Revised: RG-RV2 by Fukuhara et al.
|
|
|
- Eileen Abigail Tyler
- 5 years ago
- Views:
Transcription
1 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the deleted allele (Spns2 - ). In the targeted allele, exon 2 of the Spns2 gene (yellow box) is flanked by two loxp sites (green arrowheads). Together with the first loxp site, a neomycin selection cassette, PKG-Neo-pA (gray box), flanked by two frt sites (orange 1
2 arrowheads) has been inserted. Mice carrying the targeted Spns2 allele were crossed with transgenic mice expressing Flp recombinase under the control of cytomegalovirus early enhancer/chicken beta actin (CAG) promoter to remove the PKG-Neo-pA cassette, resulting in the Spns2 floxed mice. To generate conventional Spns2 knockout mice, the mice carrying a floxed Spns2 allele were crossed with transgenic mice expressing the Cre recombinase under the control of cytomegalovirus (CMV) promoter. To inactivate the Spns2 gene in endothelial cells, the Spns2 floxed mice were bred with Tie2-Cre mice that carry the Cre recombinase driven by the Tie2 promoter. (B) Southern blot analysis of AvrII-digested genomic DNA from tail biopsies of Spns2 f/f, Spns2 -/- and Spns2 +/+ mice were carried out using a probe indicated in A (blue line). As expected, 7.2 kb, 7.5 kb and 7.0 kb bands were detected in wild type (Spns2 +/+ ), Spns2 floxed (Spns2 f/f ) and Spns2 knockout (Spns2 -/- ) mice. (C) PCR analysis of the genomic DNA from Spns2 f/f, Spns2 -/- and Spns2 +/+ mice were performed using a primer set to amplify the exon 2 as indicated in A (red arrows). 552-bp, 842-bp and 316-bp fragments were amplified in wild type (Spns2 +/+ ), Spns2 floxed (Spns2 f/f ) and Spns2 knockout (Spns2 -/- ) mice. (D) Genomic PCR analysis of Spns2 +/+, Spns2 -/-, Spns2 f/-, Spns2 f/- ;Tie2Cre, Spns2 f/f, Spns2 f/f ;Tie2Cre mice were performed using the primer sets to amplify the exon 2 of Spns2 gene (upper panel) and Cre recombinase gene (lower panel). 2
3 Supplemental Figure 2 Spns2 KO mice are functionally disrupted for Spns2. (A) Schematic representation of the exon/intron structure of the wild type Spns2 allele and the corresponding wild type mrna (Wt mrna). A mutant mrna transcribed from the Spns2 deleted allele lacking exon 2 (Mutant mrna) is also shown at the bottom. Boxes and polylines represent exons and introns, respectively. Blue and gray boxes correspond to the coding and non-coding exons, respectively. Red boxes indicate exon 2. The size of exon is given below each exon. The mrna derived from the Spns2 deleted allele results in an in-frame deletion of exon 2. (B) RT-PCR analyses of RNAs extracted from the lungs of wild type (Spns2 +/+ ) and Spns2 KO (Spns2 -/- ) mice were performed using two sets of PCR primers as indicated in A (blue and green arrows). The size difference between the two bands is consistent with the absence of exon 2 (66 bp) in the mutant mrna. Sequencing of the PCR products showed that Spns2 KO mice express a mutant mrna transcript lacking exon 2-derived sequence encoding aa of wild type Spns2. (C) HUVECs were transfected with the plasmid expressing either wild type (Wt) or mutant (Mutant) Spns2 carboxy-terminally tagged GFP or with myristoylated 3
4 GFP-encoding plasmid (Myr-GFP). GFP and phase contrast images were obtained using an IX81 inverted microscope (Olympus). Note that mutant Spns2 protein failed to localize at the plasma membrane. (D) HEK293 cells were transfected with the plasmid expressing human Spns2 (hwt), mouse Spns2 (mwt) or mouse Spns2 mutant (mutant) together with or without the Sphk1-expressing vector as indicated at the bottom. S1P release from those cells was examined as described in Methods. Data are shown as means ± S.D. (n=4). Note that Spns2 mutant lost the ability to export S1P. 4
5 Supplemental Figure 3 Spns2 KO mice develop normally except for the symblepharon formation. (A-F) Body and organ weights of 10-week-old wild type (Spns2 +/+ ) and Spns2 KO (Spns2 -/- ) mice were measured (body weight (A), heart (B), lung (C), liver (D), spleen (E), kidney (F)). There were no differences in body and organ weights between wild type and Spns2 KO mice. Data are shown as means ± S.D. (wild type, n=11; Spns2 KO, n=10). (G) Spns2 KO mice exhibit symblepharon. 5
6 Supplemental Figure 4 The biochemical examination of the blood of Spns2 KO mice. Plasma levels of total protein (TP) (A), total bilirubin (TBIL) (B), aspartate aminotransferase (AST) (C), alanine aminotransferase (ALT) (D), triglycerol (TG) (E), glucose (GLU) (F), blood urea nitrogen (BUN) (G) and albumin (ALB) (H) in 8-week-old wild type (Spns2 +/+ ) and Spns2 KO (Spns2 -/- ) mice. There were no differences in blood biochemical parameters between wild type and Spns2 KO mice. Data are shown as means ± S.D. (n=4). 6
7 Supplemental Figure 5 The hematological profile of Spns2 KO mice. Blood was collected from 8-week-old wild type (Spns2 +/+ ) and Spns2 KO (Spns2 -/- ) mice, and analyzed for white blood cells (WBC) (A), red blood cells (RBC) (B), platelets (C), hemoglobin (HBG) (D), hematocrit (HCT) (E), mean corpuscular volume (MCV) (F), mean corpuscular hemoglobin (MCH) (G) and mean corpuscular hemoglobin concentration (MCHC) (H). Spns2 KO mice exhibited a decreased count of white blood cells compared with wild type mice. Data are shown as means ± S.D. (n=4). 7
8 Supplemental Figure 6 A representative flow cytometric analysis of peripheral blood cells from control (Spns2 +/+ ) and global Spns2 KO (Spns2 -/- ) mice. (A) The numbers represent the percentage of CD4 and CD8 single-positive (SP) T cells in total lymphocytes. (B) The numbers represent percentages of IgD + CD23 + cells in CD19 + peripheral blood lymphocytes (mature recirculating B cells; Mature rec. B) in the gate on the plots. 8
9 Supplemental Figure 7 Histological analyses of the thymi and peripheral lymph nodes (LN) of Spns2 KO mice. Representative hematoxylin and eosin stained sections of thymus and peripheral LN from wild type (Spns2 +/+ ) and Spns2 KO (Spns2 -/- ) mice. No striking structural differences of thymus and peripheral LNl between Spns2 KO and control mice were detected. 9
10 Supplemental Figure 8 Increased proportion of fully mature SP T cells and decreased proportion of semi-mature SP T cells in the thymi of Spns2 KO mice. (A) Frequencies (left) and numbers (right) of semi-mature (CD69 + CD62L lo/- ) and fully mature (CD69 lo/- CD62L + ) CD4 SP T cells in the thymi of control (Spns2 +/+ ) or Spns2 KO (Spns2 -/- ) mice (n=8). (B) Frequencies (left) and numbers (right) of semi-mature (CD69 + CD62L lo/- ) and fully mature (CD69 lo/- CD62L + ) CD8 SP T cells in thymi of control (Spns2 +/+ ) or Spns2 KO (Spns2 -/- ) mice (n=8). (C) Flow cytometric analysis for the expression of CD69 on fully mature CD62L + CD4 (left) and CD8 (right) SP T cells in the thymus of control (black line) or Spns2 KO (orange line) mice. Control antibody staining is shown shaded in grey. Data are representative for eight pairs of mice. 10
11 Supplemental Figure 9 Reduced numbers of mature recirculating B (Mature rec. B) cells in the bone marrow of Spns2 KO mice. Representative flow cytometric analysis of bone marrow derived B cells from control (Spns2 +/+ ) or Spns2 KO (Spns2 -/- ) mice. (A) The numbers indicate the percentage of IgM and IgD expressing CD19 + B cells. Mature recirculating B (Mature rec. B) cells and immature B cells were identified as CD19 + IgM + IgD + and CD19 + IgM + IgD -, respectively. (B) Frequencies (left) and numbers (right) of mature recirculating B cells and immature B cells are shown (n=11). Bars and circles indicate averages and values for individual mice, respectively. 11
12 Supplemental Figure 10 Amount of plasma S1P bound to HDL and albumin in control (Spns2 +/+ ) or Spns2 KO (Spns2 -/- ) mice. (A) Representative gel filtration profiles of plasma S1P (bars) and protein (solid line) from control (top: Spns2 +/+ ) and Spns2 KO (bottom: Spns2 -/- ) mice. 100 l of plasma was chromatographed on a Superose 12 column. Fractions were collected every 2 min (0.5 ml). For each fraction, S1P concentration was determined. Slashed and gray bars indicate the amount of S1P bound to HDL and albumin, respectively. (B) Amount of S1P associated with HDL and albumin in 100 l of plasma from control (Spns2 +/+ ) or Spns2 KO (Spns2 -/- ) mice (n=2). 12
13 Supplemental Figure 11 PCR analysis of the genomic DNA isolated from the bone marrow of wild type mouse (Spns2 +/+ ), Spns2 KO mouse (Spns2 -/- ) and 13 Spns2 KO mice reconstituted with wild type bone marrow (#1~#13) was performed as described in the legend of Supplementary Figure 1C. 552-bp and 316-bp PCR products represent the wild type and Spns2-deleted alleles, respectively. Note that the bone marrow of Spns2 KO mice was successfully reconstituted by that of wild type mice. 13
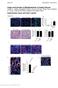
14 Supplemental Figure 12 In situ hybridization for Spns2 mrna. Antisense probe was hybridized to the sections of heart, lung, hypothalamus, olfactory bulb and kidney (Spns2: purple). Serial sections were also stained with anti-cd31 antibody (CD31: brown) to identify the endothelial cells. The scale bars represent 20 m. 14
15 Supplemental Figure 13 Plasma concentrations of S1P in control (Spns2 f/f ) or EC-Spns2 cko (Spns2 f/f ;Tie2Cre) mice. Data are shown as means ± S.D. (n=3). 15
16 Supplemental Figure 14 Reduced number of mature T cells in peripheral lymph nodes of EC-Spns2 cko mice. Frequencies (left) and numbers (right) of CD4 SP (CD4) and CD8 SP (CD8) T cells in peripheral lymph nodes from control (Spns2 f/f ) or EC-Spns2 cko (Spns2 f/f ;Tie2Cre) mice are shown (n=11). Bars and circles indicate averages and values for individual mice, respectively. 16
17 Supplemental Figure 15 Reconstitution of EC-Spns2 cko mice with control bone marrow. (A) PCR analysis of the genomic DNA isolated from the bone marrow of wild type mouse (Spns2 +/+ ), Spns2 KO mouse (Spns2 -/- ), Spns2 floxed mouse (Spns2 f/f ), 8 Spns2 floxed mice reconstituted with bone marrow from Spns2 floxed mice (Spns2 f/f Spns2 f/f ) and 9 EC-Spns2 cko 17
18 mice reconstituted with bone marrow from Spns2 floxed mice (Spns2 f/f Spns2 f/f,tie2cre) was performed as described in the legend of Supplementary Figure 1C. 552-bp, 842-bp and 316-bp PCR products represent the wild type, Spns2-floxed and Spns2-deleted alleles, respectively. Note that the bone marrow of EC-Spns2 cko was successfully reconstituted by that of Spns2 floxed mice. (B, C) Flow cytometric analyses of Spns2 floxed mice reconstituted with bone marrow from Spns2 floxed mice (Spns2 f/f Spns2 f/f ) and EC-Spns2 cko mice reconstituted with bone marrow from Spns2 floxed mice (Spns2 f/f Spns2 f/f,tie2cre). (B) Frequencies and total numbers of CD4 SP (CD4) (left) and CD8 SP (CD8) (right) T cells in the thymus are shown (Spns2 f/f Spns2 f/f, n=8; Spns2 f/f Spns2 f/f,tie2cre, n=9). (D) Frequencies and total numbers of CD4 SP (CD4) (left) and CD8 SP (CD8) (right) T cells in the peripheral blood are shown (Spns2 f/f Spns2 f/f, n=8; Spns2 f/f Spns2 f/f,tie2cre, n=9). 18
19 Supplemental Figure 16 Reduced number of mature recirculating B cells in bone marrow of EC-Spns2 cko mice. Flow cytometric analyses of bone marrow derived B cells from control (Spns2 f/f ) or EC-Spns2 cko (Spns2 f/f ;Tie2Cre) mice. (A) The numbers indicate the percentage of IgM and IgD expressing CD19 + B cells. Mature recirculating B (Mature rec. B) cells and immature B cells were identified as CD19 + IgM + IgD + and CD19 + IgM + IgD -, respectively. (B), Frequencies (left) and numbers (right) of mature recirculating B cells and immature B cells are shown in left and right panels, respectively (n=11). Bars and circles indicate averages and values for individual mice, respectively. 19
20 Supplemental Figure 17 Number of mature recirculating B cells in peripheral lymph nodes of EC-Spns2 cko mice. Frequencies (left) and numbers (right) of mature recirculating B cells (CD19 + CD23 + IgD + ) in peripheral lymph nodes from control (Spns2 f/f ) or EC-Spns2 cko (Spns2 f/f ;Tie2Cre) mice are shown (n=11). Bars and circles indicate averages and values for individual mice, respectively. 20
21 Supplemental Figure 18 Flow cytometric analyses of Spns2 floxed mice reconstituted with bone marrow from Spns2 floxed mice (Spns2 f/f Spns2 f/f ) and EC-Spns2 cko mice reconstituted with bone marrow from Spns2 floxed mice (Spns2 f/f Spns2 f/f,tie2cre). Frequencies (left) and total numbers (right) of mature recirculating B cells in the bone marrow (A) and peripheral blood (B) are shown (Spns2 f/f Spns2 f/f, n=8; Spns2 f/f Spns2 f/f,tie2cre, n=9). 21
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Table S1. Oligonucleotide primer sequences
 Table S1. Oligonucleotide primer sequences Primer Name DNA Sequence (5 -> 3 ) Description CD19c CD19d Cre7 Sfpi1lox1 Sfpi1lox2 Sfpi1lox3 5 -AACCAGTCAACACCCTTCC-3 5 -CCAGACTAGATACAGACCAG-3 5 -TCAGCTACACCAGAGACGG-3
Table S1. Oligonucleotide primer sequences Primer Name DNA Sequence (5 -> 3 ) Description CD19c CD19d Cre7 Sfpi1lox1 Sfpi1lox2 Sfpi1lox3 5 -AACCAGTCAACACCCTTCC-3 5 -CCAGACTAGATACAGACCAG-3 5 -TCAGCTACACCAGAGACGG-3
Supplemental Figure 1
 Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
doi: 1.138/nature875 a promoter firefly luciferase CNS b Supplementary Figure 1. Dual luciferase assays on enhancer activity of CNS1, 2, and 3. a. promoter sequence was inserted upstream of firefly luciferase
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Pei et al. Supplementary Figure S1
 Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
Pei et al. Supplementary Figure S1 C H-CUL9: + + + + + Myc-ROC1: - - + + + U2OS/pcDN3 U2OS/H-CUL9 U2OS/ + H-CUL9 IP: -H -myc input -H -myc 1 2 3 4 5 H-CUL9 Myc-ROC1 -H -H -H -H H-CUL9: wt RR myc-roc1:
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation
 LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
LRBA is Essential for Allogeneic Responses in Bone Marrow Transplantation Mi Young Park, 1# Raki Sudan, 1# Neetu Srivastava, 1 Sudha Neelam, 1 Christie Youngs, 1 Jia- Wang Wang, 4 Robert W. Engelman, 5,6,7
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
Table S1. Primers used in the study
 Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
Table S1. Primers used in the study Primer name Application Sequence I1F16 Genotyping GGCAAGTGAGTGAGTGCCTA I1R11 Genotyping CCCACTCGTATTGACGCTCT V19 Genotyping GGGTCTCAAAGTCAGGGTCA D18Mit184-F Genotyping
A Low salt diet. C Low salt diet + mf4-31c1 3. D High salt diet + mf4-31c1 3. B High salt diet
 A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
A Low salt diet GV [AU]:. [mmhg]: 0..... 09 9 7 B High salt diet GV [AU]:. [mmhg]: 7 8...0. 7.8 8.. 0 8 7 C Low salt diet + mf-c GV [AU]:. [mmhg]:.0.9 8.7.7. 7 8 0 D High salt diet + mf-c E Lymph capillary
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin
 Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
DRG Pituitary Cerebral Cortex
 Liver Spinal cord Pons Atg5 -/- Atg5 +/+ DRG Pituitary Cerebral Cortex WT KO Supplementary Figure S1 Ubiquitin-positive IBs accumulate in Atg5 -/- tissues. Atg5 -/- neonatal tissues were fixed and decalcified.
Liver Spinal cord Pons Atg5 -/- Atg5 +/+ DRG Pituitary Cerebral Cortex WT KO Supplementary Figure S1 Ubiquitin-positive IBs accumulate in Atg5 -/- tissues. Atg5 -/- neonatal tissues were fixed and decalcified.
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
SUPPLEMENTAL MATERIAL MATERIALS AND METHODS Generation of TSPOΔ/Δ murine embryonic fibroblasts Embryos were harvested from 13.5-day pregnant TSPOfl/fl mice. After dissection to eviscerate and remove the
Supplementary Information
 1 2 Supplementary Information 3 4 5 6 7 8 Supplementary Figure 1. Loading of R837 into PLGA nanoparticles. The loading efficiency (a) and loading capacity (b) of R837 by PLGA nanoparticles obtained at
1 2 Supplementary Information 3 4 5 6 7 8 Supplementary Figure 1. Loading of R837 into PLGA nanoparticles. The loading efficiency (a) and loading capacity (b) of R837 by PLGA nanoparticles obtained at
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Information
 Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Supplementary Information MicroRNA-212/132 family is required for epithelial stromal interactions necessary for mouse mammary gland development Ahmet Ucar, Vida Vafaizadeh, Hubertus Jarry, Jan Fiedler,
Nature Medicine doi: /nm.2548
 Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Supplementary Table 1: Genotypes of offspring and embryos from matings of Pmm2 WT/F118L mice with Pmm2 WT/R137H mice total events Pmm2 WT/WT Pmm2 WT/R137H Pmm2 WT/F118L Pmm2 R137H/F118L offspring 117 (100%)
Ramp1 EPD0843_4_B11. EUCOMM/KOMP-CSD Knockout-First Genotyping
 EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
Usp14 EPD0582_2_G09. EUCOMM/KOMP-CSD Knockout-First Genotyping
 EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
EUCOMM/KOMP-CSD Knockout-First Genotyping Introduction The majority of animals produced from the EUCOMM/KOMP-CSD ES cell resource contain the Knockout-First-Reporter Tagged Insertion allele. As well as
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Supplemental Figure 1
 Supplemental Figure 1 Supplemental Figure 1 (previous page): Heavy chain gene content of ARS/Igh66 transgenic lines. A. Tail DNA from the indicated transgenic lines was amplified for the indicated gene
Supplemental Figure 1 Supplemental Figure 1 (previous page): Heavy chain gene content of ARS/Igh66 transgenic lines. A. Tail DNA from the indicated transgenic lines was amplified for the indicated gene
Supplementary Fig. 1 Generation and genotyping of ThPOK / mice
 Supplementary Fig. 1 Generation and genotyping of ThPOK / mice (a). The strategy used to target the ThPOK locus is schematically depicted. A targeting construct was generated by inserting a loxp site (filled
Supplementary Fig. 1 Generation and genotyping of ThPOK / mice (a). The strategy used to target the ThPOK locus is schematically depicted. A targeting construct was generated by inserting a loxp site (filled
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) (b)
 Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
Supplementary Figure 1 An overview of pirna biogenesis during fetal mouse reprogramming. (a) A schematic overview of the production and amplification of a single pirna from a transposon transcript. The
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
doi:10.1038/nature10163 Supplementary Table 1 Efficiency of vector construction. Process wells recovered efficiency (%) Recombineering* 480 461 96 Intermediate plasmids 461 381 83 Recombineering efficiency
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Supplemental Material TARGETING THE HUMAN MUC1-C ONCOPROTEIN WITH AN ANTIBODY-DRUG CONJUGATE. Supplemental Tables S1 S3. and
 Supplemental Material TARGETING THE HUMAN MUC1-C ONCOPROTEIN WITH AN ANTIBODY-DRUG CONJUGATE Supplemental Tables S1 S3 and Supplemental Figures S1 S10 Supplemental Table S1. PK Parameters for MAb-3D1-MMAE
Supplemental Material TARGETING THE HUMAN MUC1-C ONCOPROTEIN WITH AN ANTIBODY-DRUG CONJUGATE Supplemental Tables S1 S3 and Supplemental Figures S1 S10 Supplemental Table S1. PK Parameters for MAb-3D1-MMAE
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Nature Immunology: doi: /ni Supplementary Figure 1. Zranb1 gene targeting.
 Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
Supplementary Figure 1 Zranb1 gene targeting. (a) Schematic picture of Zranb1 gene targeting using an FRT-LoxP vector, showing the first 6 exons of Zranb1 gene (exons 7-9 are not shown). Targeted mice
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
The RRPA knock-in allele was generated by homologous recombination in TC1 ES cells.
 Supplemental Materials Materials & Methods Generation of RRPA and RAPA Knock-in Mice The RRPA knock-in allele was generated by homologous recombination in TC1 ES cells. Targeted ES clones in which the
Supplemental Materials Materials & Methods Generation of RRPA and RAPA Knock-in Mice The RRPA knock-in allele was generated by homologous recombination in TC1 ES cells. Targeted ES clones in which the
Supplemental Information. Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis
 Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Supplemental Information Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis Yunpeng Bai ;, Lujuan Zhang #, Hongming Zhou #, Yuanshu Dong #, Qi Zeng, Weinian Shou, and Zhong-Yin Zhang
Percent survival. Supplementary fig. S3 A.
 Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Cell Selective Cardiovascular Biology of Microsomal Prostaglandin E Synthase- Chen, et al Cell specific role of mpges- Lihong Chen,,*, MD, PhD; Guangrui Yang,,*, MD, PhD; Xiufeng
SUPPLEMENTAL MATERIAL Cell Selective Cardiovascular Biology of Microsomal Prostaglandin E Synthase- Chen, et al Cell specific role of mpges- Lihong Chen,,*, MD, PhD; Guangrui Yang,,*, MD, PhD; Xiufeng
Supplementary Figures and Figure legends
 Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Supplementary Figures and Figure legends 3 Supplementary Figure 1. Conditional targeting construct for the murine Satb1 locus with a modified FLEX switch. Schematic of the wild type Satb1 locus; the conditional
Nature Biotechnology doi: /nbt Supplementary Figure 1. Substrate-linked protein evolution components.
 Supplementary Figure 1 Substrate-linked protein evolution components. (a) loxbtr subsites used for the combinational directed-evolution process to generate Brec1. Sequence differences (compared to the
Supplementary Figure 1 Substrate-linked protein evolution components. (a) loxbtr subsites used for the combinational directed-evolution process to generate Brec1. Sequence differences (compared to the
Dominguez et al., 2005
 Dominguez et al., 005 SUPPLEMENTARY INFORMATION EXPERIMENTAL PROCEDURES Generation of BACE1 targeted ES cells- For the generation of the first BACE1 line (BACE1I), a 19/ola mouse cosmid library from RZPD
Dominguez et al., 005 SUPPLEMENTARY INFORMATION EXPERIMENTAL PROCEDURES Generation of BACE1 targeted ES cells- For the generation of the first BACE1 line (BACE1I), a 19/ola mouse cosmid library from RZPD
Easi CRISPR for conditional and insertional alleles
 Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
Easi CRISPR for conditional and insertional alleles C.B Gurumurthy, University Of Nebraska Medical Center Omaha, NE cgurumurthy@unmc.edu Types of Genome edits Gene disruption/inactivation Types of Genome
MLN8237 induces proliferation arrest, expression of differentiation markers and
 Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and
 Mice We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and an Frt flanked neo cassette (Fig. S1A). Targeted BA1 (C57BL/6 129/SvEv) hybrid embryonic stem cells
Mice We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and an Frt flanked neo cassette (Fig. S1A). Targeted BA1 (C57BL/6 129/SvEv) hybrid embryonic stem cells
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase
 A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
A novel tool for monitoring endogenous alpha-synuclein transcription by NanoLuciferase tag insertion at the 3 end using CRISPR-Cas9 genome editing technique Sambuddha Basu 1, 3, Levi Adams 1, 3, Subhrangshu
Supplementary Information. Isl2b regulates anterior second heart field development in zebrafish
 Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1 Supplementary Fig. 1 shrna mediated knockdown of ZRSR2 in K562 and 293T cells. (a) ZRSR2 transcript levels in stably transduced K562 cells were determined using qrt-pcr. GAPDH was
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Generation of inducible BICD2 knock-out mice. A) The mouse BICD2 locus and gene targeting constructs. To generate an inducible Bicd2
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplemental Table S1. RT-PCR primers used in this study
 Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
doi:10.1038/nature09861 & &' -(' ()*+ ')(+,,(','-*+,&,,+ ',+' ' 23,45/0*6787*9:./09 ;78?4?@*+A786?B- &' )*+*(,-* -(' ()*+ ')(+,,(','-*+,&,,+ ',+'./)*+*(,-*..)*+*(,-*./)*+*(,-*.0)*+*(,-*..)*+*(,-*
SUPPLEMENTARY INFORMATION
 6 Relative levels of ma3 RNA 5 4 3 2 1 LN MG SG PG Spl Thy ma3 ß actin Lymph node Mammary gland Prostate gland Salivary gland Spleen Thymus MMTV target tissues Fig. S1: MMTV target tissues express ma3.
6 Relative levels of ma3 RNA 5 4 3 2 1 LN MG SG PG Spl Thy ma3 ß actin Lymph node Mammary gland Prostate gland Salivary gland Spleen Thymus MMTV target tissues Fig. S1: MMTV target tissues express ma3.
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Figure S1. Phenotypic characterization of transfected ECFC. (a) ECFC were transfected using a lentivirus with a vector encoding for either human EPO
 Figure S1. Phenotypic characterization of transfected ECFC. (a) ECFC were transfected using a lentivirus with a vector encoding for either human EPO (epoecfc) or LacZ (laczecfc) under control of a cytomegalovirus
Figure S1. Phenotypic characterization of transfected ECFC. (a) ECFC were transfected using a lentivirus with a vector encoding for either human EPO (epoecfc) or LacZ (laczecfc) under control of a cytomegalovirus
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Validation of CDK9-inhibitor treatment.
 Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
Supplementary Figure 1 Validation of CDK9-inhibitor treatment. (a) Schematic of GAPDH with the middle of the amplicons indicated in base pairs. The transcription start site (TSS) and the terminal polyadenylation
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Ricke et al., http://www.jcb.org/cgi/content/full/jcb.201205115/dc1 Figure S1. Kinetochore localization of mitotic regulators in wild-type
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Ricke et al., http://www.jcb.org/cgi/content/full/jcb.201205115/dc1 Figure S1. Kinetochore localization of mitotic regulators in wild-type
i-stop codon positions in the mcherry gene
 Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Supplementary Figure 1 i-stop codon positions in the mcherry gene The grnas (green) that can potentially generate stop codons from Trp (63 th and 98 th aa, upper panel) and Gln (47 th and 114 th aa, bottom
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Figure 1 Sox2 localizes in neutrophils of mouse spleen. (a) Microscopy analysis of Sox2 and MPO in wild-type mouse spleen. Nucl, nucleus. (b) Immunohistochemistry staining of Sox2 and MPO
Supplementary Data. Plasmid -2486CAT containing the 2.5 kb fragment of VE-cadherin promoter region (-2486
 Supplementary Data Materials and Methods Generation and Genotyping of Transgenic Mice Plasmid -2486CAT containing the 2.5 kb fragment of VE-cadherin promoter region (-2486 to +24) was a gift from P. Huber.
Supplementary Data Materials and Methods Generation and Genotyping of Transgenic Mice Plasmid -2486CAT containing the 2.5 kb fragment of VE-cadherin promoter region (-2486 to +24) was a gift from P. Huber.
TRANSGENIC ANIMALS. transient. stable. - Two methods to produce transgenic animals:
 Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
SUPPLEMENTAL METHODS. Chromatin immunoprecipitation (ChIP) assay. Cell culture. Non-ablated mouse transplantation. Hematology and flow cytometry
 SUPPLEMENTAL METHODS Cell culture EML C1 cells were cultured in IMDM 20% horse serum, 1% antibiotic-antimycotic (Gibco by Life Technologies) 15% SCF conditioned medium from BHK/ MKL cells or 50 100 ng/ml
SUPPLEMENTAL METHODS Cell culture EML C1 cells were cultured in IMDM 20% horse serum, 1% antibiotic-antimycotic (Gibco by Life Technologies) 15% SCF conditioned medium from BHK/ MKL cells or 50 100 ng/ml
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
doi:10.1038/nature11070 Supplementary Figure 1 Purification of FLAG-tagged proteins. a, Purification of FLAG-RNF12 by FLAG-affinity from nuclear extracts of wild-type (WT) and two FLAG- RNF12 transgenic
Supplemental Information. T Follicular Helper Cell-Germinal Center B Cell. Interaction Strength Regulates Entry into Plasma
 Immunity, Volume 48 Supplemental Information T Follicular Helper ell-germinal enter B ell Interaction Strength Regulates Entry into Plasma ell or Recycling Germinal enter ell Fate Wataru Ise, Kentaro Fujii,
Immunity, Volume 48 Supplemental Information T Follicular Helper ell-germinal enter B ell Interaction Strength Regulates Entry into Plasma ell or Recycling Germinal enter ell Fate Wataru Ise, Kentaro Fujii,
the tamoxifen treatment schedule used to induce recombination. Arrows indicate on which
 SUPPLEMENTAL DATA Figure S1. Characterization of Vhl / mice. (A) Left panel shows a schematic outline of the tamoxifen treatment schedule used to induce recombination. Arrows indicate on which days tamoxifen
SUPPLEMENTAL DATA Figure S1. Characterization of Vhl / mice. (A) Left panel shows a schematic outline of the tamoxifen treatment schedule used to induce recombination. Arrows indicate on which days tamoxifen
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Introducing new DNA into the genome requires cloning the donor sequence, delivery of the cloned DNA into the cell, and integration into the genome.
 Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Key Terms Chapter 32: Genetic Engineering Cloning describes propagation of a DNA sequence by incorporating it into a hybrid construct that can be replicated in a host cell. A cloning vector is a plasmid
Map-Based Cloning of Qualitative Plant Genes
 Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
Map-Based Cloning of Qualitative Plant Genes Map-based cloning using the genetic relationship between a gene and a marker as the basis for beginning a search for a gene Chromosome walking moving toward
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Persistent fibroblast growth factor 23 signalling in the parathyroid glands for secondary. hyperparathyroidism in mice with chronic kidney disease
 Supplementary Information Title: Persistent fibroblast growth factor 23 signalling in the parathyroid glands for secondary hyperparathyroidism in mice with chronic kidney disease Authors: Kazuki Kawakami
Supplementary Information Title: Persistent fibroblast growth factor 23 signalling in the parathyroid glands for secondary hyperparathyroidism in mice with chronic kidney disease Authors: Kazuki Kawakami
Supplementary Materials
 Supplementary Materials Table S1. Oligonucleotide sequences and PCR conditions used to amplify the indicated genes. TA = annealing temperature; gdna = genomic DNA; cdna = complementary DNA; c = concentration.
Supplementary Materials Table S1. Oligonucleotide sequences and PCR conditions used to amplify the indicated genes. TA = annealing temperature; gdna = genomic DNA; cdna = complementary DNA; c = concentration.
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast. Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice
 A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice Fei Chen 1,*, Guojun Dong 2,*, Limin Wu 1, Fang Wang 3, Xingzheng
Myofibroblasts in kidney fibrosis. LeBleu et al.
 Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Origin and Function of Myofibroblasts in Kidney Fibrosis Valerie S. LeBleu, Gangadhar Taduri, Joyce O Connell, Yingqi Teng, Vesselina G. Cooke, Craig Woda, Hikaru Sugimoto and Raghu Kalluri Supplementary
Supplemental Materials. Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans
 Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
Supplemental Materials Matrix Proteases Contribute to Progression of Pelvic Organ Prolapse in Mice and Humans Madhusudhan Budatha, Shayzreen Roshanravan, Qian Zheng, Cecilia Weislander, Shelby L. Chapman,
SUPPLEMENTARY INFORMATION
 Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
Supplementary Data. Flvcr1a TCTAAGGCCCAGTAGGACCC GGCCTCAACTGCCTGGGAGC AGAGGGCAACCTCGGTGTCC
 Supplementary Data Supplementary Materials and Methods Measurement of reactive oxygen species accumulation in fresh intestinal rings Accumulation of reactive oxygen species in fresh intestinal rings was
Supplementary Data Supplementary Materials and Methods Measurement of reactive oxygen species accumulation in fresh intestinal rings Accumulation of reactive oxygen species in fresh intestinal rings was
Supplementary Materials
 Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplementary Materials Construction of Synthetic Nucleoli in Human Cells Reveals How a Major Functional Nuclear Domain is Formed and Propagated Through Cell Divisision Authors: Alice Grob, Christine Colleran
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and
 1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
1 Supplemental information 2 3 Materials and Methods 4 Reagents and animals 5 8Br-cAMP was purchased from Sigma (St. Louis, MO). Silencer Negative Control sirna #1 and 6 Silencer Select Pre-designed sirna
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Rassf1a -/- Sav1 +/- mice. Supplementary Figures:
 1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
Supplementary Figure 1
 Supplementary Figure 1 Product purity of cytosine base editing for the wheat genomic loci tested. Product distributions and indels frequencies at four representative wheat genomic DNA sites in wheat protoplasts
Supplementary Figure 1 Product purity of cytosine base editing for the wheat genomic loci tested. Product distributions and indels frequencies at four representative wheat genomic DNA sites in wheat protoplasts
Before starting, write your name on the top of each page Make sure you have all pages
 Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Biology 105: Introduction to Genetics Name Student ID Before starting, write your name on the top of each page Make sure you have all pages You can use the back-side of the pages for scratch, but we will
Experimental genetics - 2 Partha Roy
 Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Partha Roy Experimental genetics - 2 Making genetically altered animal 1) Gene knock-out k from: a) the entire animal b) selected cell-type/ tissue c) selected cell-type/tissue at certain time 2) Transgenic
Loss of Cul3 in Primary Fibroblasts
 PSU McNair Scholars Online Journal Volume 5 Issue 1 Humans Being: People, Places, Perspectives and Processes Article 15 2011 Loss of Cul3 in Primary Fibroblasts Paula Hanna Portland State University Let
PSU McNair Scholars Online Journal Volume 5 Issue 1 Humans Being: People, Places, Perspectives and Processes Article 15 2011 Loss of Cul3 in Primary Fibroblasts Paula Hanna Portland State University Let
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat
 Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Supplementary Figure 1: Expression of RNF8, HERC2 and NEURL4 in the cerebellum and knockdown of RNF8 by RNAi (a) Lysates of the cerebellum from rat pups at P6, P14, P22, P30 and adult (A) rats were subjected
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure S1: (A) Schematic representation of the Jarid2 Flox/Flox
 Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Supplementary Figure S1: (A) Schematic representation of the Flox/Flox locus and the strategy used to visualize the wt and the Flox/Flox mrna by PCR from cdna. An example of the genotyping strategy is
Nature Biotechnology: doi: /nbt Supplementary Figure 1. In vitro validation of OTC sgrnas and donor template.
 Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
Supplementary Figure 1 In vitro validation of OTC sgrnas and donor template. (a) In vitro validation of sgrnas targeted to OTC in the MC57G mouse cell line by transient transfection followed by 4-day puromycin
 Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
Figure S1. Generation of HspA4 -/- mice. (A) Structures of the wild-type (Wt) HspA4 -/- allele, targeting vector and targeted allele are shown together with the relevant restriction sites. The filled rectangles
To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR
 Plasmids To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR fragments downstream of firefly luciferase (luc) in pgl3 control (Promega). pgl3- CDK6 was made by amplifying a 2,886
Plasmids To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR fragments downstream of firefly luciferase (luc) in pgl3 control (Promega). pgl3- CDK6 was made by amplifying a 2,886
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Fig.1. Over-expression of RNase L in stable polyclonal cell line
 Supplemental Data mrna Protein A kda 75 40 NEO/vector NEO/RNase L RNASE L β-actin RNASE L β-actin B % of control (neo vector), normalized 240 ** 200 160 120 80 40 0 Neo Neo/RNase L RNase L Protein Supplementary
Supplemental Data mrna Protein A kda 75 40 NEO/vector NEO/RNase L RNASE L β-actin RNASE L β-actin B % of control (neo vector), normalized 240 ** 200 160 120 80 40 0 Neo Neo/RNase L RNase L Protein Supplementary
SUPPLEMENTAL MATERIALS
 SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
SUPPLEMENL MERILS Eh-seq: RISPR epitope tagging hip-seq of DN-binding proteins Daniel Savic, E. hristopher Partridge, Kimberly M. Newberry, Sophia. Smith, Sarah K. Meadows, rian S. Roberts, Mark Mackiewicz,
