Corporate Medical Policy
|
|
|
- Bertina Simpson
- 6 years ago
- Views:
Transcription
1 Corporate Medical Policy Proteogenomic Testing for Patients with Cancer (GPS Cancer Test) File Name: Origination: Last CAP Review: Next CAP Review: Last Review: proteogenomic_testing_for_patients_with_cancer_gps_cancer_test 7/2016 3/2018 3/2019 3/2018 Description of Procedure or Service Proteogenomics The term proteome refers to the entire complement of proteins produced by an organism or cellular system, and proteomics refers to the large-scale comprehensive study of a specific proteome. Similarly, the term transcriptome refers to the entire complement of transcription products (messenger RNAs), and transcriptomics refers to the study of a specific transcriptome. Proteogenomics refers to the integration of genomic information with proteomic and transcriptomic information to provide a more complete picture of the function of the genome. A system s proteome is related to its genome and to genomic alterations. However, while the genome is relatively static over time, the proteome is more dynamic and may vary over time and/or in response to selected stressors. Proteins undergo a number of modifications as part of normal physiologic processes. Following protein translation, modifications occur by splicing events, alternative folding mechanisms, and incorporation into larger complexes and signaling networks. These modifications are linked to protein function and result in functional differences that occur by location and over time. Some of the main potential applications of proteogenomics in medicine include the following: Identifying biomarkers for diagnostic, prognostic, and predictive purposes Detecting cancer by proteomic profiles or signatures Quantitating levels of proteins and monitoring levels over time for: Cancer activity Early identification of resistance to targeted tumor therapy Correlating protein profiles with disease states. Proteogenomics is an extremely complex field due to the intricacies of protein architecture and function, the many potential proteomic targets that can be measured, and the numerous testing methods used. The types of targets currently being investigated and the testing methods used and under development are discussed next. Proteomic Targets A proteomic target can be any altered protein that results from a genetic variant. Protein alterations can occur as a result of both germline and somatic mutations. Altered protein products include mutated proteins, fusion proteins, alternative splice variants, noncoding mrnas, and posttranslational modifications (PTMs). Sequence Alterations (Mutated Protein) A mutated protein has an altered amino acid sequence that arises due to a genetic variant. A single amino acid may be replaced in a protein or multiple amino acids in sequence may be affected. Mutated proteins can arise from either germline or somatic mutations. Somatic mutations can be differentiated from germline mutations by comparison with normal and diseased tissue. Page 1 of 7
2 Fusion Proteins Fusion proteins are the product of 1 or more mutated genes that fuse together. Most fusion genes discovered to date have been oncogenic, and fusion genes have been shown to have clinical relevance in a variety of cancers. Alternative Splice Events Posttranslational enzymatic splicing of proteins results in numerous protein isoforms. Alternative splicing events can lead to abnormal protein isoforms with altered function. Some alternative splicing events have been associated with tumor-specific variants. Non-coding RNAs Non-coding portions of the genome serve as the template for non-coding RNA, which plays various roles in the regulation of gene expression. There are 2 classes of non-coding RNA (ncrna): shorter ncrnas that include micrornas and related transcript products, and longer ncrnas thought to be involved in cancer progression. Posttranslational Modifications PTMs of histone proteins occur in normal cells, and are genetically regulated. Histone proteins are found in the nuclei and play a role in gene regulation by structuring the DNA into nucleosomes. A nucleosome is composed of a histone protein core surrounded by DNA. Nucleosomes are assembled into chromatin fibers that are composed of multiple nucleosomes assembled in a specific pattern. PTMs of histone proteins include a variety of mechanisms, including methylation, acetylation, phosphorylation, glycosylation, and related modifications. Proteogenomic Testing Methods Proteogenomic testing involves isolating, separating, and characterizing proteins from biologic samples, followed by correlation with genomic and transcriptomic data. Isolation of proteins is accomplished by trypsin digestion and solubilization. The soluble mix of protein isolates is then separated into individual proteins. This is generally done in multiple stages using high-performance liquid chromatography ion-exchange chromatography, 2-dimensional gel electrophoresis and related methods. Once the individual proteins are obtained, they may be characterized using various methods and parameters, some of which are described below. Immunohistochemistry/Fluorescence in situ Hybridization Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) are standard techniques for isolating and characterizing proteins. IHC identifies proteins by using specific antibodies that bind to the protein. Therefore, this technique can only be used for known proteins and protein variants because it relies on the availability of a specific antibody. This technique also can only test a relatively small number of samples at once. There are a number of reasons why IHC and FISH are not well-suited for large-scale proteomic research. They are semiquantitative techniques and involve subjective interpretation. They are considered low-throughput assays that are time-consuming and expensive, and require a relatively large tissue sample. Some advances in IHC and FISH have addressed these limitations, including tissue microarray and reverse phase protein array. Tissue microarrays can be constructed that enable simultaneous analysis of up to 1,000 tissue samples. Reverse phase protein array, a variation on tissue microarrays, allows for a large number of proteins to be quantitated simultaneously. Mass Spectrometry Mass spectrometry (MS) separates molecules by their mass to charge ratio, and has been used as a research tool for studying proteins for many years. Development of technology that led to the application of MS to biologic samples has advanced the field of proteogenomics rapidly. However, the application of MS to clinical medicine is in its formative stages. There are currently several types of mass spectrometers and a lack of standardization in the testing methods. In addition, MS equipment is expensive and currently largely restricted to tertiary research centers. Page 2 of 7
3 The potential utility of MS lies in its ability to provide a wide range of proteomic information in an efficient manner, including: Identification of altered proteins Delineation of protein or peptide profiles for a given tissue sample Amino acid sequencing of proteins or peptides Quantitation of protein levels 3-dimensional protein structure and architecture Identification of PTMs. Top-down MS refers to identification and characterization of all proteins in a sample without prior knowledge of which proteins are present. This method provides a profile of all proteins in a system, including documentation of PMTs and other protein isoforms. This method therefore provides a protein profile or map of a specific system. Following initial analysis, intact proteins can be isolated and further analyzed to determine amino acid sequences and related information. Bottom-up MS refers to identification of known proteins in a sample. This method identifies peptide fragments that indicate the presence of a specific protein. This method depends on having peptide fragments that can reliably identify a specific protein. Selective reaction monitoring mass spectrometry (SRM-MS) is a bottom-up approach modification of MS that allows for direct quantification and specific identification of low-abundance proteins without the need for specific antibodies. This method requires the selection of a peptide fragment or signature that is used to target the specific protein. Multiplex assays have also been developed to quantitate the epidermal growth factor receptor, human epidermal growth factor receptors 2 and 3, and insulin-like growth factor-1 receptor. Bioinformatics Due to complexity of proteomic information, the multiple tests used, and the need to integrate this information with other genomic data, a bioinformatics approach is necessary to interpret proteogenomic data. Software programs are available that integrate and assist in interpretation of the vast amounts of data generated by proteogenomics research. One software platform that integrates genomic and proteomic information is PARADIGM, which is used by The Cancer Genome Atlas (TCGA) project for data analysis. Other software tools currently available include the following: The Genome Peptide Finder ( matches the amino acid sequence of peptides predicted de novo with the genome sequence. The Proteogenomic Mapping Tool ( is an academic software for mapping peptides to the genome. Peppy ( is an automated search tool that generates proteogenomic data from translated databases and integrates this information for analysis. VESPA ( is a software tool that integrates data from various platforms and provides visual display of integrated data. Ongoing Proteogenomic Database Projects Numerous ongoing databases are being constructed for proteogenomic research. Some are shown in Table 1. There are also networks of researchers coordinating their activities in this field. The Clinical Proteomic Tumor Analysis Consortium is a coordinated project among 8 analysis sites sponsored by the National Cancer Institute. This project seeks to systematically characterize the genomic and transcriptomic profiles of common cancers. As of 2014, this consortium had catalogued proteomic information for breast, colon, and ovarian cancers. All data from this project are freely available. Many existing genomic databases have begun to incorporate proteomic information. TCGA intends to profile changes in the genomes of 20 different cancers. As part of its analysis, mrna expression is used to help define signaling pathways that are either upregulated or deregulated in conjunction with genetic mutations. Currently, TCGA has published comprehensive molecular characterizations of breast, colorectal, lung, gliomas, renal, and endometrial cancers. Table 1. Proteogenomic Databases Page 3 of 7
4 Human Protein Reference Database Human Cancer Proteome Variation Database (CanProVar) Cancer Mutant Proteome Database (CMPD) ChimerDB 2.0 The Synthetic Alternative Splicing Database (SASD) NONCODE Centralized platform integrating information related to protein structure alterations, posttranslational modifications, interaction networks, and disease association. The intent is to catalog this information for each protein in the human proteome. Compiles data from published literature and publicly available databases. Protein sequence database that integrates information from various publicly available datasets into 1 platform. Contains germline and somatic mutations with an emphasis on cancer-related mutations. Protein sequence database sequencing results compiled from the exome sequencing results of the NCI-60 cell lines, The Cancer CellLine Encyclopedia (CCLE) and 5600 casses from the Cancer Genome Atlas (TCGA) network genomics studies. Contains germline and somatic mutations with an emphasis on cancer-related mutations. A comprehensive database of fusion proteins including transcript products, compiled from various publicly available datasets. A comprehensive database of alternative splicing peptides and transcript products constructed from the Integrated Pathway Analysis Database (IPAD). Database of non-coding RNAs integrating data from literature mining, specialized databases and GenBank. IncRNAtor Database of long non-coding RNA (InRNA) integrating data from multiple datasets including TCGA and ENCODE. CCLE: Cancer Cell Line Encyclopedia; TCGA: The Cancer Genome Atlas. GPS Test The GPS Cancer test is a commercially available proteogenomic test intended for patients with cancer. The test includes whole genome sequencing (20,000 genes, 3 billion base pairs), whole transcriptome (RNA) sequencing, and quantitative proteomics by mass spectrometry. The test is intended to inform personalized treatment decisions for cancer, and treatment options are listed when available, although treatment recommendations are not made. Treatment options may include Food and Drug Administration approved targeted drugs with potential for clinical benefit, active clinical trials of drugs with potential for clinical benefit, and/or available drugs to which the cancer may be resistant. Policy ***Note: This Medical Policy is complex and technical. For questions concerning the technical language and/or specific clinical indications for its use, please consult your physician. Proteogenomic testing for Patients with Cancer (GPS Cancer Test) is considered investigational for all applications. BCBSNC does not provide coverage for investigational services or procedures. Page 4 of 7
5 Benefits Application This medical policy relates only to the services or supplies described herein. Please refer to the Member's Benefit Booklet for availability of benefits. Member's benefits may vary according to benefit design; therefore member benefit language should be reviewed before applying the terms of this medical policy. When Proteogenomic testing for patients with cancer is covered Not applicable. When Proteogenomic testing for patients with cancer is not covered Proteogenomic testing of patients with cancer (including but not limited to GPS Cancer Test) is considered investigational. BCBSNC does not provide coverage for investigational services. Policy Guidelines Proteogenomic testing involves the integration of proteomic, transcriptomic, and genomic information. Proteogenomic testing can be differentiated from proteomic testing, in that proteomic testing can refer to the measurement of protein products alone, without integration of genomic and transcriptomic information. When protein products alone are tested, this is not considered proteogenomic testing. Genetic counseling is primarily aimed at patients who are at risk for inherited disorders, and experts recommend formal genetic counseling in most cases when genetic testing for an inherited condition is considered. The interpretation of the results of genetic tests and the understanding of risk factors can be very difficult and complex. Therefore, genetic counseling will assist individuals in understanding the possible benefits and harms of genetic testing, including the possible impact of the information on the individual s family. Genetic counseling may alter the utilization of genetic testing substantially and may reduce inappropriate testing. Genetic counseling should be performed by an individual with experience and expertise in genetic medicine and genetic testing methods. Proteogenomics refers to the integration of genomic data with proteomic and transcriptomic data to provide a more complete picture of the function of the genome. The current focus of proteogenomics is primarily on the diagnostic, prognostic, and predictive potential of proteogenomics in various cancers. There is one commercially available proteogenomic test, the GPS Cancer test. For individuals who have cancer and indications for genetic testing who receive proteogenomic testing, the evidence includes cross-sectional studies that correlate results with standard testing and that report comprehensive molecular characterization of various cancers, and cohort studies that use proteogenomic markers to predict outcomes and that follow quantitative levels over time. Relevant outcomes are overall survival, disease-specific survival, test accuracy and validity, and treatmentrelated mortality and morbidity. There is no published evidence on the validity or utility of the GPS Cancer test. For proteogenomic testing in general, the research is at an early stage. There is a lack of standardization of testing methods, and uncertain accuracy for most proteogenomic technologies. A few studies have described assay development and validation for proteogenomic targets, and correlation of proteogenomic testing results with standard testing methods. Other studies have used proteogenomic in conjunction with genomic testing to provide a more comprehensive molecular characterization of various cancers. A very few studies have used proteogenomic tumor markers for diagnosis or prognosis, and at least 1 study has reported following quantitative protein levels for surveillance purposes. Further research is needed to standardize and validate proteogenomic testing methods. When standardized and validated testing methods are available, the clinical validity and utility of proteogenomic testing can be adequately evaluated. The evidence is insufficient to determine the effect of the technology on health outcomes. Genetics Nomenclature Update Human Genome Variation Society (HGVS) nomenclature is used to report information on variants found in DNA and serves as an international standard in DNA diagnostics. It is being implemented for Page 5 of 7
6 genetic testing medical evidence review updates starting in HGVS nomenclature is recommended by HGVS, the Variome Project, and the Human Genome Organization (HUGO). The American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) standards and guidelines for interpretation of sequence variants represent expert opinion from ACMG, AMP, and the College of American Pathologists. These recommendations primarily apply to genetic tests used in clinical laboratories, including genotyping, single genes, panels, exomes, and genomes. Recommended standard terminology: pathogenic, likely pathogenic, uncertain significance, likely benign, and benign, to describe variants identified that cause Mendelian disorders. Billing/Coding/Physician Documentation Information This policy may apply to the following codes. Inclusion of a code in this section does not guarantee that it will be reimbursed. For further information on reimbursement guidelines, please see Administrative Policies on the Blue Cross Blue Shield of North Carolina web site at They are listed in the Category Search on the Medical Policy search page. Applicable service codes: No specific code for this service The test would likely be reported with the unlisted molecular pathology procedure code BCBSNC may request medical records for determination of medical necessity. When medical records are requested, letters of support and/or explanation are often useful, but are not sufficient documentation unless all specific information needed to make a medical necessity determination is included. Scientific Background and Reference Sources BCBSA Medical Policy Reference Manual [Electronic Version] , 6/16/2016 Medical Director review 6/2016 Specialty Matched Consultant Advisory Panel 3/2017 BCBSA Medical Policy Reference Manual [Electronic Version] , 6/8/2017 Specialty Matched Consultant Advisory Panel 3/2018 Medical Director review 4/2018 Policy Implementation/Update Information 7/26/16 New policy developed. Proteogenomic testing of patients with cancer (including but not limited to GPS Cancer Test) is considered investigational. Medical Director review 6/2016. (lpr) 4/28/17 Specialty Matched Consultant Advisory Panel review 3/29/2017. No change to policy statement. (lpr) 7/28/17 Updated Policy Guidelines with Genetics Nomenclature update. Reference added. No change to policy statement. (lpr) 4/27/18 Specialty Matched Consultant Advisory Panel review 3/28/2018. No change to policy statement. Medical Director review. (lpr) Page 6 of 7
7 Medical policy is not an authorization, certification, explanation of benefits or a contract. Benefits and eligibility are determined before medical guidelines and payment guidelines are applied. Benefits are determined by the group contract and subscriber certificate that is in effect at the time services are rendered. This document is solely provided for informational purposes only and is based on research of current medical literature and review of common medical practices in the treatment and diagnosis of disease. Medical practices and knowledge are constantly changing and BCBSNC reserves the right to review and revise its medical policies periodically. Page 7 of 7
Corporate Medical Policy Genetic Testing for Fanconi Anemia
 Corporate Medical Policy Genetic Testing for Fanconi Anemia File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_fanconi_anemia 03/2015 3/2017 3/2018 12/2017 Description
Corporate Medical Policy Genetic Testing for Fanconi Anemia File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_fanconi_anemia 03/2015 3/2017 3/2018 12/2017 Description
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics and Protein Microarrays
 Introduction to BioMEMS & Medical Microdevices Proteomics and Protein Microarrays Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
Introduction to BioMEMS & Medical Microdevices Proteomics and Protein Microarrays Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd
 QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd 1 Our current NGS & Bioinformatics Platform 2 Our NGS workflow and applications 3 QIAGEN s
QIAGEN s NGS Solutions for Biomarkers NGS & Bioinformatics team QIAGEN (Suzhou) Translational Medicine Co.,Ltd 1 Our current NGS & Bioinformatics Platform 2 Our NGS workflow and applications 3 QIAGEN s
Proteomics and Cancer
 Proteomics and Cancer Japan Society for the Promotion of Science (JSPS) Science Dialogue Program at Niitsu Senior High School Niitsu, Niigata September 4th 2006 Vladimir Valera, M.D, PhD JSPS Postdoctoral
Proteomics and Cancer Japan Society for the Promotion of Science (JSPS) Science Dialogue Program at Niitsu Senior High School Niitsu, Niigata September 4th 2006 Vladimir Valera, M.D, PhD JSPS Postdoctoral
DNA Microarray Technology
 CHAPTER 1 DNA Microarray Technology All living organisms are composed of cells. As a functional unit, each cell can make copies of itself, and this process depends on a proper replication of the genetic
CHAPTER 1 DNA Microarray Technology All living organisms are composed of cells. As a functional unit, each cell can make copies of itself, and this process depends on a proper replication of the genetic
Towards unbiased biomarker discovery
 Towards unbiased biomarker discovery High-throughput molecular profiling technologies are routinely applied for biomarker discovery to make the drug discovery process more efficient and enable personalised
Towards unbiased biomarker discovery High-throughput molecular profiling technologies are routinely applied for biomarker discovery to make the drug discovery process more efficient and enable personalised
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Microbial Metabolism Systems Microbiology
 1 Microbial Metabolism Systems Microbiology Ching-Tsan Huang ( 黃慶璨 ) Office: Agronomy Hall, Room 111 Tel: (02) 33664454 E-mail: cthuang@ntu.edu.tw MIT OCW Systems Microbiology aims to integrate basic biological
1 Microbial Metabolism Systems Microbiology Ching-Tsan Huang ( 黃慶璨 ) Office: Agronomy Hall, Room 111 Tel: (02) 33664454 E-mail: cthuang@ntu.edu.tw MIT OCW Systems Microbiology aims to integrate basic biological
PHYSICIAN RESOURCES MAPPED TO GENOMICS COMPETENCIES AND GAPS IDENTIFIED WITH CURRENT EDUCATIONAL RESOURCES AVAILABLE 06/04/14
 PHYSICIAN RESOURCES MAPPED TO GENOMICS COMPETENCIES AND GAPS IDENTIFIED WITH CURRENT EDUCATIONAL RESOURCES AVAILABLE 06/04/14 Submitted resources from physician organization representatives were mapped
PHYSICIAN RESOURCES MAPPED TO GENOMICS COMPETENCIES AND GAPS IDENTIFIED WITH CURRENT EDUCATIONAL RESOURCES AVAILABLE 06/04/14 Submitted resources from physician organization representatives were mapped
Corporate Medical Policy
 Corporate Medical Policy Genetic Testing for Statin-induced Myopathy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_statin-induced_myopathy 6/2013 7/2017 7/2018
Corporate Medical Policy Genetic Testing for Statin-induced Myopathy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_statin-induced_myopathy 6/2013 7/2017 7/2018
Comments on Use of Databases for Establishing the Clinical Relevance of Human Genetic Variants
 Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane, Rm. 1061 Rockville, MD 20852 The American Society of Human Genetics 9650 Rockville Pike Bethesda, MD 20814 24 December
Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane, Rm. 1061 Rockville, MD 20852 The American Society of Human Genetics 9650 Rockville Pike Bethesda, MD 20814 24 December
Corporate Medical Policy
 Corporate Medical Policy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: magnetic_resonance_spectroscopy 12/1997 5/2017 5/2018 5/2017 Description of Procedure or Service Magnetic
Corporate Medical Policy File Name: Origination: Last CAP Review: Next CAP Review: Last Review: magnetic_resonance_spectroscopy 12/1997 5/2017 5/2018 5/2017 Description of Procedure or Service Magnetic
Understanding life WITH NEXT GENERATION PROTEOMICS SOLUTIONS
 Innovative services and products for highly multiplexed protein discovery and quantification to help you understand the biological processes that shape life. Understanding life WITH NEXT GENERATION PROTEOMICS
Innovative services and products for highly multiplexed protein discovery and quantification to help you understand the biological processes that shape life. Understanding life WITH NEXT GENERATION PROTEOMICS
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
Chapter 18: Regulation of Gene Expression. 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer
 Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Chapter 18: Regulation of Gene Expression 1. Gene Regulation in Bacteria 2. Gene Regulation in Eukaryotes 3. Gene Regulation & Cancer Gene Regulation Gene regulation refers to all aspects of controlling
Welcome to the NGS webinar series
 Welcome to the NGS webinar series Webinar 1 NGS: Introduction to technology, and applications NGS Technology Webinar 2 Targeted NGS for Cancer Research NGS in cancer Webinar 3 NGS: Data analysis for genetic
Welcome to the NGS webinar series Webinar 1 NGS: Introduction to technology, and applications NGS Technology Webinar 2 Targeted NGS for Cancer Research NGS in cancer Webinar 3 NGS: Data analysis for genetic
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Proteomics. Manickam Sugumaran. Department of Biology University of Massachusetts Boston, MA 02125
 Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
Higher Human Biology Unit 1: Human Cells Pupils Learning Outcomes
 Higher Human Biology Unit 1: Human Cells Pupils Learning Outcomes 1.1 Division and Differentiation in Human Cells I can state that cellular differentiation is the process by which a cell develops more
Higher Human Biology Unit 1: Human Cells Pupils Learning Outcomes 1.1 Division and Differentiation in Human Cells I can state that cellular differentiation is the process by which a cell develops more
Lecture #1. Introduction to microarray technology
 Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
Target Enrichment Strategies for Next Generation Sequencing
 Target Enrichment Strategies for Next Generation Sequencing Anuj Gupta, PhD Agilent Technologies, New Delhi Genotypic Conference, Sept 2014 NGS Timeline Information burst Nearly 30,000 human genomes sequenced
Target Enrichment Strategies for Next Generation Sequencing Anuj Gupta, PhD Agilent Technologies, New Delhi Genotypic Conference, Sept 2014 NGS Timeline Information burst Nearly 30,000 human genomes sequenced
The Integrated Biomedical Sciences Graduate Program
 The Integrated Biomedical Sciences Graduate Program at the university of notre dame Cutting-edge biomedical research and training that transcends traditional departmental and disciplinary boundaries to
The Integrated Biomedical Sciences Graduate Program at the university of notre dame Cutting-edge biomedical research and training that transcends traditional departmental and disciplinary boundaries to
Functional Genomics Overview RORY STARK PRINCIPAL BIOINFORMATICS ANALYST CRUK CAMBRIDGE INSTITUTE 18 SEPTEMBER 2017
 Functional Genomics Overview RORY STARK PRINCIPAL BIOINFORMATICS ANALYST CRUK CAMBRIDGE INSTITUTE 18 SEPTEMBER 2017 Agenda What is Functional Genomics? RNA Transcription/Gene Expression Measuring Gene
Functional Genomics Overview RORY STARK PRINCIPAL BIOINFORMATICS ANALYST CRUK CAMBRIDGE INSTITUTE 18 SEPTEMBER 2017 Agenda What is Functional Genomics? RNA Transcription/Gene Expression Measuring Gene
Lecture 23: Clinical and Biomedical Applications of Proteomics; Proteomics Industry
 Lecture 23: Clinical and Biomedical Applications of Proteomics; Proteomics Industry Clinical proteomics is the application of proteomic approach to the field of medicine. Proteome of an organism changes
Lecture 23: Clinical and Biomedical Applications of Proteomics; Proteomics Industry Clinical proteomics is the application of proteomic approach to the field of medicine. Proteome of an organism changes
Chapter 15 Gene Technologies and Human Applications
 Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
Chapter Outline Chapter 15 Gene Technologies and Human Applications Section 1: The Human Genome KEY IDEAS > Why is the Human Genome Project so important? > How do genomics and gene technologies affect
The Two-Hybrid System
 Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
AP Biology Gene Expression/Biotechnology REVIEW
 AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
FINAL DOCUMENT. Global Harmonization Task Force. Title: Clinical Evidence for IVD medical devices Key Definitions and Concepts
 GHTF/SG5/N6:2012 FINAL DOCUMENT Global Harmonization Task Force Title: Clinical Evidence for IVD medical devices Key Definitions and Concepts Authoring Group: Study Group 5 of the Global Harmonization
GHTF/SG5/N6:2012 FINAL DOCUMENT Global Harmonization Task Force Title: Clinical Evidence for IVD medical devices Key Definitions and Concepts Authoring Group: Study Group 5 of the Global Harmonization
BIOCRATES Life Sciences AG Short Company Presentation
 BIOCRATES Life Sciences AG Short Company Presentation Dr. Alexandra C. Gruber MBA MMFin Director Business Development / Marketing Austria Showcase Life Science Japan, Tokyo, December 2010 BIOCRATES Life
BIOCRATES Life Sciences AG Short Company Presentation Dr. Alexandra C. Gruber MBA MMFin Director Business Development / Marketing Austria Showcase Life Science Japan, Tokyo, December 2010 BIOCRATES Life
Themes: RNA and RNA Processing. Messenger RNA (mrna) What is a gene? RNA is very versatile! RNA-RNA interactions are very important!
 Themes: RNA is very versatile! RNA and RNA Processing Chapter 14 RNA-RNA interactions are very important! Prokaryotes and Eukaryotes have many important differences. Messenger RNA (mrna) Carries genetic
Themes: RNA is very versatile! RNA and RNA Processing Chapter 14 RNA-RNA interactions are very important! Prokaryotes and Eukaryotes have many important differences. Messenger RNA (mrna) Carries genetic
TECHNOLOGIES & SERVICES FOR THERAPEUTIC ANTIBODY DEVELOPMENT
 Specialist in tissue analysis by Histology, Immunohistochemistry and In Situ Hybridization TECHNOLOGIES & SERVICES FOR THERAPEUTIC ANTIBODY DEVELOPMENT CONTENTS Histalim: who we are Our areas of expertise
Specialist in tissue analysis by Histology, Immunohistochemistry and In Situ Hybridization TECHNOLOGIES & SERVICES FOR THERAPEUTIC ANTIBODY DEVELOPMENT CONTENTS Histalim: who we are Our areas of expertise
Bioinformatics Advice on Experimental Design
 Bioinformatics Advice on Experimental Design Where do I start? Please refer to the following guide to better plan your experiments for good statistical analysis, best suited for your research needs. Statistics
Bioinformatics Advice on Experimental Design Where do I start? Please refer to the following guide to better plan your experiments for good statistical analysis, best suited for your research needs. Statistics
Introduction to Bioinformatics
 Introduction to Bioinformatics Richard Corbett Canada s Michael Smith Genome Sciences Centre Vancouver, British Columbia June 28, 2017 Our mandate is to advance knowledge about cancer and other diseases
Introduction to Bioinformatics Richard Corbett Canada s Michael Smith Genome Sciences Centre Vancouver, British Columbia June 28, 2017 Our mandate is to advance knowledge about cancer and other diseases
Introduction to Microarray Analysis
 Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Genomic Research: Issues to Consider. IRB Brown Bag August 28, 2014 Sharon Aufox, MS, LGC
 Genomic Research: Issues to Consider IRB Brown Bag August 28, 2014 Sharon Aufox, MS, LGC Outline Key genomic terms and concepts Issues in genomic research Consent models Types of findings Returning results
Genomic Research: Issues to Consider IRB Brown Bag August 28, 2014 Sharon Aufox, MS, LGC Outline Key genomic terms and concepts Issues in genomic research Consent models Types of findings Returning results
Chapter 11: Regulation of Gene Expression
 Chapter Review 1. It has long been known that there is probably a genetic link for alcoholism. Researchers studying rats have begun to elucidate this link. Briefly describe the genetic mechanism found
Chapter Review 1. It has long been known that there is probably a genetic link for alcoholism. Researchers studying rats have begun to elucidate this link. Briefly describe the genetic mechanism found
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
2. Outline the levels of DNA packing in the eukaryotic nucleus below next to the diagram provided.
 AP Biology Reading Packet 6- Molecular Genetics Part 2 Name Chapter 19: Eukaryotic Genomes 1. Define the following terms: a. Euchromatin b. Heterochromatin c. Nucleosome 2. Outline the levels of DNA packing
AP Biology Reading Packet 6- Molecular Genetics Part 2 Name Chapter 19: Eukaryotic Genomes 1. Define the following terms: a. Euchromatin b. Heterochromatin c. Nucleosome 2. Outline the levels of DNA packing
CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016
 CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016 Topics Genetic variation Population structure Linkage disequilibrium Natural disease variants Genome Wide Association Studies Gene
CS273B: Deep Learning in Genomics and Biomedicine. Recitation 1 30/9/2016 Topics Genetic variation Population structure Linkage disequilibrium Natural disease variants Genome Wide Association Studies Gene
DISCOVERY AND VALIDATION OF TARGETS AND BIOMARKERS BY MASS SPECTROMETRY-BASED PROTEOMICS. September, 2011
 DISCOVERY AND VALIDATION OF TARGETS AND BIOMARKERS BY MASS SPECTROMETRY-BASED PROTEOMICS September, 2011 1 CAPRION PROTEOMICS Leading proteomics-based service provider - Biomarker and target discovery
DISCOVERY AND VALIDATION OF TARGETS AND BIOMARKERS BY MASS SPECTROMETRY-BASED PROTEOMICS September, 2011 1 CAPRION PROTEOMICS Leading proteomics-based service provider - Biomarker and target discovery
Feature Selection of Gene Expression Data for Cancer Classification: A Review
 Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 50 (2015 ) 52 57 2nd International Symposium on Big Data and Cloud Computing (ISBCC 15) Feature Selection of Gene Expression
Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 50 (2015 ) 52 57 2nd International Symposium on Big Data and Cloud Computing (ISBCC 15) Feature Selection of Gene Expression
MICROARRAYS+SEQUENCING
 MICROARRAYS+SEQUENCING The most efficient way to advance genomics research Down to a Science. www.affymetrix.com/downtoascience Affymetrix GeneChip Expression Technology Complementing your Next-Generation
MICROARRAYS+SEQUENCING The most efficient way to advance genomics research Down to a Science. www.affymetrix.com/downtoascience Affymetrix GeneChip Expression Technology Complementing your Next-Generation
"Stratification biomarkers in personalised medicine"
 1/12 2/12 SUMMARY REPORT "Stratification biomarkers in personalised medicine" Workshop to clarify the scope for stratification biomarkers and to identify bottlenecks in the discovery and the use of such
1/12 2/12 SUMMARY REPORT "Stratification biomarkers in personalised medicine" Workshop to clarify the scope for stratification biomarkers and to identify bottlenecks in the discovery and the use of such
Accessible answers. Targeted sequencing: accelerating and amplifying answers for oncology research
 Accessible answers Targeted sequencing: accelerating and amplifying answers for oncology research Help advance precision medicine Accelerate results with Ion Torrent NGS Life without cancer. This is our
Accessible answers Targeted sequencing: accelerating and amplifying answers for oncology research Help advance precision medicine Accelerate results with Ion Torrent NGS Life without cancer. This is our
Introduction to the UCSC genome browser
 Introduction to the UCSC genome browser Dominik Beck NHMRC Peter Doherty and CINSW ECR Fellow, Senior Lecturer Lowy Cancer Research Centre, UNSW and Centre for Health Technology, UTS SYDNEY NSW AUSTRALIA
Introduction to the UCSC genome browser Dominik Beck NHMRC Peter Doherty and CINSW ECR Fellow, Senior Lecturer Lowy Cancer Research Centre, UNSW and Centre for Health Technology, UTS SYDNEY NSW AUSTRALIA
CHARTING THE COURSE FOR PRECISION MEDICINE
 A Friends of Cancer Research White Paper CHARTING THE COURSE FOR PRECISION MEDICINE ADOPTING CONSENSUS ANALYTICAL STANDARDS AND STREAMLINING APPROVAL PATHWAYS FOR POST-MARKET MODIFICATIONS FOR NGS TESTS
A Friends of Cancer Research White Paper CHARTING THE COURSE FOR PRECISION MEDICINE ADOPTING CONSENSUS ANALYTICAL STANDARDS AND STREAMLINING APPROVAL PATHWAYS FOR POST-MARKET MODIFICATIONS FOR NGS TESTS
ACCELERATING GENOMIC ANALYSIS ON THE CLOUD. Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia to analyze thousands of genomes
 ACCELERATING GENOMIC ANALYSIS ON THE CLOUD Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia to analyze thousands of genomes Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia
ACCELERATING GENOMIC ANALYSIS ON THE CLOUD Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia to analyze thousands of genomes Enabling the PanCancer Analysis of Whole Genomes (PCAWG) consortia
GUIDANCE ON THE EVALUATION OF NON ACCREDITED QUALIFICATIONS
 GUIDANCE ON THE EVALUATION OF NON ACCREDITED QUALIFICATIONS 1. Introduction 1.1 This document provides guidance notes for the assessment of academic qualifications that have not been formally accredited
GUIDANCE ON THE EVALUATION OF NON ACCREDITED QUALIFICATIONS 1. Introduction 1.1 This document provides guidance notes for the assessment of academic qualifications that have not been formally accredited
Developing an Accurate and Precise Companion Diagnostic Assay for Targeted Therapies in DLBCL
 Developing an Accurate and Precise Companion Diagnostic Assay for Targeted Therapies in DLBCL James Storhoff, Ph.D. Senior Manager, Diagnostic Test Development World Cdx, Boston, Sep. 10th Molecules That
Developing an Accurate and Precise Companion Diagnostic Assay for Targeted Therapies in DLBCL James Storhoff, Ph.D. Senior Manager, Diagnostic Test Development World Cdx, Boston, Sep. 10th Molecules That
8/21/2014. From Gene to Protein
 From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
Fundamentals of Clinical Genomics
 Fundamentals of Clinical Genomics Wellcome Genome Campus Hinxton, Cambridge, UK 17-19 January 2018 Lectures and Workshops to be held in the Rosalind Franklin Pavilion Lunch and Dinner to be held in the
Fundamentals of Clinical Genomics Wellcome Genome Campus Hinxton, Cambridge, UK 17-19 January 2018 Lectures and Workshops to be held in the Rosalind Franklin Pavilion Lunch and Dinner to be held in the
3.1.4 DNA Microarray Technology
 3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
Bridging the Gap in Precision Medicine An Informatics Solution Connecting Research, Pathology, and Clinical Care
 Bridging the Gap in Precision Medicine An Informatics Solution Connecting Research, Pathology, and Clinical Care O R A C L E W H I T E P A P E R F E B R U A R Y 2 0 1 6 Disclaimer The following is intended
Bridging the Gap in Precision Medicine An Informatics Solution Connecting Research, Pathology, and Clinical Care O R A C L E W H I T E P A P E R F E B R U A R Y 2 0 1 6 Disclaimer The following is intended
Terminology for personalized medicine
 Terminology for personalized medicine Sarah Ali-Khan 1 PhD, Stephanie Kowal 2, Westerly Luth 2, Richard Gold 1 SJD, and Tania Bubela 2 PhD JD 1. Centre for Intellectual Property Policy (CIPP), Faculty
Terminology for personalized medicine Sarah Ali-Khan 1 PhD, Stephanie Kowal 2, Westerly Luth 2, Richard Gold 1 SJD, and Tania Bubela 2 PhD JD 1. Centre for Intellectual Property Policy (CIPP), Faculty
Pathology Facilities and Potential for Collaboration. The University of Western Australia
 Pathology Facilities and Potential for Collaboration UWA Pathology and Laboratory Medicine Pathology Research Themes Experimental pathology Infection and Immunity Translational pathology Cores Core Pathology
Pathology Facilities and Potential for Collaboration UWA Pathology and Laboratory Medicine Pathology Research Themes Experimental pathology Infection and Immunity Translational pathology Cores Core Pathology
Complete automation for NGS interpretation and reporting with evidence-based clinical decision support
 Brochure Bioinformatics for Clinical Oncology Testing Complete automation for NGS interpretation and reporting with evidence-based clinical decision support Sample to Insight Powering clinical insights
Brochure Bioinformatics for Clinical Oncology Testing Complete automation for NGS interpretation and reporting with evidence-based clinical decision support Sample to Insight Powering clinical insights
Supplementary Figure 1. Utilization of publicly available antibodies in different applications.
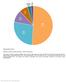 Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Proteomics: A Challenge for Technology and Information Science. What is proteomics?
 Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Integrating Genomics in Family Medicine
 Lehigh Valley Health Network LVHN Scholarly Works Department of Family Medicine Integrating Genomics in Family Medicine Brian Stello MD Lehigh Valley Health Network, Brian.Stello@lvhn.org Follow this and
Lehigh Valley Health Network LVHN Scholarly Works Department of Family Medicine Integrating Genomics in Family Medicine Brian Stello MD Lehigh Valley Health Network, Brian.Stello@lvhn.org Follow this and
PV92 PCR Bio Informatics
 Purpose of PCR Chromosome 16 PV92 PV92 PCR Bio Informatics Alu insert, PV92 locus, chromosome 16 Introduce the polymerase chain reaction (PCR) technique Apply PCR to population genetics Directly measure
Purpose of PCR Chromosome 16 PV92 PV92 PCR Bio Informatics Alu insert, PV92 locus, chromosome 16 Introduce the polymerase chain reaction (PCR) technique Apply PCR to population genetics Directly measure
Unit 6: Molecular Genetics & DNA Technology Guided Reading Questions (100 pts total)
 Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 16 The Molecular Basis of Inheritance Unit 6: Molecular Genetics
Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 16 The Molecular Basis of Inheritance Unit 6: Molecular Genetics
Next Generation Sequencing. Target Enrichment
 Next Generation Sequencing Target Enrichment Next Generation Sequencing Your Partner in Every Step from Sample to Data NGS: Revolutionizing Genetic Analysis with Single-Molecule Resolution Next generation
Next Generation Sequencing Target Enrichment Next Generation Sequencing Your Partner in Every Step from Sample to Data NGS: Revolutionizing Genetic Analysis with Single-Molecule Resolution Next generation
TheraLin. Universal Tissue Fixative Enabling Molecular Pathology
 TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
TheraLin Universal Tissue Fixative Enabling Molecular Pathology TheraLin Universal Tissue Fixative Enabling Molecular Pathology Contents Page # TheraLin Universal Tissue Fixative 3 Introduction 5 Easy
AGRO/ANSC/BIO/GENE/HORT 305 Fall, 2016 Overview of Genetics Lecture outline (Chpt 1, Genetics by Brooker) #1
 AGRO/ANSC/BIO/GENE/HORT 305 Fall, 2016 Overview of Genetics Lecture outline (Chpt 1, Genetics by Brooker) #1 - Genetics: Progress from Mendel to DNA: Gregor Mendel, in the mid 19 th century provided the
AGRO/ANSC/BIO/GENE/HORT 305 Fall, 2016 Overview of Genetics Lecture outline (Chpt 1, Genetics by Brooker) #1 - Genetics: Progress from Mendel to DNA: Gregor Mendel, in the mid 19 th century provided the
Biology 201 (Genetics) Exam #3 120 points 20 November Read the question carefully before answering. Think before you write.
 Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Chapter 16: Gene Expression from Biology by OpenStax College is licensed under a Creative Commons Attribution 3.0 Unported license.
 Chapter 16: Gene Expression from Biology by OpenStax College is licensed under a Creative Commons Attribution 3.0 Unported license. 2013, Rice University. CHAPTER 16 GENE EXPRESSION 429 16 GENE EXPRESSION
Chapter 16: Gene Expression from Biology by OpenStax College is licensed under a Creative Commons Attribution 3.0 Unported license. 2013, Rice University. CHAPTER 16 GENE EXPRESSION 429 16 GENE EXPRESSION
BIOLOGY. Chapter 16 GenesExpression
 BIOLOGY Chapter 16 GenesExpression CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 18 Gene Expression 2014 Pearson Education, Inc. Figure 16.1 Differential Gene Expression results
BIOLOGY Chapter 16 GenesExpression CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 18 Gene Expression 2014 Pearson Education, Inc. Figure 16.1 Differential Gene Expression results
First Annual Biomarker Symposium Quest Diagnostics Clinical Trials
 First Annual Biomarker Symposium Quest Diagnostics Clinical Trials Terry Robins, Ph.D. Director Biomarker R&D and Scientific Affairs Quest Diagnostics Clinical Trials Key Considerations: Biomarker Development
First Annual Biomarker Symposium Quest Diagnostics Clinical Trials Terry Robins, Ph.D. Director Biomarker R&D and Scientific Affairs Quest Diagnostics Clinical Trials Key Considerations: Biomarker Development
Preanalytical Variables in Blood Collection: Impact on Precision Medicine
 Preanalytical Variables in Blood Collection: Impact on Precision Medicine Carolyn Compton, MD, PhD Chair, Scientific Advisory Committee, Indivumed GmbH Professor of Life Sciences, ASU Professor of Laboratory
Preanalytical Variables in Blood Collection: Impact on Precision Medicine Carolyn Compton, MD, PhD Chair, Scientific Advisory Committee, Indivumed GmbH Professor of Life Sciences, ASU Professor of Laboratory
Epigenetics, Environment and Human Health
 Epigenetics, Environment and Human Health A. Karim Ahmed National Council for Science and the Environment Washington, DC May, 2015 Epigenetics A New Biological Paradigm A Question about Cells: All cells
Epigenetics, Environment and Human Health A. Karim Ahmed National Council for Science and the Environment Washington, DC May, 2015 Epigenetics A New Biological Paradigm A Question about Cells: All cells
Regulation of Gene Expression
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 15 Regulation of Gene Expression Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 15 Regulation of Gene Expression Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Fusion Gene Analysis. ncounter Elements. Molecules That Count WHITE PAPER. v1.0 OCTOBER 2014 NanoString Technologies, Inc.
 Fusion Gene Analysis NanoString Technologies, Inc. Fusion Gene Analysis ncounter Elements v1.0 OCTOBER 2014 NanoString Technologies, Inc., Seattle WA 98109 FOR RESEARCH USE ONLY. Not for use in diagnostic
Fusion Gene Analysis NanoString Technologies, Inc. Fusion Gene Analysis ncounter Elements v1.0 OCTOBER 2014 NanoString Technologies, Inc., Seattle WA 98109 FOR RESEARCH USE ONLY. Not for use in diagnostic
DNA. bioinformatics. epigenetics methylation structural variation. custom. assembly. gene. tumor-normal. mendelian. BS-seq. prediction.
 Epigenomics T TM activation SNP target ncrna validation metagenomics genetics private RRBS-seq de novo trio RIP-seq exome mendelian comparative genomics DNA NGS ChIP-seq bioinformatics assembly tumor-normal
Epigenomics T TM activation SNP target ncrna validation metagenomics genetics private RRBS-seq de novo trio RIP-seq exome mendelian comparative genomics DNA NGS ChIP-seq bioinformatics assembly tumor-normal
RNA-Sequencing analysis
 RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
RNA-Sequencing analysis Markus Kreuz 25. 04. 2012 Institut für Medizinische Informatik, Statistik und Epidemiologie Content: Biological background Overview transcriptomics RNA-Seq RNA-Seq technology Challenges
Presented by: Tess L. Crisostomo Laboratory Quality Assurance/Compliance Officer Naval Medical Center San Diego
 Presented by: Tess L. Crisostomo Laboratory Quality Assurance/Compliance Officer Naval Medical Center San Diego Discuss the difference between DNA Genotyping and Genome Sequencing What makes you who you
Presented by: Tess L. Crisostomo Laboratory Quality Assurance/Compliance Officer Naval Medical Center San Diego Discuss the difference between DNA Genotyping and Genome Sequencing What makes you who you
Outline and learning objectives. From Proteomics to Systems Biology. Integration of omics - information
 From to Systems Biology Outline and learning objectives Omics science provides global analysis tools to study entire systems How to obtain omics - What can we learn Limitations Integration of omics - In-class
From to Systems Biology Outline and learning objectives Omics science provides global analysis tools to study entire systems How to obtain omics - What can we learn Limitations Integration of omics - In-class
Gene Regulation Solutions. Microarrays and Next-Generation Sequencing
 Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Cytomics in Action: Cytokine Network Cytometry
 Cytomics in Action: Cytokine Network Cytometry Jonni S. Moore, Ph.D. Director, Clinical and Research Flow Cytometry and PathBioResource Associate Professor of Pathology & Laboratory Medicine University
Cytomics in Action: Cytokine Network Cytometry Jonni S. Moore, Ph.D. Director, Clinical and Research Flow Cytometry and PathBioResource Associate Professor of Pathology & Laboratory Medicine University
Reflection paper on co-development of pharmacogenomic biomarkers and Assays in the context of drug development
 1 2 3 24 June 2010 EMA/CHMP/641298/2008 Committee for Medicinal Products for Human Use (CHMP) 4 5 6 7 Reflection paper on co-development of pharmacogenomic biomarkers and Assays in the context of drug
1 2 3 24 June 2010 EMA/CHMP/641298/2008 Committee for Medicinal Products for Human Use (CHMP) 4 5 6 7 Reflection paper on co-development of pharmacogenomic biomarkers and Assays in the context of drug
MCDB 1041 Class 21 Splicing and Gene Expression
 MCDB 1041 Class 21 Splicing and Gene Expression Learning Goals Describe the role of introns and exons Interpret the possible outcomes of alternative splicing Relate the generation of protein from DNA to
MCDB 1041 Class 21 Splicing and Gene Expression Learning Goals Describe the role of introns and exons Interpret the possible outcomes of alternative splicing Relate the generation of protein from DNA to
Intro to Microarray Analysis. Courtesy of Professor Dan Nettleton Iowa State University (with some edits)
 Intro to Microarray Analysis Courtesy of Professor Dan Nettleton Iowa State University (with some edits) Some Basic Biology Genes are DNA sequences that code for proteins. (e.g. gene lengths perhaps 1000
Intro to Microarray Analysis Courtesy of Professor Dan Nettleton Iowa State University (with some edits) Some Basic Biology Genes are DNA sequences that code for proteins. (e.g. gene lengths perhaps 1000
BIO 101 : The genetic code and the central dogma
 BIO 101 : The genetic code and the central dogma NAME Objectives The purpose of this exploration is to... 1. design experiments to decipher the genetic code; 2. visualize the process of protein synthesis;
BIO 101 : The genetic code and the central dogma NAME Objectives The purpose of this exploration is to... 1. design experiments to decipher the genetic code; 2. visualize the process of protein synthesis;
Genomics Market Share, Size, Analysis, Growth, Trends and Forecasts to 2024 Hexa Research
 Genomics Market Share, Size, Analysis, Growth, Trends and Forecasts to 2024 Hexa Research " Increasing usage of novel genomics techniques and tools, evaluation of their benefit to patient outcome and focusing
Genomics Market Share, Size, Analysis, Growth, Trends and Forecasts to 2024 Hexa Research " Increasing usage of novel genomics techniques and tools, evaluation of their benefit to patient outcome and focusing
Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD
 Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD Department of Biopharmacy, Faculty of Pharmacy, Silpakorn University Example of critical checkpoints
Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD Department of Biopharmacy, Faculty of Pharmacy, Silpakorn University Example of critical checkpoints
LATE-PCR. Linear-After-The-Exponential
 LATE-PCR Linear-After-The-Exponential A Patented Invention of the Laboratory of Human Genetics and Reproductive Biology Lab. Director: Lawrence J. Wangh, Ph.D. Department of Biology, Brandeis University,
LATE-PCR Linear-After-The-Exponential A Patented Invention of the Laboratory of Human Genetics and Reproductive Biology Lab. Director: Lawrence J. Wangh, Ph.D. Department of Biology, Brandeis University,
Cancer Genetics Solutions
 Cancer Genetics Solutions Cancer Genetics Solutions Pushing the Boundaries in Cancer Genetics Cancer is a formidable foe that presents significant challenges. The complexity of this disease can be daunting
Cancer Genetics Solutions Cancer Genetics Solutions Pushing the Boundaries in Cancer Genetics Cancer is a formidable foe that presents significant challenges. The complexity of this disease can be daunting
If Dna Has The Instructions For Building Proteins Why Is Mrna Needed
 If Dna Has The Instructions For Building Proteins Why Is Mrna Needed if a strand of DNA has the sequence CGGTATATC, then the complementary each strand of DNA contains the info needed to produce the complementary
If Dna Has The Instructions For Building Proteins Why Is Mrna Needed if a strand of DNA has the sequence CGGTATATC, then the complementary each strand of DNA contains the info needed to produce the complementary
LECTURE: 22 IMMUNOGLOBULIN DIVERSITIES LEARNING OBJECTIVES: The student should be able to:
 LECTURE: 22 Title IMMUNOGLOBULIN DIVERSITIES LEARNING OBJECTIVES: The student should be able to: Identify the chromosome that contains the gene segments that encode the surface immunoglobulin heavy chain
LECTURE: 22 Title IMMUNOGLOBULIN DIVERSITIES LEARNING OBJECTIVES: The student should be able to: Identify the chromosome that contains the gene segments that encode the surface immunoglobulin heavy chain
PLTW Biomedical Science Medical Interventions Course Outline
 Follow the fictitious Smith family as you learn about the prevention, diagnosis, and treatment of disease. Play the role of biomedical professionals to analyze case information and diagnose and treat your
Follow the fictitious Smith family as you learn about the prevention, diagnosis, and treatment of disease. Play the role of biomedical professionals to analyze case information and diagnose and treat your
The Genetic Code and Transcription. Chapter 12 Honors Genetics Ms. Susan Chabot
 The Genetic Code and Transcription Chapter 12 Honors Genetics Ms. Susan Chabot TRANSCRIPTION Copy SAME language DNA to RNA Nucleic Acid to Nucleic Acid TRANSLATION Copy DIFFERENT language RNA to Amino
The Genetic Code and Transcription Chapter 12 Honors Genetics Ms. Susan Chabot TRANSCRIPTION Copy SAME language DNA to RNA Nucleic Acid to Nucleic Acid TRANSLATION Copy DIFFERENT language RNA to Amino
2D gel Western blotting using antibodies against ubiquitin, SUMO and acetyl PTM
 2D gel Western blotting using antibodies against ubiquitin, SUMO and acetyl PTM Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc www.kendricklabs.com Talk Outline Significance Method description
2D gel Western blotting using antibodies against ubiquitin, SUMO and acetyl PTM Nancy Kendrick, Jon Johansen & Matt Hoelter, Kendrick Labs Inc www.kendricklabs.com Talk Outline Significance Method description
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor
 Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Tumor Growth Suppression Through the Activation of p21, a Cyclin-Dependent Kinase Inhibitor Nicholas Love 11/28/01 A. What is p21? Introduction - p21 is a gene found on chromosome 6 at 6p21.2 - this gene
Introduction to Bioinformatics and Gene Expression Technology
 Vocabulary Introduction to Bioinformatics and Gene Expression Technology Utah State University Spring 2014 STAT 5570: Statistical Bioinformatics Notes 1.1 Gene: Genetics: Genome: Genomics: hereditary DNA
Vocabulary Introduction to Bioinformatics and Gene Expression Technology Utah State University Spring 2014 STAT 5570: Statistical Bioinformatics Notes 1.1 Gene: Genetics: Genome: Genomics: hereditary DNA
Tech Note. Using the ncounter Analysis System with FFPE Samples for Gene Expression Analysis. ncounter Gene Expression. Molecules That Count
 ncounter Gene Expression Tech Note Using the ncounter Analysis System with FFPE Samples for Gene Expression Analysis Introduction For the past several decades, pathologists have kept samples obtained from
ncounter Gene Expression Tech Note Using the ncounter Analysis System with FFPE Samples for Gene Expression Analysis Introduction For the past several decades, pathologists have kept samples obtained from
GNPS: Global Natural Products Social Molecular Networking Delivering data-enabled, community-driven research
 GNPS: Global Natural Products Social Molecular Networking Delivering data-enabled, community-driven research Mingxun Wang 1,2,4, Jeremy Carver 1,4, Julie Wertz 1,4, Laurence Bernstein 1,4, Seungjin Na
GNPS: Global Natural Products Social Molecular Networking Delivering data-enabled, community-driven research Mingxun Wang 1,2,4, Jeremy Carver 1,4, Julie Wertz 1,4, Laurence Bernstein 1,4, Seungjin Na
Bio11 Announcements. Ch 21: DNA Biology and Technology. DNA Functions. DNA and RNA Structure. How do DNA and RNA differ? What are genes?
 Bio11 Announcements TODAY Genetics (review) and quiz (CP #4) Structure and function of DNA Extra credit due today Next week in lab: Case study presentations Following week: Lab Quiz 2 Ch 21: DNA Biology
Bio11 Announcements TODAY Genetics (review) and quiz (CP #4) Structure and function of DNA Extra credit due today Next week in lab: Case study presentations Following week: Lab Quiz 2 Ch 21: DNA Biology
MOLECULAR BIOLOGY OF EUKARYOTES 2016 SYLLABUS
 03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
03-442 Lectures: MWF 9:30-10:20 a.m. Doherty Hall 2105 03-742 Advanced Discussion Section: Time and place to be announced Probably Mon 4-6 p.m. or 6-8p.m.? Once we establish who is taking the advanced
Introduction to Pharmacogenetics Competency
 Introduction to Pharmacogenetics Competency Updated on 6/2015 Pre-test Question # 1 Pharmacogenetics is the study of how genetic variations affect drug response a) True b) False Pre-test Question # 2 Pharmacogenetic
Introduction to Pharmacogenetics Competency Updated on 6/2015 Pre-test Question # 1 Pharmacogenetics is the study of how genetic variations affect drug response a) True b) False Pre-test Question # 2 Pharmacogenetic
Algorithms in Bioinformatics
 Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline Central Dogma of Molecular
Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline Central Dogma of Molecular
