Nature Structural & Molecular Biology: doi: /nsmb.2548
|
|
|
- Chester Preston
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference
2 electron maps. The former is colored blue and contoured at 1 σ, and the latter is black and contoured at 5 σ. Both maps were calculated for Å resolution range and were subject to three fold real space averaging. The Cα atoms of the cross-linked cysteines are emphasized as yellow spheres. (b) Stereo view of the structural superposition of single WT Glt Ph (PDB accession code 2nwl) and Glt Ph out protomers shown in ribbon representation. The superposition was generated using residues , which correspond to the structurally conserved region between outward and inward facing states. The trimerization and transport domains are colored orange and blue for the wild type and brown and black for Glt Ph out, respectively. The substrate and the cross-linking Hg 2+ ion are emphasized as spheres, and the cysteines are shown in stick representation.
3 Supplementary Figure 2. RH421 dye senses coupled Na + /Asp binding to Glt Ph and TBA inhibits transport catalyzed by Glt Ph. (a) Emission spectra of RH421 in a buffer containing in mm 10 HEPES/KOH ph 7.4, 99 KCl (black line); after addition of 0.2 mm DDM and 1 µm Glt Ph -WT (blue line); and after further addition of 10 mm NaCl and 1 mm Asp (green line). Fluorescence was excited at 532 nm. (b) Proteo-liposomes, containing WT Glt Ph at protein to lipid ratio of 1:50 were loaded with buffer containing 300 mm choline chloride and assayed in the uptake buffer containing 100 mm NaCl, 200 mm choline chloride, 100 nm 3 H-Asp and the indicated amounts of TBA (open circles) or DL-TBOA (solid circles) for 10 minutes. Averages of three measurements with standard errors are shown.
4 Supplementary Figure 3. TBA and Asp bind in a competitive manner. Binding isotherms for Glt Ph out (blue) and Glt Ph in (red) at 25 C. (a) TBA titrations in the presence of 100 mm NaCl. The solid black lines through the data are fits with the following parameters: K D = 3.8 and 1.5 µm, ΔH = -2.6 and -8.7 kcal/mol and n = 1.0 and 1.0 for Glt Ph out and Glt Ph in, respectively. (b) Asp titrations in the presence of 0.6 mm TBA and 30 mm NaCl. The solid black lines through the data are fits with the following parameters: K D = 0.13 and 0.43 µm, ΔH = and -8.3 kcal/mol and n = 0.9 and 0.5 for Glt Ph out and Glt Ph in, respectively.
5 Supplementary Figure 4. Na+ binding in the presence of coupled ligands. Na+ titrations derived from fluorescence experiments using GltPhout (a, blue), GltPhin (b, red) and the WT (c, black) at 25 C. Left panels are Na+ titrations in the presence of 1 (solid circles), 0.1 (open circles) and 0.01 (solid triangles) mm Asp. Right panels are Na+ titrations in the presence of 10 (solid circles), 1 (open circles) and 0.1 (solid triangles) mm TBA. The data were fitted to Hill equations yielding average Hill coefficients, nhill as indicated on the graphs.
6 Supplementary Figure 5. Asp bind in 1M Na+ and determination of the binding state functions. (a) Binding isotherms for GltPhout (blue, left) and GltPhin (red, right) at 25 C in the presence of 1M NaCl. Upper panels, the solid colored lines are the heat powers developed during the titration. Lower panels, the black solid lines through the data are fits with the following parameters: ΔH = and kcal/mol and n = 0.7 and 0.9 for GltPhout and GltPhin, respectively. The KD values could not be calculated accurately due to the lack of experimental points in the transition regions of the plots. (b) Schematic representation of the reactions for which thermodynamic parameters were obtained experimentally (solid lines) or calculated (dashed lines). Left panel, H0s and CPs were determined experimentally for Asp binding under three conditions: in the presence of [Na+] < 60 mm (reaction 1), in the presence of [Na+] = 1 M (reaction 2) and in the presence of [TBA] = 0.6 mm and [Na+] = 30 mm (reaction 3). The H0s and CPs for the binding of 2 Na+ ions and consequent L-TBA binding were calculated as differences between those for reactions 1 and 2, and for reactions 2 and 3, respectively. Right panel, G0s for 3Na/Asp (reaction 1) and 2Na/L-TBA (reaction 3) binding were determined from the linear extrapolations in Fig. 2c and G0 of 2 Na+ binding alone (reaction 2) measured
7 directly. The G 0 s of Asp and L-TBA binding to the transporters pre-bound to 2 Na + ions were calculated as differences between those for reactions 1 and 2, and for reactions 3 and 2, respectively. G 0 of L-TBA replacement by Asp was calculated as a difference between that of reactions 1 and 3.
8 out in Supplementary Figure 6. DL-TBOA binding. Shown are data for Glt Ph (blue) and Glt Ph (red). a, ITC data for DL-TBOA titrations in the presence of 100 mm NaCl at 25 C. The solid black lines through the data are fits with the following parameters: K D = 2.6 and 16.0 µm, ΔH = -9.8 and -8.9 kcal/mol and n = 1.4 and 1.2 for Glt out Ph and Glt in Ph, respectively. b, Temperature dependencies of DL-TBOA binding enthalpies obtained at 200 mm NaCl. Straight line fits to the data yielded ΔC P estimates of -148 and -334 cal mol -1 deg -1 for Glt out Ph and Glt in Ph, respectively.
9 Supplementary Table 1: Thermodynamic parameters of binding reactions Hg 2+ binding 1 mm Na + K D, nm ΔH, kcal/mol Glt Ph -L66C/S300C Glt Ph -K55C/A364C mm Na +, 0.2 mm Asp Glt Ph -L66C/S300C Glt Ph -K55C/A364C # Binding parameters were obtained by fitting data in Figure 1, c and d to independent binding sites model. n is the number of the binding sites. n # Competitive binding of TBA and Asp. Glt Ph out Glt Ph in ΔG Asp ΔG TBA, ΔG TBA to Asp, ΔG calc TBA to Asp, kcal/mol kcal/mol kcal/mol kcal/mol Glt Ph out Glt Ph in ΔH 0 Asp, kcal/mol ΔH 0 TBA, kcal/mol ΔH 0 TBA to Asp, kcal/mol ΔH 0 TBA to Asp calc, kcal/mol ± ± ± ± ± ± G Asp, G TBA, H 0 Asp and H 0 TBA are Asp and TBA binding free energies and standard enthalpies, respectively. The Gs were measured by ITC in the presence of 30 mm Na +. The H 0 s are averages of at least three ITC experiments performed at Na + concentrations between 10 and 100 mm (Fig. 2c). G TBA to Asp, G calc TBA to Asp, H TBA to Asp and H calc TBA to Asp are the experimental and calculated free energies and enthalpies of TBA replacement by Asp, respectively. The experimental G TBA to Asp and H 0 TBA to Asp were obtained by ITC in the presence of 0.6 mm TBA and 30 mm Na +. The enthalpies are averages of three experiments. calc G TBA to Asp was calculated using the following competition equation: K D, calc =K D,Asp (1+[TBA]/K D,TBA ), where K D, Asp and K D, TBA correspond to G Asp and G TBA, respectively. The competition equation is valid when concentration of TBA is significantly higher than that of the protein. In our experiments, TBA to protein molar ratio was at least 15. H 0 TBA to Asp calc are the differences between the binding enthalpies of Asp and TBA.
10 SUPPLEMENTARY NOTE The standard free energy of Asp and Na + binding. The coupled binding reaction is: T + Asp + "# Na + $ T# Asp# Na " +, (S1) where T represents the transporter, and α is the number of ions coupled to Asp binding. The total free energy of this reaction is: "G Total = "G 0 + RT lna TAspNa # RT lna Asp # $% RT lna Na # RT lna T, (S2) where G 0 is the standard free energy, defined as the free energy of the reaction at 25 C, 1 atm pressure and 1 M concentration of the reactants, and the a T, a TAspNa+, a Asp and a Na are the activities of the free transporter, the bound transporter, Asp and Na +, respectively. At a given Na + activity, G Total = 0 and a T = a TAspNa+ when the activity of Asp equals to apparent dissociation constant K D,app. RT lnk D,app = "G 0 # $% RT ln a Na (S3) Considering that RTlnK D,app = G app, where G app is the apparent free energy of Asp binding, and RTlna Na = µ Na, where µ Na is the chemical potential of the ion, we obtain the final expression of equation (1): "G app = "G 0 # $% µ Na Calculations of the thermodynamic parameters. The thermodynamic parameters for 3Na + /Asp binding to Glt Ph out and Glt Ph in, including H, G and Cp were measured directly by ITC. However, the thermodynamic parameters of Na + ions biding could not be determined by ITC
11 because the affinity for Na + is too low for direct measurements. Instead, H and Cp could be assessed from T + Asp + 3Na + " 1 T# Asp# Na 3 + T + 2Na + " 2 T# Na Asp + Na + " 3 T# Asp# Na 3 + (S4) In the above scheme, H and Cp values for reactions (1) and (3) were experimentally determined (experimentally accessible reactions 1 and 2, respectively, in Supplementary Fig. 7a), and the corresponding values for reaction (4) were calculated as differences between (1) and (3): H 2 = H 1 - H 3 and Cp 2 = Cp 1 - Cp 3 (S5) G for reaction (2) was measured directly using fluorescence-based assay. The Asp affinity to Na + -saturated transporter, reaction (3), is too high to measure by either ITC or the fluorescence assay, and G 3 of this reaction was instead calculated as (Supplementary Fig. 7b): G 3 = G 1 - G 2 (S6) Similarly, H and Cp for TBA binding could not be determined by ITC because of insufficient heats of binding. Gs were still accessible from the fluorescence-based affinity measurements. Instead, H and Cp were determined via a two-step process: the transporters were first saturated with TBA, and then titrated with Asp. In the scheme below, the experimentally observed reactions are (1) and (5) (experimentally accessible reactions 1 and 3, respectively, in Supplementary Fig. 7a), while reaction (4) cannot be characterized directly: T + Asp + 3Na + " 1 T# Asp# Na 3 + T + TBA + 2Na + " 4 T# TBA# Na Asp + Na + " 5 T# Asp# Na TBA (S7)
12 assuming Na + stoichiometry of 2 and 3 for TBA and Asp binding, respectively. The values of the thermodynamic parameters for reaction (4) were calculated as differences between corresponding values of reactions (1) and (5). Temperature dependence of the binding free energy. The temperature dependences of the free energies were calculated using the thermodynamic parameters either measured or calculated at T ref = 298 K as described above, and the corresponding Cp values based on the following standard thermodynamic expressions: "H(T) = "H(T ref ) + "Cp(T # T ref ) (S8) "S(T) = "S(T ref ) + "Cp# ln T T ref (S9) "G(T) = "H(T) # T$ "S(T) (S10)
Supplementary Figure 1. WT ph 4.5. γ (ion s
 Supplementary Figure 1 a WT ph 4.5 b 10 5 γ (ion s 1 ) 10 4 10 3 10 2 WT Y445A EA/YA E148A µcal s 1 Supplementary Figure 1 ph dependence of Cl transport and binding. a) Cl transport rates at ph 7.5 (white)
Supplementary Figure 1 a WT ph 4.5 b 10 5 γ (ion s 1 ) 10 4 10 3 10 2 WT Y445A EA/YA E148A µcal s 1 Supplementary Figure 1 ph dependence of Cl transport and binding. a) Cl transport rates at ph 7.5 (white)
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Materials and Methods Circular dichroism (CD) spectroscopy. Far ultraviolet (UV) CD spectra of apo- and holo- CaM and the CaM mutants were recorded on a Jasco J-715 spectropolarimeter
SUPPLEMENTARY MATERIAL Materials and Methods Circular dichroism (CD) spectroscopy. Far ultraviolet (UV) CD spectra of apo- and holo- CaM and the CaM mutants were recorded on a Jasco J-715 spectropolarimeter
SUPPLEMENTARY INFORMATION. Determinants of laminin polymerization revealed. by the structure of the α5 chain amino-terminal region
 SUPPLEMENTARY INFORMATION Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region Sadaf-Ahmahni Hussain 1, Federico Carafoli 1 & Erhard Hohenester 1 1 Department
SUPPLEMENTARY INFORMATION Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region Sadaf-Ahmahni Hussain 1, Federico Carafoli 1 & Erhard Hohenester 1 1 Department
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D
 Supplementary Information for Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D Federico Forneris a,b, B. Tom Burnley a,b,c and Piet Gros a * a Crystal
Supplementary Information for Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D Federico Forneris a,b, B. Tom Burnley a,b,c and Piet Gros a * a Crystal
Supplemental Information Molecular Cell, Volume 41
 Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
Supplementary Information Titles
 Supplementary Information Titles Please list each supplementary item and its title or caption, in the order shown below. Note that we do NOT copy edit or otherwise change supplementary information, and
Supplementary Information Titles Please list each supplementary item and its title or caption, in the order shown below. Note that we do NOT copy edit or otherwise change supplementary information, and
Automated Chemical Denaturation as a Tool to Evaluate Protein Stability and Optimize the Formulation of Biologics
 Automated Chemical Denaturation as a Tool to Evaluate Protein Stability and Optimize the Formulation of Biologics Ernesto Freire Johns Hopkins University Baltimore, MD 21218 ef@jhu.edu AVIA 2304 Automated
Automated Chemical Denaturation as a Tool to Evaluate Protein Stability and Optimize the Formulation of Biologics Ernesto Freire Johns Hopkins University Baltimore, MD 21218 ef@jhu.edu AVIA 2304 Automated
Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation
 Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation Julien Pernier, Jozsef Orban 1, Balendu Sankara Avvaru, Antoine Jégou, Guillaume Romet- Lemonne, Bérengère
Dimeric WH2 domains in Vibrio VopF promote actin filament barbed end uncapping and assisted elongation Julien Pernier, Jozsef Orban 1, Balendu Sankara Avvaru, Antoine Jégou, Guillaume Romet- Lemonne, Bérengère
Conformational changes in IgE contribute to its. uniquely slow dissociation rate from receptor FcεRI
 Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI M.D. Holdom, A.M. Davies, J.E. Nettleship, S.C. Bagby, B. Dhaliwal, E. Girardi, J. Hunt, H.J. Gould,
Colchicine. Colchicine. a b c d e
 α1-tub Colchicine Laulimalide RB3 β1-tub Vinblastine α2-tub Taxol Colchicine β2-tub Maytansine a b c d e Supplementary Figure 1 Structures of microtubule and tubulin (a)the cartoon of head to tail arrangement
α1-tub Colchicine Laulimalide RB3 β1-tub Vinblastine α2-tub Taxol Colchicine β2-tub Maytansine a b c d e Supplementary Figure 1 Structures of microtubule and tubulin (a)the cartoon of head to tail arrangement
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Functional activity of fluorescence-labeled ribosome complexes used in this study, as determined by the time-resolved puromycin assay.
 Supplementary Figure 1 Functional activity of fluorescence-labeled ribosome complexes used in this study, as determined by the time-resolved puromycin assay. Closed circles, unlabeled PRE complex; open
Supplementary Figure 1 Functional activity of fluorescence-labeled ribosome complexes used in this study, as determined by the time-resolved puromycin assay. Closed circles, unlabeled PRE complex; open
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and
 Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Supplementary Tables Supplementary Table 1: List of CH3 domain interface residues in the first chain (A) and their side chain contacting residues in the second chain (B) a Interface Res. in Contacting
Nature Structural & Molecular Biology: doi: /nsmb.3428
 Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Supplementary Figure 1 Biochemical characterization of the monou and oligou activity switch of TUT4(7). (a) Mouse TUT4 and human TUT7 were assayed for monou and Lin28-dependent oligou addition activities
Quantitative Evaluation of the Ability of Ionic Liquids to Offset the Cold- Induced Unfolding of Proteins
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2014 Supplimentary informations Quantitative Evaluation of the Ability of Ionic Liquids
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2014 Supplimentary informations Quantitative Evaluation of the Ability of Ionic Liquids
Masayoshi Honda, Jeehae Park, Robert A. Pugh, Taekjip Ha, and Maria Spies
 Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
Molecular Cell, Volume 35 Supplemental Data Single-Molecule Analysis Reveals Differential Effect of ssdna-binding Proteins on DNA Translocation by XPD Helicase Masayoshi Honda, Jeehae Park, Robert A. Pugh,
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/11/e1601625/dc1 Supplementary Materials for A molecular mechanism of chaperone-client recognition This PDF file includes: Lichun He, Timothy Sharpe, Adam Mazur,
advances.sciencemag.org/cgi/content/full/2/11/e1601625/dc1 Supplementary Materials for A molecular mechanism of chaperone-client recognition This PDF file includes: Lichun He, Timothy Sharpe, Adam Mazur,
SUPPLEMENTARY INFORMATION
 Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Results Construct purification and coupling. Two A1-GP1bα ReaLiSM constructs, with and without cysteine residues near the N and C-termini (Fig. S2a), were expressed and purified by Ni affinity chromatography
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Acceleration of protein folding by four orders of magnitude through a single amino acid substitution
 Acceleration of protein folding by four orders of magnitude through a single amino acid substitution Daniel J. A. Roderer 1, Martin A. Schärer 1, Marina Rubini 2 * and Rudi Glockshuber 1 AUTHOR ADDRESS
Acceleration of protein folding by four orders of magnitude through a single amino acid substitution Daniel J. A. Roderer 1, Martin A. Schärer 1, Marina Rubini 2 * and Rudi Glockshuber 1 AUTHOR ADDRESS
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein.
 Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Supplementary Table 1. DNA sequence synthesized to express the Zika virus NS5 protein. MR766 NS5 sequence 1 ACAGAGAACAGATTGGTGGTGGAGGTGGGACGGGAGAGACTCTGGGAGAGAAGTGGAAAG 61 CTCGTCTGAATCAGATGTCGGCCCTGGAGTTCTACTCTTATAAAAAGTCAGGTATCACTG
Accurate label-free reaction kinetics determination using initial rate heat. measurements
 Supplementary Information Accurate label-free reaction kinetics determination using initial rate heat measurements Kourosh Honarmand Ebrahimi 1 *, Peter-Leon Hagedoorn 1, Denise Jacobs 2, Wilfred R. Hagen
Supplementary Information Accurate label-free reaction kinetics determination using initial rate heat measurements Kourosh Honarmand Ebrahimi 1 *, Peter-Leon Hagedoorn 1, Denise Jacobs 2, Wilfred R. Hagen
SUPPLEMENTARY INFORMATION. Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly
 SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
SUPPLEMENTARY INFORMATION Reengineering Protein Interfaces Yields Copper-Inducible Ferritin Cage Assembly Dustin J. E. Huard, Kathleen M. Kane and F. Akif Tezcan* Department of Chemistry and Biochemistry,
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
has only one nucleotide, U20, between G19 and A21, while trna Glu CUC has two
 SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
Problem Set No. 3 Due: Thursday, 11/04/10 at the start of class
 Department of Chemical Engineering ChE 170 University of California, Santa Barbara Fall 2010 Problem Set No. 3 Due: Thursday, 11/04/10 at the start of class Objective: To understand and develop models
Department of Chemical Engineering ChE 170 University of California, Santa Barbara Fall 2010 Problem Set No. 3 Due: Thursday, 11/04/10 at the start of class Objective: To understand and develop models
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Structure and mechanism of a canonical poly(adp-ribose) glycohydrolase Mark S. Dunstan 1$, Eva Barkauskaite 2$, Pierre Lafite 3, Claire E. Knezevic 4, Amy Brassington 1, Marijan
SUPPLEMENTARY INFORMATION Structure and mechanism of a canonical poly(adp-ribose) glycohydrolase Mark S. Dunstan 1$, Eva Barkauskaite 2$, Pierre Lafite 3, Claire E. Knezevic 4, Amy Brassington 1, Marijan
A Non-Sequence-Specific DNA Binding Mode of RAG1 Is Inhibited by RAG2
 doi:10.1016/j.jmb.2009.02.020 J. Mol. Biol. (2009) 387, 744 758 Available online at www.sciencedirect.com A Non-Sequence-Specific DNA Binding Mode of RAG1 Is Inhibited by RAG2 Shuying Zhao, Lori M. Gwyn,
doi:10.1016/j.jmb.2009.02.020 J. Mol. Biol. (2009) 387, 744 758 Available online at www.sciencedirect.com A Non-Sequence-Specific DNA Binding Mode of RAG1 Is Inhibited by RAG2 Shuying Zhao, Lori M. Gwyn,
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis
 S1 of S9 Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis Victoria Steffen, Julia Otten, Susann Engelmann, Andreas Radek, Michael Limberg, Bernd
S1 of S9 Supplementary Materials: A Toolbox of Genetically Encoded FRET-Based Biosensors for Rapid L-Lysine Analysis Victoria Steffen, Julia Otten, Susann Engelmann, Andreas Radek, Michael Limberg, Bernd
Site specific immobilization of a potent antimicrobial peptide onto silicone catheters: evaluation against urinary tract infection pathogens
 Supplementary information Site specific immobilization of a potent antimicrobial peptide onto silicone catheters: evaluation against urinary tract infection pathogens Biswajit Mishra, a Anindya Basu, a
Supplementary information Site specific immobilization of a potent antimicrobial peptide onto silicone catheters: evaluation against urinary tract infection pathogens Biswajit Mishra, a Anindya Basu, a
Specificity: Induced Fit
 Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
SUPPLEMENTARY INFORMATION
 This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
This supplementary information is an extension of the letter with the same title and includes further discussion on the comparison of our designed Fe B Mb (computer model and crystal structure) with the
Final exam. Please write your name on the exam and keep an ID card ready.
 Biophysics of Macromolecules Prof. R. Jungmann and Prof. J. Lipfert SS 2017 Final exam Final exam First name: Last name: Student number ( Matrikelnummer ): Please write your name on the exam and keep an
Biophysics of Macromolecules Prof. R. Jungmann and Prof. J. Lipfert SS 2017 Final exam Final exam First name: Last name: Student number ( Matrikelnummer ): Please write your name on the exam and keep an
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron
 Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Molecular Interactions Research
 Molecular Interactions Research Group (MIRG) Satya P. Yadav, Aaron P. Yamniuk, John Newitt, Michael L. Doyle, Ed Eisenstein, Thomas A. Neubert Presented at: ABRF 2013 (RG9 session), Palm Springs, CA March
Molecular Interactions Research Group (MIRG) Satya P. Yadav, Aaron P. Yamniuk, John Newitt, Michael L. Doyle, Ed Eisenstein, Thomas A. Neubert Presented at: ABRF 2013 (RG9 session), Palm Springs, CA March
Supplementary Information. Structural basis for duplex RNA recognition and cleavage by A.
 Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the
 Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the roots of three-day-old etiolated seedlings of Col-0
Supplementary Figure 1 PZA inhibits root hair formation as well as cell elongation in the maturation zone of eto1-2 roots. (A) The PI staining of the roots of three-day-old etiolated seedlings of Col-0
The MIRG 2002 Study: Assembly State, Thermodynamic and Kinetic Analysis of an Enzyme/Inhibitor Interaction
 The MIRG 2002 Study: Assembly State, Thermodynamic and Kinetic Analysis of an Enzyme/Inhibitor Interaction Model System Carbonic Anhydrase II (CA-II = ~30 kda) 4-Carboxybenzenesulfonamide (CBS = 201.2
The MIRG 2002 Study: Assembly State, Thermodynamic and Kinetic Analysis of an Enzyme/Inhibitor Interaction Model System Carbonic Anhydrase II (CA-II = ~30 kda) 4-Carboxybenzenesulfonamide (CBS = 201.2
Protein-Protein Interactions II
 Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Supplementary Figure 1. Antibody-induced cargo release studied by native PAGE. A clear band corresponding to the cargo strand (lane 1) is visible.
 Supplementary Figure 1. Antibody-induced cargo release studied by native PAGE. A clear band corresponding to the cargo strand (lane 1) is visible. Because SYBR Gold is less sensitive to single stranded
Supplementary Figure 1. Antibody-induced cargo release studied by native PAGE. A clear band corresponding to the cargo strand (lane 1) is visible. Because SYBR Gold is less sensitive to single stranded
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
doi:10.1038/nature10016 Supplementary discussion on binding site density for protein complexes on the surface: The density of biotin sites on the chip is ~10 3 biotin-peg per µm 2. The biotin sites are
Supplementary Information. Single-molecule analysis reveals multi-state folding of a guanine. riboswitch
 Supplementary Information Single-molecule analysis reveals multi-state folding of a guanine riboswitch Vishnu Chandra 1,4,#, Zain Hannan 1,5,#, Huizhong Xu 2,# and Maumita Mandal 1,2,3,6* Department of
Supplementary Information Single-molecule analysis reveals multi-state folding of a guanine riboswitch Vishnu Chandra 1,4,#, Zain Hannan 1,5,#, Huizhong Xu 2,# and Maumita Mandal 1,2,3,6* Department of
Recovery of the Formation and Function of Oxidized G-Quadruplexes by a Pyrenemodified
 Supporting Information Recovery of the Formation and Function of Oxidized G-Quadruplexes by a Pyrenemodified Guanine Tract Shuntaro Takahashi, Ki Tae Kim, Peter Podbevsek, Janez Plavec, Byeang Hyean Kim,
Supporting Information Recovery of the Formation and Function of Oxidized G-Quadruplexes by a Pyrenemodified Guanine Tract Shuntaro Takahashi, Ki Tae Kim, Peter Podbevsek, Janez Plavec, Byeang Hyean Kim,
SPARK. Southern Illinois University Edwardsville. Drake Jensen Southern Illinois University Edwardsville,
 Southern Illinois University Edwardsville SPARK Chemistry Faculty Research, Scholarship, and Creative Activity Chemistry Spring 2-15-2015 The exchanged EF-hands in calmodulin and troponin C chimeras impair
Southern Illinois University Edwardsville SPARK Chemistry Faculty Research, Scholarship, and Creative Activity Chemistry Spring 2-15-2015 The exchanged EF-hands in calmodulin and troponin C chimeras impair
Site-specific time-resolved FRET reveals local variations in the unfolding mechanism in an apparently two-state protein unfolding transition
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 Supplementary information for Site-specific time-resolved FRET reveals local variations
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2017 Supplementary information for Site-specific time-resolved FRET reveals local variations
Supplementary Information
 Supplementary Information Minimal mechanistic model of sirna-dependent target RNA slicing by recombinant human Argonaute2 protein Andrea Deerberg*, Sarah Willkomm* and Tobias Restle Institute of Molecular
Supplementary Information Minimal mechanistic model of sirna-dependent target RNA slicing by recombinant human Argonaute2 protein Andrea Deerberg*, Sarah Willkomm* and Tobias Restle Institute of Molecular
Supplementary Information. Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly
 Supplementary Information Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly Joshua M. Karchin, Jeung-Hoi Ha, Kevin E. Namitz, Michael S. Cosgrove, and
Supplementary Information Small Molecule-Induced Domain Swapping as a Mechanism for Controlling Protein Function and Assembly Joshua M. Karchin, Jeung-Hoi Ha, Kevin E. Namitz, Michael S. Cosgrove, and
Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis
 Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Supplemental material to accompany Structure and Possible Mechanism of the CcbJ Methyltransferase from Streptomyces caelestis Jacob Bauer, a Gabriela Ondrovičová, a Lucie Najmanová, b Vladimír Pevala,
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain
 Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
Supplementatry Fig 1. Domain structure, biophysical characterisation and electron microscopy of a TD. (a) XTACC3/Maskin and XMAP215/chTOG domain architecture. Various C-terminal fragments were cloned and
TAG-LITE RECEPTOR LIGAND BINDING ASSAY
 TAG - L I T E TAG-LITE RECEPTOR LIGAND BINDING ASSAY PROTOCOL ASSAY PRINCIPLE The Tag-lite Ligand Binding Assay is a homogeneous alternative to radio ligand binding assays for HTS and compound profiling.
TAG - L I T E TAG-LITE RECEPTOR LIGAND BINDING ASSAY PROTOCOL ASSAY PRINCIPLE The Tag-lite Ligand Binding Assay is a homogeneous alternative to radio ligand binding assays for HTS and compound profiling.
Tag-lite Tachykinin NK1 labeled Cells, ready-to-use (transformed & labeled), 200 tests* (Part# C1TT1NK1)
 TAG - L I T E Tag-lite Tachykinin NK1 Receptor Ligand Binding Assay protocol Tachykinin NK1 labeled cells for: 200 tests Part#: C1TT1NK1 Lot#: 03-1 March 2016 (See expiration date on package label) Store
TAG - L I T E Tag-lite Tachykinin NK1 Receptor Ligand Binding Assay protocol Tachykinin NK1 labeled cells for: 200 tests Part#: C1TT1NK1 Lot#: 03-1 March 2016 (See expiration date on package label) Store
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a)
 Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplementary Figure 1
 Supplementary Figure 1 2 Supplementary Figure 1: Sequence alignment of HsHSD17B8 and HsCBR4 of with KAR orthologs. The secondary structure elements as calculated by DSSP and residue numbers are displayed
Supplementary Figure 1 2 Supplementary Figure 1: Sequence alignment of HsHSD17B8 and HsCBR4 of with KAR orthologs. The secondary structure elements as calculated by DSSP and residue numbers are displayed
PHF20 is an effector protein of p53 double lysine methylation
 SUPPLEMENTARY INFORMATION PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53 Gaofeng Cui 1, Sungman Park 2, Aimee I Badeaux 3, Donghwa Kim 2, Joseph Lee 1,
SUPPLEMENTARY INFORMATION PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53 Gaofeng Cui 1, Sungman Park 2, Aimee I Badeaux 3, Donghwa Kim 2, Joseph Lee 1,
Supporting Information
 Copyright WILEY-VCH Verlag GmbH & Co. KGaA, 69469 Weinheim, Germany, 2014. Supporting Information for Small, DOI: 10.1002/smll.201303558 Ultraspecific and Highly Sensitive Nucleic Acid Detection by Integrating
Copyright WILEY-VCH Verlag GmbH & Co. KGaA, 69469 Weinheim, Germany, 2014. Supporting Information for Small, DOI: 10.1002/smll.201303558 Ultraspecific and Highly Sensitive Nucleic Acid Detection by Integrating
Name with Last Name, First: BIOE111: Functional Biomaterial Development and Characterization FINAL EXAM (April 27, 2017)
 BIOE111: Functional Biomaterial Development and Characterization FINAL EXAM (April 27, 2017) Question 0: Fill in your name and student ID on each page. (1) Question 1: Briefly define the following terms
BIOE111: Functional Biomaterial Development and Characterization FINAL EXAM (April 27, 2017) Question 0: Fill in your name and student ID on each page. (1) Question 1: Briefly define the following terms
Supplementary Information
 Supplementary Information Supplemental Figure 1. VVD-III purifies in a reduced state. (a) The cell pellet of VVD-III (VVD 36 C108A:M135I:M165I) is green compared to VVD-I (wild type VVD 36) due to the
Supplementary Information Supplemental Figure 1. VVD-III purifies in a reduced state. (a) The cell pellet of VVD-III (VVD 36 C108A:M135I:M165I) is green compared to VVD-I (wild type VVD 36) due to the
Supporting Information
 Supporting Information Peroxidase vs. Peroxygenase Activity: Substrate Substituent Effects as Modulators of Enzyme Function in the Multifunctional Catalytic Globin Dehaloperoxidase Ashlyn H. McGuire, Leiah
Supporting Information Peroxidase vs. Peroxygenase Activity: Substrate Substituent Effects as Modulators of Enzyme Function in the Multifunctional Catalytic Globin Dehaloperoxidase Ashlyn H. McGuire, Leiah
Challenges to measuring intracellular Ca 2+ Calmodulin: nature s Ca 2+ sensor
 Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Calcium Signals in Biological Systems Lecture 3 (2/9/0) Measuring intracellular Ca 2+ signals II: Genetically encoded Ca 2+ sensors Henry M. Colecraft, Ph.D. Challenges to measuring intracellular Ca 2+
Oligonucleotides were purchased from Eurogentec, purified by denaturing gel electrophoresis
 SUPPLEMENRY INFORMION Purification of probes and Oligonucleotides sequence Oligonucleotides were purchased from Eurogentec, purified by denaturing gel electrophoresis and recovered by electroelution. Labelling
SUPPLEMENRY INFORMION Purification of probes and Oligonucleotides sequence Oligonucleotides were purchased from Eurogentec, purified by denaturing gel electrophoresis and recovered by electroelution. Labelling
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade
 Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Development of a quantitative fluorescence-based ligandbinding
 1 2 3 4 5 6 7 Development of a quantitative fluorescence-based ligandbinding assay Conor J. Breen 1, 2, *, Mathilde Raverdeau 2 & H. Paul Voorheis 2 1 Department of Biology, Maynooth University, Maynooth,
1 2 3 4 5 6 7 Development of a quantitative fluorescence-based ligandbinding assay Conor J. Breen 1, 2, *, Mathilde Raverdeau 2 & H. Paul Voorheis 2 1 Department of Biology, Maynooth University, Maynooth,
Supplemental Information. Single-Molecule Imaging Reveals How. Mre11-Rad50-Nbs1 Initiates DNA Break Repair
 Molecular Cell, Volume 67 Supplemental Information Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair Logan R. Myler, Ignacio F. Gallardo, Michael M. Soniat, Rajashree A. Deshpande,
Molecular Cell, Volume 67 Supplemental Information Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair Logan R. Myler, Ignacio F. Gallardo, Michael M. Soniat, Rajashree A. Deshpande,
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature08627 Supplementary Figure 1. DNA sequences used to construct nucleosomes in this work. a, DNA sequences containing the 601 positioning sequence (blue)24 with a PstI restriction site
doi: 10.1038/nature08627 Supplementary Figure 1. DNA sequences used to construct nucleosomes in this work. a, DNA sequences containing the 601 positioning sequence (blue)24 with a PstI restriction site
ABSTRACT. William J. Cressman Master of Science, Molecular mechanisms of protein allostery are not well understood,
 ABSTRACT Title of Document: PROBING ALLOSTERIC COMMUNICATION BETWEEN DISORDERED SURFACES IN A PROTEIN William J. Cressman Master of Science, 2015 Directed By: Dr. Dorothy Beckett Department of Chemistry
ABSTRACT Title of Document: PROBING ALLOSTERIC COMMUNICATION BETWEEN DISORDERED SURFACES IN A PROTEIN William J. Cressman Master of Science, 2015 Directed By: Dr. Dorothy Beckett Department of Chemistry
Expanded View Figures
 Expanded View Figures Figure EV1. AM3-CLE45 control experiments and bam3 alleles. A Relative primary root length of indicated genotypes at 9 dag, in response to increasing amounts of CLE45 in the media.
Expanded View Figures Figure EV1. AM3-CLE45 control experiments and bam3 alleles. A Relative primary root length of indicated genotypes at 9 dag, in response to increasing amounts of CLE45 in the media.
A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy
 Nature Methods A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy Luis A Campos, Jianwei Liu, Xiang Wang, Ravishankar Ramanathan, Douglas S English & Victor
Nature Methods A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy Luis A Campos, Jianwei Liu, Xiang Wang, Ravishankar Ramanathan, Douglas S English & Victor
Supplementary Figure 1. Two activation pathways and four conformations of β 2 integrins. KIM127 (red) can specifically detect
 Supplementary Figure 1 Two activation pathways and four conformations of β 2 integrins. KIM127 (red) can specifically detect integrin extension (E + ) and mab24 (green) can specifically detect headpiece-opening
Supplementary Figure 1 Two activation pathways and four conformations of β 2 integrins. KIM127 (red) can specifically detect integrin extension (E + ) and mab24 (green) can specifically detect headpiece-opening
An Ultra-sensitive Colorimetric Detection of Tetracyclines using the Shortest Aptamer with Highly Enhanced Affinity
 An Ultra-sensitive Colorimetric Detection of Tetracyclines using the Shortest Aptamer with Highly Enhanced Affinity Young Seop Kwon, Nurul Hanun Ahmad Raston, and Man Bock Gu*. School of Life Sciences
An Ultra-sensitive Colorimetric Detection of Tetracyclines using the Shortest Aptamer with Highly Enhanced Affinity Young Seop Kwon, Nurul Hanun Ahmad Raston, and Man Bock Gu*. School of Life Sciences
DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli
 Supplementary Information DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli Geraldine Fulcrand 1,2, Samantha Dages 1,2, Xiaoduo Zhi 1,2, Prem Chapagain
Supplementary Information DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli Geraldine Fulcrand 1,2, Samantha Dages 1,2, Xiaoduo Zhi 1,2, Prem Chapagain
SUPPLEMENTARY INFORMATION. Design and Characterization of Bivalent BET Inhibitors
 SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
RNP purification, components and activity.
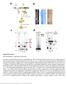 Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Supplementary Figure 1. FRET probe labeling locations in the Cas9-RNA-DNA complex.
 Supplementary Figure 1. FRET probe labeling locations in the Cas9-RNA-DNA complex. (a) Cy3 and Cy5 labeling locations shown in the crystal structure of Cas9-RNA bound to a cognate DNA target (PDB ID: 4UN3)
Supplementary Figure 1. FRET probe labeling locations in the Cas9-RNA-DNA complex. (a) Cy3 and Cy5 labeling locations shown in the crystal structure of Cas9-RNA bound to a cognate DNA target (PDB ID: 4UN3)
Use of ITC/DSC for the studies of biomolecularstability and interaction of excipients with proteins. Natalia Markova
 Use of ITC/DSC for the studies of biomolecularstability and interaction of excipients with proteins Natalia Markova MIBio2012 Outline Brief intro to the specifics of protein stability profiling and optimization
Use of ITC/DSC for the studies of biomolecularstability and interaction of excipients with proteins Natalia Markova MIBio2012 Outline Brief intro to the specifics of protein stability profiling and optimization
SUPPLEMENTAL INFORMATION. Center Hamburg-Eppendorf, Center for Molecular Neurobiology, ZMNH, Hamburg, Germany
 SUPPLEMENTAL INFORMATION Title: Inter-motif communication induces hierarchical Ca 2+ -filling of Caldendrin Authors: Uday Kiran 1, Phanindranath Regur 1, Michael R. Kreutz 3,4*, Yogendra Sharma 1,2*, Asima
SUPPLEMENTAL INFORMATION Title: Inter-motif communication induces hierarchical Ca 2+ -filling of Caldendrin Authors: Uday Kiran 1, Phanindranath Regur 1, Michael R. Kreutz 3,4*, Yogendra Sharma 1,2*, Asima
Engineering splicing factors with designed specificities
 nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
nature methods Engineering splicing factors with designed specificities Yang Wang, Cheom-Gil Cheong, Traci M Tanaka Hall & Zefeng Wang Supplementary figures and text: Supplementary Figure 1 Supplementary
Staple W100 (H) H3. Electron density maps of region C of the CDR-H3 loop in the 4E10 Fab SAH-MPER (671
 4E10/MPER (671 683KKK) 4E10/SAH-MPER (671 683KKK) (q) 4E10/SAH-MPER (671 683KKK) (q)pser W100B (H) Da-Gly-pSer Tether C-term L100C (H) P4 G100A (H) W100 (H) H3 G99 (H) T98 (H) N-term Supplementary Figure
4E10/MPER (671 683KKK) 4E10/SAH-MPER (671 683KKK) (q) 4E10/SAH-MPER (671 683KKK) (q)pser W100B (H) Da-Gly-pSer Tether C-term L100C (H) P4 G100A (H) W100 (H) H3 G99 (H) T98 (H) N-term Supplementary Figure
Supporting Information Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy
 Supporting Information Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy Jonathan R. Burns* & Stefan Howorka* 1 Contents 1. Design of DNA nanopores... 3 1.1.
Supporting Information Defined Bilayer Interactions of DNA Nanopores Revealed with a Nuclease-Based Nanoprobe Strategy Jonathan R. Burns* & Stefan Howorka* 1 Contents 1. Design of DNA nanopores... 3 1.1.
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport
 Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Supplementary materials for Structure of an open clamp type II topoisomerase-dna complex provides a mechanism for DNA capture and transport Ivan Laponogov 1,2, Dennis A. Veselkov 1, Isabelle M-T. Crevel
Electronic Supplementary Information (ESI)
 Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2018 Electronic Supplementary Information (ESI) Palladium(II) and platinum(ii) saccharinate
Electronic Supplementary Material (ESI) for Dalton Transactions. This journal is The Royal Society of Chemistry 2018 Electronic Supplementary Information (ESI) Palladium(II) and platinum(ii) saccharinate
Globins. The Backbone structure of Myoglobin 2. The Heme complex in myoglobin. Lecture 10/01/2009. Role of the Globin.
 Globins Lecture 10/01/009 The Backbone structure of Myoglobin Myoglobin: 44 x 44 x 5 Å single subunit 153 amino acid residues 11 residues are in an a helix. Helices are named A, B, C, F. The heme pocket
Globins Lecture 10/01/009 The Backbone structure of Myoglobin Myoglobin: 44 x 44 x 5 Å single subunit 153 amino acid residues 11 residues are in an a helix. Helices are named A, B, C, F. The heme pocket
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
Microscale Thermophoresis
 AN INTRODUCTION Microscale Thermophoresis A sensitive method to detect and quantify molecular interactions OCTOBER 2012 CHAPTER 1 Overview Microscale thermophoresis (MST) is a new method that enables the
AN INTRODUCTION Microscale Thermophoresis A sensitive method to detect and quantify molecular interactions OCTOBER 2012 CHAPTER 1 Overview Microscale thermophoresis (MST) is a new method that enables the
Characterization of G4-G4 crosstalk in c-kit promoter region
 Supplementary information Characterization of G4-G4 crosstalk in c-kit promoter region Riccardo Rigo #, Claudia Sissi #,* # Department of Pharmaceutical and Pharmacological Sciences, University of Padova,
Supplementary information Characterization of G4-G4 crosstalk in c-kit promoter region Riccardo Rigo #, Claudia Sissi #,* # Department of Pharmaceutical and Pharmacological Sciences, University of Padova,
A quantitative investigation of linker histone interactions with nucleosomes and chromatin
 White et al Supplementary figures and figure legends A quantitative investigation of linker histone interactions with nucleosomes and chromatin 1 Alison E. White, Aaron R. Hieb, and 1 Karolin Luger 1 White
White et al Supplementary figures and figure legends A quantitative investigation of linker histone interactions with nucleosomes and chromatin 1 Alison E. White, Aaron R. Hieb, and 1 Karolin Luger 1 White
SUPPLEMENTARY INFORMATION Figures. Supplementary Figure 1 a. Page 1 of 30. Nature Chemical Biology: doi: /nchembio.2528
 SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
