Simultaneous analysis of relative protein expression levels across multiple samples using itraq isobaric tags with 2D nano LC MS/MS
|
|
|
- Beatrix Chambers
- 6 years ago
- Views:
Transcription
1 Simultaneous analysis of relative protein expression levels across multiple samples using itraq isobaric tags with 2D nano LC MS/MS Richard D Unwin 1,2, John R Griffiths 1 & Anthony D Whetton 1 1 Stem Cell & Leukaemia Proteomics Laboratory, School of Cancer and Enabling Sciences, Faculty of Medical and Human Sciences, University of Manchester, Manchester Academic Health Science Centre, Wolfson Molecular Imaging Centre, Manchester, UK. 2 Centre for Advanced Diagnostics and Experimental Therapeutics, Manchester NIHR Biomedical Research Centre, Central Manchester University Hospitals NHS Foundation Trust, Manchester, UK. Correspondence should be addressed to R.D.U. (r.unwin@manchester.ac.uk). Published online 26 August 2010; doi: /nprot In this paper, we describe the use of itraq (isobaric Tags for Relative and Absolute Quantitation) tags for comparison of protein expression levels between multiple samples. These tags label all peptides in a protein digest before labeled samples are pooled, fractionated and analyzed using mass spectrometry (MS). As the tags are isobaric, the intensity of each peak is the sum of the intensity of this peptide from all samples, providing a moderate enhancement in sensitivity. On peptide fragmentation, amino-acid sequence ions also show this summed intensity, providing a sensitivity enhancement. However, the distinct distribution of isotopes in the tags is such that, on further fragmentation, a tag-specific reporter ion is released. The relative intensities of these ions represent the relative amount of peptide in the analytes. Integration of the relative quantification data for the peptides allows relative quantification of the protein. This protocol discusses the rationale behind design, optimization and performance of experiments, comparing protein samples using itraq chemistries combined with strong cation exchange chromatographic fractionation and MS. INTRODUCTION The incorporation of isobaric tags is a widely used strategy for obtaining relative quantification of peptides. Samples are derivatized with one of several chemically identical itraq (isobaric Tags for Relative and Absolute Quantification) tags. Critically, these have identical overall mass but vary in terms of the distribution of heavy carbon and nitrogen isotopes within their structure. Therefore, when samples are pooled, the same peptide from different samples will appear at the same mass-to-charge ratio in a mass spectrum. However, on fragmentation of labeled peptides by collision-induced dissociation, peptide fragments generate aminoacid sequence information (inferring peptide identity), whereas itraq tags fragment to release tag-specific (and sample-specific) reporter ions. The ratios of these reporter ions are representative of the proportions of each peptide in the individual samples. Up to eight samples can be analyzed in tandem using this methodology. This protocol is complementary to and extends documents supplied by AB Sciex, including the Protocol 1 ( com/literature/cms_ pdf) and Chemistry Reference Guide 2 (available from AB Sciex, part no ). The principle of using this type of chemistry for relative quantification in proteomics was demonstrated by Thompson et al. 3. They synthesized peptides containing a tandem mass tag and showed that this strategy could indeed be used to obtain relative quantification in tandem MS experiment. Ross et al. 4 then published a similar approach using itraq, in which they described a tag that contained an amine-reactive moiety enabling reaction with any peptide. This protocol deals exclusively with the itraq tagging technology, available as either four-channel or eight-channel reagents. The four-channel reagents have nominal reporter ion masses at m/z. The eight-channel contains reporters at and 121 m/z (120 is not used because of the presence of phenylalanine immonium ion at this m/z value). Practically, we have experienced little difference between the two types of reagents. The itraq technology has proved to be successful in numerous experimental contexts. The primary advantage over an in vitro labeling strategy such as SILAC (stable isotope labeling by amino acids in cell culture) is that itraq strategies are applicable to primary samples, e.g., human biofluids 5 7 and disease tissues 8 10 or primary tissues from animal models 11. Although SILAC allows sample mixing before organelle fractionation, thus decreasing handling errors in theory, by our method (using appropriately designed samplesplitting experiments in which a cell pellet is halved and each aliquot processed in an identical manner before labeling with different itraq reagents and quantifying the errors introduced during the procedure), the biochemist can fractionate biological material effectively with minor errors before itraq labeling. However, for protein enrichment methodologies that are less reproducible, the ability to pool sample before labeling, which is possible by SILAC, perhaps makes it the method of choice if the experiment is being performed in cultured cells. The multiplex nature of the reagent is also advantageous over other chemical labeling methodologies, such as labeling with 18 O during digestion 12 or with 13 C-acrylamide 13, as by their very nature these experiments are limited to binary comparisons. This ability to analyze multiple samples simultaneously facilitates time course studies 14,15, or allows a larger number of samples to be analyzed against the same control (or analysis of biological replicate samples) simultaneously, saving on sample preparation and analysis time 16 and improving experimental design. As the itraq reagents essentially label all peptides in a sample, it is ideally suited to the quantification of posttranslational modifications 17 19, and may also be used to label whole proteins before subsequent protein fractionation steps, if required 20. As with many proteomics methods for relative quantification, success in this technique is dependent on sample preparation. The peptide-reactive group on the itraq tags is a N-hydroxysuccinimide moiety, which will label peptides through free amine groups, 1574 VOL.5 NO nature protocols
2 namely, those at the N-terminus and on lysine side chains. As a result, it is important to ensure that buffers used to prepare protein before labeling do not contain free amines; hence, common reagents such as Tris and ammonium bicarbonate should be avoided. This chemistry will also react with cysteine, hence the requirement that they are blocked before labeling, and, at lower ph, can also react with tyrosine residues. As neither of these reactions is 100% efficient, they should be minimized by careful design of sample buffer systems. For example, ensuring that the buffering capacity of the labeling buffer is sufficient to maintain the ph > 7.5 during labeling will minimize reaction with tyrosine residues to a large degree. Moreover, when performing large-scale quantitation experiments, it is important to determine the technical variation introduced through the sample handling procedure. To test this, it is recommended that each experiment use all channels, with at least two of the channels occupied by technical or biological replicates. If this is not possible, it is recommended that a separate experiment be performed containing such replicates to assess interexperimental variation. Experimental design When performing itraq analyses, it is recommended that careful consideration be given to the samples that are labeled in order to obtain the highest possible confidence in the quantification data, allowing protein differences to be assigned with confidence. We routinely use all eight channels in any experiment, and include at least one replicate sample that enables the assessment of technical and/or biological variation. Analysis of the variation of ratios from such replicates, in which all proteins should theoretically be present in a 1:1 ratio, allows the investigator to determine, at a global level, how reproducible the sample handling, labeling and analysis are, and what values can be reliably used as cutoffs by determining the boundaries within which > 95% of the control data lie (therefore, a 5% probability that a protein lies outside of these limits by chance). This replicate can also be used on an individual protein basis, with proteins in which the control ratio deviates significantly from 1 being identified as having potentially poor quality quantification. In instances in which a study consists of less than eight samples, this design is relatively simple. For studies with more than eight samples, it is recommended that a reference sample be obtained by pooling equivalent amounts of each sample in the study and running replicates of this sample in the eight-plex, alongside six individual test samples. Comparison of ratios of each sample versus the pooled reference allows protein expression across different itraq experiments to be assessed. It is also important that the whole process be replicated to obtain statistical confidence in peptide/protein changes. Sample preparation and protein digestion. Before labeling, ensure that protein samples are prepared in the absence of compounds containing free amine groups such as Tris and ammonium bicarbonate. These will react with itraq-labeling reagents at a later stage in the protocol. A suitable cell lysis buffer for cultured mammalian cells consists of 1 M triethylammonium bicarbonate (TEAB) with up to 0.1% (wt/vol) SDS. If this buffer is unsuitable for the sample under investigation, alternative buffers/detergents can be used. However, if they contain components that are likely to interfere with itraq labeling (amine based) or trypsin activity, these components should be removed by buffer exchange before analysis. Alternative buffers should be tested by processing a standard protein mixture before performing a full experiment. Once the protein concentration has been determined using a suitable protein assay method, equal amounts of protein should be labeled. Each vial of itraq reagent will label up to 100 µg of peptide, although it is possible to obtain data after labeling as little as 10 µg of protein. It is important to ensure that samples are in equal volumes before digestion and labeling, and that the final concentration of SDS during digestion is 0.05% (wt/vol), as concentrations > 0.1% (wt/vol) will inhibit trypsin activity. Additional 1 M TEAB may be added to reduce the SDS concentration if necessary. Note that we recommend the use of 1 M TEAB throughout the protocol, as this buffer is easily removed at the end of the experiment and as such the increase in concentration has no deleterious effect on the protocol (while providing greater buffering capacity than the 0.5 M TEAB supplied by AB Sciex), minimizing itraqlabeling of tyrosine residues. For complex samples such as biological fluids or whole-cell lysates/subcellular fractions, pooled labeled peptides should be prefractionated off-line before analysis by liquid chromatography/ tandem mass spectrometry (LC MS/MS). In this protocol, we describe the use of strong cation exchange (SCX) fractionation for this purpose. Although other modes of chromatography are equally applicable, each should be carefully tested to allow optimal separation of the specific sample type and compatibility with downstream separation tools. When analyzing labeled peptide samples in a mass spectrometer, collision energy settings are very important in obtaining good itraq data, and a compromise often has to be made between obtaining good reporter ions and good peptide sequence ions. Similarly, it is important to ensure that LC conditions are such that all salt content is removed before injection onto the analytical column, and again these features of the method should be optimized and tested before commencement of a full experiment. MATERIALS REAGENTS Triethylammonium bicarbonate (TEAB, supplied as 1 M stock; Sigma-Aldrich, cat. no. T7408) SDS (Sigma-Aldrich, cat. no. L3771). It is dissolved at 0.1% (wt/vol) concentration in 1 M TEAB, with the remainder discarded after use. itraq reagent-labeling kit (AB Sciex, cat. nos (four-plex) and (eight-plex)). Store at 20 C CRITICAL itraq reagents are extremely susceptible to hydrolysis. Therefore, ensure that caps are tight on arrival, and take care to prevent reagent freeze/thawing when removing aliquots from the freezer. Tris-(2-carboxyethyl)phosphine (TCEP; Sigma, cat. no. C4706). Stock of 50 mm is prepared in water and stored at 20 C. Methylmethanethiosulfate (Thermo Scientific, cat. no ). Stock of 200 mm is prepared in isopropanol and stored at 20 C. Sequencing-grade modified trypsin (Promega, cat. no. V5280) Water (HPLC grade; Rathburn, cat. no. RH1020) Potassium chloride (Sigma-Aldrich, cat. no. P9541) Ethanol (Fisher, cat. no. E/0650DF/17) Isopropanol (Sigma-Aldrich, cat. no. I9516) Hydrochloric acid (supplied at 37% (vol/vol) (~4.1 M); Sigma-Aldrich, cat. no. H1758) Acetonitrile (HPLC grade; Fisher, cat. no. A062617)! CAUTION It is toxic and flammable. Use it in a fume hood. Handle using gloves and goggles. nature protocols VOL.5 NO
3 Formic acid (Fluka, cat. no )! CAUTION It is corrosive. Use it in a fume hood. Handle using gloves and goggles. EQUIPMENT Off-line LC system (LC Packings, see EQUIPMENT SETUP) Nano LC Pump (LC Packings, see EQUIPMENT SETUP) QStar Q-ToF mass spectrometer (AB Sciex, see EQUIPMENT SETUP) CRITICAL Any mass spectrometer capable of performing MS/MS analysis of peptides and resolving the itraq reporter ions (m/z ) can be used for this analysis, although data analysis workflows will be instrument dependent. Before a complete analysis, it is recommended that defined mixtures of 8 10 proteins, with 6 8 proteins at the same level and 2 4 at variable levels, be prepared and analyzed to ensure that the system is suitable and quantification accuracy is acceptable. ProteinPilot software (AB Sciex, cat. no ) CRITICAL ProteinPilot currently only supports protein identification and quantification from data generated using AB Sciex instruments (although ProteinPilot v3 will perform identification only on Mascot generic format (.mgf) files generated from non- AB Sciex platforms). For other instrument platforms, an alternative data analysis package is required. A good list of alternatives is described in reference 19. SpeedVac (Labconco, cat. no ) Electrospray emitters (New Objective, cat. no. FS CE-50) Glass sample vials with lids (Chromacol, cat. no. 03-FISV) Heater block (Scientific Laboratory Supplies, cat. no. BLO1312) Acclaim PepMap C18 (5 µm, 100 Å trapping column, 5 mm 300 µm internal diameter; LC Packings, cat. no ) Pepmap C18 (3 µm, 100 Å analytical column, 15 cm 75 µm internal diameter; LC Packings, cat. no ) SCX chromatography column ( cm polysulfoethyl A column, 5 µm beads, 200 Å pore size; PolyLC, cat. no. 102SE0502) REAGENT SETUP LC buffers Prepare buffers fresh every 1 2 d, using clean glassware retained only for this purpose. SCX chromatography buffer A (SCX-A) is composed of 20% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid (ph 2.7). SCX chromatography buffer B (SCX-B) is composed of 20% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid and 1 M KCl (ph 2.7). Reversed-phase chromatography buffer A (RP-A) is composed of 2% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid in water (97.9%), and is used as both a sample-loading buffer and as a component of the reversedphase chromatography gradient. Reversed-phase chromatography buffer B (RP-B) is composed of 80% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid in water (19.9%). Samples Samples can be prepared in advance and stored at 80 C for at least 6 months before labeling. Extracted protein should be in an amine-free buffer, ideally prepared using the conditions described above, i.e., 1 M TEAB with 0.1% (wt/vol) SDS. Should this buffer prove inappropriate for some sample types, alternative buffers can be used, e.g., urea, alternative detergents and so on, but they must be amine free and the components must not interfere with trypsin digestion. Samples that have been successfully analyzed in our laboratory include human and mouse plasma, mammalian cell lines and primary murine hematopoietic cells, along with commercially available purified proteins. It is advisable to try to label a simple protein mixture with any new buffer (alongside the same mixture in 1 M TEAB with 0.1% SDS) before using it for sample lysis and labeling. EQUIPMENT SETUP Offline LC LC Packings ISC-3000 SP pump, with a FAMOS autosampler and a UV detector. Autosampler setup consists of a 1-ml syringe and a 2-ml sample loop, loading onto a SCX chromatography column ( cm Polysulfoethyl A column, 5 µm beads, 200 Å pore size; PolyLC, cat. no. 102SE0502). Online LC Nano LC system with a FAMOS autosampler, Ultimate gradient pump and SWITCHOS loading pump, with vacuum degasser (now superseded by the Ultimate 3,000 modular LC system). System setup consists of a 25-µl volume syringe, 100-µl sample loop, Acclaim PepMap C18, 5 µm, 100 Å trapping column (5 mm 300 µm internal diameter; LC Packings, cat. no ) and a Pepmap C18, 3 µm, 100 Å analytical column (15 cm 75 µm internal diameter; LC Packings, cat. no ). Mass spectrometer QStar XL (now superseded by the QStar Elite mass spectrometer) equipped with Analyst control software and fitted with a standard NanoSpray II source with a MicroIonSpray II head. PROCEDURE Sample preparation and protein digestion TIMING 2 h, followed by overnight incubation 1 After determining the protein concentration using a suitable assay, aliquot up to 100 µg of protein into separate tubes and equalize sample volumes using 1 M TEAB. CRITICAL STEP Ensure that the protein sample is prepared in a suitable buffer containing no free amine groups. A recommended buffer is 1 M TEAB with 0.1% (wt/vol) SDS, which efficiently lyses mammalian cell lines. CRITICAL STEP Ensure that each sample for labeling contains equal amounts of protein in the same volume of buffer, up to a maximum volume of 75 µl. Ideally, this volume should be as low as possible (20 µl) to minimize downstream handling steps. 2 Reduce cysteine disulfide bonds with 0.1 volume of 50 mm TCEP. Mix on a vortex, pulse spin and incubate at 60 C for 1 h. 3 Alkylate the reduced cysteine residues by adding 0.05 volumes of 200 mm methylmethanethiosulfate. Mix on a vortex, pulse spin and incubate at room temperature (~22 C) for 10 min. 4 To each tube, add 10 µg of trypsin, which has been reconstituted in the supplied buffer and diluted in 20 µl of 1 M TEAB. CRITICAL STEP Ensure that the final concentration of SDS is not > 0.05% (wt/vol) to prevent inhibition of trypsin. TEAB (1 M) should be used to adjust the SDS concentration if necessary. 5 Mix on a vortex, pulse spin and incubate samples at 37 C overnight. Peptide labeling with itraq reagents TIMING 4 5 h 6 If necessary, reduce sample volumes using a SpeedVac at room temperature, avoiding sample drying out, and adjust the volume back up to 30 µl using 1 M TEAB. PAUSE POINT Samples may be stored at 20 C for up to 1 month, although it is recommended that they be labeled and pooled as soon as possible VOL.5 NO nature protocols
4 7 Remove required itraq reagents from the freezer and return unused vials quickly to prevent thawing and formations of condensation. For four-plex reagent experiments, note the correction factors supplied with the itraq kit. Pulse spin reagent vials to ensure that contents are collected at the bottom of the vial. CRITICAL STEP itraq reagent vials should be removed from the freezer and allowed to thaw for only 2 3 min before opening. 8 Add 60 µl of ethanol for a four-plex reagent kit or 60 µl of isopropanol for an eight-plex kit to each reagent vial. Mix and pulse spin. 9 Transfer the entire contents of the reagent vial to the appropriate sample tube and rinse with a further 10 µl of organic solvent (ethanol or isopropanol). 10 Mix each tube on a vortex mixer and pulse spin, then allow to react at room temperature for 1 h if four-plex, or for 2 h if eight-plex. 11 Reduce the volume to ~30 µl on a SpeedVac at room temperature, avoiding sample drying out. PAUSE POINT Samples may be stored at 20 C for 2 3 months with no effect on protein quantification. Sample fractionation using SCX chromatography TIMING 2 h, without sample drying after fractionation 12 Add 100 µl of SCX-A to each sample tube and pool to a single tube. CRITICAL STEP If precipitate is present, add a further 50 µl of SCX-A to dissolve. 13 Rinse each tube with a further 100 µl of SCX-A and add to pooled material. 14 If necessary, adjust ph of pooled samples to ph < 2.7 using 1 M hydrochloric acid. Check using standard laboratory ph paper. CRITICAL STEP For successful binding of peptides to the SCX column, sample ph must be below Transfer the entire sample to a glass sample vial and adjust the volume to 1.6 ml with SCX-A. 16 Load the entire sample onto the analytical column using a 2-ml injection loop and wash with 100% SCX-A at 1 ml min 1 for a minimum of 30 min. CRITICAL STEP Washing is complete once the UV signal has returned to baseline. 17 Separate peptides using a suitable gradient elution at a flow rate of 400 µl min 1. A typical gradient comprises an increase from 0 to 15% SCX-B over 35 min, 15 to 30% SCX-B in 5 min and 30 to 100% SCX-B in 5 min. Hold at 100% SCX-B for 15 min to wash the column and reequilibrate at 100% SCX-A for a final 15 min. This gradient should be sufficient to elute a single peptide (e.g., Glu-Fibrinopeptide) with a peak width of 30 s at baseline. 18 Collect fractions into 1.5-ml tubes at suitable intervals (typically 1 min). Typically, fractions are taken from 5 min to min after the start of the gradient. 19 Dry samples on a SpeedVac to ~20 µl. PAUSE POINT Samples may be stored at 20 C for 2 3 months with no effect on protein quantification. Online reversed-phase nano LC MS/MS analysis of fractions TIMING 3 h per fraction 20 Resuspend sample fractions in RP-A to a volume of 180 µl and centrifuge for 5 min at > 10,000g to remove particulates. 21 Transfer 60 µl to an autosampler vial and load the full amount onto a trapping column for desalting. 22 Wash with 100% RP-A for 40 min at 30 µl min 1. CRITICAL STEP It is important to wash for this amount of time to ensure complete removal of all salts from the sample. 23 Elute peptides over an analytical column at a typical flow rate of nl min 1 using a suitable gradient profile. A typical gradient consists of 2 40% RP-B over 80 min, followed by washing at 100% RP-B for 10 min and a reequilibration with 98% RP-A for 20 min. nature protocols VOL.5 NO
5 CRITICAL STEP The steepness of the elution gradient should be tailored to sample complexity. A typical gradient for a whole-cell lysate is given in Step 23 above. 24 Set up the mass spectrometer to acquire a time-of-flight mass spectrometry scan for 1 s duration, followed by two MS/MS scans of 1.5 s each for the two most abundant precursors. Enable each precursor to be fragmented twice before dynamic exclusion for 2 min. Typical source voltages and collision energy settings, optimized for five distinct peptides from an itraq-labeled mixture of standard proteins (described in ANTICIPATED RESULTS), are as follows: Parameter Setting Curtain gas 10 IonSpray voltage 2,300 V Source temperature 120 C Declustering potential Declustering potential 2 Focusing potential 70 V 15 V 320 V Collision energy, 2 + precursors *m/z + 7 Collision energy, 3 + precursors 0.054*m/z + 5 Collision energy, 4 + precursors 0.054*m/z + 4 MS mass range m/z 400 1,200 MS/MS mass range m/z 50 1,600 (enhance all ON) CRITICAL STEP Instrument settings should be optimized on a known standard before analysis. Settings described here are for guideline purposes only. Key parameters for optimization are collision energies, along with the duration of MS and MS/MS modes, and the number of MS/MS scans per cycle. Good end points for optimization are the number of itraq-labeled and quantified peptides identified, and the average reporter ion area of the identified peptides. Often this requires a trade-off, in which the number of identifications is slightly compromised to acquire sufficient signal from the itraq reporter ions. When using the QStar Elite, it is also important to optimize the Smart IDA criteria and determine whether this feature provides an analytical advantage. Postacquisition data analysis TIMING Variable, depending on data set size 25 Once data have been acquired, all data files are submitted to the ProteinPilot analysis software for peptide identification and quantification, peptide grouping into proteins and protein ratio calculation. Search settings are dependent on sample preparation workflow, but for a whole-proteome analysis using the procedure described above, a suitable search (Paragon) method would be as follows: Parameter Setting Sample type itraq 8-plex (peptide labeled) Cys alkylation MMTS Digestion Trypsin Instrument QSTAR ESI Special factors None Quantitate Checked Bias correction Checked Search effort Thorough ID Detected protein threshold > 0.1 (20%) False discovery rate analysis Checked 1578 VOL.5 NO nature protocols
6 These settings would need to be altered if the sample under analysis was, e.g., from an enriched phosphopeptide population. The low threshold for detected proteins is set to enable more accurate determination of false discovery rate within the experiment. These data can then be used to define an appropriate confidence cutoff for which a protein identification is likely to be correct at a required false discovery rate. If replicate samples are included in the experimental design, technical/biological variation can be assessed and cutoff values imposed for confident protein fold changes 16. CRITICAL STEP Several quality control values should be obtained from this output, such as efficiency of the labeling reaction (labeled lysine residues versus total lysine residues; it should be approaching 100%), efficiency of reduction/alkylation reaction (alkylated cysteine residues versus total cysteine residues; it should be approaching 100%) and protein loading ( Applied bias calculated in the Summary Statistics tab; it should be within 1.2- to 1.3-fold for a typical whole-cell lysate experiment). With a generic tool such as MASCOT, these can be determined using variable modifications in the search criteria. For all data manipulations and statistical calculations, data should be converted into log 2 space. TIMING Steps 1 5, Sample preparation and protein digestion: 2 h, then overnight Steps 6 11, Peptide labeling with itraq reagents: 4 5 h Steps 12 19, Strong cation exchange (SCX) chromatography: 2 h Steps 20 24, Nano LC MS/MS analysis of fractions: 3 h per fraction Step 25, Postacquisition data analysis: variable, depending on data set size Troubleshooting advice can be found in Table 1. Table 1 Troubleshooting table. Step Problem Possible reason Solution 5 Failed digestion Inappropriate trypsin concentration Inappropriate ph for trypsin activity Inhibiting substance in sample buffer Repeat digestion ensuring 10 µg trypsin per 100 µg sample protein Check that the ph of the sample before digestion is between 7.5 and 8.5. Adjust if necessary by addition of 1 M TEAB or by changing buffer (acetone precipitation or buffer exchange) Check sample buffer components and if necessary assess buffer suitability by digesting known protein(s) in solution 9, 25 Failed itraq labeling Reagent has degraded Check storage conditions and shelf life of itraq reagent. If necessary, label a single stock protein resuspended and digested in 1 M TEAB 18 No UV trace in strong cation exchange chromatography 23 Poor resolution reversed phase chromatography Inhibiting substance in sample buffer Digest failed Poor binding to column Problem with LC setup/column or chromatography method Presence of contaminants Overloading of column Problem with LC setup/column or chromatography method Check sample buffer components and if necessary assess buffer suitability by labeling known protein(s) in this buffer solution Check digest by running a proportion of the sample from before and after digest on a gel Ensure that sample ph is < 2.7 before loading Check by performing a separation of a single protein digest or a single peptide in the same buffers/volumes as are used for sample analysis Contaminants binding the column can affect quality of chromatography. Check MS trace for any sign of polymer contamination. Check column integrity postacquisition by analysis of a peptide digest standard Reduce peptide loading, or alternatively source a suitable higher capacity column Check by performing a separation of a single protein digest or a single peptide in the same buffers/volumes as are used for sample analysis (continued) nature protocols VOL.5 NO
7 Table 1 Troubleshooting table (continued). Step Problem Possible reason Solution 24 No signal in MS Poor ionization Check source and needle voltages, and check nanospray emitter. Replace emitter if required 25 Poor reproducibility between replicate samples Low number of peptide/ protein identifications Low proportion of identified peptides/proteins with quantification Wide range of applied bias values Poor peptide fragmentation Poor itraq reporter ion intensities Biological or technical variation Poor peptide fragmentation Poor chromatography Inappropriate MS settings Inefficient itraq labeling Poor detection of itraq reporter ions Unequal amounts of protein loaded at the start of the experiment Variation during sample handling Vastly different samples being analyzed Check collision energy settings and ensure that they are optimal for sample type Check collision energy and ion transmission settings and ensure they are optimal for sample type Check sample preparation and handling methods for sources of error. Minimize sample handling steps, where possible. If problem persists, identify the exact source of variation by taking a single sample through the protocol and splitting it in half at each stage up to the labeling step Check collision energy settings and ensure they are optimal for sample type. Run standard itraq-labeled sample to optimize and check Plot extracted ion chromatographs in MS to check quality of the chromatography. Run standard itraq-labeled sample to optimize and check If the same peptide is fragmented many times, check chromatography (above) and dynamic exclusion criteria. Also check instrument raw sensitivity using a low level injection of a standard protein mix Calculate efficiency of labeling on lysine residues. If poor, see TROUBLESHOOTING advice for Step 9 (above) Check and optimize ion transmission values and collision energy settings using a defined itraq-labeled peptide standard Check protein assay results for the samples and repeat if necessary Check sample handling step and ensure that the number of steps and hence variation remains at a minimum If > 50% of the proteins identified are indeed altered in their concentration, then the calculated applied bias is uninformative and inappropriate. This may be the case for studies looking at, e.g., test and control immunoprecipitations for determination of proteinprotein interactions. In this case, a set of spiked-in standard proteins may be appropriate so that experimental variability can be estimated ANTICIPATED RESULTS For method development, it is important to assess each step in the protocol before conducting a full experiment. This includes testing itraq labeling in the sample buffer, optimizing chromatography gradients for peptide fractionation and optimizing mass spectrometer setting for the acquisition of MS/MS spectra containing product ions for peptide identification and itraq reporter ions for peptide-relative quantification. A good sample to prepare for such optimization is eight simple mixtures, each containing 5 6 proteins, all at the same concentration, and two proteins the levels of which vary between samples. By generating aliquots of such samples, various alterations to the sample preparation buffers can be made relatively easily. We have previously used mixtures containing equimolar amounts of human serotransferrin, BSA, β-lactoglobulin and α-lactalbumin, chick lysozyme and E. coli β-galactosidase. This was supplemented with chick ovotransferrin at 0.5:1.5:1.8: 0.2:1.9:0.1:1, and bovine carbonic anhydrase II at 1.5:0.5:0.2:1.8:0.1:1.9:1. These ratios ensure that the total molar amount of each protein in the itraq-labeled and pooled sample is the same 14. When digested and itraq labeled, this sample can be used to assess the range of separation on the SCX off-line fractionation, although peak width should be assessed using a simple mixture of single peptides, or even a single peptide. Injection of this sample onto the MS allows assessment of the system s sensitivity (on our system, we routinely identify all these proteins at 20 fmol each on the column). To test quantification, more should be injected (200 fmol of each protein on the column). The type of data that this analysis should generate is shown in Figure 1. Here, the mixture was fractionated into eight fractions using a shortened SCX gradient (0 250 mm KCl for 10 min, 1-min fractions collected, and the first and last fractions not analyzed), and each fraction was 1580 VOL.5 NO nature protocols
8 a Intensity, counts c log 2 (ratio) : : : : : : : m/z, Da Precursor ion 300 y y9 200 b3 y b6 100 b1 y b5 y7 y Lysozyme C - Gallus gallus G T D V Q A W I R Expected ratio 1:1:1:1:1:1:1: m/z Carbonic β- anhydrase 2 Serotransferrin galactosidase Serum albumin 1,000 1,200 1,400 Ovotransferrin Intensity, counts log 2 (control:control) 1, ,500 1, fold m/z, Da b y b b y ,000 m/z Unused peptide score Transferrin - Gallus gallus S A G W N I P I G T L L H R Expected ratio 1:0.5:1.5:1.6:0.2:1.9:0.1:1 y8 b y b b6 y y5 y9 Figure 1 Expected data from a typical itraq experiment. (a) Representative spectra from a protein that does not change across eight itraq-labeled samples. (b) Representative spectra from a protein the concentration of which changes across eight itraq-labeled samples. In both spectra, note the strong b-ion series that results from the presence of the N-terminal itraq label. (c) Ratiometric data from the test mixture described in this protocol, showing anticipated results for three of the six proteins whose levels do not change across the experiments and for the two proteins, carbonic anhydrase 2 and ovotransferrin, which do (at 1:1.5:0.5:0.2:1.8:0.1:1.9:1 and 1:0.5:1.5:1.8:0.2:1.9:0.1:1, respectively). These ratios are indicated by diamonds, demonstrating that itraq generally underestimated the degree of fold change, but is successful at identifying differences in protein concentration. β-lactoglobulin, α-lactalbumin and lysozyme are omitted for reasons of clarity. A completed example of this experiment is shown in reference 14. (d) Example of data acquired from a control itraq experiment in which the same complex sample (in this case, a whole-cell lysate) is loaded into two channels. This demonstrates that accuracy of quantification generally increases with the number of peptides identified (represented by unused peptide score, a function of the number and quality of the peptides unique to each protein) and that the majority of the control:control ratios are within b d 2-fold 1.25-fold 2-fold analyzed by MS with an estimated 200 fmol on the column using the conditions described in the protocol. Representative spectra from proteins with no change and with changing levels are shown (Fig. 1a,b). Results of the quantification analysis (protein ratio with 95% confidence intervals) are shown in Figure 1c. Note that itraq quantification underestimated the amount of real fold change between samples, presumably because of low levels of almost isobaric species being fragmented simultaneously. This topic is discussed in more detail elsewhere 21. However, in reality, this does not have an effect on the final data, as those proteins defined by the analysis as changes are usually correct. In addition, this underestimation of fold change is also a feature of other methods of protein quantification, such as SILAC 22. Owing to the wide variety of sample types that can be analyzed using this method, and the wide variety of MS setups that can be used, it is inappropriate to provide a benchmark for the results from a full experiment, although for any data set certain quality control criteria can be assessed. The efficiency of trypsin hydrolysis and itraq labeling can be assessed from the ProteinPilot results file by calculating the number of labeled lysine residues versus total lysine residues. This value should be near 100% (assessing labeling on N-termini is harder, as N-terminal modification or blocking will prevent itraq labeling, plus any peptide degradation after labeling will yield unlabeled N-termini). The efficiency of the reduction/alkylation reaction can be assessed in a similar manner. These studies can be performed before pooling and fractionation if required by taking a 1 µl sample out of each labeling reaction, desalting using a StageTip or ZipTip and analyzing by LC MS/MS, if required. Protein loading and sample handling can be assessed in part by checking the applied bias (value by which all ratios need to be multiplied to generate a median protein ratio of 1.0) calculated in the Summary Statistics tab of ProteinPilot. These values should all be within 1.2- to 1.3-fold for a typical whole-cell lysate experiment. If internal replicates (technical and/or biological) are included within the experiment, these should be compared to allow assessment of ratio cutoffs outside of which a protein/peptide nature protocols VOL.5 NO
9 can be considered as changed. An example is shown in Figure 1d, with the raw data used to construct this figure provided as Supplementary Data. In this experiment, biological replicates of a control mock-transfected cell line (both harvested at the same time as the test -transfected samples) were labeled and included in the eight-plex experiment. Data were extracted and proteins with acceptable quantification properties ( > 3 quantified spectra assigned to each protein, not unique peptide sequences) selected. The log 2 ratio is calculated for each protein and plotted against the unused peptide score (in ProteinPilot, this score is a function of the number of unique peptides identified and their confidence the sum of the log 10 (% confidence) for all unique peptides for MASCOT searches, this could be substituted with the MASCOT score). It is important to check whether the distribution of ratios is approximately normal (around 0) to ensure that the normalization and statistics used during the analysis are appropriate. We typically calculate the values between which > 95% of the wellquantified data points lie, ensuring that the boundaries are set at an equal distance from unity. For a whole-cell lysate (Fig. 1d) in our hands, this routinely yields ratio fold changes of < 0.8 and > 1.25 ( 0.32 < log 2 (ratio) < 0.32). Other groups may model this distribution more effectively and enable generation of cutoffs that are specific to the number of peptides identified (wider for proteins with few peptides, tighter for proteins with many peptides in which quantification is more confident) 21. Note: Supplementary information is available via the HTML version of this article. Acknowledgments This work is funded in part by the Leukaemia and Lymphoma Research Fund, UK, and by the Manchester NIHR Biomedical Research Centre. AUTHOR CONTRIBUTIONS R.D.U. and J.R.G. acquired the data for setup experiments; R.D.U. analyzed the data; R.D.U., J.R.G. and A.D.W. wrote the paper. COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests. Published online at Reprints and permissions information is available online at reprintsandpermissions/. 1. Applied Biosystems. Applied Biosystems itraq TM Reagents: Amine-Modifying Labeling Reagents for Multiplexed Relative and Absolute Protein Quantitation Protocol (Applied Biosystems, Foster City, CA, 2004). 2. Applied Biosystems. itraq Reagents Chemical Reference Guide (Applied Biosystems, Foster City, CA, 2004). 3. Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, (2003). 4. Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, (2004). 5. Hardt, M. et al. Assessing the effects of diurnal variation on the composition of human parotid saliva: quantitative analysis of native peptides using itraq reagents. Anal. Chem. 77, (2005). 6. Ogata, Y. et al. Differential protein expression in male and female human lumbar cerebrospinal fluid using itraq reagents after abundant protein depletion. Proteomics 7, (2007). 7. Kristiansson, M.H., Bhat, V.B., Babu, I.R., Wishnok, J.S. & Tannenbaum, S.R. Comparative time-dependent analysis of potential inflammation biomarkers in lymphoma-bearing SJL mice. J. Proteome Res. 6, (2007). 8. DeSouza, L. et al. Search for cancer markers from endometrial tissues using differentially labeled tags itraq and clcat with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 4, (2005). 9. Bouchal, P. et al. Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (itraq-2dlc-ms/ms) based quantitative proteomic analysis. J. Proteome Res. 8, (2009). 10. Garbis, S.D. et al. Search for potential markers for prostate cancer diagnosis, prognosis and treatment in clinical tissue specimens using amine-specific isobaric tagging (itraq) with two-dimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 7, (2008). 11. Unwin, R.D. et al. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood 107, (2006). 12. Schnölzer, M., Jedrzejewski, P. & Lehmann, W.D. Protease-catalyzed incorporation of 18 O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis 17, (1996). 13. Faca, V. et al. Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. J. Proteome Res. 5, (2006). 14. Williamson, A.J.K. et al. Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol. Cell. Proteomics 7, (2008). 15. Lu, R. et al. Systems-level dynamic analysis of fate change in murine embryonic stem cells. Nature 462, (2009). 16. Pierce, A. et al. Eight-channel itraq enables comparison of the activity of six leukemogenic tyrosine kinases. Mol. Cell. Proteomics 7, (2008). 17. Zhang, Y. et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, (2005). 18. Trinidad, J. Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, (2007). 19. Wolf-Yadlin, A., Hautaniemi, S., Lauffenburger, D.A. & White, F.M. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. USA 104, (2007). 20. Wiese, S., Reidegeld, K.A., Meyer, H.E. & Warscheid, B. Protein labeling by itraq: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7, (2007). 21. Karp, N.A. et al. Addressing accuracy and precision issues in itraq quantitation. Mol. Cell Proteomics, published online (10 April 2010). 22. Ong, S.-E., Kratchmarova, I. & Mann, M. Properties of 13C-substituted arginine in stable isotope labelling by amino acids in cell culture (SILAC). J. Proteome Res. 2, (2003) VOL.5 NO nature protocols
Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium
 Supplementary data itraq-based Proteomics Analyses Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium supplemented with 5g L -1 glucose for 48 hours at 37 C with
Supplementary data itraq-based Proteomics Analyses Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium supplemented with 5g L -1 glucose for 48 hours at 37 C with
Investigating IκB Kinase Inhibition in Breast Cancer Cells
 Investigating IκB Kinase Inhibition in Breast Cancer Cells Biomarker Discovery on the TripleTOF 5600 System Using itraq Reagent Labeling Emma Rusilowicz 1, Thiri Zaw 2, Matthew J. McKay 2, Ted Hupp 1 and
Investigating IκB Kinase Inhibition in Breast Cancer Cells Biomarker Discovery on the TripleTOF 5600 System Using itraq Reagent Labeling Emma Rusilowicz 1, Thiri Zaw 2, Matthew J. McKay 2, Ted Hupp 1 and
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Quantitative mass spec based proteomics
 Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
ProteinPilot Report for ProteinPilot Software
 ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification
 Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification Protein Expression Analysis using the TripleTOF 5600 System and itraq Reagents Vojtech Tambor 1, Christie
Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification Protein Expression Analysis using the TripleTOF 5600 System and itraq Reagents Vojtech Tambor 1, Christie
Peptide enrichment and fractionation
 Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements
 Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Accelerating Throughput for Targeted Quantitation of Proteins/Peptides in Biological Samples
 Technical Note Accelerating Throughput for Targeted Quantitation of Proteins/Peptides in Biological Samples Biomarker Verification Studies using the QTRAP 5500 Systems and the Reagents Triplex Sahana Mollah,
Technical Note Accelerating Throughput for Targeted Quantitation of Proteins/Peptides in Biological Samples Biomarker Verification Studies using the QTRAP 5500 Systems and the Reagents Triplex Sahana Mollah,
Strategies for Quantitative Proteomics. Atelier "Protéomique Quantitative" La Grande Motte, France - June 26, 2007
 Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System
 Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
New Approaches to Quantitative Proteomics Analysis
 New Approaches to Quantitative Proteomics Analysis Chris Hodgkins, Market Development Manager, SCIEX ANZ 2 nd November, 2017 Who is SCIEX? Founded by Dr. Barry French & others: University of Toronto Introduced
New Approaches to Quantitative Proteomics Analysis Chris Hodgkins, Market Development Manager, SCIEX ANZ 2 nd November, 2017 Who is SCIEX? Founded by Dr. Barry French & others: University of Toronto Introduced
Mass Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System
 GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
Agilent Multiple Affinity Removal Spin Cartridges for the Depletion of High-Abundant Proteins from Human Proteomic Samples.
 Agilent Multiple Affinity Removal Spin Cartridges for the Depletion of High-Abundant Proteins from Human Proteomic Samples Instructions Third edition July 2005 General Information Introduction The Agilent
Agilent Multiple Affinity Removal Spin Cartridges for the Depletion of High-Abundant Proteins from Human Proteomic Samples Instructions Third edition July 2005 General Information Introduction The Agilent
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B
 Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins*
 Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
SILAC Protein Quantitation Kits
 INSTRUCTIONS SILAC Protein Quantitation Kits A33969 A33970 A33971 A33972 A33973 MAN0016245 Rev C.0 Pub. Part No. 2161996.8 Number A33969 Description SILAC Protein Quantitation Kit (LysC) DMEM DMEM for
INSTRUCTIONS SILAC Protein Quantitation Kits A33969 A33970 A33971 A33972 A33973 MAN0016245 Rev C.0 Pub. Part No. 2161996.8 Number A33969 Description SILAC Protein Quantitation Kit (LysC) DMEM DMEM for
Improving Sensitivity for an Immunocapture LC-MS Assay of Infliximab in Rat Plasma Using Trap-and-Elute MicroLC-MS
 Improving Sensitivity for an Immunocapture LC-MS Assay of in Rat Plasma Using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest
Improving Sensitivity for an Immunocapture LC-MS Assay of in Rat Plasma Using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest
Universal Solution for Monoclonal Antibody Quantification in Biological Fluids Using Trap-Elute MicroLC-MS Method
 Universal Solution for Monoclonal Antibody Quantification in Biological Fluids Using Trap-Elute MicroLC-MS Method Featuring the SCIEX QTRAP 6500+ LC-MS/MS System with OptiFlow Turbo V source and M5 MicroLC
Universal Solution for Monoclonal Antibody Quantification in Biological Fluids Using Trap-Elute MicroLC-MS Method Featuring the SCIEX QTRAP 6500+ LC-MS/MS System with OptiFlow Turbo V source and M5 MicroLC
Supporting information
 Supporting information Simple and Integrated Spintip-based Technology Applied for Deep Proteome Profiling Wendong Chen,, Shuai Wang, Subash Adhikari, Zuhui Deng, Lingjue Wang, Lan Chen, Mi Ke, Pengyuan
Supporting information Simple and Integrated Spintip-based Technology Applied for Deep Proteome Profiling Wendong Chen,, Shuai Wang, Subash Adhikari, Zuhui Deng, Lingjue Wang, Lan Chen, Mi Ke, Pengyuan
Automated platform of µlc-ms/ms using SAX trap column for
 Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Improving the Detection Limits for Highly Basic Neuropeptides Using CESI-MS
 Improving the Detection Limits for Highly Basic Neuropeptides Using CESI-MS Stephen Lock SCIEX UK Overview Who Should Read This: Senior Scientists and Lab Directors with an Interest in Protein Analysis
Improving the Detection Limits for Highly Basic Neuropeptides Using CESI-MS Stephen Lock SCIEX UK Overview Who Should Read This: Senior Scientists and Lab Directors with an Interest in Protein Analysis
Biotherapeutic Non-Reduced Peptide Mapping
 Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
SUPPLEMENTARY INFORMATION
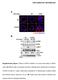 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS
 Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest and Lei Xiong SCIEX, Redwood City,
Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest and Lei Xiong SCIEX, Redwood City,
Instructions SILAC Protein Quantitation Kits
 Instructions SILAC Protein Quantitation Kits CIL Catalog No. Description DMEM LYS C SILAC Protein Quantitation Kit DMEM (Dulbecco s Modified Eagle s Medium) Kit contains: SILAC DMEM Media, 2 x 500 ml Dialyzed
Instructions SILAC Protein Quantitation Kits CIL Catalog No. Description DMEM LYS C SILAC Protein Quantitation Kit DMEM (Dulbecco s Modified Eagle s Medium) Kit contains: SILAC DMEM Media, 2 x 500 ml Dialyzed
Quantification of Host Cell Protein Impurities Using the Agilent 1290 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System
 Application Note Biotherapeutics Quantification of Host Cell Protein Impurities Using the Agilent 9 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System Authors Linfeng Wu and Yanan Yang
Application Note Biotherapeutics Quantification of Host Cell Protein Impurities Using the Agilent 9 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System Authors Linfeng Wu and Yanan Yang
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Supplemental materials and methods. The itraq labelling of the proteins was done as described previously [1]. Briefly, C2C12 myotubes
![Supplemental materials and methods. The itraq labelling of the proteins was done as described previously [1]. Briefly, C2C12 myotubes Supplemental materials and methods. The itraq labelling of the proteins was done as described previously [1]. Briefly, C2C12 myotubes](/thumbs/78/78704364.jpg) 1 Supplemental materials and methods 2 itraq labelling and nanolc-ms/ms analysis 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 The itraq labelling of the proteins was done as described previously [1]. Briefly,
1 Supplemental materials and methods 2 itraq labelling and nanolc-ms/ms analysis 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 The itraq labelling of the proteins was done as described previously [1]. Briefly,
Click Chemistry Capture Kit
 http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
Innovations for Protein Research. Protein Research. Powerful workflows built on solid science
 Innovations for Protein Research Protein Research Powerful workflows built on solid science Your partner in discovering innovative solutions for quantitative protein and biomarker research Applied Biosystems/MDS
Innovations for Protein Research Protein Research Powerful workflows built on solid science Your partner in discovering innovative solutions for quantitative protein and biomarker research Applied Biosystems/MDS
Tools and considerations to increase resolution of complex proteome samples by two-dimensional offline LC/MS. Application
 Tools and considerations to increase resolution of complex proteome samples by two-dimensional offline LC/MS Application Martin Vollmer Patric Hörth Edgar Nägele Abstract Off-line 2D-HPLC applying a continuous
Tools and considerations to increase resolution of complex proteome samples by two-dimensional offline LC/MS Application Martin Vollmer Patric Hörth Edgar Nägele Abstract Off-line 2D-HPLC applying a continuous
Proteomics and some of its Mass Spectrometric Applications
 Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE.
 User Protocol 122643 Rev. 12 May 2005 JSW Page 1 of 7 ProteoExtract Albumin/IgG Removal Kit, Maxi Cat. No. 122643 1. Introduction One of the major challenges in functional proteomics is the handling of
User Protocol 122643 Rev. 12 May 2005 JSW Page 1 of 7 ProteoExtract Albumin/IgG Removal Kit, Maxi Cat. No. 122643 1. Introduction One of the major challenges in functional proteomics is the handling of
ProteoEnrich CAT-X SEC Kit Merck: Lichrospher Composition. Silica Particle size. 25 µm particles with 6 nm pore size Ligand SO 3
 Novagen User Protocol TB428 Rev. A 1105 1 of 6 ProteoEnrich TM CAT-X SEC Kit About the Kit ProteoEnrich CAT-X SEC Kit 71539-3 Description The ProteoEnrich CAT-X SEC Kit provides a highly specific method
Novagen User Protocol TB428 Rev. A 1105 1 of 6 ProteoEnrich TM CAT-X SEC Kit About the Kit ProteoEnrich CAT-X SEC Kit 71539-3 Description The ProteoEnrich CAT-X SEC Kit provides a highly specific method
Proteomics. Proteomics is the study of all proteins within organism. Challenges
 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Algorithm for Matching Additional Spectra
 Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
NPTEL VIDEO COURSE PROTEOMICS PROF. SANJEEVA SRIVASTAVA
 LECTURE-24 Quantitative proteomics: Stable isotope labeling by amino acids in cell culture (SILAC) TRANSCRIPT Welcome to the proteomics course. In today s lecture we will talk about Quantitative proteomics:
LECTURE-24 Quantitative proteomics: Stable isotope labeling by amino acids in cell culture (SILAC) TRANSCRIPT Welcome to the proteomics course. In today s lecture we will talk about Quantitative proteomics:
Appendix. Table of contents
 Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
LC/MS/MS Solutions for Biomarker Discovery QSTAR. Elite Hybrid LC/MS/MS System. More performance, more reliability, more answers
 LC/MS/MS Solutions for Biomarker Discovery QSTAR Elite Hybrid LC/MS/MS System More performance, more reliability, more answers More is better and the QSTAR Elite LC/MS/MS system has more to offer. More
LC/MS/MS Solutions for Biomarker Discovery QSTAR Elite Hybrid LC/MS/MS System More performance, more reliability, more answers More is better and the QSTAR Elite LC/MS/MS system has more to offer. More
ProteinPilot Software for Protein Identification and Expression Analysis
 ProteinPilot Software for Protein Identification and Expression Analysis Providing expert results for non-experts and experts alike ProteinPilot Software Overview New ProteinPilot Software transforms protein
ProteinPilot Software for Protein Identification and Expression Analysis Providing expert results for non-experts and experts alike ProteinPilot Software Overview New ProteinPilot Software transforms protein
Ultra-Sensitive Quantification of Trastuzumab Emtansine in Mouse Plasma using Trap-Elute MicroLC MS Method
 Ultra-Sensitive Quantification of Trastuzumab Emtansine in Mouse Plasma using Trap-Elute MicroLC MS Method Featuring SCIEX QTRAP 6500+ System with OptiFlow Turbo V source and M5 MicroLC system Lei Xiong,
Ultra-Sensitive Quantification of Trastuzumab Emtansine in Mouse Plasma using Trap-Elute MicroLC MS Method Featuring SCIEX QTRAP 6500+ System with OptiFlow Turbo V source and M5 MicroLC system Lei Xiong,
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore, India
 Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Jet Stream Proteomics for Sensitive and Robust Standard Flow LC/MS
 Jet Stream Proteomics for Sensitive and Robust Standard Flow LC/MS Technical Overview Authors Yanan Yang, Vadiraj hat, and Christine Miller Agilent Technologies, Inc. Santa Clara, California Introduction
Jet Stream Proteomics for Sensitive and Robust Standard Flow LC/MS Technical Overview Authors Yanan Yang, Vadiraj hat, and Christine Miller Agilent Technologies, Inc. Santa Clara, California Introduction
Faster, easier, flexible proteomics solutions
 Agilent HPLC-Chip LC/MS Faster, easier, flexible proteomics solutions Our measure is your success. products applications soft ware services Phospho- Anaysis Intact Glycan & Glycoprotein Agilent s HPLC-Chip
Agilent HPLC-Chip LC/MS Faster, easier, flexible proteomics solutions Our measure is your success. products applications soft ware services Phospho- Anaysis Intact Glycan & Glycoprotein Agilent s HPLC-Chip
Solutions Guide. MX Series II Modular Automation for Nano and Analytical Scale HPLC And Low Pressure Fluid Switching Applications
 Solutions Guide MX Series II Modular Automation for Nano and Analytical Scale HPLC And Low Pressure Fluid Switching Applications Page 1 of 12 Table of Contents Sample Injection... 3 Two- Selection... 4
Solutions Guide MX Series II Modular Automation for Nano and Analytical Scale HPLC And Low Pressure Fluid Switching Applications Page 1 of 12 Table of Contents Sample Injection... 3 Two- Selection... 4
ProteinPilot Software Overview
 ProteinPilot Software Overview High Quality, In-Depth Protein Identification and Protein Expression Analysis Sean L. Seymour and Christie L. Hunter SCIEX, USA As mass spectrometers for quantitative proteomics
ProteinPilot Software Overview High Quality, In-Depth Protein Identification and Protein Expression Analysis Sean L. Seymour and Christie L. Hunter SCIEX, USA As mass spectrometers for quantitative proteomics
RayBio Genomic DNA Magnetic Beads Kit
 RayBio Genomic DNA Magnetic Beads Kit Catalog #: 801-112 User Manual Last revised January 4 th, 2017 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100
RayBio Genomic DNA Magnetic Beads Kit Catalog #: 801-112 User Manual Last revised January 4 th, 2017 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100
for water and beverage analysis
 Thermo Scientific EQuan MAX Plus Systems Automated, high-throughput LC-MS solutions for water and beverage analysis Pesticides Pharmaceuticals Personal care products Endocrine disruptors Perfluorinated
Thermo Scientific EQuan MAX Plus Systems Automated, high-throughput LC-MS solutions for water and beverage analysis Pesticides Pharmaceuticals Personal care products Endocrine disruptors Perfluorinated
Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research
 PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
Lecture 5: 8/31. CHAPTER 5 Techniques in Protein Biochemistry
 Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Antibody Array User s Guide
 Antibody Microarray User s Guide Rev. 9.1 Page 1 TABLE OF CONTENTS Introduction... 2 How it works... 3 Experimental considerations... 4 Components... 4 Additional Material Required... 6 Reagent Preparation...
Antibody Microarray User s Guide Rev. 9.1 Page 1 TABLE OF CONTENTS Introduction... 2 How it works... 3 Experimental considerations... 4 Components... 4 Additional Material Required... 6 Reagent Preparation...
ichemistry SOLUTIONS Integrated chemistries to boost mass spectrometry workflows
 ichemistry SOLUTIONS Integrated chemistries to boost mass spectrometry workflows ichemistry SOLUTIONS ichemistry solutions ichemistry solutions are the world s only reagents and consumables that are customdesigned
ichemistry SOLUTIONS Integrated chemistries to boost mass spectrometry workflows ichemistry SOLUTIONS ichemistry solutions ichemistry solutions are the world s only reagents and consumables that are customdesigned
Increasing Sensitivity In LC-MS. Simplification and prefractionation of complex protein samples
 Increasing Sensitivity In LC-MS Simplification and prefractionation of complex protein samples Protein Analysis Workflow Simplify Fractionate Separate Identify High Abundant Protein Removal reduction of
Increasing Sensitivity In LC-MS Simplification and prefractionation of complex protein samples Protein Analysis Workflow Simplify Fractionate Separate Identify High Abundant Protein Removal reduction of
DNA Purification Magnetic Beads
 DNA Purification Magnetic Beads Catalog #: 801-109 User Manual Last revised December 17 th, 2018 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
DNA Purification Magnetic Beads Catalog #: 801-109 User Manual Last revised December 17 th, 2018 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
Application Manual ProteoSpin Abundant Serum Protein Depletion Kit
 Application Manual ProteoSpin Abundant Serum Protein Depletion Kit For Use with P/N 17300 ProteoSpin Abundant Serum Protein Depletion Kit Basic Features Efficient removal of highly abundant proteins 1
Application Manual ProteoSpin Abundant Serum Protein Depletion Kit For Use with P/N 17300 ProteoSpin Abundant Serum Protein Depletion Kit Basic Features Efficient removal of highly abundant proteins 1
Introduction. Benefits of the SWATH Acquisition Workflow for Metabolomics Applications
 SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
Proteomics Training Kit
 CONTENTS I. INTRODUCTION II. STANDARD PREPARATION III. PROTEIN EXPRESSION: MASSPREP MIX 1 VS. MIX 2 (75 µm) IV. PROTEIN EXPRESSION: MASSPREP MIX 1 VS. MIX 2 (300 µm) V. ORDERING INFORMATION I. INTRODUCTION
CONTENTS I. INTRODUCTION II. STANDARD PREPARATION III. PROTEIN EXPRESSION: MASSPREP MIX 1 VS. MIX 2 (75 µm) IV. PROTEIN EXPRESSION: MASSPREP MIX 1 VS. MIX 2 (300 µm) V. ORDERING INFORMATION I. INTRODUCTION
DNA isolation from tissue DNA isolation from eukaryotic cells (max. 5 x 106 cells) DNA isolation from paraffin embedded tissue
 INDEX KIT COMPONENTS 3 STORAGE AND STABILITY 3 BINDING CAPACITY 3 INTRODUCTION 3 IMPORTANT NOTES 4 EUROGOLD TISSUE DNA MINI KIT PROTOCOLS 5 A. DNA isolation from tissue 5 B. DNA isolation from eukaryotic
INDEX KIT COMPONENTS 3 STORAGE AND STABILITY 3 BINDING CAPACITY 3 INTRODUCTION 3 IMPORTANT NOTES 4 EUROGOLD TISSUE DNA MINI KIT PROTOCOLS 5 A. DNA isolation from tissue 5 B. DNA isolation from eukaryotic
Component. Buffer RL
 GenElute FFPE RNA Purification Catalog number RNB400 Product Description Sigma s GenElute FFPE RNA Purification Kit provides a rapid method for the isolation and purification of total RNA (including microrna)
GenElute FFPE RNA Purification Catalog number RNB400 Product Description Sigma s GenElute FFPE RNA Purification Kit provides a rapid method for the isolation and purification of total RNA (including microrna)
Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection.
 Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection Application Note Christian Wenz Andreas Rüfer Abstract Agilent Equipment Agilent 3100
Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection Application Note Christian Wenz Andreas Rüfer Abstract Agilent Equipment Agilent 3100
For concentration and purification of circulating cell-free DNA for variable sample volumes
 NeoGeneStar TM Circulating DNA Purification Kit For concentration and purification of circulating cell-free DNA for variable sample volumes (Additional information and protocol suggestions for cell free
NeoGeneStar TM Circulating DNA Purification Kit For concentration and purification of circulating cell-free DNA for variable sample volumes (Additional information and protocol suggestions for cell free
MagExtactor -His-tag-
 Instruction manual MagExtractor-His-tag-0905 F0987K MagExtactor -His-tag- Contents NPK-701 100 preparations Store at Store at 4 C [1] Introduction [2] Components [3] Materials required [4] Protocol3 1.
Instruction manual MagExtractor-His-tag-0905 F0987K MagExtactor -His-tag- Contents NPK-701 100 preparations Store at Store at 4 C [1] Introduction [2] Components [3] Materials required [4] Protocol3 1.
Simplifying ImmunoAffinity Capture Workflow
 CAE TUDY MART Digest ImmunoAffinity (IA) Kit implifying ImmunoAffinity Capture Workflow Rapid, ensitive, LC-RM Quantitative Analysis of Proteins in Plasma As pharmaceuticals grow more efficacious, reporting
CAE TUDY MART Digest ImmunoAffinity (IA) Kit implifying ImmunoAffinity Capture Workflow Rapid, ensitive, LC-RM Quantitative Analysis of Proteins in Plasma As pharmaceuticals grow more efficacious, reporting
Cross Linking Immunoprecipitation
 301PR 03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Cross Linking Immunoprecipitation Utilizes Protein A/G Agarose& DSS for Antibody
301PR 03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Cross Linking Immunoprecipitation Utilizes Protein A/G Agarose& DSS for Antibody
Method Optimisation in Bottom-Up Analysis of Proteins
 Method Optimisation in Bottom-Up Analysis of Proteins M. Styles, 1 D. Smith, 2 J. Griffiths, 2 L. Pereira, 3 T. Edge 3 1 University of Manchester, UK; 2 Patterson Institute, Manchester, UK; 3 Thermo Fisher
Method Optimisation in Bottom-Up Analysis of Proteins M. Styles, 1 D. Smith, 2 J. Griffiths, 2 L. Pereira, 3 T. Edge 3 1 University of Manchester, UK; 2 Patterson Institute, Manchester, UK; 3 Thermo Fisher
GELFREE 8100 Fractionation System. Frequently Asked Questions
 INDEX Basics...3 Features...3 Sample Loading...4 Operation...5 Fraction Recovery...6 Tips...7 Troubleshooting...7 Use and Use Restrictions: The Products are sold, and deliverables of any services are provided
INDEX Basics...3 Features...3 Sample Loading...4 Operation...5 Fraction Recovery...6 Tips...7 Troubleshooting...7 Use and Use Restrictions: The Products are sold, and deliverables of any services are provided
columns PepSwift and ProSwift Capillary Monolithic Reversed-Phase Columns
 columns PepSwift and ProSwift Capillary Monolithic Reversed-Phase Columns PepSwift and ProSwift monolithic columns are specially designed for high-resolution LC/MS analysis in protein identification, biomarker
columns PepSwift and ProSwift Capillary Monolithic Reversed-Phase Columns PepSwift and ProSwift monolithic columns are specially designed for high-resolution LC/MS analysis in protein identification, biomarker
Hichrom Limited 1 The Markham Centre, Station Road, Theale, Reading, Berkshire, RG7 4PE, UK Tel: +44 (0) Fax: +44 (0)
 Hichrom Limited 1 The Markham Centre, Station Road, Theale, Reading, Berkshire, RG7 4PE, UK Tel: +44 (0)118 930 3660 Fax: +44 (0)118 932 3484 Email: www.hichrom.co.uk Table of Contents Sample Injection...
Hichrom Limited 1 The Markham Centre, Station Road, Theale, Reading, Berkshire, RG7 4PE, UK Tel: +44 (0)118 930 3660 Fax: +44 (0)118 932 3484 Email: www.hichrom.co.uk Table of Contents Sample Injection...
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Investigating Biological Variation of Liver Enzymes in Human Hepatocytes
 Investigating Biological Variation of Liver Enzymes in Human Hepatocytes SWATH Acquisition on the TripleTOF Systems Xu Wang 1, Hui Zhang 2, Christie Hunter 1 1 SCIEX, USA, 2 Pfizer, USA MS/MS ALL with
Investigating Biological Variation of Liver Enzymes in Human Hepatocytes SWATH Acquisition on the TripleTOF Systems Xu Wang 1, Hui Zhang 2, Christie Hunter 1 1 SCIEX, USA, 2 Pfizer, USA MS/MS ALL with
ProteoExtract Protein Precipitation Kit Cat. No
 User Protocol 539180 Rev. 12-October-06 JSW ProteoExtract Protein Precipitation Kit Cat. No. 539180 Note that this user protocol is not lot-specific and is representative of the current specifications
User Protocol 539180 Rev. 12-October-06 JSW ProteoExtract Protein Precipitation Kit Cat. No. 539180 Note that this user protocol is not lot-specific and is representative of the current specifications
Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS
 Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS Sensitive workflow for the detection of PTMs in biological samples from small sample volumes Klaus Faserl, 1 Bettina Sarg, 1
Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS Sensitive workflow for the detection of PTMs in biological samples from small sample volumes Klaus Faserl, 1 Bettina Sarg, 1
SERVA BluePrep Protein EndotoxinEx Macro Kit
 INSTRUCTION MANUAL SERVA BluePrep Protein EndotoxinEx Macro Kit (Cat. No. 42086.01) 1 Contents 1. SERVA BluePrep Protein EndotoxinEx Macro Kit 3 1.1. Kit Components 3 1.2. Specifications 4 1.3. Storage
INSTRUCTION MANUAL SERVA BluePrep Protein EndotoxinEx Macro Kit (Cat. No. 42086.01) 1 Contents 1. SERVA BluePrep Protein EndotoxinEx Macro Kit 3 1.1. Kit Components 3 1.2. Specifications 4 1.3. Storage
PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%)
 1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
Fast and reliable purification of up to 100 µg of transfection-grade plasmid DNA using a spin-column.
 INSTRUCTION MANUAL ZymoPURE Plasmid Miniprep Kit Catalog Nos. D4209, D4210, D4211 & D4212 (Patent Pending) Highlights Fast and reliable purification of up to 100 µg of transfection-grade plasmid DNA using
INSTRUCTION MANUAL ZymoPURE Plasmid Miniprep Kit Catalog Nos. D4209, D4210, D4211 & D4212 (Patent Pending) Highlights Fast and reliable purification of up to 100 µg of transfection-grade plasmid DNA using
ProteoSpin Total Protein Concentration, Detergent Clean-Up and Endotoxin Removal Mini Kit Product Insert Product # 22800
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com ProteoSpin Total Protein Concentration, Detergent Clean-Up and
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com ProteoSpin Total Protein Concentration, Detergent Clean-Up and
SERVA BluePrep Major Serum Protein Removal Kit
 INSTRUCTION MANUAL SERVA BluePrep Major Serum Protein Removal Kit (Cat. No. 42079.01) 1 Contents 1. SERVA BluePrep Major Serum Protein Removal Kit 3 2. Procedure 4 2.1. Notes prior to use 4 2.2. Column
INSTRUCTION MANUAL SERVA BluePrep Major Serum Protein Removal Kit (Cat. No. 42079.01) 1 Contents 1. SERVA BluePrep Major Serum Protein Removal Kit 3 2. Procedure 4 2.1. Notes prior to use 4 2.2. Column
FavorPrep Blood / Cultured Cell Genomic DNA Extraction Maxi Kit. User Manual
 TM FavorPrep Blood / Cultured Cell Genomic DNA Extraction Maxi Kit User Manual Cat. No.: FABGK 003 (10 Preps) FABGK 003-1 (24 Preps) For Research Use Only v.1005 Introduction TM FavorPrep Genomic DNA Extraction
TM FavorPrep Blood / Cultured Cell Genomic DNA Extraction Maxi Kit User Manual Cat. No.: FABGK 003 (10 Preps) FABGK 003-1 (24 Preps) For Research Use Only v.1005 Introduction TM FavorPrep Genomic DNA Extraction
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis
 Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Computing with large data sets
 Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Rapid Peptide Catabolite ID using the SCIEX Routine Biotransform Solution
 Rapid Peptide Catabolite ID using the SCIEX Routine Biotransform Solution Rapidly Identify Major Catabolites with the SCIEX X500R QTOF System and MetabolitePilot TM 2.0 Software Ian Moore and Jinal Patel
Rapid Peptide Catabolite ID using the SCIEX Routine Biotransform Solution Rapidly Identify Major Catabolites with the SCIEX X500R QTOF System and MetabolitePilot TM 2.0 Software Ian Moore and Jinal Patel
Biotherapeutic Peptide Mapping Information Dependent Acquisition (IDA) Method
 Biotherapeutic Peptide Mapping Information Dependent Acquisition (IDA) Method Routine biotherapeutic accurate mass peptide mapping analysis on the X500B QTOF System Method details for the routine peptide
Biotherapeutic Peptide Mapping Information Dependent Acquisition (IDA) Method Routine biotherapeutic accurate mass peptide mapping analysis on the X500B QTOF System Method details for the routine peptide
foodproof Sample Preparation Kit III
 For food testing purposes FOR IN VITRO USE ONLY Version 1, June 2015 For isolation of DNA from raw material and food products of plant and animal origin for PCR analysis Order No. S 400 06.1 Kit for 50
For food testing purposes FOR IN VITRO USE ONLY Version 1, June 2015 For isolation of DNA from raw material and food products of plant and animal origin for PCR analysis Order No. S 400 06.1 Kit for 50
GenElute FFPE DNA Purification Kit. Catalog number DNB400 Storage temperature -20 ºC TECHNICAL BULLETIN
 GenElute FFPE DNA Purification Kit Catalog number DNB400 Storage temperature -20 ºC TECHNICAL BULLETIN Product Description GenElute FFPE DNA Purification Kit provides a rapid method for the isolation and
GenElute FFPE DNA Purification Kit Catalog number DNB400 Storage temperature -20 ºC TECHNICAL BULLETIN Product Description GenElute FFPE DNA Purification Kit provides a rapid method for the isolation and
Analysis of the Massachusetts Cannabis Pesticides List Using the SCIEX QTRAP System
 Analysis of the Massachusetts Cannabis Pesticides List Using the SCIEX QTRAP 6500+ System Evoking the power of MS3 to reduce matrix background in cannabis extracts Craig M. Butt 1, Robert Di Lorenzo 2,
Analysis of the Massachusetts Cannabis Pesticides List Using the SCIEX QTRAP 6500+ System Evoking the power of MS3 to reduce matrix background in cannabis extracts Craig M. Butt 1, Robert Di Lorenzo 2,
基于质谱的蛋白质药物定性定量分析技术及应用
 基于质谱的蛋白质药物定性定量分析技术及应用 蛋白质组学质谱数据分析 单抗全蛋白测序及鉴定 Bioinformatics Solutions Inc. Waterloo, ON, Canada 上海 中国 2017 1 3 PEAKS Studio GUI Introduction Overview of PEAKS Studio GUI Project View Tasks, running info,
基于质谱的蛋白质药物定性定量分析技术及应用 蛋白质组学质谱数据分析 单抗全蛋白测序及鉴定 Bioinformatics Solutions Inc. Waterloo, ON, Canada 上海 中国 2017 1 3 PEAKS Studio GUI Introduction Overview of PEAKS Studio GUI Project View Tasks, running info,
NanoLC-Ultra system. data sheet. Introducing the NanoLC-Ultra family of high pressure HPLCs for proteomics research
 data sheet NanoLC-Ultra system The new NanoLC-Ultra is Eksigent s third generation system, delivering superior gradient precision at pressures up to 10,000 psi. Introducing the NanoLC-Ultra family of high
data sheet NanoLC-Ultra system The new NanoLC-Ultra is Eksigent s third generation system, delivering superior gradient precision at pressures up to 10,000 psi. Introducing the NanoLC-Ultra family of high
High-resolution Analysis of Charge Heterogeneity in Monoclonal Antibodies Using ph-gradient Cation Exchange Chromatography
 High-resolution Analysis of Charge Heterogeneity in Monoclonal Antibodies Using ph-gradient Cation Exchange Chromatography Agilent 1260 Infinity Bio-inert Quaternary LC System with Agilent Bio Columns
High-resolution Analysis of Charge Heterogeneity in Monoclonal Antibodies Using ph-gradient Cation Exchange Chromatography Agilent 1260 Infinity Bio-inert Quaternary LC System with Agilent Bio Columns
Liver Mitochondria Proteomics Employing High-Resolution MS Technology
 Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Thermo Scientific EASY-Spray LC Columns Optimization - Red flags to watch for and how to troubleshoot your workflow Estee Toole
 Thermo Scientific EASY-Spray LC Columns Optimization - Red flags to watch for and how to troubleshoot your workflow Estee Toole The world leader in serving science What Makes a Thermo Scientific EASY-Spray
Thermo Scientific EASY-Spray LC Columns Optimization - Red flags to watch for and how to troubleshoot your workflow Estee Toole The world leader in serving science What Makes a Thermo Scientific EASY-Spray
TaKaRa MiniBEST Plasmid Purification Kit Ver.4.0
 Cat. # 9760 For Research Use TaKaRa MiniBEST Plasmid Purification Kit Ver.4.0 Product Manual Table of Contents I. Description... 3 II. Kit Components... 3 III. Shipping and Storage... 4 IV. Preparation
Cat. # 9760 For Research Use TaKaRa MiniBEST Plasmid Purification Kit Ver.4.0 Product Manual Table of Contents I. Description... 3 II. Kit Components... 3 III. Shipping and Storage... 4 IV. Preparation
Identification of human serum proteins detectable after Albumin removal with Vivapure Anti-HSA Kit
 Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Mammalian Chromatin Extraction Kit
 Mammalian Chromatin Extraction Kit Catalog Number KA3925 100 extractions Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3
Mammalian Chromatin Extraction Kit Catalog Number KA3925 100 extractions Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3
Agilent 3100 OFFGEL Fractionator pi-based fractionation of proteins and peptides with liquid-phase recovery
 Agilent 3100 OFFGEL Fractionator pi-based fractionation of proteins and peptides with liquid-phase recovery Achieve unprecedented sensitivity in LC/MS-based proteomics experiments Sample complexity and
Agilent 3100 OFFGEL Fractionator pi-based fractionation of proteins and peptides with liquid-phase recovery Achieve unprecedented sensitivity in LC/MS-based proteomics experiments Sample complexity and
Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics
 Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics INTRODUCTION Peptide sequencing by nano LC-ESI-MS/MS is a widespread technique used for protein identification in proteomics.
Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics INTRODUCTION Peptide sequencing by nano LC-ESI-MS/MS is a widespread technique used for protein identification in proteomics.
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins
 Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Agilent Human 14 Multiple Affinity Removal System Spin Cartridges for the Depletion of High-Abundant Proteins from Human Proteomic Samples
 Agilent Human 14 Multiple Affinity Removal System Spin Cartridges for the Depletion of High-Abundant Proteins from Human Proteomic Samples Instructions Second edition October 2008 General Information Introduction
Agilent Human 14 Multiple Affinity Removal System Spin Cartridges for the Depletion of High-Abundant Proteins from Human Proteomic Samples Instructions Second edition October 2008 General Information Introduction
Genomic DNA Mini Kit (Blood/Cultured Cell) For research use only
 Genomic DNA Mini Kit (Blood/Cultured Cell) For research use only Sample : up to 300 µl of whole blood, up to 200 µl of frozen blood, up to 200 µl of buffy coat, cultured animal cells (up to 1 x 107), 9
Genomic DNA Mini Kit (Blood/Cultured Cell) For research use only Sample : up to 300 µl of whole blood, up to 200 µl of frozen blood, up to 200 µl of buffy coat, cultured animal cells (up to 1 x 107), 9
