Knob-Hole Temperature ( oc) Supplementary Figure 4
11 (a) Not Secreted + WT Fc WT Fc HC LC-Fc HC/ LC-Fc/ HC/ LC-Fc LC-Fc HC (b) Not Secreted + >> K392D K409D D399K E356K HC LC-Fc HC/ LC-Fc/ HC/ LC-Fc LC-Fc HC Supplementary Figure 5
12 (a) D399K / K409 D:K392 D D399K K409 D K392 D K392 K409 D399 (b) D399K:E356K / K409 D:K392 D E356 D399K K409 D K409 K392 D K392 E356K D399 Supplementary Figure 6
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION A human XRCC4-XLF complex bridges DNA ends. Sara N. Andres 1, Alexandra Vergnes 2, Dejan Ristic 3, Claire Wyman 3, Mauro Modesti 2,4, and Murray Junop 2,4 1 Department of Biochemistry
SUPPLEMENTARY INFORMATION A human XRCC4-XLF complex bridges DNA ends. Sara N. Andres 1, Alexandra Vergnes 2, Dejan Ristic 3, Claire Wyman 3, Mauro Modesti 2,4, and Murray Junop 2,4 1 Department of Biochemistry
11 questions for a total of 120 points
 Your Name: BYS 201, Final Exam, May 3, 2010 11 questions for a total of 120 points 1. 25 points Take a close look at these tables of amino acids. Some of them are hydrophilic, some hydrophobic, some positive
Your Name: BYS 201, Final Exam, May 3, 2010 11 questions for a total of 120 points 1. 25 points Take a close look at these tables of amino acids. Some of them are hydrophilic, some hydrophobic, some positive
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 29 September 2005
 Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 9 September 005 Focus concept Purification of a novel seed storage protein allows sequence analysis and
Case 7 A Storage Protein From Seeds of Brassica nigra is a Serine Protease Inhibitor Last modified 9 September 005 Focus concept Purification of a novel seed storage protein allows sequence analysis and
SUPPLEMENTAL FIGURE LEGENDS. Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
Packing of Secondary Structures
 7.88 Lecture Notes - 5 7.24/7.88J/5.48J The Protein Folding and Human Disease Packing of Secondary Structures Packing of Helices against sheets Packing of sheets against sheets Parallel Orthogonal Table:
7.88 Lecture Notes - 5 7.24/7.88J/5.48J The Protein Folding and Human Disease Packing of Secondary Structures Packing of Helices against sheets Packing of sheets against sheets Parallel Orthogonal Table:
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Structural bioinformatics
 Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
7.013 Problem Set 3 FRIDAY October 8th, 2004
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert. Weinberg, Dr. laudette ardel Name: T: 7.013 Problem Set 3 FRIDY October 8th, 2004 Problem
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert. Weinberg, Dr. laudette ardel Name: T: 7.013 Problem Set 3 FRIDY October 8th, 2004 Problem
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
Nature Structural & Molecular Biology: doi: /nsmb.2548
 Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Fundamentals of Protein Structure
 Outline Fundamentals of Protein Structure Yu (Julie) Chen and Thomas Funkhouser Princeton University CS597A, Fall 2005 Protein structure Primary Secondary Tertiary Quaternary Forces and factors Levels
Outline Fundamentals of Protein Structure Yu (Julie) Chen and Thomas Funkhouser Princeton University CS597A, Fall 2005 Protein structure Primary Secondary Tertiary Quaternary Forces and factors Levels
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
1. DNA replication. (a) Why is DNA replication an essential process?
 ame Section 7.014 Problem Set 3 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68120 by 5:00pm on Friday
ame Section 7.014 Problem Set 3 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68120 by 5:00pm on Friday
DNA.notebook March 08, DNA Overview
 DNA Overview Deoxyribonucleic Acid, or DNA, must be able to do 2 things: 1) give instructions for building and maintaining cells. 2) be copied each time a cell divides. DNA is made of subunits called nucleotides
DNA Overview Deoxyribonucleic Acid, or DNA, must be able to do 2 things: 1) give instructions for building and maintaining cells. 2) be copied each time a cell divides. DNA is made of subunits called nucleotides
Jürgen Fichtl, Development Characterization Pharma Technical Development, Penzberg, Germany Mass Spec 2013, September 23-26, Boston
 Analytical Strategies for Assessment of Disulfide Linkages in Biopharmaceuticals Jürgen Fichtl, Development Characterization Pharma Technical Development, Penzberg, Germany Mass Spec 2013, September 23-26,
Analytical Strategies for Assessment of Disulfide Linkages in Biopharmaceuticals Jürgen Fichtl, Development Characterization Pharma Technical Development, Penzberg, Germany Mass Spec 2013, September 23-26,
Zwitterion Chromatography ZIC
 Zwitterion Chromatography ZIC A novel technique, with unique selectivity, suitable for preparative scale separations? PhD Einar Pontén What is Zwitterion Chromatography? Our definition: Liquid chromatography
Zwitterion Chromatography ZIC A novel technique, with unique selectivity, suitable for preparative scale separations? PhD Einar Pontén What is Zwitterion Chromatography? Our definition: Liquid chromatography
Virtual bond representation
 Today s subjects: Virtual bond representation Coordination number Contact maps Sidechain packing: is it an instrumental way of selecting and consolidating a fold? ASA of proteins Interatomic distances
Today s subjects: Virtual bond representation Coordination number Contact maps Sidechain packing: is it an instrumental way of selecting and consolidating a fold? ASA of proteins Interatomic distances
Supporting Online Material. Av1 and Av2 were isolated and purified under anaerobic conditions according to
 Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Structure formation and association of biomolecules. Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München
 Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Problem Set Unit The base ratios in the DNA and RNA for an onion (Allium cepa) are given below.
 Problem Set Unit 3 Name 1. Which molecule is found in both DNA and RNA? A. Ribose B. Uracil C. Phosphate D. Amino acid 2. Which molecules form the nucleotide marked in the diagram? A. phosphate, deoxyribose
Problem Set Unit 3 Name 1. Which molecule is found in both DNA and RNA? A. Ribose B. Uracil C. Phosphate D. Amino acid 2. Which molecules form the nucleotide marked in the diagram? A. phosphate, deoxyribose
Chapter 8. One-Dimensional Structural Properties of Proteins in the Coarse-Grained CABS Model. Sebastian Kmiecik and Andrzej Kolinski.
 Chapter 8 One-Dimensional Structural Properties of Proteins in the Coarse-Grained CABS Model Abstract Despite the significant increase in computational power, molecular modeling of protein structure using
Chapter 8 One-Dimensional Structural Properties of Proteins in the Coarse-Grained CABS Model Abstract Despite the significant increase in computational power, molecular modeling of protein structure using
MOLEBIO LAB #3: Electrophoretic Separation of Proteins
 MOLEBIO LAB #3: Electrophoretic Separation of Proteins Introduction: Proteins occupy a central position in the structure and function of all living organisms. Some proteins serve as structural components
MOLEBIO LAB #3: Electrophoretic Separation of Proteins Introduction: Proteins occupy a central position in the structure and function of all living organisms. Some proteins serve as structural components
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Introduction to Protein Purification
 Introduction to Protein Purification 1 Day 1) Introduction to Protein Purification. Input for Purification Protocol Development - Guidelines for Protein Purification Day 2) Sample Preparation before Chromatography
Introduction to Protein Purification 1 Day 1) Introduction to Protein Purification. Input for Purification Protocol Development - Guidelines for Protein Purification Day 2) Sample Preparation before Chromatography
Protein 3D Structure Prediction
 Protein 3D Structure Prediction Michael Tress CNIO ?? MREYKLVVLGSGGVGKSALTVQFVQGIFVDE YDPTIEDSYRKQVEVDCQQCMLEILDTAGTE QFTAMRDLYMKNGQGFALVYSITAQSTFNDL QDLREQILRVKDTEDVPMILVGNKCDLEDER VVGKEQGQNLARQWCNCAFLESSAKSKINVN
Protein 3D Structure Prediction Michael Tress CNIO ?? MREYKLVVLGSGGVGKSALTVQFVQGIFVDE YDPTIEDSYRKQVEVDCQQCMLEILDTAGTE QFTAMRDLYMKNGQGFALVYSITAQSTFNDL QDLREQILRVKDTEDVPMILVGNKCDLEDER VVGKEQGQNLARQWCNCAFLESSAKSKINVN
Cristian Micheletti SISSA (Trieste)
 Cristian Micheletti SISSA (Trieste) michelet@sissa.it Mar 2009 5pal - parvalbumin Calcium-binding protein HEADER CALCIUM-BINDING PROTEIN 25-SEP-91 5PAL 5PAL 2 COMPND PARVALBUMIN (ALPHA LINEAGE) 5PAL 3
Cristian Micheletti SISSA (Trieste) michelet@sissa.it Mar 2009 5pal - parvalbumin Calcium-binding protein HEADER CALCIUM-BINDING PROTEIN 25-SEP-91 5PAL 5PAL 2 COMPND PARVALBUMIN (ALPHA LINEAGE) 5PAL 3
Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words).
 1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
Thr Gly Tyr. Gly Lys Asn
 Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Your unique body characteristics (traits), such as hair color or blood type, are determined by the proteins your body produces. Proteins are the building blocks of life - in fact, about 45% of the human
Ligand immobilization using thiol-disulphide exchange
 A P P L I C A T I T E 9 Ligand immobilization using thiol-disulphide exchange Abstract This Application ote describes an immobilization procedure based on thioldisulphide exchange, providing a valuable
A P P L I C A T I T E 9 Ligand immobilization using thiol-disulphide exchange Abstract This Application ote describes an immobilization procedure based on thioldisulphide exchange, providing a valuable
CFSSP: Chou and Fasman Secondary Structure Prediction server
 Wide Spectrum, Vol. 1, No. 9, (2013) pp 15-19 CFSSP: Chou and Fasman Secondary Structure Prediction server T. Ashok Kumar Department of Bioinformatics, Noorul Islam College of Arts and Science, Kumaracoil
Wide Spectrum, Vol. 1, No. 9, (2013) pp 15-19 CFSSP: Chou and Fasman Secondary Structure Prediction server T. Ashok Kumar Department of Bioinformatics, Noorul Islam College of Arts and Science, Kumaracoil
Zool 3200: Cell Biology Exam 3 3/6/15
 Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
7.014 Quiz II 3/18/05. Write your name on this page and your initials on all the other pages in the space provided.
 7.014 Quiz II 3/18/05 Your Name: TA's Name: Write your name on this page and your initials on all the other pages in the space provided. This exam has 10 pages including this coversheet. heck that you
7.014 Quiz II 3/18/05 Your Name: TA's Name: Write your name on this page and your initials on all the other pages in the space provided. This exam has 10 pages including this coversheet. heck that you
Disease and selection in the human genome 3
 Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Biomolecules: lecture 6
 Biomolecules: lecture 6 - to learn the basics on how DNA serves to make RNA = transcription - to learn how the genetic code instructs protein synthesis - to learn the basics on how proteins are synthesized
Biomolecules: lecture 6 - to learn the basics on how DNA serves to make RNA = transcription - to learn how the genetic code instructs protein synthesis - to learn the basics on how proteins are synthesized
has only one nucleotide, U20, between G19 and A21, while trna Glu CUC has two
 SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
MIT Department of Biology 7.28, Spring Molecular Biology
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology Question 1. You are interested in understanding the residues of a specific group I intron that function specifically in catalysis of the
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology Question 1. You are interested in understanding the residues of a specific group I intron that function specifically in catalysis of the
Dynamic Programming Algorithms
 Dynamic Programming Algorithms Sequence alignments, scores, and significance Lucy Skrabanek ICB, WMC February 7, 212 Sequence alignment Compare two (or more) sequences to: Find regions of conservation
Dynamic Programming Algorithms Sequence alignments, scores, and significance Lucy Skrabanek ICB, WMC February 7, 212 Sequence alignment Compare two (or more) sequences to: Find regions of conservation
Rajan Sankaranarayanan
 Proofreading during translation of the genetic code Rajan Sankaranarayanan Centre for Cellular and Molecular Biology (CCMB) Council for Scientific and Industrial Research (CSIR) Hyderabad, INDIA Accuracy
Proofreading during translation of the genetic code Rajan Sankaranarayanan Centre for Cellular and Molecular Biology (CCMB) Council for Scientific and Industrial Research (CSIR) Hyderabad, INDIA Accuracy
 Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Linkage preference of USP30 and USP domain architecture.
 Supplementary Figure 1 Linkage preference of USP30 and USP domain architecture. a, Catalytic efficiencies determined from Coomassie-stained gel-based kinetics for all eight diub linkages (see Supplementary
Supplementary Figure 1 Linkage preference of USP30 and USP domain architecture. a, Catalytic efficiencies determined from Coomassie-stained gel-based kinetics for all eight diub linkages (see Supplementary
Homology Modelling. Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen
 Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Introduction to Proteins
 Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Protein Synthesis. Application Based Questions
 Protein Synthesis Application Based Questions MRNA Triplet Codons Note: Logic behind the single letter abbreviations can be found at: http://www.biology.arizona.edu/biochemistry/problem_sets/aa/dayhoff.html
Protein Synthesis Application Based Questions MRNA Triplet Codons Note: Logic behind the single letter abbreviations can be found at: http://www.biology.arizona.edu/biochemistry/problem_sets/aa/dayhoff.html
Purification: Step 1. Lecture 11 Protein and Peptide Chemistry. Cells: Break them open! Crude Extract
 Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1. Protein and Peptide Chemistry. Lecture 11. Big Problem: Crude extract is not the natural environment. Cells: Break them open!
 Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L-
 36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
36. The double bonds in naturally-occuring fatty acids are usually isomers. A. cis B. trans C. both cis and trans D. D- E. L- 37. The essential fatty acids are A. palmitic acid B. linoleic acid C. linolenic
1. DNA, RNA structure. 2. DNA replication. 3. Transcription, translation
 1. DNA, RNA structure 2. DNA replication 3. Transcription, translation DNA and RNA are polymers of nucleotides DNA is a nucleic acid, made of long chains of nucleotides Nucleotide Phosphate group Nitrogenous
1. DNA, RNA structure 2. DNA replication 3. Transcription, translation DNA and RNA are polymers of nucleotides DNA is a nucleic acid, made of long chains of nucleotides Nucleotide Phosphate group Nitrogenous
Green Fluorescent Protein. Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani
 Green Fluorescent Protein Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani Introduction The Green fluorescent protein (GFP) was first isolated from the Jellyfish Aequorea victoria,
Green Fluorescent Protein Avinash Bayya Varun Maturi Nikhileswar Reddy Mukkamala Aravindh Subhramani Introduction The Green fluorescent protein (GFP) was first isolated from the Jellyfish Aequorea victoria,
Residue Contact Prediction for Protein Structure using 2-Norm Distances
 Residue Contact Prediction for Protein Structure using 2-Norm Distances Nikita V Mahajan Department of Computer Science &Engg GH Raisoni College of Engineering, Nagpur LGMalik Department of Computer Science
Residue Contact Prediction for Protein Structure using 2-Norm Distances Nikita V Mahajan Department of Computer Science &Engg GH Raisoni College of Engineering, Nagpur LGMalik Department of Computer Science
MATF Antigen Submission Details and Standard Project Deliverables
 MATF Antigen Submission Details and Standard Project Deliverables What we require when you submit your antigen: Proteins For a recombinant protein target, we require a minimum of 400µg soluble recombinant
MATF Antigen Submission Details and Standard Project Deliverables What we require when you submit your antigen: Proteins For a recombinant protein target, we require a minimum of 400µg soluble recombinant
Purification Kits. Fast and Convenient PROSEP -A and PROSEP-G Spin Column Kits for Antibody Purification DATA SHEET
 Â Montage Antibody Purification Kits Fast and Convenient PROSEP -A and PROSEP-G Spin Column Kits for Antibody Purification DATA SHEET Available with immobilized Protein A or Protein G Easy-to-use Antibody
 Montage Antibody Purification Kits Fast and Convenient PROSEP -A and PROSEP-G Spin Column Kits for Antibody Purification DATA SHEET Available with immobilized Protein A or Protein G Easy-to-use Antibody
Denis V. Kurek, Sergey A. Lopatin, *Vladimir E. Tikhonov, Valery P. Varlamov
 NEW AFFINITY SORBENTS FOR PURIFICATION OF RECOMBINANT PROTEINS WITH THE USE OF CHITIN-BINDING DOMAIN AS AN AFFINITY TAG Denis V. Kurek, Sergey A. Lopatin, *Vladimir E. Tikhonov, Valery P. Varlamov Centre
NEW AFFINITY SORBENTS FOR PURIFICATION OF RECOMBINANT PROTEINS WITH THE USE OF CHITIN-BINDING DOMAIN AS AN AFFINITY TAG Denis V. Kurek, Sergey A. Lopatin, *Vladimir E. Tikhonov, Valery P. Varlamov Centre
ARBRE-P4EU Consensus Protein Quality Guidelines for Biophysical and Biochemical Studies Minimal information to provide
 ARBRE-P4EU Consensus Protein Quality Guidelines for Biophysical and Biochemical Studies Minimal information to provide Protein name and full primary structure, by providing a NCBI (or UniProt) accession
ARBRE-P4EU Consensus Protein Quality Guidelines for Biophysical and Biochemical Studies Minimal information to provide Protein name and full primary structure, by providing a NCBI (or UniProt) accession
Examining the components of your peptide sample with AccuPep QC. Lauren Lu, Ph.D. October 29, 2015, 9:00-10:00 AM EST
 Examining the components of your peptide sample with AccuPep QC Lauren Lu, Ph.D. October 29, 2015, 9:00-10:00 AM EST When do I need custom peptides? Custom peptides play an important role in many research
Examining the components of your peptide sample with AccuPep QC Lauren Lu, Ph.D. October 29, 2015, 9:00-10:00 AM EST When do I need custom peptides? Custom peptides play an important role in many research
Homology Modelling. Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen
 Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Protein Structure and Function! Lecture 4: ph, pka and pi!
 Protein Structure and Function! Lecture 4: ph, pka and pi! Definition of ph and pk a! ph is a measure of the concentration of H +.! + ph = log 10[H ] For a weak acid,! HA #!!"! H + + A!, K a = [H + ][A!
Protein Structure and Function! Lecture 4: ph, pka and pi! Definition of ph and pk a! ph is a measure of the concentration of H +.! + ph = log 10[H ] For a weak acid,! HA #!!"! H + + A!, K a = [H + ][A!
Supplementary Figures
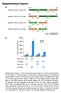 Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
Supplementary Figures a HindIII XbaI pcdna3.1/v5-his A_Nfluc-VVD 1-398 37-186 Nfluc Linker 1 VVD HindIII XhoI pcdna3.1/v5-his A_VVD-Nfluc 37-186 1-398 VVD Linker 2 Nfluc HindIII XbaI pcdna3.1/v5-his A_Cfluc-VVD
42 fl organelles = 34.5 fl (1) 3.5X X 0.93 = 78,000 (2)
 SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
SUPPLEMENTAL DATA Supplementary Experimental Procedures Fluorescence Microscopy - A Zeiss Axiovert 200M microscope equipped with a Zeiss 100x Plan- Apochromat (1.40 NA) DIC objective and Hamamatsu Orca
Site-directed Mutagenesis
 Site-directed Mutagenesis Applications Subtilisin (Met à Ala mutation resistant to oxidation) Fluorescent proteins Protein structure-function Substrate trapping mutants Identify regulatory regions/sequences
Site-directed Mutagenesis Applications Subtilisin (Met à Ala mutation resistant to oxidation) Fluorescent proteins Protein structure-function Substrate trapping mutants Identify regulatory regions/sequences
Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2
 Structural evidence for consecutive -like modules in the spliceosomal ATPase Brr2 Lingdi Zhang, Tao Xu, Corina Maeder, Laura-Oana Bud, James Shanks, Jay Nix, Christine Guthrie, Jeffrey A. Pleiss, Rui Zhao
Structural evidence for consecutive -like modules in the spliceosomal ATPase Brr2 Lingdi Zhang, Tao Xu, Corina Maeder, Laura-Oana Bud, James Shanks, Jay Nix, Christine Guthrie, Jeffrey A. Pleiss, Rui Zhao
Bivalirudin Purification:
 Bivalirudin Purification: Sorbent Screening and Overload Experiments Marc Jacob, Joshua Heng, and Tivadar Farkas Phenomenex, Inc., 411 Madrid Ave., Torrance, CA 90501 USA PO94190412_W Abstract In this
Bivalirudin Purification: Sorbent Screening and Overload Experiments Marc Jacob, Joshua Heng, and Tivadar Farkas Phenomenex, Inc., 411 Madrid Ave., Torrance, CA 90501 USA PO94190412_W Abstract In this
 www.lessonplansinc.com Topic: Gene Mutations WS Summary: Students will learn about frame shift mutations and base substitution mutations. Goals & Objectives: Students will be able to demonstrate how mutations
www.lessonplansinc.com Topic: Gene Mutations WS Summary: Students will learn about frame shift mutations and base substitution mutations. Goals & Objectives: Students will be able to demonstrate how mutations
The study of protein secondary structure and stability at equilibrium ABSTRACT
 The study of protein secondary structure and stability at equilibrium Michelle Planicka Dept. of Physics, North Georgia College and State University, Dahlonega, GA REU, Dept. of Physics, University of
The study of protein secondary structure and stability at equilibrium Michelle Planicka Dept. of Physics, North Georgia College and State University, Dahlonega, GA REU, Dept. of Physics, University of
Purification of antibody fragments and single domain antibodies with Amsphere A3 Protein A resin
 Purification of antibody fragments and single domain antibodies with Amsphere A3 Protein A resin Platteau, Gerald 1 *; Ströhlein, Guido 1 ; Gaspariunaite, Vaiva 2 ; Vincke, Cécile 2 ; Sterckx, Yann 2 ;
Purification of antibody fragments and single domain antibodies with Amsphere A3 Protein A resin Platteau, Gerald 1 *; Ströhlein, Guido 1 ; Gaspariunaite, Vaiva 2 ; Vincke, Cécile 2 ; Sterckx, Yann 2 ;
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Daily Agenda. Warm Up: Review. Translation Notes Protein Synthesis Practice. Redos
 Daily Agenda Warm Up: Review Translation Notes Protein Synthesis Practice Redos 1. What is DNA Replication? 2. Where does DNA Replication take place? 3. Replicate this strand of DNA into complimentary
Daily Agenda Warm Up: Review Translation Notes Protein Synthesis Practice Redos 1. What is DNA Replication? 2. Where does DNA Replication take place? 3. Replicate this strand of DNA into complimentary
1. Introduction. 2. Materials and Method
 American Journal of Pharmacological Sciences, 2016, Vol. 4, No. 1, 1-6 Available online at http://pubs.sciepub.com/ajps/4/1/1 Science and Education Publishing DOI:10.12691/ajps-4-1-1 In-silico Designing
American Journal of Pharmacological Sciences, 2016, Vol. 4, No. 1, 1-6 Available online at http://pubs.sciepub.com/ajps/4/1/1 Science and Education Publishing DOI:10.12691/ajps-4-1-1 In-silico Designing
Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
 SUPPLEMENTARY INFORMATION Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs Malka Kitayner 1, 3, Haim Rozenberg 1, 3, Remo Rohs 2, 3, Oded Suad 1, Dov Rabinovich
SUPPLEMENTARY INFORMATION Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs Malka Kitayner 1, 3, Haim Rozenberg 1, 3, Remo Rohs 2, 3, Oded Suad 1, Dov Rabinovich
In Silico Analysis Shows That Single Aminoacid Variations In Rhesus Macacque Fcγreceptor Affect Protein Stability And Binding Affinity To IgG1
 Georgia State University ScholarWorks @ Georgia State University Biology Theses Department of Biology 4-24-2013 In Silico Analysis Shows That Single Aminoacid Variations In Rhesus Macacque Fcγreceptor
Georgia State University ScholarWorks @ Georgia State University Biology Theses Department of Biology 4-24-2013 In Silico Analysis Shows That Single Aminoacid Variations In Rhesus Macacque Fcγreceptor
Gene Expression Translation U C A G A G
 Why? ene Expression Translation How do cells synthesize polypeptides and convert them to functional proteins? The message in your DN of who you are and how your body works is carried out by cells through
Why? ene Expression Translation How do cells synthesize polypeptides and convert them to functional proteins? The message in your DN of who you are and how your body works is carried out by cells through
Folding simulation: self-organization of 4-helix bundle protein. yellow = helical turns
 Folding simulation: self-organization of 4-helix bundle protein yellow = helical turns Protein structure Protein: heteropolymer chain made of amino acid residues R + H 3 N - C - COO - H φ ψ Chain of amino
Folding simulation: self-organization of 4-helix bundle protein yellow = helical turns Protein structure Protein: heteropolymer chain made of amino acid residues R + H 3 N - C - COO - H φ ψ Chain of amino
The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR
 The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR S. Thomas, C. S. Fernando, J. Roach, U. DeSilva and C. A. Mireles DeWitt The objective
The Expression of Recombinant Sheep Prion Protein (RecShPrPC) and its Detection Using Western Blot and Immuno-PCR S. Thomas, C. S. Fernando, J. Roach, U. DeSilva and C. A. Mireles DeWitt The objective
Promix TM. Enter a New Era in Biomolecule Analysis with. Columns. Unsurpassed Selectivity and Peak Capacity for Peptides and Proteins
 Promix TM Enter a New Era in Biomolecule Analysis with Promix TM Columns Unsurpassed electivity and Peak Capacity for Peptides and Proteins Applications: Proteomics Peptide/Protein Analysis Peptide/Protein
Promix TM Enter a New Era in Biomolecule Analysis with Promix TM Columns Unsurpassed electivity and Peak Capacity for Peptides and Proteins Applications: Proteomics Peptide/Protein Analysis Peptide/Protein
GFP CCD2 GFP IP:GFP
 D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
D1 D2 1 75 95 148 178 492 GFP CCD1 CCD2 CCD2 GFP D1 D2 GFP D1 D2 Beclin 1 IB:GFP IP:GFP Supplementary Figure 1: Mapping domains required for binding to HEK293T cells are transfected with EGFP-tagged mutant
Lecture 11: Gene Prediction
 Lecture 11: Gene Prediction Study Chapter 6.11-6.14 1 Gene: A sequence of nucleotides coding for protein Gene Prediction Problem: Determine the beginning and end positions of genes in a genome Where are
Lecture 11: Gene Prediction Study Chapter 6.11-6.14 1 Gene: A sequence of nucleotides coding for protein Gene Prediction Problem: Determine the beginning and end positions of genes in a genome Where are
Aims: -Purification of a specific protein. -Study of protein-protein interactions
 Aims: -Purification of a specific protein -Study of protein-protein interactions This is a reliable method for purifying total IgG from crude protein mixtures such as serum. Protein A (linked to resin
Aims: -Purification of a specific protein -Study of protein-protein interactions This is a reliable method for purifying total IgG from crude protein mixtures such as serum. Protein A (linked to resin
DATA. mrna CODON CHART
 Alien Encounters! Background: In the year 2050, an incredible archeological discovery was made in the middle of a remote area of South America. It was an area where a large meteor had struck Earth. A pile
Alien Encounters! Background: In the year 2050, an incredible archeological discovery was made in the middle of a remote area of South America. It was an area where a large meteor had struck Earth. A pile
Hmwk # 8 : DNA-Binding Proteins : Part II
 The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
Worksheet: Mutations Practice
 Worksheet: Mutations Practice There are three ways that DNA can be altered when a mutation (change in DNA sequence) occurs. 1. Substitution one base-pairs is replaced by another: Example: G to C or A to
Worksheet: Mutations Practice There are three ways that DNA can be altered when a mutation (change in DNA sequence) occurs. 1. Substitution one base-pairs is replaced by another: Example: G to C or A to
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
In Vitro Monitoring of the Formation of Pentamers from the Monomer of GST Fused HPV 16 L1
 This journal is The Royal Society of Chemistry 213 In Vitro Monitoring of the Formation of Pentamers from the Monomer of GST Fused HPV 16 L1 Dong-Dong Zheng, a Dong Pan, a Xiao Zha, ac Yuqing Wu,* a Chunlai
This journal is The Royal Society of Chemistry 213 In Vitro Monitoring of the Formation of Pentamers from the Monomer of GST Fused HPV 16 L1 Dong-Dong Zheng, a Dong Pan, a Xiao Zha, ac Yuqing Wu,* a Chunlai
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Application Note USD Purification of Mouse IgM from Cell Culture Supernatant by Cation Exchange Chromatography on CM Ceramic HyperD F Sorbent
 Application Note USD 241 Purification of Mouse IgM from Cell Culture Supernatant by Cation Exchange Chromatography on CM Ceramic HyperD F Sorbent What this Study Demonstrates T h i s s t u d y o n C a
Application Note USD 241 Purification of Mouse IgM from Cell Culture Supernatant by Cation Exchange Chromatography on CM Ceramic HyperD F Sorbent What this Study Demonstrates T h i s s t u d y o n C a
1. Bloomsbury BBSRC Centre for Structural Biology, Birkbeck College and University College London.
 Purification/Polishing of His-tagged proteins - Application of Centrifugal Vivapure Ion-exchange Membrane Devices to the Purification/Polishing of Histagged Background Multi-milligram quantities of highly
Purification/Polishing of His-tagged proteins - Application of Centrifugal Vivapure Ion-exchange Membrane Devices to the Purification/Polishing of Histagged Background Multi-milligram quantities of highly
Supplemental Information. The structural basis of R Spondin recognition by LGR5 and RNF43
 Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
High-resolution structure of a papaya plant-defence barwin-like protein solved by in-house sulphur-sad phasing
 High-resolution structure of a papaya plant-defence barwin-like protein solved by in-house sulphur-sad phasing Joëlle Huet, Emmanuel Jean Teinkela Mbosso, Sameh Soror, Franck Meyer, Yvan Looze, René Wintjens,
High-resolution structure of a papaya plant-defence barwin-like protein solved by in-house sulphur-sad phasing Joëlle Huet, Emmanuel Jean Teinkela Mbosso, Sameh Soror, Franck Meyer, Yvan Looze, René Wintjens,
7.88J Protein Folding Problem Fall 2007
 MIT OpenCourseWare http://ocw.mit.edu 7.88J Protein Folding Problem Fall 2007 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. 7.88 Lecture Notes - 8 7.24/7.88J/5.48J
MIT OpenCourseWare http://ocw.mit.edu 7.88J Protein Folding Problem Fall 2007 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. 7.88 Lecture Notes - 8 7.24/7.88J/5.48J
Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D
 Supplementary Information for Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D Federico Forneris a,b, B. Tom Burnley a,b,c and Piet Gros a * a Crystal
Supplementary Information for Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D Federico Forneris a,b, B. Tom Burnley a,b,c and Piet Gros a * a Crystal
Sasidhar N. Nirudodhi, Lin Tzihsuan, Justin Sperry, Jason C. Rouse, James A. Carroll
 Impact of Sequence Variants on the Higher-Order Structure (HOS) of Monoclonal Antibodies Using an Optimized HDX- MS Method with a Dual-Protease Co-Immobilized Column Format Sasidhar N. Nirudodhi, Lin Tzihsuan,
Impact of Sequence Variants on the Higher-Order Structure (HOS) of Monoclonal Antibodies Using an Optimized HDX- MS Method with a Dual-Protease Co-Immobilized Column Format Sasidhar N. Nirudodhi, Lin Tzihsuan,
Algorithm for Matching Additional Spectra
 Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
NPTEL VIDEO COURSE PROTEOMICS PROF. SANJEEVA SRIVASTAVA
 LECTURE-06 PROTEIN PURIFICATION AND PEPTIDE ISOLATION USING CHROMATOGRAPHY TRANSCRIPT Welcome to the proteomics course. Today, we will talk about protein purification and peptide isolation using chromatography
LECTURE-06 PROTEIN PURIFICATION AND PEPTIDE ISOLATION USING CHROMATOGRAPHY TRANSCRIPT Welcome to the proteomics course. Today, we will talk about protein purification and peptide isolation using chromatography
Protein Purification Products. Complete Solutions for All of Your Protein Purification Applications
 Protein Purification Products Complete Solutions for All of Your Protein Purification Applications FLAG-Tagged Protein Products EXPRESS with the pcmv-dykddddk Vector Set Fuse your protein of interest to
Protein Purification Products Complete Solutions for All of Your Protein Purification Applications FLAG-Tagged Protein Products EXPRESS with the pcmv-dykddddk Vector Set Fuse your protein of interest to
Supplemental Material for Xue et al. List. Supplemental Figure legends. Figure S1. Related to Figure 1. Figure S2. Related to Figure 3
 Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
Examination of Protein G stability and Binding Characteristics Using Four Body Nearest Neighbor Contact Potentials
 Examination of Protein G stability and Binding Characteristics Using Four Body Nearest Neighbor Contact Potentials Abstract Gregory M. Reck George Mason University A computational geometry technique employing
Examination of Protein G stability and Binding Characteristics Using Four Body Nearest Neighbor Contact Potentials Abstract Gregory M. Reck George Mason University A computational geometry technique employing
Protein Structure Prediction by Constraint Logic Programming
 MPRI C2-19 Protein Structure Prediction by Constraint Logic Programming François Fages, Constraint Programming Group, INRIA Rocquencourt mailto:francois.fages@inria.fr http://contraintes.inria.fr/ Molecules
MPRI C2-19 Protein Structure Prediction by Constraint Logic Programming François Fages, Constraint Programming Group, INRIA Rocquencourt mailto:francois.fages@inria.fr http://contraintes.inria.fr/ Molecules
Types of chromatography
 Chromatography Physical separation method based on the differential migration of analytes in a mobile phase as they move along a stationary phase. Mechanisms of Separation: Partitioning Adsorption Exclusion
Chromatography Physical separation method based on the differential migration of analytes in a mobile phase as they move along a stationary phase. Mechanisms of Separation: Partitioning Adsorption Exclusion
ASPP1 Fw GGTTGGGAATCCACGTGTTG ASPP1 Rv GCCATATCTTGGAGCTCTGAGAG
 Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Materials and Methods Plasmids: the following plasmids were used in the supplementary data: pwzl-myc- Lats2 (Aylon et al, 2006), pretrosuper-vector and pretrosuper-shp53 (generous gift of
Supplemental Information for:
 Supplemental Information for: Antibody-induced dimerization of FGFR1 promotes receptor endocytosis independently of its kinase activity Łukasz Opaliński*, Aleksandra Sokołowska-Wędzina, Martyna Szczepara,
Supplemental Information for: Antibody-induced dimerization of FGFR1 promotes receptor endocytosis independently of its kinase activity Łukasz Opaliński*, Aleksandra Sokołowska-Wędzina, Martyna Szczepara,
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
