Supporting information
|
|
|
- Colin Watson
- 5 years ago
- Views:
Transcription
1 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc f (gcatgggcaagcttgaggacggcgtcatgt) and cc f (gaggccgtggtaccatagaggcgggcg), that includes 512 bp of cc2570 coding sequence and 162 bp upstream of its ATG codon, was cut with HindIII-KpnI and inserted between the same sites of pbgent (Gent R ). Conjugation of strain PC7167 (CB15 diva305) with strain CJW2130 (E. coli S17-1 carrying ptc67) resulted in the insertion of ptc67 at the cc2570 locus of PC7167, which is about 10-kb downstream the ftsi gene (previously called diva), maintaining a full copy of the coding sequence of cc2570 and yielding strain CJW2142. A similar strain construction was obtained by insertion of ptc67 into wild-type CB15N by conjugation, creating CJW2138. CJW2138 cells were indistinguishable from wild-type cells with respect to cell morphology and growth rate, verifying that insertion of ptc67 does not interfere with cell division. To move diva305 (ftsi(ts)) mutation into a synchronizable cell background, CB15N was transduced with a ΦCR30 phage lysate prepared from strain CJW2142, and transductants were selected for gentamycin resistance, creating strain CJW2141. The presence of diva305 (ftsi(ts)) mutation in CJW2141 was confirmed by DNA sequencing. Strain CJW2577 expressing ftsz-yfp under the control of Pvan was obtained by transduction of CJW2141 with a ΦCR30 phage lysate prepared from strain CJW1438. To construct ptc34-3, the ftsi coding region (1770-bp) was PCR amplified with primers egfp-pbp3d (ccgctcaagcttcgatgagcctctcgaacctgggtccc) and egfp-pbp3r (gttgaacaggtaccacagtcttttggtcatagg), cleaved with HindIII and KpnI, and cloned into pxgfp4c1 (Kan R ) using the same sites. ptc34-3 was then inserted into the xylx locus of CB15N by 1
2 transformation to create strain CJW1822 carrying a gfp-ftsi fusion under the control of the xylose-inducible promoter (Pxyl) as a second ftsi copy on the chromosome. To obtain ptc49, a DNA fragment containing the entire ftsi coding region was first obtained by PCR with primers egfp-pbp3d and egfp-pbp3r (see above) and cloned into the TOPO pcr2.1 vector (Invitrogen). This plasmid was used as template to create a S296A mutation by site-directed mutagenesis with primers pbp3-s296af (gtctacgagatgggcgcgaccttcaaggccttc) and pbp3-s296ar (gaaggccttgaaggtcgcgcccatctcgtagac), digested with HindIII and KpnI, and the ftsi S296A fragment cloned between the same sites of pxgfp4c1. ptc49 was then inserted into the xylx locus of CB15N by transformation to generate strain CJW2136 carrying a gfp-ftsi S296A fusion under the control of Pxyl. Strains CJW1823 and CJW2137 were obtained by transduction of strains CJW1822 and CJW2136, respectively, with a ΦCR30 phage lysate prepared from strain CJW1550 to insert the ftsz-mcherry fusion under the control of Pvan at the vana chromosomal locus. Strains CJW2235 and CJW2236 were obtained by mating CB15N carrying the ftsi(ts) mutation (CJW2141) with E. coli strains CJW1821 (Pxyl-gfp-ftsI) and CJW2127 (Pxyl-gfp-ftsI S296A ), respectively. To create ptc71, the cc2561 coding region, corresponding to the region upstream the ftsi gene locus, was PCR amplified with primers cc f (ctggactagtgggccggcaaggcacccg) and cc r (gaggctcatatgaacgccccctggac), digested with SpeI and NdeI, and a gfp-ftsi DNA fragment was released from ptc34-3 by digestion with NdeI and PstI. A triple ligation of the two fragments was then performed into pnpts138 digested with SpeI and PstI. ptc71 was inserted into CB15N by transformation and selection for Kan R. Transformants were then incubated in PYE medium without antibiotics and streaked on PYE plates containing 3% sucrose in order to select for those in which the pnpts138 vector has been excised, maintaining a gfp- 2
3 ftsi fusion under control of the native ftsi promoter, yielding strain CJW2144. Strain CJW2144 was confirmed by PCR and DNA sequencing. To create strain CJW2143, a ΦCR30 phage lysate prepared from strain YB1585 was used to replace native ftsz with ftsz under the control of Pxyl by transduction. A gfp-ftsi 1-90 fragment was PCR amplified from plasmid ptc34-3 using primers gfp+1f (ggcatatggtgagcaagggcgaggagc) and cc r (ccgggatcctcaatcgccccgcgcgccttcgac) and cloned into pxgfp4c1 using Nde1 and Kpn1 restriction sites. The plasmid prp3 was then inserted into the xylx locus of CB15N by transformation to create strain CJW2851 (CB15N Pxyl::Pxyl-gfp-ftsI 1-90 ) carrying the gfp-ftsi 1-90 fusion under the control of Pxyl. A ΦCR30 phage lysate was prepared from strain CJW2851 and used to introduce gfp-ftsi 1-90 under the control of Pxyl by transduction into CJW2141 (CB15N ftsi(ts)) to obtain strain CJW2852 (CB15N ftsi(ts) Pxyl::Pxyl-gfp-ftsI 1-90 ). Legends of supporting figures Fig. S1. Immunoblot analysis of GFP-PBP3 or GFP-PBP3 S296A in different backgrounds and conditions. Various strains (as indicated) were grown to log phase at 30 C. When indicated, the cultures contained xylose or glucose and were incubated at 37 C for 1.5 h. Whole cell extracts of log-phase cultures were prepared by sonicating cells producing GFP-PBP3 or GFP-PBP3 S296A under various conditions. Thirty micrograms of these extracts were analyzed by immunoblotting using anti-gfp antibody. The asterisk shows a degradation product that may result from the known C-terminal proteolytic processing of PBP3 (Hara et al., 1989; Nagasawa et al., 1989; Nakamura et al., 1983). The arrowheads mark the position of GFP-PBP3 and free GFP 3
4 (produced from the CJW2144 control strain) as indicated. Molecular weight markers (MW) are also shown. Fig. S2. The dynamic behavior of PBP3 also occurs under conditions of endogenous production and in the absence of de novo protein synthesis. The FRAP experiments were performed as described as in Fig. 5 except that only the fluorescence intensities of a bleached region (closed diamonds) is shown over time. The solid red line is the predicted recovery curve determined by least-square fitting of the data as described in the Experimental Procedures (r ). A. Polar GFP-PBP3 signals of CJW1822 cells (CB15N Pxyl::Pxyl::gfp-ftsI) grown overnight in the presence of xylose to accumulate GFP-PBP3 were photobleached in time-lapse sequences. B. Same as (A) except that CJW1822 cells (CB15N Pxyl::Pxyl::gfp-ftsI) grown in the presence of xylose were washed in medium containing glucose and incubated in the same glucose medium for 30 min to stop synthesis of GFP-PBP3 from Pxyl. Cells were then incubated with 20 µg/ml chloramphenicol (CAM) for 20 min to block protein synthesis before being placed on an agarose-padded slide that contained glucose and CAM. Whole cell intensity did not increase and remained constant for as long as 60 min under these conditions (data not shown), verifying that GFP-PBP3 synthesis was effectively blocked. C. Same as (A) except that GFP-PBP3 was produced at the native level as the only source of PBP3 from CJW2144 cells (CB15N ftsi::gfp-ftsi). D. Same as (B) except that the medial GFP-PBP3 signals from predivisional CJW1822 cells were photobleached. E. Same as (C) except that the medial GFP-PBP3 signals from predivisional CJW2144 cells, which produce GFP-PBP3 at endogenous levels, were photobleached. 4
5 Fig. S3. Cephalexin binds to several PBPs and fosfomycin affects PBP3 localization. A. The specificity of cephalexin binding to PBP3 was tested using a BOCILLIN-FL competition binding assay. The membrane-enriched fractions of CB15N Δbla cells (lacking the β-lactamaseencoding gene) were incubated in a buffer containing increasing amounts of cephalexin or an excess of Penicillin G (+). They were then incubated with 10µM of the fluorescent penicillin derivative, BOCILLIN-FL. Lane 1 and 2, samples incubated with 0 (-) or 10 mm (+) Penicillin G, respectively. Lane 3 to 7, samples incubated with 0, 0.001, 0.01, 0.1 and 1 mm cephalexin, respectively. Fluorescent bands that disappeared in the presence of a large excess of penicillin identify PBPs whereas bands that remain present correspond to unspecific binding of the BOCILLIN-FL (indicated by the bracket on the right). Incubation with 0.01 mm or higher of cephalexin (lanes 5-7) results in the concomitant disappearance of several specific PBP fluorescent bands, suggesting that cephalexin is not specific for a particular PBP (such as PBP3). Molecular weight markers (MW) are shown. B. Quantitative analysis of GFP-PBP3 and FtsZ-mCherry localization upon treatment with fosfomycin. At least 114 cells from 3 separate microscopy experiments were analyzed. Polar GFP-PBP3 localization pattern was analyzed in cells showing no visible constriction by DIC microscopy, while medial GFP-PBP3 localization was considered in terms of colocalization with FtsZ-mCherry in cells showing clear constriction. Movie legend Movie S1. Time-lapse microscopy of E. coli cells producing a PBP3 N361S mutant. E. coli K-12 cells (LMC502 leu + pbpb r1 ) producing PBP3 N361S (Taschner et al., 1988) were placed on LB- 5
6 containing agarose-padded slide and imaged every 5 min during the course of several cell cycles at 37ºC. Representative images of phase contrast are shown. Division produces pointed poles. Scale bar, 1µm. 6
7 7
8 8
9 9
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of
 Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Supplemental Information: Table S1, Plasmid and Strain Construction, Supplemental Figures, and Movie Legends
 Supplemental Information: Table S1, Plasmid and Strain Construction, Supplemental Figures, and Movie Legends Supplemental Table Table S1: Strains and plasmids Na me Genotype/description Reference/source
Supplemental Information: Table S1, Plasmid and Strain Construction, Supplemental Figures, and Movie Legends Supplemental Table Table S1: Strains and plasmids Na me Genotype/description Reference/source
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Laloux and Jacobs-Wagner, http://www.jcb.org/cgi/content/full/jcb.201303036/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Supporting data on the PopZ variants. Related
Supplemental material Laloux and Jacobs-Wagner, http://www.jcb.org/cgi/content/full/jcb.201303036/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Supporting data on the PopZ variants. Related
Supporting Information
 Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Supporting Information Development of a 2,4-Dinitrotoluene-Responsive Synthetic Riboswitch in E. coli cells Molly E. Davidson, Svetlana V. Harbaugh, Yaroslav G. Chushak, Morley O. Stone, Nancy Kelley-
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
The fusion used in most of these studies (MinC4-GFP) contains GFP and
 Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Gregory, Becker and Pogliano, Supplemental Materials page 1 Supplemental Methods Description of the MinC and MinD GFP insertions The fusion used in most of these studies (MinC4-GFP) contains GFP and flanking
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Chapter 10 (Part II) Gene Isolation and Manipulation
 Biology 234 J. G. Doheny Chapter 10 (Part II) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. What does PCR stand for? 2. What does the
Biology 234 J. G. Doheny Chapter 10 (Part II) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. What does PCR stand for? 2. What does the
Table S1. List of DNA constructs and primers, part 1 Construct
 SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
SUPPLEMENTARY TABLES: Table S1. List of DNA constructs and primers, part 1 Construct Comment (Expressed protein) Vector, PCR primers and template or source of Restriction Sites name Resistance construct
YG1 Control. YG1 PstI XbaI. YG5 PstI XbaI Water Buffer DNA Enzyme1 Enzyme2
 9/9/03 Aim: Digestion and gel extraction of YG, YG3, YG5 and 8/C. Strain: E. coli DH5α Plasmid: Bba_J600, psbc3 4,, 3 6 7, 8 9 0, 3, 4 8/C 8/C SpeI PstI 3 3 x3 YG YG PstI XbaI.5 3.5 x YG3 YG3 PstI XbaI
9/9/03 Aim: Digestion and gel extraction of YG, YG3, YG5 and 8/C. Strain: E. coli DH5α Plasmid: Bba_J600, psbc3 4,, 3 6 7, 8 9 0, 3, 4 8/C 8/C SpeI PstI 3 3 x3 YG YG PstI XbaI.5 3.5 x YG3 YG3 PstI XbaI
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis
 FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G. Bernhardt, David
FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis Jeffrey Meisner, Paula Montero Llopis, Lok-To Sham, Ethan Garner, Thomas G. Bernhardt, David
Figure legends for supplement
 Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Figure legends for supplement Supplemental Figure 1 Characterization of purified and recombinant proteins Relevant fractions related the final stage of the purification protocol(bingham et al., 1998; Toba
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1.
 Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
Supplemental figures Supplemental Figure 1: Fluorescence recovery for FRAP experiments depicted in Figure 1. Percent of original fluorescence was plotted as a function of time following photobleaching
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I) We have made considerable progress in our analysis of the gene for
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I) We have made considerable progress in our analysis of the gene for
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive
 Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive to ionizing radiation. However, why these mutants are
Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive to ionizing radiation. However, why these mutants are
Supplemental Information
 Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplemental Information ATP-dependent unwinding of U4/U6 snrnas by the Brr2 helicase requires the C-terminus of Prp8 Corina Maeder 1,3, Alan K. Kutach 1,2,3, and Christine Guthrie 1 1 Department of Biochemistry
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Conversion of plasmids into Gateway compatible cloning
 Conversion of plasmids into Gateway compatible cloning Rafael Martinez 14072011 Overview: 1. Select the right Gateway cassette (A, B or C). 2. Design primers to amplify the right Gateway cassette from
Conversion of plasmids into Gateway compatible cloning Rafael Martinez 14072011 Overview: 1. Select the right Gateway cassette (A, B or C). 2. Design primers to amplify the right Gateway cassette from
Biotechnology Project VI lesson. 1 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO into the expression vector pegfp-c3
 26-11-2018 - Biotechnology Project VI lesson 1 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO into the expression vector pegfp-c3 2 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO or pegfp-c3 into the
26-11-2018 - Biotechnology Project VI lesson 1 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO into the expression vector pegfp-c3 2 - Subcloning NRG1-III-β3 from pcr-blunt II-TOPO or pegfp-c3 into the
transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected with the wwp-luc reporter, and FLAG-tagged FHL1,
 Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
Supplementary Data Supplementary Figure Legends Supplementary Figure 1 FHL-mediated TGFβ-responsive reporter transcription and the promoter occupancy of Smad proteins. (A) HepG2 cells were co-transfected
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators
 Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh,
Supplemental Information for: A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators Nick T. Peters, Thuy Dinh,
Journal of Experimental Microbiology and Immunology (JEMI) Vol. 6:20-25 Copyright December 2004, M&I UBC
 Preparing Plasmid Constructs to Investigate the Characteristics of Thiol Reductase and Flavin Reductase With Regard to Solubilizing Insoluble Proteinase Inhibitor 2 in Bacterial Protein Overexpression
Preparing Plasmid Constructs to Investigate the Characteristics of Thiol Reductase and Flavin Reductase With Regard to Solubilizing Insoluble Proteinase Inhibitor 2 in Bacterial Protein Overexpression
Creating pentr vectors by BP reaction
 Creating pentr vectors by BP reaction Tanya Lepikhova and Rafael Martinez 15072011 Overview: 1. Design primers to add the attb sites to gene of interest 2. Perform PCR with a high fidelity DNA polymerase
Creating pentr vectors by BP reaction Tanya Lepikhova and Rafael Martinez 15072011 Overview: 1. Design primers to add the attb sites to gene of interest 2. Perform PCR with a high fidelity DNA polymerase
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supporting Information
 Supporting Information Risser and Callahan 10.1073/pnas.0909152106 SI Text Plasmid Construction. Plasmid pdr211 is a mobilizable shuttle vector containing P pete -pats. A fragment containing P pete was
Supporting Information Risser and Callahan 10.1073/pnas.0909152106 SI Text Plasmid Construction. Plasmid pdr211 is a mobilizable shuttle vector containing P pete -pats. A fragment containing P pete was
Figure 1. Map of cloning vector pgem T-Easy (bacterial plasmid DNA)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
2014 Pearson Education, Inc. CH 8: Recombinant DNA Technology
 CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity.
 Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Supplemental Figure 1 HDA18 has an HDAC domain and therefore has concentration dependent and TSA inhibited histone deacetylase activity. (A) Amino acid alignment of HDA5, HDA15 and HDA18. The blue line
Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role
 Supporting Information Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role Micah T. Nelp, Anthony P. Young, Branden M. Stepanski, Vahe Bandarian* Department
Supporting Information Human Viperin Causes Radical SAM Dependent Elongation of E. coli Hinting at its Physiological Role Micah T. Nelp, Anthony P. Young, Branden M. Stepanski, Vahe Bandarian* Department
Supplementary information. Interplay between Penicillin-binding proteins and SEDS proteins promotes. bacterial cell wall synthesis
 Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
Supplementary information Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis Sophie Leclercq 1, Adeline Derouaux 1, Samir Olatunji 1, Claudine Fraipont
A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Journal of Plant Research A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana Linna Leng 1 Qianqian Liang
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Mcbio 316: Exam 3. (10) 2. Compare and contrast operon vs gene fusions.
 Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
SUPPLEMENTARY EXPEMENTAL PROCEDURES
 SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
SUPPLEMENTARY EXPEMENTAL PROCEDURES Plasmids- Total RNAs were extracted from HeLaS3 cells and reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) to obtain DNA template for the
Polymerase Chain Reaction
 Polymerase Chain Reaction Amplify your insert or verify its presence 3H Taq platinum PCR mix primers Ultrapure Water PCR tubes PCR machine A. Insert amplification For insert amplification, use the Taq
Polymerase Chain Reaction Amplify your insert or verify its presence 3H Taq platinum PCR mix primers Ultrapure Water PCR tubes PCR machine A. Insert amplification For insert amplification, use the Taq
Molecular cloning: 6 RBS+ arac/ laci
 Molecular cloning: 6 RBS+ arac/ laci Resource: 6 RBS: from the parts: B0030, B0032, B0034, J61100, J61107, J61127; renamed as RBS1 RBS2 RBS6; arac: C0080 laci: C0012 July 6 th Plasmid mini prep: 6 RBS;
Molecular cloning: 6 RBS+ arac/ laci Resource: 6 RBS: from the parts: B0030, B0032, B0034, J61100, J61107, J61127; renamed as RBS1 RBS2 RBS6; arac: C0080 laci: C0012 July 6 th Plasmid mini prep: 6 RBS;
EZ Vision DNA Dye as Loading Buffer
 EZ Vision DNA Dye as Loading Buffer Code Description Size N472-1ML N472-KIT N650-1ML N650-KIT N313-1ML N313-KIT N473-2PK N473-3PK EZ Vision One DNA Dye as Loading Buffer, 6X EZ Vision Two, DNA Dye as Loading
EZ Vision DNA Dye as Loading Buffer Code Description Size N472-1ML N472-KIT N650-1ML N650-KIT N313-1ML N313-KIT N473-2PK N473-3PK EZ Vision One DNA Dye as Loading Buffer, 6X EZ Vision Two, DNA Dye as Loading
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Contents. 1 Basic Molecular Microbiology of Bacteria... 1 Exp. 1.1 Isolation of Genomic DNA Introduction Principle...
 Contents 1 Basic Molecular Microbiology of Bacteria... 1 Exp. 1.1 Isolation of Genomic DNA... 1 Introduction... 1 Principle... 1 Reagents Required and Their Role... 2 Procedure... 3 Observation... 4 Result
Contents 1 Basic Molecular Microbiology of Bacteria... 1 Exp. 1.1 Isolation of Genomic DNA... 1 Introduction... 1 Principle... 1 Reagents Required and Their Role... 2 Procedure... 3 Observation... 4 Result
Supplemental Data. Bacterial Birth Scar Proteins Mark Future Flagellum Assembly Site
 Supplemental Data Bacterial Birth Scar Proteins Mark Future Flagellum Assembly Site Edgar Huitema, Sean Pritchard, David Matteson, Sunish Kumar Radhakrishnan, and Patrick H. Viollier - 1 - Figure S1. Z-Ring
Supplemental Data Bacterial Birth Scar Proteins Mark Future Flagellum Assembly Site Edgar Huitema, Sean Pritchard, David Matteson, Sunish Kumar Radhakrishnan, and Patrick H. Viollier - 1 - Figure S1. Z-Ring
CH 8: Recombinant DNA Technology
 CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
CH 8: Recombinant DNA Technology Biotechnology the use of microorganisms to make practical products Recombinant DNA = DNA from 2 different sources What is Recombinant DNA Technology? modifying genomes
Step 1: Digest vector with Reason for Step 1. Step 2: Digest T4 genomic DNA with Reason for Step 2: Step 3: Reason for Step 3:
 Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Construct Design and Cloning Guide for Cas9-triggered homologous recombination
 Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Construct Design and Cloning Guide for Cas9-triggered homologous recombination Written by Dan Dickinson (ddickins@live.unc.edu) and last updated December 2013. Reference: Dickinson DJ, Ward JD, Reiner
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Regulation of ARE transcript 3 end processing by the. yeast Cth2 mrna decay factor
 Regulation of ARE transcript 3 end processing by the yeast Cth2 mrna decay factor Manoël Prouteau, Marie-Claire Daugeron and Bertrand Séraphin Supplementary Information Material and Methods Plasmid construction
Regulation of ARE transcript 3 end processing by the yeast Cth2 mrna decay factor Manoël Prouteau, Marie-Claire Daugeron and Bertrand Séraphin Supplementary Information Material and Methods Plasmid construction
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
The Central Dogma. 1) A simplified model. 2) Played out across life. 3) Many points for control.
 Molecular Biology The Central Dogma 1) A simplified model. 2) Played out across life. 3) Many dis@nct points for control. Molecular Biology DNA: four nucleo@de bases (GC,AT) (2 bits) gene@c code in 3 base
Molecular Biology The Central Dogma 1) A simplified model. 2) Played out across life. 3) Many dis@nct points for control. Molecular Biology DNA: four nucleo@de bases (GC,AT) (2 bits) gene@c code in 3 base
P52A/R67A. The plasmids expressing AD-fused Atg1 variants were constructed as follows: DNA
 SUPPLEMENTAL DATA The Autophagy-Related Protein Kinase Atg1 Interacts with the Ubiquitin-Like Protein Atg8 via the Atg8 Family Interacting Motif to Facilitate Autophagosome Formation Hitoshi Nakatogawa,
SUPPLEMENTAL DATA The Autophagy-Related Protein Kinase Atg1 Interacts with the Ubiquitin-Like Protein Atg8 via the Atg8 Family Interacting Motif to Facilitate Autophagosome Formation Hitoshi Nakatogawa,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending)
 peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
Journal of Experimental Microbiology and Immunology (JEMI) Vol. 6:26-34 Copyright December 2004, M&I UBC
 Cloning EDTA monooxygenase as a Model Protein to Characterize the Effects of Flavin oxidoreductase on Solubility of Proteins in Protein Overexpression Systems MIKAMEH KAZEM Department of Microbiology and
Cloning EDTA monooxygenase as a Model Protein to Characterize the Effects of Flavin oxidoreductase on Solubility of Proteins in Protein Overexpression Systems MIKAMEH KAZEM Department of Microbiology and
SUPPLEMENTARY INFORMATION
 a Pxyl ms2d mgfp b Pxyl ms2(dlfg) mgfp Pvan mcherry Transcription STOP codon bs48 + Xylose + Vanillic Acid transcriptional terminator Pvan mcherry bs48 STOP transcriptional codon terminator + Xylose Transcription
a Pxyl ms2d mgfp b Pxyl ms2(dlfg) mgfp Pvan mcherry Transcription STOP codon bs48 + Xylose + Vanillic Acid transcriptional terminator Pvan mcherry bs48 STOP transcriptional codon terminator + Xylose Transcription
Materials and methods. by University of Washington Yeast Resource Center) from several promoters, including
 Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
The Biotechnology Toolbox
 Chapter 15 The Biotechnology Toolbox Cutting and Pasting DNA Cutting DNA Restriction endonuclease or restriction enzymes Cellular protection mechanism for infected foreign DNA Recognition and cutting specific
Chapter 15 The Biotechnology Toolbox Cutting and Pasting DNA Cutting DNA Restriction endonuclease or restriction enzymes Cellular protection mechanism for infected foreign DNA Recognition and cutting specific
Diagnosis Sanger. Interpreting and Troubleshooting Chromatograms. Volume 1: Help! No Data! GENEWIZ Technical Support
 Diagnosis Sanger Interpreting and Troubleshooting Chromatograms GENEWIZ Technical Support DNAseq@genewiz.com Troubleshooting This troubleshooting guide is based on common issues seen from samples within
Diagnosis Sanger Interpreting and Troubleshooting Chromatograms GENEWIZ Technical Support DNAseq@genewiz.com Troubleshooting This troubleshooting guide is based on common issues seen from samples within
An antibiotic selection marker for nematode transgenesis
 nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
nature methods An antibiotic selection marker for nematode transgenesis Rosina Giordano-Santini, Stuart Milstein, Nenad Svrzikapa, Domena Tu, Robert Johnsen, David Baillie, Marc Vidal & Denis Dupuy Supplementary
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplemental Data. Hu et al. Plant Cell (2017) /tpc
 1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
1 2 3 4 Supplemental Figure 1. DNA gel blot analysis of homozygous transgenic plants. (Supports Figure 1.) 5 6 7 8 Rice genomic DNA was digested with the restriction enzymes EcoRⅠ and BamHⅠ. Lanes in the
-7 ::(teto) ::(teto) 120. GFP membranes merge. overexposed. Supplemental Figure 1 (Marquis et al.)
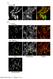 P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
P spoiie -gfp -7 ::(teto) 120-91 ::(teto) 120 GFP membranes merge overexposed Supplemental Figure 1 (Marquis et al.) Figure S1 TetR-GFP is stripped off the teto array in sporulating cells that contain
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Craft et al., http://www.jcb.org/cgi/content/full/jcb.201409036/dc1 Figure S1. GFP -tubulin interacts with endogenous -tubulin. Ciliary
Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe:
 1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability
 Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/315/5819/1709/dc1 Supporting Online Material for RISPR Provides Acquired Resistance Against Viruses in Prokaryotes Rodolphe Barrangou, Christophe Fremaux, Hélène Deveau,
www.sciencemag.org/cgi/content/full/315/5819/1709/dc1 Supporting Online Material for RISPR Provides Acquired Resistance Against Viruses in Prokaryotes Rodolphe Barrangou, Christophe Fremaux, Hélène Deveau,
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection
 Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
Protocol for cloning SEC-based repair templates using Gibson assembly and ccdb negative selection Written by Dan Dickinson (daniel.dickinson@austin.utexas.edu) and last updated January 2018. A version
TrueORF TM cdna Clones and PrecisionShuttle TM Vector System
 TrueORF TM cdna Clones and PrecisionShuttle TM Vector System Application Guide Table of Contents Package Contents and Storage Conditions... 2 Related, Optional Reagents... 2 Related Products... 2 Available
TrueORF TM cdna Clones and PrecisionShuttle TM Vector System Application Guide Table of Contents Package Contents and Storage Conditions... 2 Related, Optional Reagents... 2 Related Products... 2 Available
mod-1::mcherry unc-47::gfp RME ser-4::gfp vm2 vm2 vm2 vm2 VNC
 A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
A mod-1::mcherry unc-47::gfp RME B ser-4::gfp vm2 vm2 VNC vm2 vm2 VN Figure S1 Further details of mod- 1 and ser- 4 reporter expression patterns. (A) Adult head region showing mod- 1::mCherry and unc-
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
pri 201 DNA Series (High-Expression Vectors for Plant Transformation)
 For Research Use Cat #. 3264 3265 3266 3267 pri 201 DNA Series (High-Expression Vectors for Plant Transformation) Product Manual Table of Contents I. Description... 3 II. Product Information... 3 III.
For Research Use Cat #. 3264 3265 3266 3267 pri 201 DNA Series (High-Expression Vectors for Plant Transformation) Product Manual Table of Contents I. Description... 3 II. Product Information... 3 III.
Supplemental Data. Sethi et al. (2014). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Supplemental Data Supplemental Figure 1. MYC2 Binds to the E-box but not the E1-box of the MPK6 Promoter. (A) E1-box and E-box (wild type) containing MPK6 promoter fragment. The region shown in red denotes
Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin
 Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin Godefroid Charbon, 1,4 Matthew T. Cabeen, 1 and Christine Jacobs-Wagner 1,2,3,5 1 Department of Molecular, Cellular
Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin Godefroid Charbon, 1,4 Matthew T. Cabeen, 1 and Christine Jacobs-Wagner 1,2,3,5 1 Department of Molecular, Cellular
Recombinant DNA Technology
 Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
Recombinant DNA Technology Common General Cloning Strategy Target DNA from donor organism extracted, cut with restriction endonuclease and ligated into a cloning vector cut with compatible restriction
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Recombinant DNA Libraries and Forensics
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 A. Library construction Recombinant DNA Libraries and Forensics Recitation Section 18 Answer Key April 13-14, 2005 Recall that earlier
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 A. Library construction Recombinant DNA Libraries and Forensics Recitation Section 18 Answer Key April 13-14, 2005 Recall that earlier
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and
 Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts. (A) Shoot morphology of 19-day-old nla mutants under Pi-sufficient conditions. (B) [ 33 P]Pi uptake
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Recombinant DNA Technology. The Role of Recombinant DNA Technology in Biotechnology. yeast. Biotechnology. Recombinant DNA technology.
 PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
GenBuilder TM Plus Cloning Kit User Manual
 GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
GenBuilder TM Plus Cloning Kit User Manual Cat. No. L00744 Version 11242017 Ⅰ. Introduction... 2 I.1 Product Information... 2 I.2 Kit Contents and Storage... 2 I.3 GenBuilder Cloning Kit Workflow... 2
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rainero et al., http://www.jcb.org/cgi/content/full/jcb.201109112/dc1 Figure S1. The expression of DGK- is reduced upon transfection
