Supporting Information
|
|
|
- Emily Henderson
- 6 years ago
- Views:
Transcription
1 Supporting Information Nishimura et al /pnas SI Text SI Materials and Methods. Plasmid construction. The cdnas of human Gα q were amplified and subcloned into pcmv5. Mutants of Gα q (R60K, L78N, V184S, I190N, and P193C) were generated by site-directed PCR mutagenesis. A soluble and functional chimeric protein, Gα i/q, which comprises the N-terminal helix (residues 1 28) of rat Gα i1, the Arg-Ser linker, and the core domain of mouse Gα q (residues ) was generated by overlapping PCR as previously described (1). X-ray data collection. For X-ray data collection, the crystal was mounted on a rayon loop and flash-frozen in liquid nitrogen. The X-ray data were collected at SPring-8 on beamline BL41XU. The crystal was maintained at 100 K in a nitrogen stream during exposure to the X-ray beam with a wavelength of 1.00 Å. Diffraction from a total oscillation range of 180 was recorded by the charge-coupled device detector (ADSC Quantum 315). The dataset was indexed and merged using the HKL2000 program suite (2). Structure determination. The phases were determined by the molecular replacement (MR) method using the GDP þ AlF 4 form of the Gα i/q subunit (PDB ID: 2BCJ) (1) and the Gβγ subunits (PDB ID: 1GP2) (3) as a search model. Two independent MR calculations using the Gα i/q and Gβγ coordinates were performed by the PHASER (4) program, which successfully gave solutions corresponding to the known architecture of the heterotrimer. One complex was included in the asymmetric unit of the crystal. In the model building, the structure of the heterotrimer was at first refined by CNS (5). The inhibitor model was then built into the residual electron-density map using the graphic program Coot (6). In this process, the ring-shaped densities with an extended body (corresponding to the cyclic backbone with the YM subregion) were traced by two different orientations of an inhibitor structure, a clockwise one and an anticlockwise one, as well as with several different conformations of the subregion. Only the present structure fully fitted into the densities accompanied with the lowest R-values. The structure was finally refined by REFMAC with TLS parameterization (7). The figures were prepared using PyMOL (DeLano Scientific). Trypsin protection assay. 293Tcells were transfected with each Gα. At 48 h posttransfection, the cells were washed with PBS and lysed with a lysis buffer (20 mm Hepes-NaOH ph 8.0, 100 mm NaCl, 3 mm MgCl 2,1mMEDTA,1mMDTT, 10 μm GDP, and 0.5% Lubrol-PX). Lysates were cleared by centrifugation at 14,000 g for 10 min, and supernatants were pretreated with or without 10 μm YM for 15 min at 30 C. Next, supernatants (1.5 mg ml) were incubated with or without AlF 4 (10 mm MgCl 2, 5 mm NaF, and 50 μm AlCl 3 ) for 15 min at 30 C and then treated with 5 μg ml TPCK-trypsin for 20 min at 30 C. The reactions were stopped by the addition of Laemmli sample buffer. The digestion of Gα q or Gα 14 was detected using an anti-gα q/11 antibody (Santa Cruz Biotechnology) or anti-gα 14 antibody (Gramsch Laboratories) that recognizes the C-terminal tail of the corresponding protein. AlF 4 -induced Gα dissociation from Gβγ. The assay was performed as previously described by Yamaguchi et al. (8) with some modification. Briefly, 293T cells were transfected with each Gα, Gβ 1, and FLAG-Gγ 2. At 48 h posttransfection, the cells were pretreated with or without 10 μm YM for 30 min and incubated with or without AlF 4 for an additional 2 h. The cells were then washed with PBS and lysed with lysis buffer with or without 10 μm YM and AlF 4. Lysates were cleared by centrifugation at 14,000 g for 10 min, and the supernatants were then incubated with the anti-flag antibody and protein G-Sepharose for 1 h. Immunoprecipitates were washed three times with lysis buffer and treated with Laemmli sample buffer. 1. Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ (2005) Snapshot of activated G proteins at the membrane: The Galphaq GRK2 Gbetagamma complex. Science 310: Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: Wall MA, et al. (1995) The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83: McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D 61: Brunger AT, et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D 54: Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D 60: Winn MD, Murshudov GN, Papiz MZ (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 374: Yamaguchi Y, Katoh H, Negishi M (2003) N-terminal short sequences of alpha subunits of the G12 family determine selective coupling to receptors. J Biol Chem 278: of5
2 Fig. S1. The α subunit selectivity of YM Purified Gα s (A), Gα i1 (B), Gα o (C), or Gα 13 (D) at a concentration of 100 nm was preincubated with (open circles) or without (filled circles) 10 μm YM for 3 min, and GTPγS binding assay was performed. Each value represents the mean S:D: from three independent experiments. Fig. S2. YM sensitivity of Gα 14 and Gα 16. The AlF 4 -induced conformational changes in Gα 14 and Gα 16 were evaluated with the trypsin protection assay (A) or the dissociation from Gβγ (B), respectively. (A) 293T cells were transfected with Gα q or Gα 14 plasmid. Cell lysate was pretreated with or without 10 μm YM and then incubated with or without AlF 4.Gαin the lysate was digested with trypsin. The trypsin sensitivities of Gα q and Gα 14 were analyzed by immunoblotting using anti-gα q and anti-gα 14 antibodies, respectively. Partially protected 38 kda products of Gα q and Gα 14 are shown as Protected Gα q and Protected Gα 14, respectively. (B) 293T cells transfected with Gα, Gβ 1, and FLAG-Gγ 2 were preincubated with or without 10 μm YM After 30 min, 293T cells were treated with or without AlF 4 for an additional 2 h. The whole-cell lysate (WCL) was prepared and incubated with an anti-flag antibody and protein G-Sepharose. Coimmunoprecipitation of Gα q or Gα 16 with FLAG-Gβ 1γ 2 was detected with anti-gα q and anti-gα 16 antibodies, respectively. 2of5
3 Fig. S3. Comparison of YM sensitivity between Gα i/q monomer and Gα i/qβγ heterotrimer. (A) Inhibition of GTPγS binding to His-Gα i/qβγ heterotrimer. Purified His-Gα i/qβγ (100 nm) was preincubated with (open circles) or without (filled circles) 10 μm YM for 3 min, and the GTPγS binding assay was performed. (B) Comparison of dose-dependent inhibition of GTPγS binding to His-Gα i/q and His-Gα i/qβγ. Purified His-Gα i/q (filled circles) or His-Gα i/qβγ (open circles) was preincubated with the indicated concentration of YM , and the GTPγS binding assay was performed for 120 min. Each value represents the mean S:D: from three independent experiments. Fig. S4. Comparison of overall structure of Gα i/qβγ-ym with those of other Gαβγ. (A) Stereo view of the Gα i/qβ 1γ 2 (red), which is bound to GDP (purple) and YM (cyan), superimposed on Gα i1β 1γ 2 (yellow) (1) and Gα t/iβ 1γ 1 (gray) (2). The helical domain of Gα i/q may be rotated through the hinge regions (Linker 1 and Switch I) compared with those of Gα i1 and Gα t/i.(b) Stereo view of the Gα i/q (red), which is bound to GDP and YM , superimposed on the Gα i/q bound to GDP and AlF 4 from Gα i/q-grk2-gβγ complex (yellow) (3) and Gα i/q-p63rhogef-rhoa complex (gray) (4). (C) Close-up view of the three switch regions of Gα i/q. The conformations of three switch regions of Gα i/qβγ bound to YM are essentially the same as those of Gα i1βγ and Gα t/iβγ, which display the typical inactive GDP-bound conformation. 1 Wall MA, et al. (1995) The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83: Lambright DG, et al. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379: Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ (2005) Snapshot of activated G proteins at the membrane: The Galphaq GRK2 Gbetagamma complex. Science 310: Lutz S, et al. (2007) Structure of Galphaq p63rhogef RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318: of5
4 Fig. S5. Stereo view of the structure model of YM bound to the heterotrimer. The jf o j-jf c j omit electron-density map contoured at 3.3σ (gray mesh) is superimposed. The map was calculated without the contribution of YM Oxygen and nitrogen are shown in red and blue, respectively. The hydrogen bonds within a distance of 3.3 Å are shown as dashed lines. Fig. S6. Trypsin protection assay of each Gα q mutant. 293T cells were transfected with each Gα q mutant plasmid. Cell lysate was preincubated with or without AlF 4, and then Gα q was digested with trypsin. Fig. S7. Stereo view of the interaction between YM and Gα i/qβγ.gα i/qβγ-ym complex depicted as a stick model is shown in the same orientation as Fig. 3E. GTPase domain, helical domain, and two linkers of Gα i/q are shown in yellow, green, and light red, respectively. Gβ and YM are light blue and cyan, respectively. Oxygen and nitrogen are shown in red and blue, respectively. Hydrogen bonds are shown as dashed lines. 4of5
5 Table S1. Crystallographic data for the Gα i/qβγ-ym complex X-ray data Space group I4 1 Cell parameters [Å] a ¼ b ¼ 173.3, c ¼ 60.9 Resolution [Å] * ( ) Mosaicity V M ½Å 3 DaŠ V solvent [%] Reflections, total/unique 80;301 19;018 Completeness [%] 94.1 (76.9) hi σ I i 11.1 (2.4) Redundancy 4.2 (4.1) R merge [%] 5.9 (52.6) Refinement Number of residues included Gα q 342 (of 355) Gβ 330 (of 340) Gγ 50 (of 78) Number of atoms 5,829 R work R free [%] Ramachandran plot [%] Most favored 81.6 Additionally allowed 16.4 Generously allowed 1.8 Disallowed 0.2 (Thr87 of Gβ) Average B-factor [Å 2 ] Gα q 91.1 Gβ 91.2 Gγ 93.1 GDP 90.7 YM R.m.s. bond length [Å], angles [ ] 0.011, 1.2 *Numbers in parentheses refer to statistics for the outer resolution shell. R work ¼ ΣjjF obs j jf calc jj ΣjF obs j. R free is the same as R work except for a 5% subset of all reflections that were never used in the crystallographic refinement. 5of5
Supporting Information
 Supporting Information Slep et al. 10.1073/pnas.0801569105 SI Materials and Methods Mouse RGS16, residues 53 180, was subcloned into a modified pgex-2t vector (GE Healthcare) to yield a thrombin-cleavable
Supporting Information Slep et al. 10.1073/pnas.0801569105 SI Materials and Methods Mouse RGS16, residues 53 180, was subcloned into a modified pgex-2t vector (GE Healthcare) to yield a thrombin-cleavable
SUPPLEMENTARY INFORMATION
 Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
Molecular basis of RNA-dependent RNA polymerase II activity Elisabeth Lehmann, Florian Brueckner, and Patrick Cramer Gene Center Munich and Center for integrated Protein Science CiPS M, Department of Chemistry
The YTH domain (residues ) of human YTHDF2 (NP_ ) was subcloned
 Supplementary information, Data S1 Materials and Methods Protein Expression, Purification and Crystallization The YTH domain (residues 383-553) of human YTHDF2 (NP_057342.2) was subcloned into a modified
Supplementary information, Data S1 Materials and Methods Protein Expression, Purification and Crystallization The YTH domain (residues 383-553) of human YTHDF2 (NP_057342.2) was subcloned into a modified
The Skap-hom Dimerization and PH Domains Comprise
 Molecular Cell, Volume 32 Supplemental Data The Skap-hom Dimerization and PH Domains Comprise a 3 -Phosphoinositide-Gated Molecular Switch Kenneth D. Swanson, Yong Tang, Derek F. Ceccarelli, Florence Poy,
Molecular Cell, Volume 32 Supplemental Data The Skap-hom Dimerization and PH Domains Comprise a 3 -Phosphoinositide-Gated Molecular Switch Kenneth D. Swanson, Yong Tang, Derek F. Ceccarelli, Florence Poy,
Supporting Online Material. Av1 and Av2 were isolated and purified under anaerobic conditions according to
 Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Supporting Online Material Materials and Methods Av1 and Av2 were isolated and purified under anaerobic conditions according to published protocols (S1). Crystals of nf-, pcp- and adp-av2:av1 complexes
Appendix B Dansyl probe syntheses and characterization and D-8-Ad:P450cam structure determination
 201 Appendix B Dansyl probe syntheses and characterization and D-8-Ad:P450cam structure determination Acknowlegements. The structure of the D-8-Ad:P450cam conjugate was determined by Anna-Maria A. Hays.
201 Appendix B Dansyl probe syntheses and characterization and D-8-Ad:P450cam structure determination Acknowlegements. The structure of the D-8-Ad:P450cam conjugate was determined by Anna-Maria A. Hays.
Six genes, Lsm1, Lsm2, Lsm3, Lsm5, Lsm6, and Lsm7, were amplified from the
 Supplementary information, Data S1 Methods Clones and protein preparation Six genes, Lsm1, Lsm2, Lsm3, Lsm5, Lsm6, and Lsm7, were amplified from the Saccharomyces cerevisiae genomic DNA by polymerase chain
Supplementary information, Data S1 Methods Clones and protein preparation Six genes, Lsm1, Lsm2, Lsm3, Lsm5, Lsm6, and Lsm7, were amplified from the Saccharomyces cerevisiae genomic DNA by polymerase chain
Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and
 Materials and Methods Immunoprecipitation and immunoblot analysis Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and immunoblot analyses with corresponding
Materials and Methods Immunoprecipitation and immunoblot analysis Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and immunoblot analyses with corresponding
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
Supplemental Information
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supplemental Information Experimental Procedures Cloning, expression, purification and mutagenesis
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supplemental Information Experimental Procedures Cloning, expression, purification and mutagenesis
Nature Structural & Molecular Biology: doi: /nsmb.1969
 Supplementary Methods Structure determination All the diffraction data sets were collected on BL-41XU (using ADSC Quantum 315 HE CCD detector) at SPring8 (Harima, Japan) or on BL5A (using ADSC Quantum
Supplementary Methods Structure determination All the diffraction data sets were collected on BL-41XU (using ADSC Quantum 315 HE CCD detector) at SPring8 (Harima, Japan) or on BL5A (using ADSC Quantum
Gα i Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
Supplemental Information. The structural basis of R Spondin recognition by LGR5 and RNF43
 Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
Supplemental Information The structural basis of R Spondin recognition by LGR5 and RNF43 Po Han Chen 1, Xiaoyan Chen 1, Deyu Fang 2, Xiaolin He 1* 1 Department of Molecular Pharmacology and Biological
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport
 Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
doi:10.1038/nature10963 Supplementary Table 1 Data collection, phasing and refinement statistics. Crystal Native Derivative-1 (OsO 4 ) Derivative-2 (Orange-Pt) Data collection Space group C2 C2 C2 Cell
Conformational changes in IgE contribute to its. uniquely slow dissociation rate from receptor FcεRI
 Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
Supplementary Information For. A genetically encoded tool for manipulation of NADP + /NADPH in living cells
 Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
Supplementary Information For A genetically encoded tool for manipulation of NADP + /NADPH in living cells Valentin Cracan 1,2,3, Denis V. Titov 1,2,3, Hongying Shen 1,2,3, Zenon Grabarek 1* and Vamsi
Figure S2, related to Figure 1. Stereo images of the CarD/RNAP complex and. electrostatic potential surface representation of the CarD/RNAP interface
 Structure, Volume 21 Supplemental Information Structure of the Mtb CarD/RNAP -Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD Gulcin Gulten
Structure, Volume 21 Supplemental Information Structure of the Mtb CarD/RNAP -Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD Gulcin Gulten
Nature Structural & Molecular Biology: doi: /nsmb.3428
 Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supporting Information
 Supporting Information Xiao et al. 10.1073/pnas.1309211110 SI Methods Protein Expression and Purification. Bacmid was generated and recombinant baculovirus amplified using standard procedures (Bac-to-Bac;
Supporting Information Xiao et al. 10.1073/pnas.1309211110 SI Methods Protein Expression and Purification. Bacmid was generated and recombinant baculovirus amplified using standard procedures (Bac-to-Bac;
SUPPLEMENTARY INFORMATION Figures. Supplementary Figure 1 a. Page 1 of 30. Nature Chemical Biology: doi: /nchembio.2528
 SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
ab G alpha i Activation Assay Kit
 ab173234 G alpha i Activation Assay Kit Instructions for Use For the simple and fast measurement of G alpha i activation. This product is for research use only and is not intended for diagnostic use. Version
ab173234 G alpha i Activation Assay Kit Instructions for Use For the simple and fast measurement of G alpha i activation. This product is for research use only and is not intended for diagnostic use. Version
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table.
 Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Supplementary Table 1. The Q-PCR primer sequence is summarized in the following table. Name Sequence (5-3 ) Application Flag-u ggactacaaggacgacgatgac Shared upstream primer for all the amplifications of
Colchicine. Colchicine. a b c d e
 α1-tub Colchicine Laulimalide RB3 β1-tub Vinblastine α2-tub Taxol Colchicine β2-tub Maytansine a b c d e Supplementary Figure 1 Structures of microtubule and tubulin (a)the cartoon of head to tail arrangement
α1-tub Colchicine Laulimalide RB3 β1-tub Vinblastine α2-tub Taxol Colchicine β2-tub Maytansine a b c d e Supplementary Figure 1 Structures of microtubule and tubulin (a)the cartoon of head to tail arrangement
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression
 Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
Supplemental figures Supplemental Figure. 1. Silencing expression of Celsr3 by shrna. To examine the silencing effects of Celsr3 shrna, we co transfected 293T cells with expression plasmids for the shrna
absorption spectra were measured on a Hewlett Packard 8452A diode array spectrophotometer.
 S1 Experimental General: P450cam was expressed and purified as previously described. 1 Steady state UV-visible absorption spectra were measured on a Hewlett Packard 8452A diode array spectrophotometer.
S1 Experimental General: P450cam was expressed and purified as previously described. 1 Steady state UV-visible absorption spectra were measured on a Hewlett Packard 8452A diode array spectrophotometer.
SUPPLEMENTARY INFORMATION. Design and Characterization of Bivalent BET Inhibitors
 SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
RhoC Activation Assay Kit
 Product Manual RhoC Activation Assay Kit Catalog Number STA-403-C 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family
Product Manual RhoC Activation Assay Kit Catalog Number STA-403-C 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family
has only one nucleotide, U20, between G19 and A21, while trna Glu CUC has two
 SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
Structural basis of transferrin sequestration by transferrin-binding protein B
 Supplementary Information for Structural basis of transferrin sequestration by transferrin-binding protein B Charles Calmettes, Joenel Alcantara, Rong-Hua Yu, Anthony B. Schryvers and Trevor F. Moraes*
Supplementary Information for Structural basis of transferrin sequestration by transferrin-binding protein B Charles Calmettes, Joenel Alcantara, Rong-Hua Yu, Anthony B. Schryvers and Trevor F. Moraes*
SUPPLEMENTARY INFORMATION
 Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Structure of a tyrosyl-trna synthetase splicing factor bound to a group I intron RNA Paul J. Paukstelis 1, Jui-Hui Chen 2, Elaine Chase 2, Alan M. Lambowitz 1,*, and Barbara L. Golden 2,*,. 1 Institute
Supporting Information
 Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
Supplemental Material
 Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
Supplemental Material Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase Shuang Yang, 1, 2 Xiangdong Zheng, 1, 2, 3 Chao Lu, 4 Guo-Min Li, 2 C. David Allis, 4 1, 2, 3, 5* and Haitao
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
DOI:.38/ncb327 a b Sequence coverage (%) 4 3 2 IP: -GFP isoform IP: GFP IP: -GFP IP: GFP Sequence coverage (%) 4 3 2 IP: -GFP IP: GFP 33 52 58 isoform 2 33 49 47 IP: Control IP: Peptide Sequence Start
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein
 Paper Presentation PLoS ONE 2011 Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein Debanu Das, Mireille Herve, Julie Feuerhelm, etc. and Dominique
Paper Presentation PLoS ONE 2011 Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein Debanu Das, Mireille Herve, Julie Feuerhelm, etc. and Dominique
Gα 13 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
Supplementary information
 Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
SUPPLEMENTARY INFORMATION doi:10.1038/nature10258 Supplementary Figure 1 Reconstitution of the CENP-A nucleosome with recombinant human histones H2A, H2B, H4, and CENP-A. a, Purified recombinant human
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron
 Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I
 BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
 SUPPLEMENTARY INFORMATION Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs Malka Kitayner 1, 3, Haim Rozenberg 1, 3, Remo Rohs 2, 3, Oded Suad 1, Dov Rabinovich
SUPPLEMENTARY INFORMATION Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs Malka Kitayner 1, 3, Haim Rozenberg 1, 3, Remo Rohs 2, 3, Oded Suad 1, Dov Rabinovich
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Not The Real Exam Just for Fun! Chemistry 391 Fall 2018
 Not The Real Exam Just for Fun! Chemistry 391 Fall 2018 Do not open the exam until ready to begin! Rules of the Game: This is a take-home Exam. The exam is due on Thursday, October 11 th at 9 AM. Otherwise,
Not The Real Exam Just for Fun! Chemistry 391 Fall 2018 Do not open the exam until ready to begin! Rules of the Game: This is a take-home Exam. The exam is due on Thursday, October 11 th at 9 AM. Otherwise,
Supplemental Online Material
 Supplemental Online Material Supplemental Figures Supplemental Figure 1. A) Crosslinking efficiencies of 5 -biotin tagged oligonucleotides screened with the FASTDXL method 1. Reactive and nonreactive cysteine
Supplemental Online Material Supplemental Figures Supplemental Figure 1. A) Crosslinking efficiencies of 5 -biotin tagged oligonucleotides screened with the FASTDXL method 1. Reactive and nonreactive cysteine
Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplemental material JCB Hoffmann et al., http ://www.jcb.org /cgi /content /full /jcb.201506071 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. TRA IP harbors a PCNA-binding PIP box required for its recruitment
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA
 Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Cdc42 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
SUPPLEMENTAL FIGURE LEGENDS. Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Homology alignment of DDR2 amino acid sequence. Shown are the amino acid sequences of human DDR2, mouse DDR2 and the closest homologs in zebrafish and C. Elegans.
CSE : Computational Issues in Molecular Biology. Lecture 19. Spring 2004
 CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
Attenuation of synaptic toxicity and MARK4/PAR1-mediated Tau phosphorylation by
 Supplementary Methods and Figures Attenuation of synaptic toxicity and MARK4/PAR1-mediated Tau phosphorylation by methylene blue for Alzheimer s disease treatment Wenchao Sun 1, Seongsoo Lee 1,2, Xiaoran
Supplementary Methods and Figures Attenuation of synaptic toxicity and MARK4/PAR1-mediated Tau phosphorylation by methylene blue for Alzheimer s disease treatment Wenchao Sun 1, Seongsoo Lee 1,2, Xiaoran
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a)
 Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Protein expression and purification from Hi5 insect cells is as described (1, 2).
 Materials & methods Protein expression and purification Protein expression and purification from Hi5 insect cells is as described (1, 2). The purified complex exhibited a 2:2:2 stoichiometry as determined
Materials & methods Protein expression and purification Protein expression and purification from Hi5 insect cells is as described (1, 2). The purified complex exhibited a 2:2:2 stoichiometry as determined
Rab5 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells
 SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
Supplemental Table 1: Sequences of real time PCR primers. Primers were intronspanning
 Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
Symbol Accession Number Sense-primer (5-3 ) Antisense-primer (5-3 ) T a C ACTB NM_001101.3 CCAGAGGCGTACAGGGATAG CCAACCGCGAGAAGATGA 57 HSD3B2 NM_000198.3 CTTGGACAAGGCCTTCAGAC TCAAGTACAGTCAGCTTGGTCCT 60
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
SUPPLEMENTARY INFORMATION
 This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
SUPPLEMENTAL DATA Experimental procedures
 SUPPLEMENTAL DATA Experimental procedures Cloning, protein production and purification. The DNA sequences corresponding to residues R10-S250 (DBD) and E258-T351 (IPT/TIG) in human EBF1 (gi:31415878), and
SUPPLEMENTAL DATA Experimental procedures Cloning, protein production and purification. The DNA sequences corresponding to residues R10-S250 (DBD) and E258-T351 (IPT/TIG) in human EBF1 (gi:31415878), and
SUPPLEMENTARY INFORMATION. Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly
 SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the
 Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the roots of three-day-old etiolated seedlings of Col-0
Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the roots of three-day-old etiolated seedlings of Col-0
Figure S1
 Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Supporting Information
 Title Structure of bacterial cellulose synthase subunit D Hu, Song-Qing; Gao, Yong-Gui; Tajima, Kenji; Sunagaw Author(s) Yoda, Takanori; Shimura, Daisuke; Satoh, Yasuharu; M CitationProceedings of the
Title Structure of bacterial cellulose synthase subunit D Hu, Song-Qing; Gao, Yong-Gui; Tajima, Kenji; Sunagaw Author(s) Yoda, Takanori; Shimura, Daisuke; Satoh, Yasuharu; M CitationProceedings of the
Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity
 Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity Stanley G. Kimani 1 *, Sushil Kumar 1 *, Nitu Bansal 2 *, Kamalendra Singh 3, Vladyslav Kholodovych
Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity Stanley G. Kimani 1 *, Sushil Kumar 1 *, Nitu Bansal 2 *, Kamalendra Singh 3, Vladyslav Kholodovych
Nature Structural & Molecular Biology: doi: /nsmb.2307
 A novel locally-closed conformation of a bacterial pentameric proton-gated ion channel Marie S. Prevost*, Ludovic Sauguet*, Hugues Nury, Catherine Van Renterghem, Christèle Huon, Frederic Poitevin, Marc
A novel locally-closed conformation of a bacterial pentameric proton-gated ion channel Marie S. Prevost*, Ludovic Sauguet*, Hugues Nury, Catherine Van Renterghem, Christèle Huon, Frederic Poitevin, Marc
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade
 Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1
 Supplemental information for: Structure determination and activity manipulation of the turfgrass AA receptor FePYR1 Zhizhong Ren 1, 2,#, Zhen Wang 1, 2,#, X Edward Zhou 3, Huazhong Shi 4, Yechun Hong 1,
Supplemental information for: Structure determination and activity manipulation of the turfgrass AA receptor FePYR1 Zhizhong Ren 1, 2,#, Zhen Wang 1, 2,#, X Edward Zhou 3, Huazhong Shi 4, Yechun Hong 1,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Arf6 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
Supporting Information
 Supporting Information Battisti et al. 10.1073/pnas.1210275109 SI Text SI Materials and Methods. Virus propagation and purification. Egggrown Newcastle disease virus (NDV; strain B1), harvested at 42 h
Supporting Information Battisti et al. 10.1073/pnas.1210275109 SI Text SI Materials and Methods. Virus propagation and purification. Egggrown Newcastle disease virus (NDV; strain B1), harvested at 42 h
SUPPLEMENTARY INFORMATION
 Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Anti-HB-EGF (Human) mab
 Page 1 For Research Use Only. Not for use in diagnostic procedures. CODE No. D308-3 Anti-HB-EGF (Human) mab CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 3H4 Mouse IgG1
Page 1 For Research Use Only. Not for use in diagnostic procedures. CODE No. D308-3 Anti-HB-EGF (Human) mab CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 3H4 Mouse IgG1
human Cdc45 Figure 1c. (c)
 1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
1 Details of the refined crystallographic model of human Cdc45 and comparison of its active-site region with that of bacterial RecJ. (a) Stereo view of a representative example of the final 2F o -F c electron
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
RheB Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
Supporting Information
 Supporting Information Drugs Modulate Interactions Between the First Nucleotide-Binding Domain and the Fourth Cytoplasmic Loop of Human P-glycoprotein Tip W. Loo and David M. Clarke Department of Medicine
Supporting Information Drugs Modulate Interactions Between the First Nucleotide-Binding Domain and the Fourth Cytoplasmic Loop of Human P-glycoprotein Tip W. Loo and David M. Clarke Department of Medicine
A(+1) A( 7) RNA hairpin
 PP7 CP dimer A(+1) A( 7) RNA hairpin 5 3 Supplemental Figure 1. Sample electron density of RNA hairpin. Cartoon showing PP7 FG CP (wheat) bound to RNA hairpin (violet) showing 2Fo-Fc electron density (blue)
PP7 CP dimer A(+1) A( 7) RNA hairpin 5 3 Supplemental Figure 1. Sample electron density of RNA hairpin. Cartoon showing PP7 FG CP (wheat) bound to RNA hairpin (violet) showing 2Fo-Fc electron density (blue)
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
SUPPLEMENTARY INFORMATION
 Supplementary Methods Protein expression and in vitro binding studies. Recombinant baculovirus carrying GST, GST-mCC, GST-mSec, GST-mSecA and GST-mSecD were generated according to the manufacturer s instructions
Supplementary Methods Protein expression and in vitro binding studies. Recombinant baculovirus carrying GST, GST-mCC, GST-mSec, GST-mSecA and GST-mSecD were generated according to the manufacturer s instructions
Figure S1 Alpha-carbon backbones of components from FSH-FSHRHB complex. a, Stereo view of FSHRHB (red) with every 10 th residue marked.
 a 230 80 80 130 110 230 110 180 60 180 130 60 210 200160 40 210 200 160 40 90 20 90 20 250 150 150 190 100 250 50 30 100 220 140 220 190 140 50 30 240 240 120 70 120 70 170 170 b 70 70 60 60 20 20 50 50
a 230 80 80 130 110 230 110 180 60 180 130 60 210 200160 40 210 200 160 40 90 20 90 20 250 150 150 190 100 250 50 30 100 220 140 220 190 140 50 30 240 240 120 70 120 70 170 170 b 70 70 60 60 20 20 50 50
Supplementary Information for. Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction
 Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure
 ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
A Brief Introduction to Structural Biology and Protein Crystallography
 A Brief Introduction to Structural Biology and Protein Crystallography structural biology of H2O http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/water-structure.jpg Protein polymers fold up into
A Brief Introduction to Structural Biology and Protein Crystallography structural biology of H2O http://courses.cm.utexas.edu/jrobertus/ch339k/overheads-1/water-structure.jpg Protein polymers fold up into
Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from
 Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
SUPPLEMENTARY INFORMATION
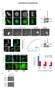 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
Ral Activation Assay Kit
 Product Manual Ral Activation Assay Kit Catalog Number STA-408 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family of
Product Manual Ral Activation Assay Kit Catalog Number STA-408 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family of
Anti-CLOCK (Mouse) mab
 Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-CLOCK (Mouse) mab CODE No. D349-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal CLSP4 Mouse IgG1
Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-CLOCK (Mouse) mab CODE No. D349-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal CLSP4 Mouse IgG1
Learning to Use PyMOL (includes instructions for PS #2)
 Learning to Use PyMOL (includes instructions for PS #2) To begin, download the saved PyMOL session file, 4kyz.pse from the Chem 391 Assignments web page: http://people.reed.edu/~glasfeld/chem391/assign.html
Learning to Use PyMOL (includes instructions for PS #2) To begin, download the saved PyMOL session file, 4kyz.pse from the Chem 391 Assignments web page: http://people.reed.edu/~glasfeld/chem391/assign.html
Supplemental Material Liu et al.
 Supplemental Material Liu et al. Supplemental Methods Protein expression, purification Recombinant etud11 (a.a 2344-2515) and Tud7-11 (a.a. 1617-2515) proteins (Tud7-11) were expressed in the Rosetta strain
Supplemental Material Liu et al. Supplemental Methods Protein expression, purification Recombinant etud11 (a.a 2344-2515) and Tud7-11 (a.a. 1617-2515) proteins (Tud7-11) were expressed in the Rosetta strain
Supplemental Online Material. The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/-
 #1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
#1074683s 1 Supplemental Online Material Materials and Methods Cell lines and tissue culture The mouse embryonic fibroblast cell line #10 derived from β-arrestin1 -/- -β-arrestin2 -/- knock-out animals
Crystal Structure of a Self-Splicing Group I Intron with Both Exons
 Supplementary Data for: Crystal Structure of a Self-Splicing Group I Intron with Both Exons Peter L. Adams, Mary R. Stahley, Anne B. Kosek, Jimin Wang and Scott A. Strobel The supplementary material includes
Supplementary Data for: Crystal Structure of a Self-Splicing Group I Intron with Both Exons Peter L. Adams, Mary R. Stahley, Anne B. Kosek, Jimin Wang and Scott A. Strobel The supplementary material includes
