Developing a novel inhibitor for Cdc14 in the fungus Aspergillus niger. Stephanie Zhang
|
|
|
- Anna Lindsey
- 5 years ago
- Views:
Transcription
1 Developing a novel inhibitor for Cdc14 in the fungus Aspergillus niger Stephanie Zhang ABSTRACT Fungal pathogens are a major cause of crop damage. In these fungi, Cdc14 could be a critical phosphatase for regulating cell division by ending the mitotic process, as demonstrated by studies in Saccharomyces cerevisiae. Due to Cdc14 s absence in higher plants and noncritical role in animals, an inhibitor which can reduce its activity could function as a potential fungicide. In this project, I developed an inhibitor for Cdc14 in the fungal pathogen Aspergillus niger (AnCdc14), a pathogen that causes black mold on crops. After characterizing the catalytic specificity of AnCdc14, I designed multiple inhibitors that modeled the specificity. A combination of bioinformatic and biochemical approaches was used to determine the most effective inhibitor: one that contained four benzene rings and functioned through irreversible inhibition. Through computer modeling, I optimized this inhibitor to improve its affinity for AnCdc14. This modified inhibitor can be tested in vitro and in vivo for its efficiency in preventing A. niger growth and eliminating its pathogenicity. Due to high conservation of Cdc14 among fungi species, the inhibitor I created could be tested in multiple fungi, such as F. graminearum. Eventually, these results could be developed into a new antifungal compound that broadly prevents plant fungal infections.
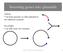
2 INTRODUCTION Every year, fungal pathogens destroy more than 125 million tons of food, enough to feed 600 million people (1). Fungicides are a common method to control these fungal damages, but fungi inevitably evolve resistance to these chemicals. Therefore, there exists a long felt yet unfulfilled need to find new, effective, and economic fungicides. One path to find new fungicides is to find molecules that inhibit a biological process necessary for fungi growth and pathogenesis. Molecules that inhibit the enzyme Cdc14 are great candidates for such a purpose because Cdc14 is essential for fungal development. Cdc14 in Saccharomyces cerevisiae (S. cerevisiae) has been researched extensively and shown to dephosphorylate its substrates. It plays a major role in the Mitotic Exit Network by inactivating cyclin-dependent kinases (CDKs), thereby ending mitosis and regulating cytokinesis (2,3) (Figure 1). When ScCdc14 is inhibited or not present, the cell is unable to finish mitosis and therefore unable to develop properly (4). Studies also demonstrate that ScCdc14 helps to coordinate DNA repair and genome stability (5). Figure 1. Regulation of mitosis in S. cerevisiae: Cdc14 promotes mitotic exit by antagonizing mitotic CDK activity. The activity of Cdc14 is regulated by two signaling pathways, the Cdc14 early anaphase release (FEAR) network and the mitotic exit network (MEN). Diagram from Source 4. 1
3 Cdc14 is an attractive target for antifungal drug development for several other important reasons. First, genetic deletion of Cdc14 severely retards growth and eliminates pathogenicity of Fusarium graminearum (F. graminearum) and Magnaporthe oryzae (M. oryzae) (6,7). Second, catalytic sequences of all Cdc14 orthologs in different fungi species are highly conserved, indicating that these orthologs had similar substrate specificity (8). This makes design of competitive inhibitors for a broad family of fungi achievable. Third, Cdc14 phosphatase genes are conserved in fungi but are absent from higher plants (9). Genetic studies indicate its function in animal and human cells is not critical, contradicting its function in fungal cells (10). Thus, plants or consuming animals and humans are likely to be unaffected by compounds that specifically inhibit Cdc14. Aspergillus niger (A. niger), is a fungus that causes black mold on certain crops, fruits, and vegetables such as rice, grapes, apricots, onions, and peanuts. A. niger produces Ochratoxin A, a toxin that can pose a major health hazard to both humans and animals (11). Therefore, developing a fungicide that can control the A. niger pathogenic infection has both economic and health values. In this study, I characterized the catalytic specificity for Cdc14 in A. niger (AnCdc14) and found that optimal substrates for AnCdc14 contain the amino acid sequence phosphorylated serine (Ser(P))-proline (P)-X (any amino acid)-lysine or arginine (Lys/Arg). This is similar to the specificity for other Cdc14 orthologs, such as Cdc14 in S. cerevisiae (ScCdc14), Cdc14 in F. graminearum (FgCdc14), and Cdc14 in M. oryzae (MoCdc14), suggesting that inhibitors that are efficient on AnCdc14 could also inhibit its orthologs and vice versa. An initial screening of 10,000 potential Cdc14 inhibitors at the Bindley Bioscience Center has identified 13 lead inhibitors of ScCdc14 (see Table S2 in the appendix). To look for 2
4 competitive inhibitors that are highly specific for the Cdc14 family but broadly reactive across fungal species, I tested these 13 inhibitors and selected the most efficient one for AnCdc14 using computer modeling as well as a standard phosphatase assay and an IC50 curve. The IC50 value generated from this curve is the concentration of an inhibitor needed to reduce biological activity by half; it is a measurement for drug efficiency and is commonly used in pharmacological research. With a combination of in vitro and in silico techniques, I modified and optimized this inhibitor to ultimately create an improved inhibitor for AnCdc14. This generation two molecule has the potential to inhibit AnCdc14 and broadly act against all fungi orthologs of Cdc14, thus making it a good candidate for a future fungicide. 3
5 MATERIALS AND METHODS Transformation and Purification Purified AnCdc14 DNA was inserted into the plasmid pet15b (produced by Genscript) between the NdeI and BamHI restriction sites. This created a fusion on the 5 end of the AnCdc14 with the 6xHis tag sequence. This sequence ultimately allowed for isolation and purification of the target protein by nickel affinity chromatography. The genetically modified plasmid was then transformed into competent Escherichia coli (E. coli) BL21 AI cells with the heat shock method. DNA was mixed with the bacteria cells and placed in 42 C for one minute, then in ice, then in 37 C incubation for 30 minutes (12). The transformed bacteria were selected by ampicillin (as the original plasmid contained an ampicillin-resistant maker) and then amplified overnight. Harvested bacteria were lysed on ice for 30 min in lysis buffer (25mM HEPES ph7.5, 500 mm NaCl, 0.1% (v/v)triton x-100, 10 mm imidazole, 10% (v/v) glycerol) supplemented with 50 mg/ml Lysozyme,10 μm Leupeptin, 1 μm Pepstatin, and universal Nuclease. The bacteria were sonicated to reduce viscosity. After centrifugation at 35,000 g for 30 min at 4 C, the resulting soluble extract was used to run AKTA chromatography. During the chromatography, proteins with a Histidine tag bound to the nickel column and were then washed with imidazole. The imidazole was later dialyzed away, leaving just purified proteins. A Bradford Assay was performed with the purified protein to measure the concentration using a standard curve. SDS-Page gel electrophoresis was run for one hour at 180 volts with a running gel containing 30% acrylamide with a cross-linking ratio of 29:1. This gel was set in Coomassie blue staining solution before being imaged with Bio-Rad Gel Doc EZ imaging 4
6 system and analyzed with ImageLab. The molecular weight of the protein confirmed that the purified protein was AnCdc14. Phosphatase Assays To confirm that AnCdc14 had tyrosine phosphatase activity, phosphatase reactions were conducted. Sodium orthovanadate inhibits all tyrosine phosphatases and was added at three concentrations (0, 500, and 1000 μm) to a solution of 0.3 μm of AnCdc14 and 300 mm of artificial substrate pnpp. The amount of dephosphorylated pnpp (p-nitrophenol, or pnp) produced from each of the three reactions was calculated using a standard pnp curve. To determine necessary concentrations and time needed to achieve steady state conditions, 30 mm of pnpp and 0.3 μm of AnCdc14 were incubated for 60 minutes at 24 C. Absorbance values at 405 nm determined the amount of pnp produced and were measured every minute. The results indicated that 20 minutes at 30 C was sufficient to reach steady state with 0.3 μm of AnCdc14 and 30 mm of pnpp. A Michaelis-Menten assay was then performed to ensure that future assays would be under steady-state conditions, to determine the average initial velocity of the reaction, and to find the values for k cat and K m in AnCdc14. This assay was performed with various concentrations of the substrate pnpp (1, 3, 6, 12.5, 30, and 50 mm) and 0.3 μm AnCdc14 for 20 minutes at 30 C. The results were graphed on ImageLab and fitted to the Michaelis-Menten equation. Catalytic Specificity AnCdc14 s catalytic ability to dephosphorylate 24 artificial substrates (Table S1) with different amino acid sequences was tested to determine AnCdc14 s catalytic specificity (13). 100 μm of each substrate were mixed with 0.13 μm of AnCdc14 for 20 minutes at 30 C (an additional phosphatase assay determined these conditions necessary to reach steady state). The 5
7 amount of phosphate released from each reaction was calculated using a standard curve for BioMol Green, which reacts with free phosphate, and the results were plotted on a bar graph to determine AnCdc14 s catalytic specificity. In silico Inhibition of AnCdc14 A model of AnCdc14 s protein structure was made based on the corresponding amino acid sequence and the known crystal structure of ScCdc14: 5XW4. This homology model was built using Phyre2, a suite of software tools to predict and analyze protein structure, function, and mutations (14). 3D Ligand Site, a specific tool of Phyre2, was used to find the binding site of the homology model (15). Specific inhibitor molecules (Table S2) were docked into the model s catalytic site using MCule s 1-Click Docking tool, a website that docks inhibitors and generates values that represent binding affinity ( S scores). The inhibitors were docked at the position Arg188_C1279, which corresponded to the point X: , Y: , Z: in the homology model. The S values were graphed in a bar graph to predict results for the in vitro inhibition assays. In vitro Inhibition of AnCdc14 The same 13 inhibitors (Table S2) were tested in vitro to find the accuracy and validity of the homology model and to determine the most efficient inhibitor for AnCdc μm of each inhibitor were mixed with 30 mm of pnpp and 0.3 μm AnCdc14. From this assay, three inhibitors were determined to be the most effective and have the highest binding affinity for AnCdc14, thus producing less product. Each of these three inhibitors were used to plot an IC 50 curve at concentrations of 0, 0.25, 0.5, 1, 2, 5, 10, 25, 50, 100, and 200 μm with 30 mm pnpp and 0.3 μm AnCdc14. The 6
8 IC 50 value and K i value were determined for each of these inhibitors. From these assays, the best inhibitor was identified and was used in further experiments. Mechanism of Inhibition To determine the mechanism of inhibition that the top inhibitor used, Michaelis-Menten curves were created for three different inhibitor concentrations: 0, 2.5, and 10 μm. These inhibitors were tested at pnpp concentrations of 1, 2, 5, 10, 25, 50, 75, and 100 mm with 0.6 μm AnCdc14 for 20 minutes at 30 C. The three curves were graphed on ImageLab and fitted to the Michaelis-Menten equation. To test for reversibility of inhibition, 10 μm of inhibitor and 0.6 μm of enzyme were incubated at 25 C for 0, 15, and 30 minutes. Following the incubation time, 30 mm of pnpp was added and incubated for 20 minutes at 30 C. Optimization of Inhibitor in silico The identified best inhibitor was optimized in MCule s 1-Click Docking. The site s construction tool was used to add structures to the original inhibitor to increase its binding affinity for AnCdc14 and thus produce a better inhibitor. 7
9 RESULTS AND DISCUSSION Transformation and Purification After inducing expression of AnCdc14 in E. coli, I extracted proteins from the transformed cells and ran nickel affinity chromatography to purify the AnCdc14 enzyme. The UV absorbance trace from the AKTA purification system showed a large peak where there was a high imidazole concentration, suggesting reaction tubes 20 through 30 contained the protein (Figure 2A). To ensure that the purified protein was AnCdc14, I ran gel electrophoresis using a fraction of purified protein sample. A thick band at 51.0 kda, very close to calculated size of 52.2 kda based on amino acid sequence, suggested that I isolated the correct protein (Figure 2B). Figure 2. Purification of AnCdc14: A. The UV absorbance signal during the AKTA purification run for each volume of sample that passed through the column suggests successful purification of protein. The large spike in absorbance signifies which samples from the AKTA system contained the highest concentration of imidazole. B. SDS-Page gel analysis using Coomassie blue staining demonstrates a thick band of AnCdc14 at 51.0 kda. 8
10 With purified AnCdc14, I performed a Bradford assay (data not shown) to find that the AnCdc14 concentration was µm. Phosphatase Assays Phosphatase activity of AnCdc14 was confirmed by an assay using sodium orthovanadate (a protein tyrosine phosphatase inhibitor) as an inhibitor and pnpp (an artificial small molecule) as the substrate. The results demonstrated that at inhibitor concentration of 500 µm, percent inhibition increased slightly, from 0% to 7.3%; at inhibitor concentration of 1000 µm, the percent inhibition increased at a much greater magnitude, from 7.3% to 40.5% (Figure 3A). The result indicated that increased concentrations of phosphatase inhibitor decreased AnCdc14 activity. The sodium orthovanadate assay confirmed AnCdc14 s phosphatase activity, and I found the enzyme s specific activity to be 6.51 min -1. In the reaction of AnCdc14 with pnpp, I found that the graph was linear for the first 40 minutes (Figure 3B). Because the linear portion of the graph represented when my enzyme was in the steady state, I determined that all further experiments conducted with steady state properties would have to be conducted in less than 40 minutes. Figure 3. AnCdc14 Phosphatase Activity and Steady State Conditions: A. Sodium orthovanadate, a protein tyrosine phosphatase inhibitor, was added to AnCdc14 at various concentrations. Percent inhibition increased with increased concentration of the inhibitor. The error bars shown are standard deviations for the three trials I completed. B. Shown is absorbance at nm over 60 minutes as AnCdc14 reacted with the substrate pnpp. The absorbance is a measure representative of the amount of phosphate produced from the AnCdc14 catalyzed reaction. 9
11 With the steady-state time parameter determined, I calculated k cat and K m values for AnCdc14 using a Michaelis-Menten curve. The curve yielded that the K m value was mm, v max was µm/min, and k cat was 0.92 sec -1 (Figure 4). These values were used in future experiments to calculate k cat /K m and K i values. Figure 4. Michaelis-Menten Curve with the Substrate pnpp: The average velocity of product produced in µm per minute for each of the substrate concentrations were fit to a Michaelis-Menten curve. The velocity of the phosphate produced was calculated using the absorbance values of the reactions after 20 minutes of reacting at 30 C. The error bars shown represent standard deviation over the three conducted trials, and the curve was generated by ImageLab and is fit by the Michaelis-Menten equation: velocity=(v max *[substrate])/(k m +[substrate]). Catalytic Specificity To find the AnCdc14 substrate specificity, I selected 24 substrates (Table S1) and tested catalytic efficiency of AnCdc14 on them. These 24 substrates were selected based on previous studies with ScCdc14 and other Cdc14 family members (2). I measured the amount of phosphate that was produced by each substrate reaction catalyzed by AnCdc14. These values were used to calculate the k cat /K m values for each substrate. Higher quotients indicate substrates with higher binding affinity to AnCdc14. Substrates #1, #7, and #24 were found to be the best substrates and #8, #9, #14, and #17 were shortly behind (Figure 5). Upon comparison of the corresponding sequences, I found that these sequences shared a phosphorylated serine group followed by a proline in the +1 position and a lysine in the 10
12 Average k cat /K m (M -1 sec -1 ) +3 position (Table S1). This sequence was absent at the substrates showing low AnCdc14 binding affinity, such as #10, #11, #12, #13, and it also matched the highly conserved substrate specificity among other Cdc14 family members, including ScCdc14 (16) Figure 5. Substrate Specificity of AnCdc14: The average k cat /K m values for each substrate were used to compare substrate sequences and determine catalytic specificity of AnCdc14. The error bars on the graph indicate the standard deviations among the three trials. The substrates corresponding to the numbers on the x-axis can be found in Table S1 (in the appendix). In silico Inhibition of AnCdc14 Substrate Number Due to limited research on AnCdc14 s protein structure, I had to develop a model of it based on ScCdc14 s crystal structure and AnCdc14 s gene sequence (Figure 6A). With my homology model, I tested 13 pre-screened inhibitors to predict their efficiency. These inhibitors were designed and selected based on the specificity of ScCdc14. I docked each of these inhibitors into AnCdc14 s active site via the computer software program 1-Click Docking. This procedure produced an S score that represented the binding affinity of the inhibitor for the enzyme. S scores with higher absolute values represented higher binding affinity. Though many of the inhibitors had similar S scores, inhibitors #2 and #4 (Table S2) were marginally better than the others in terms of S score, with inhibitors #1, #3, and #6 following (Figure 6C). Inhibitor #4 11
13 is shown docked in Figure 6B at the active site, Arg188, which was determined by 3D Ligand Site. Figure 6. Docking Inhibitors for AnCdc14: A. The homology model of AnCdc14 was developed based on the structure of ScCdc14. Shown are the predicted alpha helices and beta sheets. The section highlighted in red at the front of the model represents the active site, Arg188. B. Inhibitor #4, which was deemed to be the best inhibitor by comparison, docked to the homology model at the active site. C. The absolute values of the S values generated by 1- Click Docking are graphed for each inhibitor to compare binding affinity for the 13 inhibitors. The inhibitor molecules corresponding to the numbers on the x-axis can be found in Table S2 (in the appendix). In vitro Inhibition of AnCdc14 To validate the prediction made by 1-Click Docking, in vitro assays with each inhibitor were performed and the inhibition percentage was calculated for each inhibitor. In line with the predictions, inhibitors #1, #3, and #4 were AnCdc14 s top inhibitors, with number 4 being the most effective (Figure 7A). 12
14 From the IC 50 curve equations generated from the assays, I found that the IC 50 values for inhibitors #1, #3, and #4 were 31.9, 9.1, and 7.5 µm (Figures 7B, 7C, 7D). Inhibitor #4 had the smallest IC 50 value, indicating that it required the smallest concentration to inhibit AnCdc14 by 50%, thus it is the most efficient inhibitor. These results also supported the inhibitor assay results discussed in the previous paragraph (Figure 7A). Figure 7. In vitro Inhibition of AnCdc14: A. The percent inhibition for each of the compounds was used to determine the best inhibitors. The error bars represent standard deviation among the three trials. The compounds associated with each inhibitor number on the x-axis can be found in Table S2 (in the appendix). B-D. IC 50 curves for inhibitors #1, #3, and #4 (respectively). The R 2 values of these curves were , 0.964, and , respectively. The data were analyzed via nonlinear curve fitting in the My Curve Fit software, using the IC 50 formula: y=100/(1+(x/ IC 50 )^cooperativity). Using inhibitor #4 s IC 50 value as well as AnCdc14 s K m value, K i was calculated to be 5.3 µm, the inhibitor concentration required to bind half of the enzyme molecules. In addition, from the IC50 curves, I found that the cooperativity values for inhibitor #3 and #4 were 2.18 and 2.17, respectively, suggesting that AnCdc14 acted as a dimer (two 13
15 catalytically active sites). This was further supported by the fact that the R 2 values increased considerably when the cooperativity factor was added into the IC50 equation. This is also consistent with studies of other Cdc14 family members, as ScCdc14 is also a homo-dimer. Mechanism of Inhibition To determine which method of inhibition the inhibitor #4 actually employed, two concentrations of inhibitor #4 were used in the mechanism assay (Figure 8A, Table 1). I found that both K m and v max changed, suggesting the mechanism of inhibition to be mixed: both competitive (higher K m ) and noncompetitive (lower v max ). Figure 8. AnCdc14 is Inhibited Irreversibly: A. The average initial velocity of phosphate produced for different substrate concentrations from two trials created Michaelis-Menten curves. Inhibitor concentrations used for the different curves were 0 µm (black), 2.5 µm (blue), and 10 µm (green). The curves were generated by ImageLab and are fit by the Michaelis-Menten equation: velocity=(v max *[substrate])/(k m +[substrate]).b. Percent inhibition graphed for different amounts enzyme and inhibitor incubation times shows that inhibitor #4 is an irreversible inhibitor. The line of best fit shown has an equation of y=0.5741x , and the R 2 value is , representing a strong correlation. Table 1. v max and K m Results for Mechanism of Inhibition Assay: The v max and K m values were calculated from the Michaelis-Menten curves and their equations for each concentration of inhibitor. [Inhibitor] (µm) v max (µm/min) K m (mm)
16 However, another potential explanation for these data was that AnCdc14 was inhibited irreversibly. Irreversible inhibition occurs when the inhibitor remains covalently bound to the enzyme s active site, making it unable to bind more substrates and thereby lose its catalytic function (Figure 9). One criterion used to identify irreversible inhibitors is showing that percentage inhibition increases over time. By incubating inhibitor #4 with AnCdc14, I received results that showed 55.5% inhibition at 0 minute, 65.32% inhibition at 15 minute, and 72.71% inhibition at 30 minute (Figure 8B). These data suggested that longer periods of time increased the percent inhibition of AnCdc14, demonstrating that AnCdc14 was inhibited irreversibly by inhibitor #4. The irreversible inhibition mechanism was further supported by two sets of previously discussed results: the binding affinity for inhibitor #4 on AnCdc14 was high (Figure 7A) and inhibitor #4 s molecular structure fit well into my homology model s binding site (Figure 6B). Both suggested that inhibitor #4 might be able to covalent bonds as an irreversible inhibitor rather than hydrogen bonds as a reversible inhibitor. Figure 9. Diagrams of Inhibition Mechanisms: A. A reversible inhibitor binds to an enzyme in either the active site (competitive inhibition) or another site (noncompetitive inhibition). This temporarily blocks the substrate from binding to the enzyme but does not permanently disable enzyme activity. B. An irreversible inhibitor binds to the active site of an enzyme and creates covalent bonds, thus permanently disabling the enzyme and not allowing substrates to bind. Diagrams from Source
17 Optimization of Inhibitor in silico Both in silico and in vitro test suggested that inhibitor #4 was the best inhibitor. Using 1- Click Docking, I was able to change and optimize inhibitor #4 for increased binding affinity for AnCdc14. Such changes involved adding one more benzene ring, adding chlorine to create more intermolecular bonds with the hydrogens in the catalytic site, and adding a hydroxyl group to increase the number of hydrogen bonds (Figure 10A). Specifically, the binding affinity of inhibitor to AnCdc14 increased, with a change in absolute value of S value from 8.8 to 9.8. The optimized inhibitor can be seen docked into the AnCdc14 homology model in Figure 10B. Figure 10. Optimized Inhibitor #4: A. The structure of inhibitor #4 was slightly changed in order to increase binding affinity for the active site of AnCdc14. A benzene ring and a Chlorine group were added to the original structure. B. The optimized inhibitor docked in the active site had a higher binding affinity, with a new absolute value of S value as 9.8, compared to the original S value of 8.8 Arg188, the binding site, is labeled in the figure. 16
18 CONCLUSIONS AND FUTURE WORK In this project, I demonstrated that AnCdc14 conserved the catalytic specificity of the Cdc14 family, and I developed an inhibitor that can broadly act against all fungi with a Cdc14 ortholog. The inhibitor I developed in this project could be developed into a pesticide to act against A. niger and other fungal pathogens such as F. graminearum and M. oryzae. However, further studies both in vitro and in vivo are needed to confirm its ability to inhibit AnCdc14 and other Cdc14 orthologs, such as ScCdc14, FgCdc14, and MoCdc14. A plant infection assay using this new inhibitor could be conducted to demonstrate that it can actually prevent infection from pathogens containing Cdc14 orthologs. In addition, research conducted on whether the new fungicide causes any health problem in plants and animals, especially humans, will be needed before the new inhibitor can be considered as a real fungicide candidate. 17
19 APPENDIX Table S1. Substrate Naming Convention: The {p} represents a phosphorylated amino acid. The letters represent amino acid groups. Table S2. Inhibitor Naming Convention. 1 HT{pS}PIKSIG 2 HT{pT}PIKSIG 3 HT{pY}PIKSIG 4 HT{pS}AIKSIG 5 HT{pS}PIASIG 6 HT{pS}PIRSIG 7 HT{pS}PIKKIG 8 HT{pS}PKKSIG 9 HT{pS}PIKSKG 10 HT{pS}PKASIG 11 HT{pS}PIAKIG 12 HT{pS}PIASKG 13 HT{pS}PIASIK 14 AT{pS}PIKSIG 15 HA{pS}PIKSIG 16 HT{pS}KIKSIG 17 HT{pS}PEKSIG 18 HT{pS}PIKEIG 19 HT{pS}PIKSEG 20 HT{pS}PIKSIE 21 HT{pS}PIKSIK 22 KT{pS}PIKSIG 23 HK{pS}PIKSIG 24 HT{pS}PIKKRG 18
20 REFERENCES 1. Fisher, M., Henk, D., Briggs, C., Brownstein, J., Madoff, L., McCraw, S., and Gurr, S. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484, Bremmer, S., Hall, H., Martinez, J., Eissler, C., Hinrichsen, T., Rossie, S., Parker, L., Hall, M., and Charbonneau, H. (2011) Cdc14 Phosphatases Preferentially Dephosphorylate a Subset of Cyclin-dependent kinase (Cdk) Sites Containing Phosphoserine. Journal of Biological Chemistry 287, Visintin, R., Craig, K., Hwang, E., Prinz, S., Tyers, M., and Amon, A. (1998) The Phosphatase Cdc14 Triggers Mitotic Exit by Reversal of Cdk-Dependent Phosphorylation. Molecular Cell 2, Stegmeier, F., and Amon, A. (2004) Closing Mitosis: The Functions of the Cdc14 Phosphatase and Its Regulation. Annual Review of Genetics 38, Eissler, C., Mazón, G., Powers, B., Savinov, S., Symington, L., and Hall, M. (2014) The Cdk/Cdc14 Module Controls Activation of the Yen1 Holliday Junction Resolvase to Promote Genome Stability. Molecular Cell 54, Li, C., Melesse, M., Zhang, S., Hao, C., Wang, C., Zhang, H., Hall, M., and Xu, J. (2015) FgCDC14 regulates cytokinesis, morphogenesis, and pathogenesis in Fusarium graminearum. Molecular Microbiology 98, Li, C., Cao, S., Zhang, C., Zhang, Y., Zhang, Q., Xu, J., and Wang, C. (2017) MoCDC14 is important for septation during conidiation and appressorium formation in Magnaporthe oryzae. Molecular Plant Pathology. 8. Mocciaro, A., and Schiebel, E. (2010) Cdc14: a highly conserved family of phosphatases with nonconserved functions?. Journal of Cell Science 123, Kerk, D., Templeton, G., and Moorhead, G. (2007) Evolutionary Radiation Pattern of Novel Protein Phosphatases Revealed by Analysis of Protein Data from the Completely Sequenced Genomes of Humans, Green Algae, and Higher Plants. PLANT PHYSIOLOGY 146, Ah-Fong, A., and Judelson, H. (2011) New Role for Cdc14 Phosphatase: Localization to Basal Bodies in the Oomycete Phytophthora and Its Evolutionary Coinheritance with Eukaryotic Flagella. PLoS ONE 6, e Abarca, M. L., Bragulat, M. R., Castella, G., & Cabanes, F. J. (1994). Ochratoxin A production by strains of Aspergillus niger var. niger. Applied and environmental microbiology, 60(7), Froger, A., & Hall, J. E. (2007). Transformation of Plasmid DNA into E. coli Using the Heat Shock Method. Journal of Visualized Experiments : JoVE, (6), Powers, B., Melesse, M., Eissler, C., Charbonneau, H., and Hall, M. (2016) Measuring Activity and Specificity of Protein Phosphatases. Methods in Molecular Biology, Kelley, LA, et al. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, Wass M.N., Kelley L.A. and Sternberg M.J. (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. NAR 38, W Gray, C. (2003) The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. The EMBO Journal 22, Stahl, S. (2011). Stahl's Essential Psychopharmacology: Neuroscientific Basis and Practical Applications. 4th ed. Cambridge: Cambridge University Press. All figures and images were generated and created by Stephanie Zhang unless otherwise specified.
BACTERIAL PRODUCTION EXPRESSION METHOD OVERVIEW: PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT kda (full-length) 34.
 BACTERIAL PRODUCTION PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT 2015-XXXX XXXX pet-32a 50.9 kda (full-length) 34.0 kda (cleaved) EXPRESSION METHOD OVERVIEW: Plasmid DNA was transformed into BL21
BACTERIAL PRODUCTION PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT 2015-XXXX XXXX pet-32a 50.9 kda (full-length) 34.0 kda (cleaved) EXPRESSION METHOD OVERVIEW: Plasmid DNA was transformed into BL21
His-Spin Protein Miniprep
 INSTRUCTIONS His-Spin Protein Miniprep Catalog No. P2001 (10 purifications) and P2002 (50 purifications). Highlights Fast 5 minute protocol to purify His-tagged proteins from cell-free extracts Screen
INSTRUCTIONS His-Spin Protein Miniprep Catalog No. P2001 (10 purifications) and P2002 (50 purifications). Highlights Fast 5 minute protocol to purify His-tagged proteins from cell-free extracts Screen
5.36 Biochemistry Laboratory Spring 2009
 MIT OpenCourseWare http://ocw.mit.edu 5.36 Biochemistry Laboratory Spring 2009 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. Laboratory Manual for URIECA
MIT OpenCourseWare http://ocw.mit.edu 5.36 Biochemistry Laboratory Spring 2009 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. Laboratory Manual for URIECA
6 Enzymes II W. H. Freeman and Company
 6 Enzymes II 2017 W. H. Freeman and Company The role of an enzyme in an enzyme-catalyzed reaction is to: A. bind a transition state intermediate, such that it cannot be converted back to substrate. B.
6 Enzymes II 2017 W. H. Freeman and Company The role of an enzyme in an enzyme-catalyzed reaction is to: A. bind a transition state intermediate, such that it cannot be converted back to substrate. B.
Purification and Kinetic Phosphate Inhibition Analysis of Bacterial Alkaline Phosphatase
 Purification and Kinetic Phosphate Inhibition Analysis of Bacterial Alkaline Phosphatase Roshan Chikarmane CH 467 University of Oregon June 7, 26 Introduction Background Information. Alkaline Phosphatase
Purification and Kinetic Phosphate Inhibition Analysis of Bacterial Alkaline Phosphatase Roshan Chikarmane CH 467 University of Oregon June 7, 26 Introduction Background Information. Alkaline Phosphatase
Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005
 INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005 Highlights Fast protocol to purify Strep-tagged proteins from cell-free extracts Screen your recombinant colonies directly for
INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005 Highlights Fast protocol to purify Strep-tagged proteins from cell-free extracts Screen your recombinant colonies directly for
pt7ht vector and over-expressed in E. coli as inclusion bodies. Cells were lysed in 6 M
 Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions
 Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Supporting Information Stabilization of a virus-like particle and its application as a nanoreactor at physiological conditions Lise Schoonen, b Sjors Maassen, b Roeland J. M. Nolte b and Jan C. M. van
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
OPPF-UK Standard Protocols: Insect Cell Purification
 OPPF-UK Standard Protocols: Insect Cell Purification Last Updated 6 th October 2016 Joanne Nettleship joanne@strubi.ox.ac.uk OPPF-UK SOP: Insect Cell Purification Table of Contents Suggested Schedule...
OPPF-UK Standard Protocols: Insect Cell Purification Last Updated 6 th October 2016 Joanne Nettleship joanne@strubi.ox.ac.uk OPPF-UK SOP: Insect Cell Purification Table of Contents Suggested Schedule...
SERVA Ni-NTA Magnetic Beads
 INSTRUCTION MANUAL SERVA Ni-NTA Magnetic Beads Magnetic beads for Affinity Purification of His-Tag Fusion Proteins (Cat. No. 42179) SERVA Electrophoresis GmbH - Carl-Benz-Str. 7-69115 Heidelberg Phone
INSTRUCTION MANUAL SERVA Ni-NTA Magnetic Beads Magnetic beads for Affinity Purification of His-Tag Fusion Proteins (Cat. No. 42179) SERVA Electrophoresis GmbH - Carl-Benz-Str. 7-69115 Heidelberg Phone
Structural bases for N-glycan processing by mannosidephosphorylase
 Supporting information Volume 71 (2015) Supporting information for article: Structural bases for N-glycan processing by mannosidephosphorylase Simon Ladevèze, Gianluca Cioci, Pierre Roblin, Lionel Mourey,
Supporting information Volume 71 (2015) Supporting information for article: Structural bases for N-glycan processing by mannosidephosphorylase Simon Ladevèze, Gianluca Cioci, Pierre Roblin, Lionel Mourey,
Strep-Spin Protein Miniprep Kit Catalog No. P2004 & P2005
 INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004 & P2005 Highlights Fast & Simple: Purify Strep-tagged proteins from cell-free extracts using a spin-column in 7 minutes High-Quality:
INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004 & P2005 Highlights Fast & Simple: Purify Strep-tagged proteins from cell-free extracts using a spin-column in 7 minutes High-Quality:
MagExtactor -His-tag-
 Instruction manual MagExtractor-His-tag-0905 F0987K MagExtactor -His-tag- Contents NPK-701 100 preparations Store at Store at 4 C [1] Introduction [2] Components [3] Materials required [4] Protocol3 1.
Instruction manual MagExtractor-His-tag-0905 F0987K MagExtactor -His-tag- Contents NPK-701 100 preparations Store at Store at 4 C [1] Introduction [2] Components [3] Materials required [4] Protocol3 1.
Supplemental Experimental Procedures
 Supplemental Experimental Procedures Cloning, expression and purification of untagged P C, Q D -N 0123, Q D- N 012 and Q D -N 01 - The DNA sequence encoding non tagged P C (starting at the Arginine 57
Supplemental Experimental Procedures Cloning, expression and purification of untagged P C, Q D -N 0123, Q D- N 012 and Q D -N 01 - The DNA sequence encoding non tagged P C (starting at the Arginine 57
Prac Results: Alkaline Phosphatase
 The Steady State Assumption BMED282 U2L3 Enzyme s and Regulation [conc] Product Substrate Time (min) The Steady State Assumption Prac Results: Alkaline Phosphatase [conc] Total Enzyme added = [] + [Efree]
The Steady State Assumption BMED282 U2L3 Enzyme s and Regulation [conc] Product Substrate Time (min) The Steady State Assumption Prac Results: Alkaline Phosphatase [conc] Total Enzyme added = [] + [Efree]
Roche Molecular Biochemicals Technical Note No. LC 12/2000
 Roche Molecular Biochemicals Technical Note No. LC 12/2000 LightCycler Absolute Quantification with External Standards and an Internal Control 1. General Introduction Purpose of this Note Overview of Method
Roche Molecular Biochemicals Technical Note No. LC 12/2000 LightCycler Absolute Quantification with External Standards and an Internal Control 1. General Introduction Purpose of this Note Overview of Method
GST Fusion Protein Purification Kit
 Glutathione Resin GST Fusion Protein Purification Kit Cat. No. L00206 Cat. No. L00207 Technical Manual No. TM0185 Version 01042012 Index 1. Product Description 2. Related Products 3. Purification Procedure
Glutathione Resin GST Fusion Protein Purification Kit Cat. No. L00206 Cat. No. L00207 Technical Manual No. TM0185 Version 01042012 Index 1. Product Description 2. Related Products 3. Purification Procedure
Synthetic Biology for
 Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost
 Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen University,
Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen University,
AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA
 AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA Encodes hereditary information Used to specify the amino
AP Biology Book Notes Chapter 3 v Nucleic acids Ø Polymers specialized for the storage transmission and use of genetic information Ø Two types DNA Encodes hereditary information Used to specify the amino
RISE Program Workshop in Protein Purification
 RISE Program Workshop in Protein Purification Objectives: The purpose of this workshop is to introduce students to the principles and practice of protein purification. Each afternoon session will consist
RISE Program Workshop in Protein Purification Objectives: The purpose of this workshop is to introduce students to the principles and practice of protein purification. Each afternoon session will consist
HOOK 6X His Protein Purification (Bacteria)
 182PR-02 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Purification (Bacteria) For The Purification Of His-Tagged Proteins
182PR-02 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Purification (Bacteria) For The Purification Of His-Tagged Proteins
nature methods Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates
 nature methods Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates Audur Magnusdottir, Ida Johansson, Lars-Göran Dahlgren, Pär Nordlund & Helena Berglund Supplementary
nature methods Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates Audur Magnusdottir, Ida Johansson, Lars-Göran Dahlgren, Pär Nordlund & Helena Berglund Supplementary
Ni-NTA Agarose. User Manual. 320 Harbor Way South San Francisco, CA Phone: 1 (888) MCLAB-88 Fax: 1 (650)
 Ni-NTA Agarose User Manual 320 Harbor Way South San Francisco, CA 94080 Phone: 1 (888) MCLAB-88 Fax: 1 (650) 871-8796 www. Contents Introduction -----------------------------------------------------------------------
Ni-NTA Agarose User Manual 320 Harbor Way South San Francisco, CA 94080 Phone: 1 (888) MCLAB-88 Fax: 1 (650) 871-8796 www. Contents Introduction -----------------------------------------------------------------------
Supplemental Information
 FOOT-AND-MOUTH DISEASE VIRUS PROTEIN 2C IS A HEXAMERIC AAA+ PROTEIN WITH A COORDINATED ATP HYDROLYSIS MECHANISM. Trevor R. Sweeney 1, Valentina Cisnetto 1, Daniel Bose 2, Matthew Bailey 3, Jon R. Wilson
FOOT-AND-MOUTH DISEASE VIRUS PROTEIN 2C IS A HEXAMERIC AAA+ PROTEIN WITH A COORDINATED ATP HYDROLYSIS MECHANISM. Trevor R. Sweeney 1, Valentina Cisnetto 1, Daniel Bose 2, Matthew Bailey 3, Jon R. Wilson
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids.
 Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Supplementary Figure S1 Purification of deubiquitinases HEK293 cells were transfected with the indicated DUB-expressing plasmids. The cells were harvested 72 h after transfection. FLAG-tagged deubiquitinases
Protocol for in vitro transcription
 Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%)
 1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
Aspergillus niger End-Point PCR Kit Product# EP32900
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Aspergillus niger End-Point PCR Kit Product# EP32900 Product Insert
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Aspergillus niger End-Point PCR Kit Product# EP32900 Product Insert
Inserting genes into plasmids
 Inserting genes into plasmids GENE cut from genome or other plasmid w/ two different enzymes PLASMID cut with same two enzymes BTEC 120 - Molecular & Cell Biotechnology 18 Inserting genes into plasmids
Inserting genes into plasmids GENE cut from genome or other plasmid w/ two different enzymes PLASMID cut with same two enzymes BTEC 120 - Molecular & Cell Biotechnology 18 Inserting genes into plasmids
Lumazine synthase protein cage nanoparticles as modular delivery platforms for targeted drug delivery
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information for Lumazine synthase protein cage nanoparticles as modular delivery
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Supporting Information for Lumazine synthase protein cage nanoparticles as modular delivery
HOOK 6X His Protein Purification (Yeast)
 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Purification (Yeast) For The Purification of His Tagged Proteins from
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Purification (Yeast) For The Purification of His Tagged Proteins from
Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and
 Materials and Methods Immunoprecipitation and immunoblot analysis Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and immunoblot analyses with corresponding
Materials and Methods Immunoprecipitation and immunoblot analysis Proteins were extracted from cultured cells using a modified buffer, and immunoprecipitation and immunoblot analyses with corresponding
Nucleic Acids, Proteins, and Enzymes
 3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
HOOK 6X His Protein Spin Purification (Bacteria)
 222PR 03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Spin Purification (Bacteria) For the Purification of His Tagged
222PR 03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Spin Purification (Bacteria) For the Purification of His Tagged
Refold SK Protein Refolding Kit
 Refold SK Protein Refolding Kit 2.5mg SK v5. 2 Table of Contents Introduction...4 Kit Content...5 Instruction for Use...6 Troubleshooting Guide...9 Application Notes...16 Introduction Overexpression of
Refold SK Protein Refolding Kit 2.5mg SK v5. 2 Table of Contents Introduction...4 Kit Content...5 Instruction for Use...6 Troubleshooting Guide...9 Application Notes...16 Introduction Overexpression of
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Chapter 5: Proteins: Primary Structure
 Instant download and all chapters Test Bank Fundamentals of Biochemistry Life at the Molecular Level 4th Edition Donald Voet https://testbanklab.com/download/test-bank-fundamentals-biochemistry-life-molecular-level-
Instant download and all chapters Test Bank Fundamentals of Biochemistry Life at the Molecular Level 4th Edition Donald Voet https://testbanklab.com/download/test-bank-fundamentals-biochemistry-life-molecular-level-
SUPPLEMENTAL MATERIAL BIOCHEMICAL AND STRUCTURAL STUDIES ON THE M. TUBERCULOSIS O 6 -METHYLGUANINE METHYLTRANSFERASE AND MUTATED VARIANTS
 SUPPLEMENTAL MATERIAL BIOCHEMICAL AND STRUCTURAL STUDIES ON THE M. TUBERCULOSIS O 6 -METHYLGUANINE METHYLTRANSFERASE AND MUTATED VARIANTS Riccardo Miggiano 1, Valentina Casazza 1, Silvia Garavaglia 1,
SUPPLEMENTAL MATERIAL BIOCHEMICAL AND STRUCTURAL STUDIES ON THE M. TUBERCULOSIS O 6 -METHYLGUANINE METHYLTRANSFERASE AND MUTATED VARIANTS Riccardo Miggiano 1, Valentina Casazza 1, Silvia Garavaglia 1,
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Nickel Chelating Resin Spin Columns
 326PR-02 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Nickel Chelating Resin Spin Columns A Ni-IDA IMAC resin for 6X-His Tagged Protein
326PR-02 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Nickel Chelating Resin Spin Columns A Ni-IDA IMAC resin for 6X-His Tagged Protein
Chapter 3 Nucleic Acids, Proteins, and Enzymes
 3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
3 Nucleic Acids, Proteins, and Enzymes Chapter 3 Nucleic Acids, Proteins, and Enzymes Key Concepts 3.1 Nucleic Acids Are Informational Macromolecules 3.2 Proteins Are Polymers with Important Structural
ALP (alkaline phosphatase) calibrators were analyzed manually in microtiter plates to find the linearity range by following this protocol:
 Exam Mol 3008 May 2009 Subject 1 (15p) ALP (alkaline phosphatase) calibrators were analyzed manually in microtiter plates to find the linearity range by following this protocol: Reaction solutions: 50
Exam Mol 3008 May 2009 Subject 1 (15p) ALP (alkaline phosphatase) calibrators were analyzed manually in microtiter plates to find the linearity range by following this protocol: Reaction solutions: 50
Proteomics. Manickam Sugumaran. Department of Biology University of Massachusetts Boston, MA 02125
 Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
SPHK1 Assay Kit. Catalog Number KA assays Version: 05. Intended for research use only.
 SPHK1 Assay Kit Catalog Number KA0906 400 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials Supplied...
SPHK1 Assay Kit Catalog Number KA0906 400 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials Supplied...
For Research Use Only. Not for use in diagnostic procedures.
 Printed December 13, 2011 Version 1.0 For Research Use Only. Not for use in diagnostic procedures. DDDDK-tagged Protein PURIFICATION GEL with Elution Peptide (MoAb. clone FLA-1) CODE No. 3326 / 3327 PURIFICATION
Printed December 13, 2011 Version 1.0 For Research Use Only. Not for use in diagnostic procedures. DDDDK-tagged Protein PURIFICATION GEL with Elution Peptide (MoAb. clone FLA-1) CODE No. 3326 / 3327 PURIFICATION
Analysing protein protein interactions using a GST-fusion protein to pull down the interacting target from the cell lysate Hong Wang and Xin Zeng
 Analysing protein protein interactions using a GST-fusion protein to pull down the interacting target from the cell lysate Hong Wang and Xin Zeng Department of Molecular Genetics, Biochemistry and Microbiology,
Analysing protein protein interactions using a GST-fusion protein to pull down the interacting target from the cell lysate Hong Wang and Xin Zeng Department of Molecular Genetics, Biochemistry and Microbiology,
PI3 Kinase Assay Kit. Catalog Number KA assays Version: 05. Intended for research use only.
 PI3 Kinase Assay Kit Catalog Number KA0904 400 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials Supplied...
PI3 Kinase Assay Kit Catalog Number KA0904 400 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials Supplied...
Cambridge International Examinations Cambridge International Advanced Subsidiary and Advanced Level
 Cambridge International Examinations Cambridge International Advanced Subsidiary and Advanced Level *2249654089* BIOLOGY 9700/21 Paper 2 AS Level Structured Questions October/November 2016 1 hour 15 minutes
Cambridge International Examinations Cambridge International Advanced Subsidiary and Advanced Level *2249654089* BIOLOGY 9700/21 Paper 2 AS Level Structured Questions October/November 2016 1 hour 15 minutes
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Comparative functional proteomics of MAP Kinase signalling. in Magnaporthe oryzae
 Comparative functional proteomics of MAP Kinase signalling in Magnaporthe oryzae Submitted by Romain Huguet to the University of Exeter as a thesis for the degree of Doctor of Philosophy, in Biological
Comparative functional proteomics of MAP Kinase signalling in Magnaporthe oryzae Submitted by Romain Huguet to the University of Exeter as a thesis for the degree of Doctor of Philosophy, in Biological
Technical tips Session 4
 Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
Technical tips Session 4 Biotinylation assay: Streptavidin is a small bacterial protein that binds with high affinity to the vitamin biotin. This streptavidin-biotin combination can be used to link molecules
7.06 Problem Set Four, 2006
 7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
NHS-Activated Agarose (Dry Form)
 560PR-01R G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name NHS-Activated Agarose (Dry Form) For covalent binding of primary amine containing
560PR-01R G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name NHS-Activated Agarose (Dry Form) For covalent binding of primary amine containing
EpiQuik Global Histone H3 Phosphorylation (Ser28) Assay Kit (Colorimetric)
 EpiQuik Global Histone H3 Phosphorylation (Ser28) Assay Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H3 Phosphorylation (Ser28) Assay Kit (Colorimetric)
EpiQuik Global Histone H3 Phosphorylation (Ser28) Assay Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Global Histone H3 Phosphorylation (Ser28) Assay Kit (Colorimetric)
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
AP Biology: Unit 5: Development. Forensic DNA Fingerprinting: Using Restriction Enzymes Bio-Rad DNA Fingerprinting Kit
 Forensic DNA Fingerprinting: Using Restriction Enzymes Bio-Rad DNA Fingerprinting Kit Background: Scientists working in forensic labs are often asked to perform DNA profiling or fingerprinting to analyze
Forensic DNA Fingerprinting: Using Restriction Enzymes Bio-Rad DNA Fingerprinting Kit Background: Scientists working in forensic labs are often asked to perform DNA profiling or fingerprinting to analyze
High-Affinity Ni-NTA Resin
 High-Affinity Ni-NTA Resin Technical Manual No. 0217 Version 20070418 I Description.... 1 II Key Features... 1 III His-Tagged Fusion Protein Purification Procedure.. 1 IV Resin Regeneration. 4 V Troubleshooting...
High-Affinity Ni-NTA Resin Technical Manual No. 0217 Version 20070418 I Description.... 1 II Key Features... 1 III His-Tagged Fusion Protein Purification Procedure.. 1 IV Resin Regeneration. 4 V Troubleshooting...
AminTRAP HIS Prepacked Column
 INDEX Ordering Information... 3 Intended Use... 3 Product Description... 3 Purification Procedure... 4 Sample Preparation... 5 Sample Purification... 6 Analysis... 6 Regeneration Procedure... 6 Use and
INDEX Ordering Information... 3 Intended Use... 3 Product Description... 3 Purification Procedure... 4 Sample Preparation... 5 Sample Purification... 6 Analysis... 6 Regeneration Procedure... 6 Use and
GENE(S) Carried by transposon
 ANSWER KEY Microbial Genetics BIO 510/610 Fall Quarter 2009 Final Exam 1.) Some Insertion Sequences transpose through a Replicative mechanism of transposition. Other Insertion Sequences utilize a Cut and
ANSWER KEY Microbial Genetics BIO 510/610 Fall Quarter 2009 Final Exam 1.) Some Insertion Sequences transpose through a Replicative mechanism of transposition. Other Insertion Sequences utilize a Cut and
Enzymes and Enzymatic Reactions Adapted with permission from the original author, Charles Hoyt
 Biol. 261 Enzymes and Enzymatic Reactions Adapted with permission from the original author, Charles Hoyt Introduction All living things use energy, give off waste, reproduce and interact with the environment.
Biol. 261 Enzymes and Enzymatic Reactions Adapted with permission from the original author, Charles Hoyt Introduction All living things use energy, give off waste, reproduce and interact with the environment.
BC 367, Exam 2 November 13, Part I. Multiple Choice (3 pts each)- Please circle the single best answer.
 Name BC 367, Exam 2 November 13, 2008 Part I. Multiple Choice (3 pts each)- Please circle the single best answer. 1. The enzyme pyruvate dehydrogenase catalyzes the following reaction. What kind of enzyme
Name BC 367, Exam 2 November 13, 2008 Part I. Multiple Choice (3 pts each)- Please circle the single best answer. 1. The enzyme pyruvate dehydrogenase catalyzes the following reaction. What kind of enzyme
Peptide deformylase from superbacteria
 Peptide deformylase from superbacteria Antibiotics Most antibiotics were originally isolated from soil-derived actinomycetes between 1940s and 1960s (Golden era of antibiotic discovery) Natural product
Peptide deformylase from superbacteria Antibiotics Most antibiotics were originally isolated from soil-derived actinomycetes between 1940s and 1960s (Golden era of antibiotic discovery) Natural product
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Strep-Tactin XT Spin Column
 Strep-Tactin XT Spin Column Purification Protocol Last date of revision Last June date 2017 of revision June 2017 Version PR90-0001 Version PR90-0001 For research use only Important licensing information
Strep-Tactin XT Spin Column Purification Protocol Last date of revision Last June date 2017 of revision June 2017 Version PR90-0001 Version PR90-0001 For research use only Important licensing information
High-Affinity Ni-NTA Resin
 High-Affinity Ni-NTA Resin Technical Manual No. 0237 Version 20070418 I Description.. 1 II Key Features... 1 III His-Tagged Fusion Protein Purification Procedure 1 IV Resin Regeneration. 4 V Troubleshooting...
High-Affinity Ni-NTA Resin Technical Manual No. 0237 Version 20070418 I Description.. 1 II Key Features... 1 III His-Tagged Fusion Protein Purification Procedure 1 IV Resin Regeneration. 4 V Troubleshooting...
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2015-2016 FUNDAMENTALS OF MOLECULAR BIOLOGY AND GENETICS BIO-4003A Time allowed: 2 hours Answer ALL questions in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2015-2016 FUNDAMENTALS OF MOLECULAR BIOLOGY AND GENETICS BIO-4003A Time allowed: 2 hours Answer ALL questions in Section
BIOC 463A Expt. 4: Column Chromatographic Methods Column Chromatography
 Column Chromatography Chromatography is the process use to separate molecules based on SOME physical property of the molecule: Mass (i.e. size) Charge Affinity for ligands or substrates Hydrophobic interactions
Column Chromatography Chromatography is the process use to separate molecules based on SOME physical property of the molecule: Mass (i.e. size) Charge Affinity for ligands or substrates Hydrophobic interactions
Fig. 1. A schematic illustration of pipeline from gene to drug : integration of virtual and real experiments.
 1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
ProteIndex Chemical-Tolerant Ni-Penta Agarose. Prepacked Cartridge. 6 FF Prepacked Cartridge, 5 x 1 ml settled resin
 Store product at 2 C 8 C. Do not freeze. The product is shipped at ambient temperature. ProteIndex TM Chemical-Tolerant Ni-Penta Agarose 6 FF Prepacked Cartridge Cat. No. 11-0229-5x1ML 11-0229-1x5ML 11-0229-5x5ML
Store product at 2 C 8 C. Do not freeze. The product is shipped at ambient temperature. ProteIndex TM Chemical-Tolerant Ni-Penta Agarose 6 FF Prepacked Cartridge Cat. No. 11-0229-5x1ML 11-0229-1x5ML 11-0229-5x5ML
Index 1. Product Description 2. Purification Procedure 3. Troubleshooting 4. Ordering Information
 High Affinity Ni-Charged Resin Cat. No. L00223 Technical Manual No. TM0217 Version 07132010 Index 1. Product Description 2. Purification Procedure 3. Troubleshooting 4. Ordering Information 1. Product
High Affinity Ni-Charged Resin Cat. No. L00223 Technical Manual No. TM0217 Version 07132010 Index 1. Product Description 2. Purification Procedure 3. Troubleshooting 4. Ordering Information 1. Product
Bacterial PE LB. Bacterial Protein Extraction Lysis Buffer. (Cat. # , , , , , )
 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Bacterial PE LB Bacterial Protein Extraction Lysis Buffer (Cat. # 786-176, 786-177, 786-185,
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Bacterial PE LB Bacterial Protein Extraction Lysis Buffer (Cat. # 786-176, 786-177, 786-185,
SUPPLEMENTARY INFORMATION. The nucleotide binding dynamics of MSH2/MSH3 are lesion-dependent.
 SUPPLEMENTARY INFORMATION The nucleotide binding dynamics of /MSH are lesion-dependent. Barbara A. L. Owen, Walter H. Lang, and Cynthia T. McMurray* mau 1 8 6 4 2 mau 8 6 4 2 mau 8 6 4 2 mau 8 7 6 5 4
SUPPLEMENTARY INFORMATION The nucleotide binding dynamics of /MSH are lesion-dependent. Barbara A. L. Owen, Walter H. Lang, and Cynthia T. McMurray* mau 1 8 6 4 2 mau 8 6 4 2 mau 8 6 4 2 mau 8 7 6 5 4
Supplementary Note 1. Enzymatic properties of the purified Syn BVR
 Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Supplementary Note 1. Enzymatic properties of the purified Syn BVR The expression vector pet15b-syn bvr allowed us to routinely prepare 15 mg of electrophoretically homogenous Syn BVR from 2.5 L of TB-medium
Enzyme kinetics. Irreversible inhibition inhibitor is bound tightly to enzyme - very slow dissociation can be covalent or non covalently bound
 Enzymes can be regulated by acceleration and inhibition inhibition very common - several different mechanisms competitive / non competitive reversible / irreversible Irreversible inhibition inhibitor is
Enzymes can be regulated by acceleration and inhibition inhibition very common - several different mechanisms competitive / non competitive reversible / irreversible Irreversible inhibition inhibitor is
6 pts. B) State two mechanisms to explain the effect shown in the figure.
 Question 1. (10 points) The rate of uptake of glucose into rat adipocytes is increased by addition of insulin to the medium. The increase reaches a maximum within 30 seconds at 37 C and is energy dependent.
Question 1. (10 points) The rate of uptake of glucose into rat adipocytes is increased by addition of insulin to the medium. The increase reaches a maximum within 30 seconds at 37 C and is energy dependent.
Polymerase Chain Reaction
 Polymerase Chain Reaction Amplify your insert or verify its presence 3H Taq platinum PCR mix primers Ultrapure Water PCR tubes PCR machine A. Insert amplification For insert amplification, use the Taq
Polymerase Chain Reaction Amplify your insert or verify its presence 3H Taq platinum PCR mix primers Ultrapure Water PCR tubes PCR machine A. Insert amplification For insert amplification, use the Taq
Bio Course Syllabus Spring Biology Spring Biology Second Year Laboratory
 Biology 216 - Spring 2015 - Biology Second Year Laboratory Laboratory Sections. CRN 55840-2 W 2:00 pm 4:50 pm ISC 107, 302, 304 Instructors. Dr. Robert. D. Simon, Room ISC 358 Office Hours: W 10:00-12:00,
Biology 216 - Spring 2015 - Biology Second Year Laboratory Laboratory Sections. CRN 55840-2 W 2:00 pm 4:50 pm ISC 107, 302, 304 Instructors. Dr. Robert. D. Simon, Room ISC 358 Office Hours: W 10:00-12:00,
Your first goal is to determine the order of assembly of the 3 DNA polymerases during initiation.
 MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
MIT Department of Biology 7.28, Spring 2005 - Molecular Biology 1 Question 1 You are studying the mechanism of DNA polymerase loading at eukaryotic DNA replication origins. Unlike the situation in bacteria,
SUMOstar Gene Fusion Technology
 Gene Fusion Technology NEW METHODS FOR ENHANCING FUNCTIONAL PROTEIN EXPRESSION AND PURIFICATION IN INSECT CELLS White Paper June 2007 LifeSensors Inc. 271 Great Valley Parkway Malvern, PA 19355 www.lifesensors.com
Gene Fusion Technology NEW METHODS FOR ENHANCING FUNCTIONAL PROTEIN EXPRESSION AND PURIFICATION IN INSECT CELLS White Paper June 2007 LifeSensors Inc. 271 Great Valley Parkway Malvern, PA 19355 www.lifesensors.com
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Ver AccuPrep His-tagged Protein Purification kit. Safety Warnings and Precautions. Warranty and Liability. Cat. No. K-7200
 Ver. 1.0 Safety Warnings and Precautions AccuPrep His-tagged protein purification kit is developed and sold for research purposes only. It is not recommended for human or animal diagnostic use, unless
Ver. 1.0 Safety Warnings and Precautions AccuPrep His-tagged protein purification kit is developed and sold for research purposes only. It is not recommended for human or animal diagnostic use, unless
CHAPTER 4 Cloning, expression, purification and preparation of site-directed mutants of NDUFS3 and NDUFS7
 CHAPTER 4 Cloning, expression, purification and preparation of site-directed mutants of NDUFS3 and NDUFS7 subunits of human mitochondrial Complex-I Q module N DUFS2, 3, 7 and 8 form the core subunits of
CHAPTER 4 Cloning, expression, purification and preparation of site-directed mutants of NDUFS3 and NDUFS7 subunits of human mitochondrial Complex-I Q module N DUFS2, 3, 7 and 8 form the core subunits of
Cambridge International Examinations Cambridge International Advanced Subsidiary and Advanced Level
 Cambridge International Examinations Cambridge International Advanced Subsidiary and Advanced Level *4506784563* BIOLOGY 9700/22 Paper 2 AS Level Structured Questions February/March 2017 1 hour 15 minutes
Cambridge International Examinations Cambridge International Advanced Subsidiary and Advanced Level *4506784563* BIOLOGY 9700/22 Paper 2 AS Level Structured Questions February/March 2017 1 hour 15 minutes
HOUR EXAM I BIOLOGY 422 FALL, In the spirit of the honor code, I pledge that I have neither given nor received help on this exam.
 Name First Last (Please Print) PID Number - HOUR EXAM I BIOLOGY 422 FALL, 2011 In the spirit of the honor code, I pledge that I have neither given nor received help on this exam. 1 Signature 2 3 4 5 6
Name First Last (Please Print) PID Number - HOUR EXAM I BIOLOGY 422 FALL, 2011 In the spirit of the honor code, I pledge that I have neither given nor received help on this exam. 1 Signature 2 3 4 5 6
Lecture 22: Questions from Quiz 2 and the Trypsin Digestion Experiment
 Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2018 Lecture 22: Questions from Quiz 2 and the Trypsin Digestion Experiment 29 March 2018 c David P. Goldenberg University of Utah goldenberg@biology.utah.edu
Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2018 Lecture 22: Questions from Quiz 2 and the Trypsin Digestion Experiment 29 March 2018 c David P. Goldenberg University of Utah goldenberg@biology.utah.edu
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Technical tips Session 5
 Technical tips Session 5 Chromatine Immunoprecipitation (ChIP): This is a powerful in vivo method to quantitate interaction of proteins associated with specific regions of the genome. It involves the immunoprecipitation
Technical tips Session 5 Chromatine Immunoprecipitation (ChIP): This is a powerful in vivo method to quantitate interaction of proteins associated with specific regions of the genome. It involves the immunoprecipitation
BIOTECHNOLOGY : PRINCIPLES AND PROCESSES
 CHAPTER 11 BIOTECHNOLOGY : PRINCIPLES AND PROCESSES POINTS TO REMEMBER Bacteriophage : A virus that infects bacteria. Bioreactor : A large vessel in which raw materials are biologically converted into
CHAPTER 11 BIOTECHNOLOGY : PRINCIPLES AND PROCESSES POINTS TO REMEMBER Bacteriophage : A virus that infects bacteria. Bioreactor : A large vessel in which raw materials are biologically converted into
Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from
 Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
Supplemental Figure 1. Secondary structure of hvps4b (above) and sequence alignments of VPS4 proteins from different species as well as four other representative members of the meiotic clade of AAA ATPases.
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Purification and biochemical characterization of acid phosphatase-i from seeds of Nelumbo nucifera Sanaullah Khan a*, Shahnaz Asmat c, Sajida Batool a, Mushtaq Ahmed b a Department
SUPPLEMENTARY MATERIAL Purification and biochemical characterization of acid phosphatase-i from seeds of Nelumbo nucifera Sanaullah Khan a*, Shahnaz Asmat c, Sajida Batool a, Mushtaq Ahmed b a Department
TECHNICAL BULLETIN. HIS-Select HF Nickel Affinity Gel. Catalog Number H0537 Storage Temperature 2 8 C
 HIS-Select HF Nickel Affinity Gel Catalog Number H0537 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description HIS-Select High Flow (HF) is an immobilized metal-ion affinity chromatography (IMAC)
HIS-Select HF Nickel Affinity Gel Catalog Number H0537 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description HIS-Select High Flow (HF) is an immobilized metal-ion affinity chromatography (IMAC)
HELINI Hepatitis B virus [HBV] Real-time PCR Kit (Genotype A to H)
![HELINI Hepatitis B virus [HBV] Real-time PCR Kit (Genotype A to H) HELINI Hepatitis B virus [HBV] Real-time PCR Kit (Genotype A to H)](/thumbs/87/95094490.jpg) HELINI Hepatitis B virus [HBV] Real-time PCR Kit (Genotype A to H) Quantitative In vitro diagnostics Instruction manual Cat. No: 8001-25/50/100 tests Compatible with: Agilent, Bio-Rad, Applied Bio systems
HELINI Hepatitis B virus [HBV] Real-time PCR Kit (Genotype A to H) Quantitative In vitro diagnostics Instruction manual Cat. No: 8001-25/50/100 tests Compatible with: Agilent, Bio-Rad, Applied Bio systems
Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further. (Promega) and DpnI (New England Biolabs, Beverly, MA).
 175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
175 Appendix III Chapter 4 Methods General. Unless otherwise noted, reagents were purchased from the commercial suppliers Fisher (Fairlawn, NJ) and Sigma-Aldrich (St. Louis, MO) and were used without further
Glutathione Resin. (Cat. # , , , ) think proteins! think G-Biosciences
 191PR 05 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Glutathione Resin (Cat. # 786 280, 786 310, 786 311, 786 312) think proteins! think
191PR 05 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Glutathione Resin (Cat. # 786 280, 786 310, 786 311, 786 312) think proteins! think
1. A scientist has identified a new membrane receptor. He wishes to determine the hormone-binding
 1. A scientist has identified a new membrane receptor. He wishes to determine the hormone-binding specificity of the receptor. Three different hormones, X, Y, and Z, were mixed with the receptor in separate
1. A scientist has identified a new membrane receptor. He wishes to determine the hormone-binding specificity of the receptor. Three different hormones, X, Y, and Z, were mixed with the receptor in separate
Biotechnology. EXAM INFORMATION Items. Points. Prerequisites. Course Length. Career Cluster EXAM BLUEPRINT. Performance Standards
 EXAM INFORMATION Items 71 Points 71 Prerequisites BIOLOGY OR CHEMISTRY Course Length DESCRIPTION Biotechnology is designed to create an awareness of career possibilities in the field of biotechnology.
EXAM INFORMATION Items 71 Points 71 Prerequisites BIOLOGY OR CHEMISTRY Course Length DESCRIPTION Biotechnology is designed to create an awareness of career possibilities in the field of biotechnology.
