Edinburgh Research Explorer
|
|
|
- Augustus Carr
- 6 years ago
- Views:
Transcription
1 Edinburgh Research Explorer Genetic prediction of complex traits: integrating infinitesimal and marked genetic effects Citation for published version: Carré, C, Gamboa, F, Cros, D, Hickey, JM, Gorjanc, G & Manfredi, E 2013, 'Genetic prediction of complex traits: integrating infinitesimal and marked genetic effects' Genetica, vol 141, pp DOI: /s Digital Object Identifier (DOI): /s Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Genetica Publisher Rights Statement: Available under Open Access General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact openaccess@ed.ac.uk providing details, and we will remove access to the work immediately and investigate your claim. Download date: 01. May. 2018
2 Genetica (2013) 141: DOI /s Genetic prediction of complex traits: integrating infinitesimal and marked genetic effects Clément Carré Fabrice Gamboa David Cros John Michael Hickey Gregor Gorjanc Eduardo Manfredi Received: 16 January 2013 / Accepted: 20 May 2013 / Published online: 30 May 2013 Ó The Author(s) This article is published with open access at Springerlink.com Abstract Genetic prediction for complex traits is usually based on models including individual (infinitesimal) or marker effects. Here, we concentrate on models including both the individual and the marker effects. In particular, we develop a Mendelian segregation model combining infinitesimal effects for base individuals and realized Mendelian sampling in descendants described by the available DNA data. The model is illustrated with an example and the analyses of a public simulated data file. Further, the potential contribution of such models is assessed by simulation. Accuracy, measured as the correlation between true (simulated) and predicted genetic values, was similar for all models compared under different genetic backgrounds. As expected, the segregation model is worthwhile when markers capture a low fraction of total genetic variance. Keywords Genetic prediction Genomic selection SNP Mendelian sampling C. Carré E. Manfredi (&) UR 631 SAGA, INRA Toulouse, B.P , Auzeville, Castanet-Tolosan Cedex, France Eduardo.Manfredi@toulouse.inra.fr C. Carré F. Gamboa IMT, Université Paul Sabatier, Toulouse, France D. Cros AGAP, CIRAD, Montpellier, France J. M. Hickey Biometrics and Statistics Unit, International Maize and Wheat Improvement Center (CIMMYT), Mexico, D.F., Mexico G. Gorjanc Animal Science Department, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia Introduction In recent years, new knowledge on molecular genetics and the rapid evolution of sequencing and genotyping technology has renewed the interest on genetic prediction of complex traits. It should be recalled, however, that genetic prediction of complex traits has been a traditional field in animal and plant breeding since the 40 s in the framework of the Selection Index (SI) theory (e.g., Hazel 1943), extended later to the best linear unbiased prediction (BLUP; Henderson 1975). These genetic prediction methods, without DNA data, were based on the individual model where covariances amongst phenotypes of related individuals are translated into unobserved covariances amongst genetic values, via theoretical relatedness coefficients amongst individuals. Anticipating the availability of low-cost whole genome DNA data, Meuwissen et al. (2001) proposed marker models where many markers genotypes represent genetic effects, while the individuals are not explicitly specified in the model. We concentrate here on a third group of models including both marker and individual effects. We first recall the families of models proposed for genetic prediction and then we develop a novel model, which is illustrated with an example. Then, we assess the relative performance of the novel model in relation to the marker model for different genetic scenarios, and we report results of the analyses of a public simulated sample. Finally, originality, limits and possible extensions of the model are discussed. Individual models for genetic prediction Both SI and BLUP are applied to the infinitesimal (or polygenic) genetic model which in its simplest version is
3 240 Genetica (2013) 141: phenotype = mean? additive genetic value? residual. This model has been called polygenic or infinitesimal since the additive genetic value is the sum of the effects, assumed to be small and homogeneous, of numerous genes on the phenotype. In the statistical model, built from the genetic model, individual effects are used to represent additive genetic effects, and they are assumed random because genotype configurations of individuals arise through random processes: y ¼ l þ Zu þ e ð1þ y is a vector of phenotypes l is a constant vector (assumed known in SI and estimated in BLUP) Z is an incidence matrix of order N y ðphenotypesþ N i ðindividualsþ; relating each of the N y phenotypes to each of the measured individuals. For simplicity, we assume only one measure per individual. In standard BLUP technology Z ¼ ½0 I 0Š, i.e., null columns for base individuals without phenotypes, the identity matrix for individuals with phenotypes (when there is a single measure for each individual), and null columns for descendants without phenotype, the usual target of prediction. In this context of genetic prediction, base individuals are defined for a given genealogy as the most distant known ancestors of individuals with recorded phenotypes, i.e., they do not have phenotypes and their parents are unknown. u is a vector of additive genetic effects, with VarðuÞ ¼ Ar 2 u, with A being the relationship matrix amongst individuals. e is a vector of residuals, with VarðÞ¼Ir e 2 e, with I being an identity matrix A further usual assumption is Covðe; uþ ¼0. The only information available to distinguish genetic effects from residuals are the structures of the (co)variance matrices of u and e. In other words, the model describes a network of phenotypic covariances (observed) which are translated into genetic covariances (unobserved) via the theoretical genetic model, in particular the relatedness coefficients in the relationship matrix A. Marker and individual models With molecular data available, prediction models evolved to include this new information (e.g., Fernando and Grossman 1989; Meuwissen et al. 2001). Fernando and Grossman (1989) proposed a prediction model which included several genetic effects: an infinitesimal effect u plus haplotype effects of maternal and paternal origin at marked quantitative trait loci (QTL) positions. Their model was reasonably conservative, given the genomic tools available by that time (say, 500 microsatellites to cover the entire genome in farm animals). In this context, they assumed that a marker allele may mark different QTL alleles in different families. Later, with many more markers (10,000 multi-allelic markers), Meuwissen et al. (2001) switched from the previous conservative model to marker models exploiting linkage disequilibrium at the population level: y ¼ l þ ZWm þ e ð2þ where: m is a vector of marked genetic effects (usually termed marker effects, although the usual hypothesis is that markers do not have a true effect per se on the phenotype) W is a matrix of marker genotypes of order N i ðindividualsþn m ðmarkersþ: With biallelic markers such as SNP, usual elements of W are 0, 1 or 2, the number of, say, the allele 1 of the marker genotype. Usually assumed (co)variances are: VarðmÞ ¼ I Nm r 2 m Covðe; mþ ¼0 where I Nm is an identity matrix of order N m. If we further assume that u ¼ Wm and VarðuÞ ¼ WW 0 r 2 m, it is possible to compute predictions for u with the individual model (1), amended such that the relationship matrix A is replaced by the realized genomic relationship matrix G ¼ WW 0 (VanRaden 2008; Goddard 2009). Application of BLUP to this model has been termed genomic BLUP and improvements have been proposed to make assumptions more realistic (departures from the homogeneous variances for marked effects in model (2)) and practical implementations when only part of the individuals are genotyped making necessary to mix the A and the G matrices for the combined analyses of individuals with or without genotypes (e.g. Aguilar et al. 2011). Marker plus individual model Alternative assumptions in an outbred population are u 6¼ Wm and VarðuÞ 6¼ WW 0 r 2 m. There are theoretical reasons and experimental results to support this point of view. Theoretically, in a Bayesian context, Gianola et al. (2009) claimed that the functional relationship between r 2 u and r2 m is elusive. They did propose simple approximations under Hardy Weinberg and linkage equilibria (LE) to relate the marked genetic variance and the additive genetic variance as r 2 u ¼ 2 P N m i¼1 p iq i r 2 m, where p i and q i are the allelic frequencies for marker i. However, assuming LE is not compatible with the essential assumption of linkage disequilibrium in the context of genome-wide analysis. Furthermore, in most experimental studies, the sum of
4 Genetica (2013) 141: variances due to marker associations does not add up to the additive genetic variance due to individual infinitesimal effects raising the problem of the hidden heritability (e.g., Yang et al. 2011). The unknown vector m represents the effects of unobserved genes that should be marked by observed markers. This model should fit all genome-wide additive effects simultaneously. However, it is not warranted that all the actual additive genetic effects in the studied genome will be effectively traced by the available markers (Yang et al. 2011). Potential problems are poor marker coverage (low density but also insufficient representation of independent DNA segments), rare alleles, small (infinitesimal) gene effects, multi-allelic genes having additive effects that are poorly traced by bi-allelic markers, or other molecular genetics mechanisms. The main assumption is that each marker allele or haplotype is associated with each unobserved QTL allele in identical way for each individual in the studied population. This may be true in some cases but it is not true in general. While an association between a marker and the QTL may be stable within parents and progeny, open populations over several generations are built up by subpopulations, each one with its own QTL allele-marker allele association. Reintroduction of infinitesimal effects in the prediction model is one of the recommended ways to control partially the lack of perfect association between marker alleles and causative alleles (Goddard and Hayes 2009). The model becomes: y ¼ l þ Zu þ ZWm þ e ð3þ with additional assumptions: VarðuÞ ¼ Rr 2 u ; and Covðe; uþ ¼Covðu; mþ ¼0; where Rr 2 u is the symmetric (co)-variance matrix of individual effects of order N i. Usually, as in model (1), R = A, the additive relationship matrix computed theoretically from genealogy data. Note that the terms in model (3) are redundant if it is assumed that u = Wm. The idea in model (3) is to include residual genetic values not taken into account by the marked effects m. In applications, this model gave better predictions than the marker model (2) (e.g., De los Campos et al. 2009; Duchemin et al. 2012). Mendelian segregation model Here, we develop a model where the genetic value of an individual is a function of infinitesimal effects of ancestors (individuals in the base, with unknown parents) and Mendelian sampling which can be traced by DNA data. In the following it is assumed that all individuals have complete genotype data and all descendants have known parents. We then discuss the departures from this complete data situation. The model starts as in (3): y ¼ l þ Zu þ ZWm þ e It is convenient to separate individuals in two groups: the base ancestors with unknown parents (indexed by b) and the descendants (indexed by d). We can now expand and decompose the vector of infinitesimal values u as: u ¼ u b u d Let P be a N i 9 N i matrix with two 1 s in each row, indicating the parents of each individual (rows of P for base individuals are null). We define the matrix M as: M ¼ I 1 2 P The matrix M is interpretable in biology (each row of M represents the individual minus half the sum of parents) and in mathematics since M has the form of a Laplacian matrix, representing the pedigree graph, with P being the adjacency matrix with elements equal to 1 at the intersection of adjacent nodes (parent and progeny nodes) or 0 otherwise. Let / be a vector of infinitesimal mendelian sampling effects which are deviations of individual genetic values from their respective parental averages. Then, the matrix operator M 21 can be used to construct additive genetic values u as linear combinations of ancestor genetic values u b and mendelian sampling / of their descendants, as illustrated in part (a) of Fig. 1, so we can write: u ¼ M 1 u b / where u can be found by partitioning the M matrix in M bb, M dd,m db and M bd blocks, as: M ¼ M bb M bd ; withm M db M bb ¼ I; and M bd ¼ 0: dd Using known results about the inverse of a lower triangular matrix, we obtain: u ¼ ¼ I 0 M 1 dd M db u b M 1 dd ub M 1 dd M dbu b þ M 1 dd / / ¼ u b u d ð4þ Equation (4) uses standard results under infinitesimal models developed when it was impossible to observe DNA, and a theoretical distribution was assigned to the unknown / (see Quaas 1976). Availability of genotypes for progeny and parents gives a realized molecular mendelian
5 242 Genetica (2013) 141: (a) /2 1/ /4 1/4 1/2 1/2 1 5 For an individual in the base, e.g. 1, For a descendant, e.g. 5, (b) /2-1/ /2-1/2 1 Snp1 Snp2 Snp For an individual, e.g. 4, + In general, for the ith individual,, with,, and representing the j-th marker genotype for the individual, the father, and the mother, respectively. Fig. 1 Genetic transmission and Mendelian sampling effects in the prediction model. a Transmission: genetic values of descendants are a function of genetic values of base individuals u b and Mendelian sampling effects predicted by s. b Observed Mendelian sampling effects sampling s, a predictor of / which can be approached as a function of marked gene effects m: W s ¼ ½M db M dd Š b m ð5þ W d where matrices W b and W d contain the marker genotypes of base and descendant individuals, respectively. Figure 1b illustrates how expression (5) represents individual deviations from parental means, in terms of marked genetic effects, for a hypothetical genealogy of 5 individuals and 3 markers. Then, replacing / by s in (4), and using (5) in(4), with D ¼ M 1 dd M db, we get: u d ¼ Du b DW b m þ W d m ¼ Du b þðw d DW b Þm ð6þ And the model for phenotypes is then: y ¼ l þ Z d Du b þ Z d ð2w d DW b Þm þ e ð7þ In the term Z d Du b, Z d (of order Ny 9 Nd) relates records to individuals (descendants d) and D relates individual genetic values to ancestors genetic values u b via simple coefficients of genome sharing (including consanguinity, i.e., multiple contributions of an ancestor to an individual). So this term in (7) concentrates all phenotype information of descendants to estimate the ancestors infinitesimal values. The term Z d (2W d - DW b ) m in (7) groups two parts: Z d (W d - DW b ) m, the molecular mendelian sampling effects where individual marked effects deviate from ancestors marked effects, and Z d W d m which represents the direct relations between markers and phenotypes. Assumptions of the model A set of possible assumptions is: u b N0; Ir 2 u m N0; Ir 2 m Covðu b ; mþ ¼ 0 The assumption of independent base individuals is usual in quantitative genetics. With DNA information and complete data it would be possible to make more general
6 Genetica (2013) 141: assumptions like u b N l u ; Hr 2 u ; where H represents a genomic matrix, thus recognizing that individuals in the base populations may share genes. Again, the model is redundant if it is assumed that u b = W b m and H ¼ W b W 0 b r2 m. Alternatively, model (7) can also accommodate fixed genetic values for individuals in the base population. Distribution of marked effects m is assumed normal but other distributions such as the Gamma may be chosen, to take into account experimental results indicating few loci with large effects and many more loci with small effects (Goddard and Hayes 2009). Analyses of data Firstly, repeated simulations were conducted to assess the predictive ability of the Mendelian segregation model MS (Eq. 7) relative to the marker model M (Eq. 2). Then, we analyzed a public sample simulated for the 12th European QTLMAS workshop by Lund et al. (2009), using several models including individual and marked genetic effects. We preferred to use simulated data at this exploratory stage to understand the behavior of the compared models. Also, to simplify interpretation at this stage, estimation and prediction were limited to the unknowns in the models (l, the vector of marked effects m and the vector of individual genetic values u) by applying known variances used to simulate the data. We used the same statistical method BLUP to all models compared, which have either one (Eqs. 1 and 2) or two (Eqs. 3 and 7) random effects in addition to random residuals. BLUP of random effects were computed as detailed in the Appendix. Relative predictive performance of the Mendelian segregation (MS) model Data were simulated using the QMSim software (Sargolzaei and Schenkel 2009). The simulated population had 1 base generation (25 individuals), 3 training generations (120 individuals) and the last generation (40 individuals) taken as prediction target. Mating was at random and the family size was 1. The simulated genome had 2 chromosomes of 1 Morgan each and 10 biallelic QTL/chromosome were responsible for the QTL fraction of genetic variance. Number of SNP markers used was either 2,000 or 200 per chromosome. Phenotypes in the base and target generations were simulated but not used to predict genetic values of the target generation. The phenotypes had variance 1 and overall heritability (infinitesimal? QTL effects) was 0.4. Three genetic scenarios were replicated 200 times: high Fig. 2 Accuracy of the marker (M) and Mendelian segregation (MS) models for the three simulation scenarios with 10, 50, or 90 % of the total genetic variance explained by QTL (90 %), intermediate (50 %), or low (10 %) proportion of genetic variance explained by QTL. Mean accuracies over 200 replicates when using 2,000 SNP markers are presented in Fig. 2 for 10, 50 and 90 % of total genetic variance explained by QTL. Accuracies were highest (0.76 for model M and 0.74 for model MS) in the training data when the genetic variance explained by QTL was high (90 %). The lowest correlations occurred for the test data under scenario 10 % (0.36 for M vs for MS). The MS model gave the best predictions when the infinitesimal effects were important (scenario 10 %) and model M gave the best predictions when QTL effects represented 90 % of genetic variance. Differences between mean accuracies of two models were small and non-significant (P \ 0.05). When fewer markers were used (200 SNP per chromosome), all accuracies were lower but the methods ranked as when using more (2,000 SNP per chromosome) markers (Table 1). The accuracy of the MS model was 12 % higher than that of the M model for the scenario with the 10 % of genetic variance explained by QTL and 5 % lower when the QTL explained the 90 % of total variance. Analyses of a public simulated sample In the data simulated for the 12th European QTLMAS workshop (Lund et al. 2009), the simulated phenotypes were influenced by 50 loci, including 15 major effect loci and 35 minor effect loci with a total heritability of 0.3. Marker information was available for 6,000 SNP (only 5,925 were polymorphic and used in our analyses) on 6 chromosomes. The population was simulated under random mating and the absence of selection. Each male was mated to 10 females and each mating pair produced 10 offspring. A data set of 4,665 individuals was split into a training set (3,165 individuals) and a test set (1,500). In the
7 244 Genetica (2013) 141: Table 1 Performance of the Mendelian segregation model: relative accuracies in the training and the test data Simulated scenario Training data (%) a Test data(%) a QTL variance 10 % 200 SNP markers ,000 SNP markers QTL variance 50 % 200 SNP markers ,000 SNP markers QTL variance 90 % 200 SNP markers ,000 SNP markers a (%) is 100 times the ratio between the average accuracy under the Mendelian segregation model and the average accuracy under the marker model training set, the base population (generation 0) included 165 individuals with unknown parents. The remaining 3,000 individuals had known parents and were born in generations 1 and 2. The test set had 1,500 individuals born in generation 3 with complete genealogy. The targets of prediction were the simulated genetic values and phenotypes of the test individuals. The data used in prediction were the phenotypes of 3,000 individuals of generations 1 and 2, and the marker genotypes of all individuals. Four models were compared using the known variances used for the simulation: the marker model (M) as in (2), the marker plus individual model (MI) as in (3), the marker plus mendelian effects model (MS) given in (7), and the individual model where the (co)-variance matrix of individual effects was the additive relationship A (individual infinitesimal model; II). The method to estimate the unknowns of all the models was BLUP. The known variances were given by Lund et al. (2009): r 2 e ¼ 3:15 and r2 u ¼ 1:35. The variance of marker effects was computed as r 2 m ¼ r2 u = 2 P j p jð1 p j Þ. Correlations between predicted values and simulated genetic values and phenotypes for the training and Table 2 Correlations between the predicted genetic values ðbu Þ, simulated genetic values (u), and simulated phenotypes (y) in the training and test data Model a M MI MS II Training data rðbu; uþ rðbu; yþ Test data rðbu; uþ rðbu; yþ a Models. M: marker model (Eq. 2); MI: marker plus individual effect model (Eq. 3); MS: Mendelian segregation model (Eq. 7); II: individual infinitesimal model based on pedigree (Eq. 1) test populations are given in Table 2. The goodness of fit of model (7) for the training data was moderate rððbu; yþ ¼ 0:53Þ but it yielded the best predictions for genetic values rððbu; uþ ¼ 0:94Þ and phenotypes rðbu; yþ ¼ 0:55 in the test sample. Model [7] was also the best to estimate the marked effects m: the correlations between estimates of m and the simulated allele substitution effects, in absolute values, were 0.69 for Model [7] and 0.56 for both the marker model and the marker? individual model. Discussion As reviewed in the Introduction, there are plausible arguments to combine marked effects models with other individual effects when analyzing complex traits. To do so, the strategy used in the MS model [7] is to decompose the individual genetic value into two terms: a contribution from base individuals, weighted by the transmission matrix D, and a contribution from mendelian sampling occurring at several meiosis from base individuals to their descendants, instead of attempting to fit twice the additive genetic value of an individual as in model [3]. In traditional infinitesimal models, mendelian sampling is an unknown theoretical random term, so predictions of future phenotypes (of future progeny) are based on ancestor phenotypes and random terms. At present, with the availability of numerous markers, mendelian sampling is realized for each individual and it can be used to improve predictions. Model [7] builds on very well-known results in quantitative genetics. Early work described how genetic transmission operates in the additive relationship matrix A (e.g., Quaas 1976 and Henderson 1976, who presented detailed factorizations of the A matrix). Subsequent models included genetic transmission at unobserved segregating QTL (e.g., Fernando and Grossman 1989; Meuwissen and Goddard 2000; Legarra and Fernando 2009) and combined within family and between family marker effects in the context of methodology for QTL search (e.g., Abecasis et al. 2000). In animal breeding, efforts have focused on combining genotype data with genealogy data in individual genomic models, as reviewed by Meuwissen et al. (2011). The model [7] developed here builds on previous work by the simultaneous inclusion of infinitesimal and marked genetic effects. In this way the model might capitalize on two advantages of molecular information: the improvement of the infinitesimal prediction by the estimation of realized mendelian sampling in descendant individuals, and by capturing marked gene effects without bias due to family structure, i.e., to predict marked effects and infinitesimal effects simultaneously and without redundancy. Here, marked effects are estimated at the level of the population (marked effects m in model MS
8 Genetica (2013) 141: [7] are not defined within family) but the family structure is taken into account in the estimation model. Results of simulations indicate that the predictive ability of the MS model is comparable to that of the marker model. On one hand, the accuracies obtained in different genetic scenarios suggest that the MS model might be useful when markers are not adequate to fully explain the genetic background (low QTL variances with high infinitesimal variance, or low marker density). On the other hand, the marker model M yielded slightly higher predictive ability than MS when QTL were important and marker density was high. This result might reflect sub-optimality of the MS model to exploit favorable situations where markers do effectively capture much of total genetic variance. This might be explained by the simple distributional assumptions that we assumed at this exploratory stage for the base individuals and the marked effects of model MS in [7] and accompanying assumptions. In particular, the marker model [2], and, more explicitly, its equivalent model Genomic BLUP, capitalizes the complete data setting studied here by estimating covariances among base individuals, and covariances between base individuals and descendants. So, for the MS model to be fully competitive, its distributional assumptions should be extended to take into account those relationships. Results for the QTLMAS example are encouraging but unique and different from those of replicated simulations. At least two reasons may be advanced to explain these different results: the more complicated genetic background and the large family size, a full-sib design, simulated in the QTLMAS data set. But the impact of such factors on predictive ability needs further investigation. Further investigation is also needed on variance component estimation of models including marker and individual effects. Duchemin et al. (2012) were able to estimate both components of variance from real data using model [3], i.e., the variance of individual effects and the variance of marker effects. We are currently studying variance components estimation for model [7], with infinitesimal effects defined only for the base individuals and variance structure designed to avoid identifiability problems. Also, at this stage of model development, we are assuming complete data, in particular genotypes of base individuals. In some situations, it may possible to impute missing data. Also, if genealogy is unknown and if all individuals are in the genotyped sample, parent-progeny pairs can be easily identified using DNA data (Rohlfs et al. 2012). However, to cover many variable situations in real life, it should be necessary to expand model [7] to include heterogeneous variances where mendelian sampling is observed for some individuals but it remains a random value for individuals without genotyped parents. Another potential improvement of the MS model in [7] is the representation of genetic transmission (as in expression [5]) and marked genetic effects (as in [2] and [7]) which may be certainly improved. Haplotypes can be used instead of single non-phased SNP. The model is also compatible with approaches where some QTL are known, markers are preselected or markers are weighted by their effects during prediction (e.g. Zhang et al. 2011). Conclusions According to the literature on prediction of complex traits, it is justified to keep, both, individual (infinitesimal) and marked gene effects in the statistical predictive model. We gave a formal derivation of a mendelian sampling MS model where individual effects are a function of infinitesimal effects of base individuals and mendelian sampling in descendants, traced using available DNA data. At this stage of research, we are assuming complete data, simple distributional assumptions for individual and marked genetic effects, and known variances. First simulation results suggest that these simplifying assumptions should be extended to render the MS model fully competitive. Acknowledgments The first author benefits from financial support from INRA (Animal Genetics Department) and the Midi-Pyrenees Region (France). We thank the computing support of the bio-informatics platform Genotoul ( Toulouse, France) and financial support from the Cost Action TD1101 of the European Union. Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited. Appendix: Computation of individual and marked genetic effects using BLUP Let r 2 i, r2 M, and r2 e be the variance of infinitesimal effects, the genetic variance due to all QTL, and the residual variance, respectively. Also the variance of individual markers is r 2 m ¼ r2 M =k, with k ¼ 2 P j p j 1 p j. Then: a u ¼ r 2 e = r2 i þ r 2 M a m ¼ r 2 e =r2 m a i ¼ r 2 e =r2 i Solutions for models compared were: For model [1]:
9 246 Genetica (2013) 141: bl bu Z 1 0 y ¼ 10 Z 0 1 Z 0 Z þ a u A 1 Z 0 y where 1 is a vector of 1 and Z is the incidence matrix. bl is the BLUE (best linear unbiased estimator) of the general mean, and bu is the solution for individual effects. For model [2]: bl bm X 1 0 y ¼ 10 X 0 1 X 0 X þ a m I X 0 y where X = ZW, i.e., the incidence matrix times the matrix of genotypes, centered by column. bm is the solution for marked effects. Predictions from model [2] can be also obtained with the individual model: bl bu Z 1 0 y ¼ 10 Z 0 1 Z 0 Z þ a u G 1 Z 0 y where G ¼ WW 0 =k For model [3]: bl X X 2 4 bu 5 ¼ 4 X X0 1 X 1 þ a u A 1 X 0 1 X 5 2 bm X X0 2 X 1 X 0 2 X 2 þ a m I y 4 X 0 1 y 5 X 0 2 y where X 1 ¼ Z and X 2 ¼ ZW. bu is the solution for individual effects and bm is the solution for marked effects. For model [7]: bl X X 2 4 cu b 5 ¼ 4 X X0 1 X 1 þ a i I X 0 1 X 2 bm X X0 2 X 1 X 0 2 X 2 þ a m I y 4 X 0 1 y 5 X 0 2 y where X 1 ¼ Z d D and X 2 ¼ Z d 2ðW d DW b Þ; and bu b is the solution for base individuals and bm is the solution for marked effects. References Abecasis GR, Cardon RL, Cookson WO (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66: Aguilar I, Misztal I, Legarra A, Tsuruta S (2011) Efficient computation of the genomic relationship matrix and other matrices used in single-step evaluation. J Animal Breed Genet 128: De Los campos G, Naya H, Gianola D, Crossa J, Legarra A et al (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182: Duchemin SI, Colombani C, Legarra A, Baloche G, Larroque H et al (2012) Genomic selection in the French Lacaune dairy sheep breed. J Dairy Sci 95: Fernando RL, Grossman M (1989) Marker assisted selection using best linear unbiased prediction. Genet Sel Evol 21: Gianola D, De Los Campos G, Hill WG, Manfredi E, Fernando R (2009) Additive genetic variability and the Bayesian alphabet. Genetics 183: Goddard ME (2009) Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136: Goddard ME, Hayes BJ (2009) Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet 10: Hazel LN (1943) The genetic basis for constructing selection indexes. Genetics 28: Henderson CR (1975) Best linear unbiased estimation and prediction under a selection model. Biometrics 31: Henderson CR (1976) Simple method for computing inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 32:69 83 Legarra A, Fernando RL (2009) Linear models for joint association and linkage QTL mapping. Genet Sel Evol 41:43 Lund MS, Sahana G, De Koning DJ, Su G, Carlborg O (2009) Comparison of analyses of the QTLMAS XII common dataset. I: genomic selection. BMC Proc 3 (Suppl 1):S1 Meuwissen TH, Goddard ME (2000) Fine mapping of quantitative trait loci using linkage disequilibria with closely linked marker loci. Genetics 155: Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: Meuwissen TH, Luan T, Woolliams JA (2011) The unified approach to the use of genomic and pedigree information in genomic evaluations revisited. J Anim Breed Genet 128: Quaas RL (1976) Computing diagonal elements and inverse of a large numerator relationship matrix. Biometrics 32: Rohlfs RV, Fullerton SM, Weir BS (2012) Familial identification: population structure and relationship distinguishability. PLoS Genet 8:e Sargolzaei M, Schenkel FS (2009) QMSim: a large-scale genome simulator for livestock. Bioinformatics 25: First published January 28, 2009, doi: /bioinformatics/btp045 Vanraden P (2008) Efficient method to compute genomic predictions. J Dairy Sci 91: Yang JT, Manolio A, Pasquale LR, Boerwinkle E, Caporaso N et al (2011) Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 43: Zhang Z, Ding X, Liu J, De Koning DJ, Zhang Q (2011) Genomic selection for QTL-MAS data using a trait-specific relationship matrix. BMC Proc 5(Suppl 3):S15
The Dimensionality of Genomic Information and Its Effect on Genomic Prediction
 GENOMIC SELECTION The Dimensionality of Genomic Information and Its Effect on Genomic Prediction Ivan Pocrnic,*,1 Daniela A. L. Lourenco,* Yutaka Masuda,* Andres Legarra, and Ignacy Misztal* *Department
GENOMIC SELECTION The Dimensionality of Genomic Information and Its Effect on Genomic Prediction Ivan Pocrnic,*,1 Daniela A. L. Lourenco,* Yutaka Masuda,* Andres Legarra, and Ignacy Misztal* *Department
Genomic Selection Using Low-Density Marker Panels
 Copyright Ó 2009 by the Genetics Society of America DOI: 10.1534/genetics.108.100289 Genomic Selection Using Low-Density Marker Panels D. Habier,*,,1 R. L. Fernando and J. C. M. Dekkers *Institute of Animal
Copyright Ó 2009 by the Genetics Society of America DOI: 10.1534/genetics.108.100289 Genomic Selection Using Low-Density Marker Panels D. Habier,*,,1 R. L. Fernando and J. C. M. Dekkers *Institute of Animal
General aspects of genome-wide association studies
 General aspects of genome-wide association studies Abstract number 20201 Session 04 Correctly reporting statistical genetics results in the genomic era Pekka Uimari University of Helsinki Dept. of Agricultural
General aspects of genome-wide association studies Abstract number 20201 Session 04 Correctly reporting statistical genetics results in the genomic era Pekka Uimari University of Helsinki Dept. of Agricultural
Practical integration of genomic selection in dairy cattle breeding schemes
 64 th EAAP Meeting Nantes, 2013 Practical integration of genomic selection in dairy cattle breeding schemes A. BOUQUET & J. JUGA 1 Introduction Genomic selection : a revolution for animal breeders Big
64 th EAAP Meeting Nantes, 2013 Practical integration of genomic selection in dairy cattle breeding schemes A. BOUQUET & J. JUGA 1 Introduction Genomic selection : a revolution for animal breeders Big
By the end of this lecture you should be able to explain: Some of the principles underlying the statistical analysis of QTLs
 (3) QTL and GWAS methods By the end of this lecture you should be able to explain: Some of the principles underlying the statistical analysis of QTLs Under what conditions particular methods are suitable
(3) QTL and GWAS methods By the end of this lecture you should be able to explain: Some of the principles underlying the statistical analysis of QTLs Under what conditions particular methods are suitable
Genetics of dairy production
 Genetics of dairy production E-learning course from ESA Charlotte DEZETTER ZBO101R11550 Table of contents I - Genetics of dairy production 3 1. Learning objectives... 3 2. Review of Mendelian genetics...
Genetics of dairy production E-learning course from ESA Charlotte DEZETTER ZBO101R11550 Table of contents I - Genetics of dairy production 3 1. Learning objectives... 3 2. Review of Mendelian genetics...
Strategy for applying genome-wide selection in dairy cattle
 J. Anim. Breed. Genet. ISSN 0931-2668 ORIGINAL ARTICLE Strategy for applying genome-wide selection in dairy cattle L.R. Schaeffer Department of Animal and Poultry Science, Centre for Genetic Improvement
J. Anim. Breed. Genet. ISSN 0931-2668 ORIGINAL ARTICLE Strategy for applying genome-wide selection in dairy cattle L.R. Schaeffer Department of Animal and Poultry Science, Centre for Genetic Improvement
Accuracy of whole genome prediction using a genetic architecture enhanced variance-covariance matrix
 G3: Genes Genomes Genetics Early Online, published on February 9, 2015 as doi:10.1534/g3.114.016261 1 2 Accuracy of whole genome prediction using a genetic architecture enhanced variance-covariance matrix
G3: Genes Genomes Genetics Early Online, published on February 9, 2015 as doi:10.1534/g3.114.016261 1 2 Accuracy of whole genome prediction using a genetic architecture enhanced variance-covariance matrix
Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus 1
 Published June 25, 2015 Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus 1 D. A. L. Lourenco,* 2 S. Tsuruta,* B. O. Fragomeni,* Y. Masuda,* I. Aguilar, A. Legarra,
Published June 25, 2015 Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus 1 D. A. L. Lourenco,* 2 S. Tsuruta,* B. O. Fragomeni,* Y. Masuda,* I. Aguilar, A. Legarra,
Conifer Translational Genomics Network Coordinated Agricultural Project
 Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 4 Quantitative Genetics Nicholas Wheeler & David Harry Oregon
Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 4 Quantitative Genetics Nicholas Wheeler & David Harry Oregon
Sequencing Millions of Animals for Genomic Selection 2.0
 Proceedings, 10 th World Congress of Genetics Applied to Livestock Production Sequencing Millions of Animals for Genomic Selection 2.0 J.M. Hickey 1, G. Gorjanc 1, M.A. Cleveland 2, A. Kranis 1,3, J. Jenko
Proceedings, 10 th World Congress of Genetics Applied to Livestock Production Sequencing Millions of Animals for Genomic Selection 2.0 J.M. Hickey 1, G. Gorjanc 1, M.A. Cleveland 2, A. Kranis 1,3, J. Jenko
POPULATION GENETICS Winter 2005 Lecture 18 Quantitative genetics and QTL mapping
 POPULATION GENETICS Winter 2005 Lecture 18 Quantitative genetics and QTL mapping - from Darwin's time onward, it has been widely recognized that natural populations harbor a considerably degree of genetic
POPULATION GENETICS Winter 2005 Lecture 18 Quantitative genetics and QTL mapping - from Darwin's time onward, it has been widely recognized that natural populations harbor a considerably degree of genetic
Quantitative Genetics
 Quantitative Genetics Polygenic traits Quantitative Genetics 1. Controlled by several to many genes 2. Continuous variation more variation not as easily characterized into classes; individuals fall into
Quantitative Genetics Polygenic traits Quantitative Genetics 1. Controlled by several to many genes 2. Continuous variation more variation not as easily characterized into classes; individuals fall into
Linkage & Genetic Mapping in Eukaryotes. Ch. 6
 Linkage & Genetic Mapping in Eukaryotes Ch. 6 1 LINKAGE AND CROSSING OVER! In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even
Linkage & Genetic Mapping in Eukaryotes Ch. 6 1 LINKAGE AND CROSSING OVER! In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even
Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace populations
 Gorjanc et al. BMC Genomics (2016) 17:30 DOI 10.1186/s12864-015-2345-z RESEARCH ARTICLE Open Access Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace
Gorjanc et al. BMC Genomics (2016) 17:30 DOI 10.1186/s12864-015-2345-z RESEARCH ARTICLE Open Access Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace
Genomic Selection using low-density marker panels
 Genetics: Published Articles Ahead of Print, published on March 18, 2009 as 10.1534/genetics.108.100289 Genomic Selection using low-density marker panels D. Habier 1,2, R. L. Fernando 2 and J. C. M. Dekkers
Genetics: Published Articles Ahead of Print, published on March 18, 2009 as 10.1534/genetics.108.100289 Genomic Selection using low-density marker panels D. Habier 1,2, R. L. Fernando 2 and J. C. M. Dekkers
Managing genetic groups in single-step genomic evaluations applied on female fertility traits in Nordic Red Dairy cattle
 Abstract Managing genetic groups in single-step genomic evaluations applied on female fertility traits in Nordic Red Dairy cattle K. Matilainen 1, M. Koivula 1, I. Strandén 1, G.P. Aamand 2 and E.A. Mäntysaari
Abstract Managing genetic groups in single-step genomic evaluations applied on female fertility traits in Nordic Red Dairy cattle K. Matilainen 1, M. Koivula 1, I. Strandén 1, G.P. Aamand 2 and E.A. Mäntysaari
A strategy for multiple linkage disequilibrium mapping methods to validate additive QTL. Abstracts
 Proceedings 59th ISI World Statistics Congress, 5-30 August 013, Hong Kong (Session CPS03) p.3947 A strategy for multiple linkage disequilibrium mapping methods to validate additive QTL Li Yi 1,4, Jong-Joo
Proceedings 59th ISI World Statistics Congress, 5-30 August 013, Hong Kong (Session CPS03) p.3947 A strategy for multiple linkage disequilibrium mapping methods to validate additive QTL Li Yi 1,4, Jong-Joo
Comparison of genomic predictions using genomic relationship matrices built with different weighting factors to account for locus-specific variances
 J. Dairy Sci. 97 :6547 6559 http://dx.doi.org/ 0.368/jds.04-80 American Dairy Science Association, 04. Open access under CC BY-NC-ND license. Comparison of genomic predictions using genomic relationship
J. Dairy Sci. 97 :6547 6559 http://dx.doi.org/ 0.368/jds.04-80 American Dairy Science Association, 04. Open access under CC BY-NC-ND license. Comparison of genomic predictions using genomic relationship
Daniela Lourenco, Un. of Georgia 6/1/17. Pedigree. Individual 1. Individual 2
 Traditional evaluation The promise of genomics for beef improvement Daniela Lourenco Keith Bertrand Pedigree Performance EPD BLUP Progeny Heather Bradford, Steve Miller, Ignacy Misztal BIF - Athens, GA
Traditional evaluation The promise of genomics for beef improvement Daniela Lourenco Keith Bertrand Pedigree Performance EPD BLUP Progeny Heather Bradford, Steve Miller, Ignacy Misztal BIF - Athens, GA
Impact of Genotype Imputation on the Performance of GBLUP and Bayesian Methods for Genomic Prediction
 Impact of Genotype Imputation on the Performance of GBLUP and Bayesian Methods for Genomic Prediction Liuhong Chen 1 *, Changxi Li 1,2, Mehdi Sargolzaei 3, Flavio Schenkel 4 * 1 Department of Agricultural,
Impact of Genotype Imputation on the Performance of GBLUP and Bayesian Methods for Genomic Prediction Liuhong Chen 1 *, Changxi Li 1,2, Mehdi Sargolzaei 3, Flavio Schenkel 4 * 1 Department of Agricultural,
Fundamentals of Genomic Selection
 Fundamentals of Genomic Selection Jack Dekkers Animal Breeding & Genetics Department of Animal Science Iowa State University Past and Current Selection Strategies selection Black box of Genes Quantitative
Fundamentals of Genomic Selection Jack Dekkers Animal Breeding & Genetics Department of Animal Science Iowa State University Past and Current Selection Strategies selection Black box of Genes Quantitative
Implementation of Genomic Selection in Pig Breeding Programs
 Implementation of Genomic Selection in Pig Breeding Programs Jack Dekkers Animal Breeding & Genetics Department of Animal Science Iowa State University Dekkers, J.C.M. 2010. Use of high-density marker
Implementation of Genomic Selection in Pig Breeding Programs Jack Dekkers Animal Breeding & Genetics Department of Animal Science Iowa State University Dekkers, J.C.M. 2010. Use of high-density marker
Genomic selection and its potential to change cattle breeding
 ICAR keynote presentations Genomic selection and its potential to change cattle breeding Reinhard Reents - Chairman of the Steering Committee of Interbull - Secretary of ICAR - General Manager of vit,
ICAR keynote presentations Genomic selection and its potential to change cattle breeding Reinhard Reents - Chairman of the Steering Committee of Interbull - Secretary of ICAR - General Manager of vit,
Genome-wide association studies (GWAS) Part 1
 Genome-wide association studies (GWAS) Part 1 Matti Pirinen FIMM, University of Helsinki 03.12.2013, Kumpula Campus FIMM - Institiute for Molecular Medicine Finland www.fimm.fi Published Genome-Wide Associations
Genome-wide association studies (GWAS) Part 1 Matti Pirinen FIMM, University of Helsinki 03.12.2013, Kumpula Campus FIMM - Institiute for Molecular Medicine Finland www.fimm.fi Published Genome-Wide Associations
Chapter 14: Mendel and the Gene Idea
 Chapter 4: Mendel and the Gene Idea. The Experiments of Gregor Mendel 2. Beyond Mendelian Genetics 3. Human Genetics . The Experiments of Gregor Mendel Chapter Reading pp. 268-276 TECHNIQUE Parental generation
Chapter 4: Mendel and the Gene Idea. The Experiments of Gregor Mendel 2. Beyond Mendelian Genetics 3. Human Genetics . The Experiments of Gregor Mendel Chapter Reading pp. 268-276 TECHNIQUE Parental generation
An introduction to genetics and molecular biology
 An introduction to genetics and molecular biology Cavan Reilly September 5, 2017 Table of contents Introduction to biology Some molecular biology Gene expression Mendelian genetics Some more molecular
An introduction to genetics and molecular biology Cavan Reilly September 5, 2017 Table of contents Introduction to biology Some molecular biology Gene expression Mendelian genetics Some more molecular
Genome-Wide Association Studies (GWAS): Computational Them
 Genome-Wide Association Studies (GWAS): Computational Themes and Caveats October 14, 2014 Many issues in Genomewide Association Studies We show that even for the simplest analysis, there is little consensus
Genome-Wide Association Studies (GWAS): Computational Themes and Caveats October 14, 2014 Many issues in Genomewide Association Studies We show that even for the simplest analysis, there is little consensus
Reliabilities of genomic prediction using combined reference data of the Nordic Red dairy cattle populations
 J. Dairy Sci. 94 :4700 4707 doi: 10.3168/jds.2010-3765 American Dairy Science Association, 2011. Open access under CC BY-NC-ND license. Reliabilities of genomic prediction using combined reference data
J. Dairy Sci. 94 :4700 4707 doi: 10.3168/jds.2010-3765 American Dairy Science Association, 2011. Open access under CC BY-NC-ND license. Reliabilities of genomic prediction using combined reference data
LECTURE 5: LINKAGE AND GENETIC MAPPING
 LECTURE 5: LINKAGE AND GENETIC MAPPING Reading: Ch. 5, p. 113-131 Problems: Ch. 5, solved problems I, II; 5-2, 5-4, 5-5, 5.7 5.9, 5-12, 5-16a; 5-17 5-19, 5-21; 5-22a-e; 5-23 The dihybrid crosses that we
LECTURE 5: LINKAGE AND GENETIC MAPPING Reading: Ch. 5, p. 113-131 Problems: Ch. 5, solved problems I, II; 5-2, 5-4, 5-5, 5.7 5.9, 5-12, 5-16a; 5-17 5-19, 5-21; 5-22a-e; 5-23 The dihybrid crosses that we
DESIGNS FOR QTL DETECTION IN LIVESTOCK AND THEIR IMPLICATIONS FOR MAS
 DESIGNS FOR QTL DETECTION IN LIVESTOCK AND THEIR IMPLICATIONS FOR MAS D.J de Koning, J.C.M. Dekkers & C.S. Haley Roslin Institute, Roslin, EH25 9PS, United Kingdom, DJ.deKoning@BBSRC.ac.uk Iowa State University,
DESIGNS FOR QTL DETECTION IN LIVESTOCK AND THEIR IMPLICATIONS FOR MAS D.J de Koning, J.C.M. Dekkers & C.S. Haley Roslin Institute, Roslin, EH25 9PS, United Kingdom, DJ.deKoning@BBSRC.ac.uk Iowa State University,
Association Mapping in Plants PLSC 731 Plant Molecular Genetics Phil McClean April, 2010
 Association Mapping in Plants PLSC 731 Plant Molecular Genetics Phil McClean April, 2010 Traditional QTL approach Uses standard bi-parental mapping populations o F2 or RI These have a limited number of
Association Mapping in Plants PLSC 731 Plant Molecular Genetics Phil McClean April, 2010 Traditional QTL approach Uses standard bi-parental mapping populations o F2 or RI These have a limited number of
Human linkage analysis. fundamental concepts
 Human linkage analysis fundamental concepts Genes and chromosomes Alelles of genes located on different chromosomes show independent assortment (Mendel s 2nd law) For 2 genes: 4 gamete classes with equal
Human linkage analysis fundamental concepts Genes and chromosomes Alelles of genes located on different chromosomes show independent assortment (Mendel s 2nd law) For 2 genes: 4 gamete classes with equal
Supplementary Note: Detecting population structure in rare variant data
 Supplementary Note: Detecting population structure in rare variant data Inferring ancestry from genetic data is a common problem in both population and medical genetic studies, and many methods exist to
Supplementary Note: Detecting population structure in rare variant data Inferring ancestry from genetic data is a common problem in both population and medical genetic studies, and many methods exist to
MAS refers to the use of DNA markers that are tightly-linked to target loci as a substitute for or to assist phenotypic screening.
 Marker assisted selection in rice Introduction The development of DNA (or molecular) markers has irreversibly changed the disciplines of plant genetics and plant breeding. While there are several applications
Marker assisted selection in rice Introduction The development of DNA (or molecular) markers has irreversibly changed the disciplines of plant genetics and plant breeding. While there are several applications
CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016
 CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016 Topics Genetic variation Population structure Linkage disequilibrium Natural disease variants Genome Wide Association Studies Gene
CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016 Topics Genetic variation Population structure Linkage disequilibrium Natural disease variants Genome Wide Association Studies Gene
Human SNP haplotypes. Statistics 246, Spring 2002 Week 15, Lecture 1
 Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Genotype AA Aa aa Total N ind We assume that the order of alleles in Aa does not play a role. The genotypic frequencies follow as
 N µ s m r - - - - Genetic variation - From genotype frequencies to allele frequencies The last lecture focused on mutation as the ultimate process introducing genetic variation into populations. We have
N µ s m r - - - - Genetic variation - From genotype frequencies to allele frequencies The last lecture focused on mutation as the ultimate process introducing genetic variation into populations. We have
Genomic prediction for Nordic Red Cattle using one-step and selection index blending
 J. Dairy Sci. 95 :909 917 doi: 10.3168/jds.011-4804 American Dairy Science Association, 01. Open access under CC BY-NC-ND license. Genomic prediction for Nordic Red Cattle using one-step and selection
J. Dairy Sci. 95 :909 917 doi: 10.3168/jds.011-4804 American Dairy Science Association, 01. Open access under CC BY-NC-ND license. Genomic prediction for Nordic Red Cattle using one-step and selection
BLUP and Genomic Selection
 BLUP and Genomic Selection Alison Van Eenennaam Cooperative Extension Specialist Animal Biotechnology and Genomics University of California, Davis, USA alvaneenennaam@ucdavis.edu http://animalscience.ucdavis.edu/animalbiotech/
BLUP and Genomic Selection Alison Van Eenennaam Cooperative Extension Specialist Animal Biotechnology and Genomics University of California, Davis, USA alvaneenennaam@ucdavis.edu http://animalscience.ucdavis.edu/animalbiotech/
Gene Linkage and Genetic. Mapping. Key Concepts. Key Terms. Concepts in Action
 Gene Linkage and Genetic 4 Mapping Key Concepts Genes that are located in the same chromosome and that do not show independent assortment are said to be linked. The alleles of linked genes present together
Gene Linkage and Genetic 4 Mapping Key Concepts Genes that are located in the same chromosome and that do not show independent assortment are said to be linked. The alleles of linked genes present together
Basic Concepts of Human Genetics
 Basic Concepts of Human Genetics The genetic information of an individual is contained in 23 pairs of chromosomes. Every human cell contains the 23 pair of chromosomes. One pair is called sex chromosomes
Basic Concepts of Human Genetics The genetic information of an individual is contained in 23 pairs of chromosomes. Every human cell contains the 23 pair of chromosomes. One pair is called sex chromosomes
DO NOT OPEN UNTIL TOLD TO START
 DO NOT OPEN UNTIL TOLD TO START BIO 312, Section 1, Spring 2011 February 21, 2011 Exam 1 Name (print neatly) Instructor 7 digit student ID INSTRUCTIONS: 1. There are 11 pages to the exam. Make sure you
DO NOT OPEN UNTIL TOLD TO START BIO 312, Section 1, Spring 2011 February 21, 2011 Exam 1 Name (print neatly) Instructor 7 digit student ID INSTRUCTIONS: 1. There are 11 pages to the exam. Make sure you
SNPs - GWAS - eqtls. Sebastian Schmeier
 SNPs - GWAS - eqtls s.schmeier@gmail.com http://sschmeier.github.io/bioinf-workshop/ 17.08.2015 Overview Single nucleotide polymorphism (refresh) SNPs effect on genes (refresh) Genome-wide association
SNPs - GWAS - eqtls s.schmeier@gmail.com http://sschmeier.github.io/bioinf-workshop/ 17.08.2015 Overview Single nucleotide polymorphism (refresh) SNPs effect on genes (refresh) Genome-wide association
Experimental Design and Sample Size Requirement for QTL Mapping
 Experimental Design and Sample Size Requirement for QTL Mapping Zhao-Bang Zeng Bioinformatics Research Center Departments of Statistics and Genetics North Carolina State University zeng@stat.ncsu.edu 1
Experimental Design and Sample Size Requirement for QTL Mapping Zhao-Bang Zeng Bioinformatics Research Center Departments of Statistics and Genetics North Carolina State University zeng@stat.ncsu.edu 1
Conifer Translational Genomics Network Coordinated Agricultural Project
 Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 3 Population Genetics Nicholas Wheeler & David Harry Oregon
Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 3 Population Genetics Nicholas Wheeler & David Harry Oregon
Mapping and Mapping Populations
 Mapping and Mapping Populations Types of mapping populations F 2 o Two F 1 individuals are intermated Backcross o Cross of a recurrent parent to a F 1 Recombinant Inbred Lines (RILs; F 2 -derived lines)
Mapping and Mapping Populations Types of mapping populations F 2 o Two F 1 individuals are intermated Backcross o Cross of a recurrent parent to a F 1 Recombinant Inbred Lines (RILs; F 2 -derived lines)
Genetic Equilibrium: Human Diversity Student Version
 Genetic Equilibrium: Human Diversity Student Version Key Concepts: A population is a group of organisms of the same species that live and breed in the same area. Alleles are alternate forms of genes. In
Genetic Equilibrium: Human Diversity Student Version Key Concepts: A population is a group of organisms of the same species that live and breed in the same area. Alleles are alternate forms of genes. In
Answers to additional linkage problems.
 Spring 2013 Biology 321 Answers to Assignment Set 8 Chapter 4 http://fire.biol.wwu.edu/trent/trent/iga_10e_sm_chapter_04.pdf Answers to additional linkage problems. Problem -1 In this cell, there two copies
Spring 2013 Biology 321 Answers to Assignment Set 8 Chapter 4 http://fire.biol.wwu.edu/trent/trent/iga_10e_sm_chapter_04.pdf Answers to additional linkage problems. Problem -1 In this cell, there two copies
Conifer Translational Genomics Network Coordinated Agricultural Project
 Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 2 Genes, Genomes, and Mendel Nicholas Wheeler & David Harry
Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 2 Genes, Genomes, and Mendel Nicholas Wheeler & David Harry
Question. In the last 100 years. What is Feed Efficiency? Genetics of Feed Efficiency and Applications for the Dairy Industry
 Question Genetics of Feed Efficiency and Applications for the Dairy Industry Can we increase milk yield while decreasing feed cost? If so, how can we accomplish this? Stephanie McKay University of Vermont
Question Genetics of Feed Efficiency and Applications for the Dairy Industry Can we increase milk yield while decreasing feed cost? If so, how can we accomplish this? Stephanie McKay University of Vermont
AP BIOLOGY Population Genetics and Evolution Lab
 AP BIOLOGY Population Genetics and Evolution Lab In 1908 G.H. Hardy and W. Weinberg independently suggested a scheme whereby evolution could be viewed as changes in the frequency of alleles in a population
AP BIOLOGY Population Genetics and Evolution Lab In 1908 G.H. Hardy and W. Weinberg independently suggested a scheme whereby evolution could be viewed as changes in the frequency of alleles in a population
Molecular markers in plant breeding
 Molecular markers in plant breeding Jumbo MacDonald et al., MAIZE BREEDERS COURSE Palace Hotel Arusha, Tanzania 4 Sep to 16 Sep 2016 Molecular Markers QTL Mapping Association mapping GWAS Genomic Selection
Molecular markers in plant breeding Jumbo MacDonald et al., MAIZE BREEDERS COURSE Palace Hotel Arusha, Tanzania 4 Sep to 16 Sep 2016 Molecular Markers QTL Mapping Association mapping GWAS Genomic Selection
11.1 Genetic Variation Within Population. KEY CONCEPT A population shares a common gene pool.
 11.1 Genetic Variation Within Population KEY CONCEPT A population shares a common gene pool. 11.1 Genetic Variation Within Population Genetic variation in a population increases the chance that some individuals
11.1 Genetic Variation Within Population KEY CONCEPT A population shares a common gene pool. 11.1 Genetic Variation Within Population Genetic variation in a population increases the chance that some individuals
User s Guide Version 1.10 (Last update: July 12, 2013)
 User s Guide Version 1.10 (Last update: July 12, 2013) Centre for Genetic Improvement of Livestock Department of Animal and Poultry Science University of Guelph Guelph, Canada Mehdi Sargolzaei and Flavio
User s Guide Version 1.10 (Last update: July 12, 2013) Centre for Genetic Improvement of Livestock Department of Animal and Poultry Science University of Guelph Guelph, Canada Mehdi Sargolzaei and Flavio
Genome-wide association mapping using single-step GBLUP!!!!
 Genome-wide association mapping using single-step GBLUP!!!! Ignacy'Misztal,'Joy'Wang'University!of!Georgia Ignacio'Aguilar,'INIA,!Uruguay! Andres'Legarra,!INRA,!France! Bill'Muir,!Purdue!University! Rohan'Fernando,!Iowa!State!!!'
Genome-wide association mapping using single-step GBLUP!!!! Ignacy'Misztal,'Joy'Wang'University!of!Georgia Ignacio'Aguilar,'INIA,!Uruguay! Andres'Legarra,!INRA,!France! Bill'Muir,!Purdue!University! Rohan'Fernando,!Iowa!State!!!'
Genome-wide prediction of maize single-cross performance, considering non-additive genetic effects
 Genome-wide prediction of maize single-cross performance, considering non-additive genetic effects J.P.R. Santos 1, H.D. Pereira 2, R.G. Von Pinho 2, L.P.M. Pires 2, R.B. Camargos 2 and M. Balestre 3 1
Genome-wide prediction of maize single-cross performance, considering non-additive genetic effects J.P.R. Santos 1, H.D. Pereira 2, R.G. Von Pinho 2, L.P.M. Pires 2, R.B. Camargos 2 and M. Balestre 3 1
Exam 1 Answers Biology 210 Sept. 20, 2006
 Exam Answers Biology 20 Sept. 20, 2006 Name: Section:. (5 points) Circle the answer that gives the maximum number of different alleles that might exist for any one locus in a normal mammalian cell. A.
Exam Answers Biology 20 Sept. 20, 2006 Name: Section:. (5 points) Circle the answer that gives the maximum number of different alleles that might exist for any one locus in a normal mammalian cell. A.
Lecture WS Evolutionary Genetics Part I - Jochen B. W. Wolf 1
 N µ s m r - - - Mutation Effect on allele frequencies We have seen that both genotype and allele frequencies are not expected to change by Mendelian inheritance in the absence of any other factors. We
N µ s m r - - - Mutation Effect on allele frequencies We have seen that both genotype and allele frequencies are not expected to change by Mendelian inheritance in the absence of any other factors. We
7-1. Read this exercise before you come to the laboratory. Review the lecture notes from October 15 (Hardy-Weinberg Equilibrium)
 7-1 Biology 1001 Lab 7: POPULATION GENETICS PREPARTION Read this exercise before you come to the laboratory. Review the lecture notes from October 15 (Hardy-Weinberg Equilibrium) OBECTIVES At the end of
7-1 Biology 1001 Lab 7: POPULATION GENETICS PREPARTION Read this exercise before you come to the laboratory. Review the lecture notes from October 15 (Hardy-Weinberg Equilibrium) OBECTIVES At the end of
Computational Workflows for Genome-Wide Association Study: I
 Computational Workflows for Genome-Wide Association Study: I Department of Computer Science Brown University, Providence sorin@cs.brown.edu October 16, 2014 Outline 1 Outline 2 3 Monogenic Mendelian Diseases
Computational Workflows for Genome-Wide Association Study: I Department of Computer Science Brown University, Providence sorin@cs.brown.edu October 16, 2014 Outline 1 Outline 2 3 Monogenic Mendelian Diseases
MSc specialization Animal Breeding and Genetics at Wageningen University
 MSc specialization Animal Breeding and Genetics at Wageningen University The MSc specialization Animal Breeding and Genetics focuses on the development and transfer of knowledge in the area of selection
MSc specialization Animal Breeding and Genetics at Wageningen University The MSc specialization Animal Breeding and Genetics focuses on the development and transfer of knowledge in the area of selection
Bio 6 Natural Selection Lab
 Bio 6 Natural Selection Lab Overview In this laboratory you will demonstrate the process of evolution by natural selection by carrying out a predator/prey simulation. Through this exercise you will observe
Bio 6 Natural Selection Lab Overview In this laboratory you will demonstrate the process of evolution by natural selection by carrying out a predator/prey simulation. Through this exercise you will observe
A journey: opportunities & challenges of melding genomics into U.S. sheep breeding programs
 A journey: opportunities & challenges of melding genomics into U.S. sheep breeding programs Presenter: Dr. Ron Lewis, Department of Animal Science, University of Nebraska-Lincoln Host/Moderator: Dr. Jay
A journey: opportunities & challenges of melding genomics into U.S. sheep breeding programs Presenter: Dr. Ron Lewis, Department of Animal Science, University of Nebraska-Lincoln Host/Moderator: Dr. Jay
 http://genemapping.org/ Epistasis in Association Studies David Evans Law of Independent Assortment Biological Epistasis Bateson (99) a masking effect whereby a variant or allele at one locus prevents
http://genemapping.org/ Epistasis in Association Studies David Evans Law of Independent Assortment Biological Epistasis Bateson (99) a masking effect whereby a variant or allele at one locus prevents
Topic 11. Genetics. I. Patterns of Inheritance: One Trait Considered
 Topic 11. Genetics Introduction. Genetics is the study of how the biological information that determines the structure and function of organisms is passed from one generation to the next. It is also concerned
Topic 11. Genetics Introduction. Genetics is the study of how the biological information that determines the structure and function of organisms is passed from one generation to the next. It is also concerned
Midterm 1 Results. Midterm 1 Akey/ Fields Median Number of Students. Exam Score
 Midterm 1 Results 10 Midterm 1 Akey/ Fields Median - 69 8 Number of Students 6 4 2 0 21 26 31 36 41 46 51 56 61 66 71 76 81 86 91 96 101 Exam Score Quick review of where we left off Parental type: the
Midterm 1 Results 10 Midterm 1 Akey/ Fields Median - 69 8 Number of Students 6 4 2 0 21 26 31 36 41 46 51 56 61 66 71 76 81 86 91 96 101 Exam Score Quick review of where we left off Parental type: the
Solve Mendelian Genetics Problems
 Solve Problems Free PDF ebook Download: Solve Problems Download or Read Online ebook solve mendelian genetics problems in PDF Format From The Best User Guide Database AP Biology I ' Cate. PRACTICE 1: BASIC.
Solve Problems Free PDF ebook Download: Solve Problems Download or Read Online ebook solve mendelian genetics problems in PDF Format From The Best User Guide Database AP Biology I ' Cate. PRACTICE 1: BASIC.
POPULATION GENETICS: The study of the rules governing the maintenance and transmission of genetic variation in natural populations.
 POPULATION GENETICS: The study of the rules governing the maintenance and transmission of genetic variation in natural populations. DARWINIAN EVOLUTION BY NATURAL SELECTION Many more individuals are born
POPULATION GENETICS: The study of the rules governing the maintenance and transmission of genetic variation in natural populations. DARWINIAN EVOLUTION BY NATURAL SELECTION Many more individuals are born
Use of sib-pair linkage methods for the estimation of the genetic variance at a quantitative trait locus
 Original article Use of sib-pair linkage methods for the estimation of the genetic variance at a quantitative trait locus H Hamann, KU Götz Bayerische Landesanstalt f3r Tierzv,cht, Prof Diirrwaechter-Platz
Original article Use of sib-pair linkage methods for the estimation of the genetic variance at a quantitative trait locus H Hamann, KU Götz Bayerische Landesanstalt f3r Tierzv,cht, Prof Diirrwaechter-Platz
-Genes on the same chromosome are called linked. Human -23 pairs of chromosomes, ~35,000 different genes expressed.
 Linkage -Genes on the same chromosome are called linked Human -23 pairs of chromosomes, ~35,000 different genes expressed. - average of 1,500 genes/chromosome Following Meiosis Parental chromosomal types
Linkage -Genes on the same chromosome are called linked Human -23 pairs of chromosomes, ~35,000 different genes expressed. - average of 1,500 genes/chromosome Following Meiosis Parental chromosomal types
Conifer Translational Genomics Network Coordinated Agricultural Project
 Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 11 Association Genetics Nicholas Wheeler & David Harry Oregon
Conifer Translational Genomics Network Coordinated Agricultural Project Genomics in Tree Breeding and Forest Ecosystem Management ----- Module 11 Association Genetics Nicholas Wheeler & David Harry Oregon
Introduction to quantitative genetics
 8 Introduction to quantitative genetics Purpose and expected outcomes Most of the traits that plant breeders are interested in are quantitatively inherited. It is important to understand the genetics that
8 Introduction to quantitative genetics Purpose and expected outcomes Most of the traits that plant breeders are interested in are quantitatively inherited. It is important to understand the genetics that
Melding genomics and quantitative genetics in sheep breeding programs: opportunities and limits
 Melding genomics and quantitative genetics in sheep breeding programs: opportunities and limits Ron Lewis Genetics Stakeholders Committee ASI Convention, Scottsdale, AZ Jan. 28, 2016 My talk Genomics road
Melding genomics and quantitative genetics in sheep breeding programs: opportunities and limits Ron Lewis Genetics Stakeholders Committee ASI Convention, Scottsdale, AZ Jan. 28, 2016 My talk Genomics road
University of Groningen. The value of haplotypes Vries, Anne René de
 University of Groningen The value of haplotypes Vries, Anne René de IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document
University of Groningen The value of haplotypes Vries, Anne René de IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document
Random Allelic Variation
 Random Allelic Variation AKA Genetic Drift Genetic Drift a non-adaptive mechanism of evolution (therefore, a theory of evolution) that sometimes operates simultaneously with others, such as natural selection
Random Allelic Variation AKA Genetic Drift Genetic Drift a non-adaptive mechanism of evolution (therefore, a theory of evolution) that sometimes operates simultaneously with others, such as natural selection
Runs of Homozygosity Analysis Tutorial
 Runs of Homozygosity Analysis Tutorial Release 8.7.0 Golden Helix, Inc. March 22, 2017 Contents 1. Overview of the Project 2 2. Identify Runs of Homozygosity 6 Illustrative Example...............................................
Runs of Homozygosity Analysis Tutorial Release 8.7.0 Golden Helix, Inc. March 22, 2017 Contents 1. Overview of the Project 2 2. Identify Runs of Homozygosity 6 Illustrative Example...............................................
Basic Concepts of Human Genetics
 Basic oncepts of Human enetics The genetic information of an individual is contained in 23 pairs of chromosomes. Every human cell contains the 23 pair of chromosomes. ne pair is called sex chromosomes
Basic oncepts of Human enetics The genetic information of an individual is contained in 23 pairs of chromosomes. Every human cell contains the 23 pair of chromosomes. ne pair is called sex chromosomes
Dr. Mallery Biology Workshop Fall Semester CELL REPRODUCTION and MENDELIAN GENETICS
 Dr. Mallery Biology 150 - Workshop Fall Semester CELL REPRODUCTION and MENDELIAN GENETICS CELL REPRODUCTION The goal of today's exercise is for you to look at mitosis and meiosis and to develop the ability
Dr. Mallery Biology 150 - Workshop Fall Semester CELL REPRODUCTION and MENDELIAN GENETICS CELL REPRODUCTION The goal of today's exercise is for you to look at mitosis and meiosis and to develop the ability
HARDY-WEINBERG EQUILIBRIUM
 29 HARDY-WEINBERG EQUILIBRIUM Objectives Understand the Hardy-Weinberg principle and its importance. Understand the chi-square test of statistical independence and its use. Determine the genotype and allele
29 HARDY-WEINBERG EQUILIBRIUM Objectives Understand the Hardy-Weinberg principle and its importance. Understand the chi-square test of statistical independence and its use. Determine the genotype and allele
1999 American Agricultural Economics Association Annual Meeting
 Producer Prediction of Optimal Sire Characteristics Impacting Farm Profitibility In a Stochastic Bio-Economic Decision Framework William Herring, and Vern Pierce 1 Copyright 1999 by Vern Pierce. All rights
Producer Prediction of Optimal Sire Characteristics Impacting Farm Profitibility In a Stochastic Bio-Economic Decision Framework William Herring, and Vern Pierce 1 Copyright 1999 by Vern Pierce. All rights
Genetic Analysis of Cow Survival in the Israeli Dairy Cattle Population
 Genetic Analysis of Cow Survival in the Israeli Dairy Cattle Population Petek Settar 1 and Joel I. Weller Institute of Animal Sciences A. R. O., The Volcani Center, Bet Dagan 50250, Israel Abstract The
Genetic Analysis of Cow Survival in the Israeli Dairy Cattle Population Petek Settar 1 and Joel I. Weller Institute of Animal Sciences A. R. O., The Volcani Center, Bet Dagan 50250, Israel Abstract The
LAB. POPULATION GENETICS. 1. Explain what is meant by a population being in Hardy-Weinberg equilibrium.
 Period Date LAB. POPULATION GENETICS PRE-LAB 1. Explain what is meant by a population being in Hardy-Weinberg equilibrium. 2. List and briefly explain the 5 conditions that need to be met to maintain a
Period Date LAB. POPULATION GENETICS PRE-LAB 1. Explain what is meant by a population being in Hardy-Weinberg equilibrium. 2. List and briefly explain the 5 conditions that need to be met to maintain a
Course Overview. Interacting genes. Complementation. Complementation. February 15
 Course Overview Interacting genes http://www.erin.utoronto.ca/~w3bio/bio207/index.htm February 15 Outline Week Topic Chapter 1 Course objectives and Introduction to genetics Ch. 1 & Ch. 2 2 Human Pedigrees
Course Overview Interacting genes http://www.erin.utoronto.ca/~w3bio/bio207/index.htm February 15 Outline Week Topic Chapter 1 Course objectives and Introduction to genetics Ch. 1 & Ch. 2 2 Human Pedigrees
H3A - Genome-Wide Association testing SOP
 H3A - Genome-Wide Association testing SOP Introduction File format Strand errors Sample quality control Marker quality control Batch effects Population stratification Association testing Replication Meta
H3A - Genome-Wide Association testing SOP Introduction File format Strand errors Sample quality control Marker quality control Batch effects Population stratification Association testing Replication Meta
Why can GBS be complicated? Tools for filtering, error correction and imputation.
 Why can GBS be complicated? Tools for filtering, error correction and imputation. Edward Buckler USDA-ARS Cornell University http://www.maizegenetics.net Many Organisms Are Diverse Humans are at the lower
Why can GBS be complicated? Tools for filtering, error correction and imputation. Edward Buckler USDA-ARS Cornell University http://www.maizegenetics.net Many Organisms Are Diverse Humans are at the lower
Concepts of Genetics Ninth Edition Klug, Cummings, Spencer, Palladino
 PowerPoint Lecture Presentation for Concepts of Genetics Ninth Edition Klug, Cummings, Spencer, Palladino Chapter 5 Chromosome Mapping in Eukaryotes Copyright Copyright 2009 Pearson 2009 Pearson Education,
PowerPoint Lecture Presentation for Concepts of Genetics Ninth Edition Klug, Cummings, Spencer, Palladino Chapter 5 Chromosome Mapping in Eukaryotes Copyright Copyright 2009 Pearson 2009 Pearson Education,
Population Genetics. Lab Exercise 14. Introduction. Contents. Objectives
 Lab Exercise Population Genetics Contents Objectives 1 Introduction 1 Activity.1 Calculating Frequencies 2 Activity.2 More Hardy-Weinberg 3 Resutls Section 4 Introduction Unlike Mendelian genetics which
Lab Exercise Population Genetics Contents Objectives 1 Introduction 1 Activity.1 Calculating Frequencies 2 Activity.2 More Hardy-Weinberg 3 Resutls Section 4 Introduction Unlike Mendelian genetics which
Introduction to Indexes
 Introduction to Indexes Robert L. (Bob) Weaber, Ph.D. University of Missouri, Columbia, MO 65211 Why do we need indexes? The complications of multiple-trait selection and animal breeding decisions may
Introduction to Indexes Robert L. (Bob) Weaber, Ph.D. University of Missouri, Columbia, MO 65211 Why do we need indexes? The complications of multiple-trait selection and animal breeding decisions may
The Making of the Fittest: Natural Selection and Adaptation
 ALLELE AND PHENOTYPE FREQUENCIES IN ROCK POCKET MOUSE POPULATIONS INTRODUCTION The tiny rock pocket mouse weighs just 15 grams, about as much as a handful of paper clips. A typical rock pocket mouse is
ALLELE AND PHENOTYPE FREQUENCIES IN ROCK POCKET MOUSE POPULATIONS INTRODUCTION The tiny rock pocket mouse weighs just 15 grams, about as much as a handful of paper clips. A typical rock pocket mouse is
Genomic selection in American chestnut backcross populations
 Genomic selection in American chestnut backcross populations Jared Westbrook The American Chestnut Foundation TACF Annual Meeting Fall 2017 Portland, ME Selection against blight susceptibility in seed
Genomic selection in American chestnut backcross populations Jared Westbrook The American Chestnut Foundation TACF Annual Meeting Fall 2017 Portland, ME Selection against blight susceptibility in seed
Inheritance Biology. Unit Map. Unit
 Unit 8 Unit Map 8.A Mendelian principles 482 8.B Concept of gene 483 8.C Extension of Mendelian principles 485 8.D Gene mapping methods 495 8.E Extra chromosomal inheritance 501 8.F Microbial genetics
Unit 8 Unit Map 8.A Mendelian principles 482 8.B Concept of gene 483 8.C Extension of Mendelian principles 485 8.D Gene mapping methods 495 8.E Extra chromosomal inheritance 501 8.F Microbial genetics
Genomic Selection in R
 Genomic Selection in R Giovanny Covarrubias-Pazaran Department of Horticulture, University of Wisconsin, Madison, Wisconsin, Unites States of America E-mail: covarrubiasp@wisc.edu. Most traits of agronomic
Genomic Selection in R Giovanny Covarrubias-Pazaran Department of Horticulture, University of Wisconsin, Madison, Wisconsin, Unites States of America E-mail: covarrubiasp@wisc.edu. Most traits of agronomic
Mapping and selection of bacterial spot resistance in complex populations. David Francis, Sung-Chur Sim, Hui Wang, Matt Robbins, Wencai Yang.
 Mapping and selection of bacterial spot resistance in complex populations. David Francis, Sung-Chur Sim, Hui Wang, Matt Robbins, Wencai Yang. Mapping and selection of bacterial spot resistance in complex
Mapping and selection of bacterial spot resistance in complex populations. David Francis, Sung-Chur Sim, Hui Wang, Matt Robbins, Wencai Yang. Mapping and selection of bacterial spot resistance in complex
Exploiting genomic information on purebred and crossbred pigs
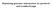 Exploiting genomic information on purebred and crossbred pigs ISSN: 1652-6880 ISBN (print version): 978-91-576-8438-7 ISBN (electronic version): 978-91-576-8439-4 ISBN: 678-94-6257-601-8 Thesis committee
Exploiting genomic information on purebred and crossbred pigs ISSN: 1652-6880 ISBN (print version): 978-91-576-8438-7 ISBN (electronic version): 978-91-576-8439-4 ISBN: 678-94-6257-601-8 Thesis committee
Using the Trio Workflow in Partek Genomics Suite v6.6
 Using the Trio Workflow in Partek Genomics Suite v6.6 This user guide will illustrate the use of the Trio/Duo workflow in Partek Genomics Suite (PGS) and discuss the basic functions available within the
Using the Trio Workflow in Partek Genomics Suite v6.6 This user guide will illustrate the use of the Trio/Duo workflow in Partek Genomics Suite (PGS) and discuss the basic functions available within the
Chapter 14. Mendel and the Gene Idea
 Chapter 14 Mendel and the Gene Idea Gregor Mendel Gregor Mendel documented a particular mechanism for inheritance. Mendel developed his theory of inheritance several decades before chromosomes were observed
Chapter 14 Mendel and the Gene Idea Gregor Mendel Gregor Mendel documented a particular mechanism for inheritance. Mendel developed his theory of inheritance several decades before chromosomes were observed
An introductory overview of the current state of statistical genetics
 An introductory overview of the current state of statistical genetics p. 1/9 An introductory overview of the current state of statistical genetics Cavan Reilly Division of Biostatistics, University of
An introductory overview of the current state of statistical genetics p. 1/9 An introductory overview of the current state of statistical genetics Cavan Reilly Division of Biostatistics, University of
LAB ACTIVITY ONE POPULATION GENETICS AND EVOLUTION 2017
 OVERVIEW In this lab you will: 1. learn about the Hardy-Weinberg law of genetic equilibrium, and 2. study the relationship between evolution and changes in allele frequency by using your class to represent
OVERVIEW In this lab you will: 1. learn about the Hardy-Weinberg law of genetic equilibrium, and 2. study the relationship between evolution and changes in allele frequency by using your class to represent
Video Tutorial 9.1: Determining the map distance between genes
 Video Tutorial 9.1: Determining the map distance between genes Three-factor linkage questions may seem daunting at first, but there is a straight-forward approach to solving these problems. We have described
Video Tutorial 9.1: Determining the map distance between genes Three-factor linkage questions may seem daunting at first, but there is a straight-forward approach to solving these problems. We have described
