Aaron A. Goodarzi, Angela T. Noon, Dorothee Deckbar, Yael Ziv, Yosef Shiloh, Markus Löbrich, and Penny A. Jeggo
|
|
|
- Peter Price
- 6 years ago
- Views:
Transcription
1 Molecular Cell, Volume 31 Supplemental Data ATM Signaling Facilitates Repair of DNA Double-Strand Breaks Associated with Heterochromatin Aaron A. Goodarzi, Angela T. Noon, Dorothee Deckbar, Yael Ziv, Yosef Shiloh, Markus Löbrich, and Penny A. Jeggo Supplemental Experimental Procedures Reagents and Irradiation: The ATM inhibitor (ATMi) KU was a gift from KuDos Pharmaceuticals and was used at a final concentration of 10 µm. Micrococcal nuclease (MNase) and neocarzinostatin were obtained from Sigma-Aldrich (Poole, UK). Calicheamicin was a provided by Ola Hammarsten. Irradiation with low energy (278 ev) Carbon-K characteristic X-rays was performed as previously described (Kuhne et al., 2005). Immunofluorescence Cells plated on glass slides were fixed for 10 min with fixative (3% (w/v) PFA, 2% (w/v) sucrose, 1X PBS) and permeablized for 1 min with 0.2 % Triton X-100 in PBS. Cells were rinsed with PBS and incubated with primary antibody diluted in PBS + 2% (w/v) BSA for 1 hour at room temperature (RT). Cells were washed three times, incubated with secondary antibody (diluted in PBS + 2% (w/v) BSA) for 30 min at RT in the dark, incubated with 4,6-diamidino-2-phenylindole (DAPI) for 10 min and washed three times with PBS. Slides were mounted using Vectasheild and visualized using a Zeiss Axioplan microscope and Simple-PCI software. For high resolution 3D imaging of deconvolved Z-stacks, slides were analyzed using an Applied Precision DeltaVision RT Olympus IX70 deconvolution microscope and softworx Suite software. Antibodies and Immunoblotting The following antibodies were used at indicated concentrations for immunofluorescence: Anti-KAP-1 ab10484 (1:1000, Abcam, UK), anti-γh2ax (1:800, Upstate Technology, UK), anti-tri-me K9 Histone H3 ab8898 (1:800, Abcam, UK), anti- CENP-A M-85 (1:100, Santa Cruz, CA), anti-hp1 (non isoform-specific antibody, i.e. detects HP1 α, β and γ) Fl-191 (1:100, Santa Cruz, CA, CA), anti-hdac1 ab19895 (1:300, Abcam, UK), anti-hdac2 H-54 (1:300, Santa Cruz, CA), 53BP1 ab21083 (1:800, Abcam UK), acetyl K9 histone H3 ab12179 (1:1000, Abcam UK), anti-hmg1 (1:400, Pharmingen, CA), anti-e2f1 E-20 (1:800, Santa Cruz, CA), anti-ku70 M19 (1:100, Santa Cruz, CA). Appropriate secondary antibodies were used at a 1:200 (green = FITC; red = Cy3 or TRITC conjugated antibodies). For immunoblotting, clarified
2 extracts were resolved on 7.5% denaturing PAGE and transferred to nitrocellulose (100V, 60 minutes in 48 mm Tris-base, 39 mm glycine and 20% (v/v) methanol). Chromosomal break analysis Confluent GM00157 cells were treated with 20 µm ATMi 30 min, then irradiated with 2 Gy, incubated for 24 hours and sub-cultured in the presence of ATMi for a further 32 hours. At this time point, maximum numbers of G2 phase cells were obtained as determined by flow cytometry. 50 ng/ml Calyculin A was added 30 minutes prior to harvest to induce premature chromosome condensation of G2 cells. Fixation and preparation of chromosome spreads was as described previously (Deckbar et al., 2007). Slides were analyzed by FISH using probes against chromosomes 7, 8 and X, according to the manufacturer s instructions (Metasystems, Germany). Semi-automated microscopy was performed using a Zeiss Axioplan2 microscope and Metafer and ISIS software (Metasystems, Germany). Expression of GFP-KAP-1 in cells with KAP-1 knockdown 1BRneo cells were transfected with 100 pmol KAP-1 B sirna per 2 x 10 5 cells, using Metafectene -Pro (according to manufacturer s instructions). 48 hr later, cells were trypsinized and simultaneously transfected with both 100 pmol KAP-1 B sirna and 1 µg pegfp-kap1-wt, S824A or S824D (or an equivalent volume of water) per 2 x 10 5 cells using Metafectene -Pro as before. 2 x10 5 transfected cells (per condition) were then plated onto glass slides in 2 ml media. 16 hours later, cells were treated ± ATMi and irradiated as normal. Nucleosome Size Control 1 x 10 7 cells were washed with PBS and once with LSB. Pelleted cells were resuspended in 5X PCV of LSB µm MC-LR and 1X protease inhibitor cocktail (Sigma-Aldrich, UK) and snap-frozen in liquid nitrogen. Cells were quick-thawed and immediately centrifuged for 10 min at rpm. The resulting supernatant was discarded and the pellet resuspended in 2X the original PCV of nuclease buffer containing 100 U/mL MNase. Samples were incubated at 37 o C for 0, 5, 10, 20, 40 or 80 min before addition of an equal volume of denaturing buffer and storage on ice. When all samples were collected, an equal volume of TE buffer was added followed by 1.5 V of 1:1 Phenol:Chloroform. Samples were inverted five times and centrifuged rpm for 10 min. The phenol (lower) phase was then removed, followed by the addition of Acetic Acid to 450 mm and then 7.5 V of ice-cold Ethanol containing 15 µg/ml of glycogen. Samples were inverted, stored on ice 5 minutes and centrifuged rpm for 10 min. The supernatant was then discarded and the DNA pellet washed with 0.5 ml 70% Ethanol. Pellets were air-dried for 10 min before being resuspended in 100 µl water. 1.5 µg DNA was resolved on a 2% (w/v) agarose gel and visualized by Ethidium Bromide staining. Pulsed Field Gel Electrophoresis (PFGE) For PFGE analysis, WT or ATM -/- MEFs were irradiated 48 hr after transfection with 30 nm mouse KAP-1 C sirna or mouse ATM sirna (Qiagen) using Metafectene -Pro. Transfection efficiency was greater than 75% (assessed under the microscope by the 2
3 number of cells showing loss of KAP1 immunofluorescent signal). At the time of irradiation, cultures were nearly confluent (>80% in G1, assessed by flow cytometry). PFGE analysis was performed according to Kühne et al (Cancer Res 64, ) with slight modifications. In short, cells were harvested either immediately after irradiation with 10 Gy (to assess DSB induction) or at 24 and 48 hr after irradiation with 80 Gy (to assess DSB repair). Cells irradiated to assess DSB induction were kept on ice 10 min before and during the irradiation procedure. Cells were then harvested on ice, swiftly embedded into 0.75% agarose plugs and immediately transferred back to 0 C for 1 h. Cells were then lysed at 50 C for 24 hr. The fraction of DNA released from the gel plug into the gel (the FAR value) was quantified by evaluating the ethidium bromidestained PFGE gel (Figure S8C). In contrast to previous studies involving less stringent cooling conditions to assess DSB induction (Deckbar et al., 2007, J Cell Biol 176, ), higher FAR values were obtained for the 10 Gy induction samples in this study (Figure 5F,G). This may have been the result of the generation of heat-labile sites during irradiation which are converted into DSBs during the lysis step of PFGE and which can lead to an over-estimation of the true DSB induction value by a factor of nearly two (Rydberg, 2000, Radiat Res 153, ). Figure S1: A maximum of ~25% of DSBs induced by NCS require ATM-signalling for repair. 1BR3 (wildtype) and AT1BR (ATM-/-) primary cells were treated with either 2 Gy IR or 50 ng/µl NCS. Cells were fixed at the indicated times post treatment and stained for H2AX (green) and DAPI (blue). 3
4 Figure S2: γh2ax foci remaining un-repaired in cells treated with ATMi are visibly associated with heterochromatin. Panel A: DMSO or 10 µm KU55933 ATM inhibitor (ATMi) was added to confluent, stationary-phase NIH 3T3 cells and, 30 minutes later, cells were irradiated with 2 Gy IR and harvested 0.5, 2, 6, 16 and 24 hours post-ir. Cells were fixed and immunostained for γh2ax (red) and DAPI (green). All images shown are overlays and are representative of the total cell population. Panel B: NIH3T3 cells were treated with ATMi, irradiated with 3 Gy IR, harvested 24 hr later and immunostained for 53BP1 (red), γh2ax (green) and DAPI (blue). Panel C: Numbers of 53BP1 foci were scored for NIH3T3 cells treated ± ATMi, irradiated with 3 Gy IR and harvested as indicated. Panel D: ATM -/- MEFs were irradiated with 3 Gy IR and harvested 24 hr later. Cells were immunostained with TriMe K9 H3 (red), γh2ax (green) and DAPI (blue). A high contrast image of the indicated cell is shown, with arrows pointing to foci scored as heterochromatic. 4
5 Figure S3: High resolution imaging of γh2ax foci remaining un-repaired in cells treated with ATMi. Panel A: NIH3T3 cells were treated with ATMi, irradiated with 3 Gy IR and harvested 24 hr later. Cells were stained with γh2ax (green) and DAPI (blue). High resolution Z-stack images were captured and deconvolved using the softworx Suite software. Regions of intense DAPI staining (purple outlines) and γh2ax foci (green outlines) were isolated for the whole stack. A single plane of view is shown. The right panel shows a close-up of the region outlined in white. Regions of isolated overlap can clearly be seen (indicated by white arrows), where γh2ax spreads into the surface of the DAPI chromocenters. Panel B: Signal intensity chromatographs are shown for three separate slices through the indicated nucleus. Green lines indicate γh2ax signal, blue lines indicate DAPI signal. Light blue dashed lines indicate the DAPI signal thresholds of euchromatin and heterochromatin. 5
6 6
7 Figure S4: >70% of γh2ax foci remaining un-repaired in cells treated with ATMi are visibly associated with heterochromatin. Panel A: DMSO or ATMi was added to stationary-phase NIH 3T3 cells and, 30 minutes later, cells were irradiated with 0.25, 0.5, 0.75, 1 or 2 Gy IR and harvested 0.5 hour later. Cells were fixed and immunostained for γh2ax (red) and DAPI (green). All images shown are overlays and are representative of the total cell population (there was no difference between DMSO and ATMi at this time point). The total number of γh2ax foci and γh2ax foci associated with regions of heterochromatin were counted for a minimum of 20 cells and the average of three experiments is shown. Panel B: DMSO or ATMi was added to stationary-phase NIH 3T3 cells and, 30 minutes later, cells were irradiated with 1, 2 or 3 Gy IR and harvested 24 hours later. Cells were fixed, immunostained and counted as above. 7
8 Figure S5: DNA size control for nucleosome preparations. Left Panel: Untreated NIH3T3 cells were processed for nucleosome solublization as described in the Experimental Procedures. Samples were incubated for the indicated times with 100 U/mL MNase, then extracted with Phenol:Chloroform and ethanol precipitated to isolate DNA. 1.5 µg of DNA was loaded on a 2% (w/v) agarose gel and visualized by Ethidium Bromide staining. Right Panel: NIH3T3 cells were processed for chromatin segregation as described in the Experimental Procedures. Samples at each stage of the segregation were extracted with Phenol:Chloroform and ethanol precipitated to isolate DNA. 1.5 µg of DNA was loaded on a 2% (w/v) agarose gel and visualized by Ethidium Bromide staining. 8
9 Figure S6: ATM-phosphorylation site mutants of KAP-1 affect the ATM-dependent DSB repair defect of 1BRneo cells. 1BRneo cells were transfected with sirna duplexes to human KAP-1 (KAP-1 B ) or scrambled sirna (mock). 48 hr later, cells were re-transfected with sirna with or without pegfp vectors encoding wildtype, S824A or S824D human KAP-1 sirna-resistant cdna. 24 hr later, cells were treated ±ATMi, irradiated and harvested as indicated. Cells were fixed and stained for 53BP1 (red) and DAPI (blue). GFP expression = green. 9
10 Figure S7: Knockdown of heterochromatin building factors in 1BRneo cells using sirna oligos. 2.3 x 105 1BRneo cells were transfected with either scrambled sirna (mock) or 20 µmol of sirna directed towards either KAP-1 (panel A), HP1α, β & γ isoform pan-specific antibody (panel B) or HDAC1 and HDAC2 (panel C). Cells were harvested and fixed 48 or 72 hours relative to the transfection procedure, as indicated. Fixed cells were immunostained for KAP-1 (panel A, red), HP1αβγ (panel B, red) or HDAC1 and HDAC2 (panel C, red). All cells were counterstained with DAPI (blue). Overlaid images (overlay signal, purple) appear in the top left-hand corner of each image. 10
11 Figure S8: KAP-1 sirna does not affect DAPI chromocenter integrity in NIH3T3 cells, but does alter histone H3 trimethylation and alleviates the requirement for ATM in DSB assessed by PFGE. Panel AB: NIH3T3 cells were transfected with scrambled control sirna (Mock) or 100 pmol KAP-1 C sirna using Metafectene-Pro. 48 hrs post transfection, cells were harvested and stained for either KAP-1 (red, Panel A) or TriMe K9 H3 (red, Panel B). Panel C: Representative pulsed-field gels for WT or ATM -/- MEFs transfected with either ATM or KAP-1 sirna and irradiated as described in the Experimental Procedures. 11
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells
 Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells
 Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
Evaluation of Metafectene-Pro mediated transfection of sirna and/or plasmids into human and murine cells Aaron A. Goodarzi and Penny A. Jeggo Genome Damage and Stability Centre, University of Sussex, East
supplementary information
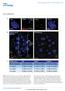 DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
DOI: 10.1038/ncb2017 Figure S1 53BP1 sirna results in a heterochromatic DSB repair defect. Panel A: NIH3T3 cells were transfected with murine 53BP1 sirna. 48 hrs later, cells were irradiated with 3 Gy
Supplementary Discussion and Figures
 Supplementary Discussion and Figures Differences in chromatin retention between the two CHD3 isoforms The pan-nuclear loss of the KAP-1-interacting CHD3 isoform from chromatin (observed in Fig. 2A) is
Supplementary Discussion and Figures Differences in chromatin retention between the two CHD3 isoforms The pan-nuclear loss of the KAP-1-interacting CHD3 isoform from chromatin (observed in Fig. 2A) is
The preparation of native chromatin from cultured human cells.
 Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
FIGURE S1. Representative images to illustrate RAD51 foci induction in FANCD2 wild-type (wt) and
 Supplementary Figures FIGURE S1. Representative images to illustrate RAD51 foci induction in FANCD2 wild-type (wt) and mutant (mut) cells. Higher magnification is shown in Figure 1A. Arrows indicate cisplatin-induced
Supplementary Figures FIGURE S1. Representative images to illustrate RAD51 foci induction in FANCD2 wild-type (wt) and mutant (mut) cells. Higher magnification is shown in Figure 1A. Arrows indicate cisplatin-induced
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
SUPPLEMENTARY INFORMATION
 Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Figure S1: Activation of the ATM pathway by I-PpoI. A. HEK293T cells were either untransfected, vector transfected, transfected with an I-PpoI expression vector, or subjected to 2Gy γ-irradiation. 24 hrs
Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and
 SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
SUPPLEMENTARY MATERIALS AND METHODS Chromatin Immunoprecipitation for qpcr analysis Four different active promoter genes were chosen, ATXN7L2, PSRC1, CELSR2 and IL24, all located on chromosome 1. Primer
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
SUPPLEMENTARY INFORMATION FIGURE LEGENDS
 SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
SUPPLEMENTARY INFORMATION FIGURE LEGENDS Fig. S1. Radiation-induced phosphorylation of Rad50 at a specific site. A. Rad50 is an in vitro substrate for ATM. A series of Rad50-GSTs covering the entire molecule
Standard Operating Procedure
 Standard Operating Procedure Title Subtitle NANoREG Work package/task: Owner and co-owner(s) HTS Comet Assay with and without FPG - 20 wells Comet assay with and without FPG using Trevigen 20-well slides
Standard Operating Procedure Title Subtitle NANoREG Work package/task: Owner and co-owner(s) HTS Comet Assay with and without FPG - 20 wells Comet assay with and without FPG using Trevigen 20-well slides
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde
 Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplementary Methods
 Supplementary Methods Antibodies For immunocytochemistry, the following antibodies were used: mouse anti-γ-h2ax (Upstate), rabbit anti-γ-h2ax (Abcam), rabbit anti-53bp1 (Novus), mouse anti-atm-phosphoserine1981
Supplementary Methods Antibodies For immunocytochemistry, the following antibodies were used: mouse anti-γ-h2ax (Upstate), rabbit anti-γ-h2ax (Abcam), rabbit anti-53bp1 (Novus), mouse anti-atm-phosphoserine1981
Confocal immunofluorescence microscopy
 Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Confocal immunofluorescence microscopy HL-6 and cells were cultured and cytospun onto glass slides. The cells were double immunofluorescence stained for Mt NPM1 and fibrillarin (nucleolar marker). Briefly,
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Ren Lab ENCODE in situ HiC Protocol for Tissue
 Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Replication-independent chromatin loading of Dnmt1 during G2 and M phases
 Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
Replication-independent chromatin loading of Dnmt1 during G2 and M phases Hariharan P. Easwaran 1, Lothar Schermelleh 2, Heinrich Leonhardt 1,2,* and M. Cristina Cardoso 1,* 1 Max Delbrück Center for Molecular
Cdc42 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining.
 Supplementary materials and methods Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining. Cells were analyzed for phosphatidylserine exposure by an annexin-v FITC/propidium
Supplementary materials and methods Flow cytometric determination of apoptosis by annexin V/propidium iodide double staining. Cells were analyzed for phosphatidylserine exposure by an annexin-v FITC/propidium
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Yeast Nuclei Isolation Kit
 Yeast Nuclei Isolation Kit Catalog Number KA3951 50 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Yeast Nuclei Isolation Kit Catalog Number KA3951 50 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Supplementary Figure 1 A green: cytokeratin 8
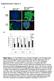 Supplementary Figure 1 A green: cytokeratin 8 green: α-sma red: α-sma blue: DAPI blue: DAPI Panc-1 Panc-1 Panc-1+hPSC Panc-1+hPSC monoculture coculture B Suppl. Figure 1: A, Immunofluorescence staining
Supplementary Figure 1 A green: cytokeratin 8 green: α-sma red: α-sma blue: DAPI blue: DAPI Panc-1 Panc-1 Panc-1+hPSC Panc-1+hPSC monoculture coculture B Suppl. Figure 1: A, Immunofluorescence staining
Arf6 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
Like use other ChIP kits, before handle ChIP assay please choose a good antibody suitable for precipitation the crosslinked protein / DNA complexes.
 ChIP Assay Kit Cat:RK20100 Like use other ChIP kits, before handle ChIP assay please choose a good antibody suitable for precipitation the crosslinked protein / DNA complexes. Manufactured by Global Headquarters
ChIP Assay Kit Cat:RK20100 Like use other ChIP kits, before handle ChIP assay please choose a good antibody suitable for precipitation the crosslinked protein / DNA complexes. Manufactured by Global Headquarters
OPPF-UK Standard Protocols: Mammalian Expression
 OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
RheB Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based RheB Activation Assay Kit Catalog Number: 81201 20 assays NewEast Biosciences 1 FAX: 610-945-2008 Table
SUPPLEMENTARY INFORMATION
 The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
The Supplementary Information (SI) Methods Cell culture and transfections H1299, U2OS, 293, HeLa cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. H1299 and 293 cells were
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Rab5 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
ab Comet Assay Kit (3- well slides)
 Version 1 Last updated 2 November 2018 ab238544 Comet Assay Kit (3- well slides) For the measurement of cellular DNA damage. This product is for research use only and is not intended for diagnostic use.
Version 1 Last updated 2 November 2018 ab238544 Comet Assay Kit (3- well slides) For the measurement of cellular DNA damage. This product is for research use only and is not intended for diagnostic use.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2579 Figure S1 Incorporation of heavy isotope-labeled amino acids and enrichment of di-glycine modified peptides. The incorporation of isotopelabeled amino acids in peptides was calculated
DOI: 10.1038/ncb2579 Figure S1 Incorporation of heavy isotope-labeled amino acids and enrichment of di-glycine modified peptides. The incorporation of isotopelabeled amino acids in peptides was calculated
Chromatin Immunoprecipitation (ChIP) Assay*
 Chromatin Immunoprecipitation (ChIP) Assay* Chromatin Immunoprecipitation (ChIP) assays are used to evaluate the association of proteins with specific DNA regions. The technique involves crosslinking of
Chromatin Immunoprecipitation (ChIP) Assay* Chromatin Immunoprecipitation (ChIP) assays are used to evaluate the association of proteins with specific DNA regions. The technique involves crosslinking of
EZ-Zyme Chromatin Prep Kit
 Instruction Manual for EZ-Zyme Chromatin Prep Kit Catalog # 17 375 Sufficient reagents for 22 enzymatic chromatin preparations per kit. Contents Page I. INTRODUCTION 2 II. EZ-Zyme KIT COMPONENTS 3 A. Provided
Instruction Manual for EZ-Zyme Chromatin Prep Kit Catalog # 17 375 Sufficient reagents for 22 enzymatic chromatin preparations per kit. Contents Page I. INTRODUCTION 2 II. EZ-Zyme KIT COMPONENTS 3 A. Provided
IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only
 IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only Introduction The IgG TrueBlot for mouse, rabbit, or goat-derived antibodies represents unique series of respective
IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only Introduction The IgG TrueBlot for mouse, rabbit, or goat-derived antibodies represents unique series of respective
Supplemental Information
 Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
Supplemental Information Intrinsic protein-protein interaction mediated and chaperonin assisted sequential assembly of a stable Bardet Biedl syndome protein complex, the BBSome * Qihong Zhang 1#, Dahai
A RRM1 H2AX DAPI. RRM1 H2AX DAPI Merge. Cont. sirna RRM1
 A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
A H2AX DAPI H2AX DAPI Merge Cont sirna Figure S1: Accumulation of RRM1 at DNA damage sites (A) HeLa cells were subjected to in situ detergent extraction without IR irradiation, and immunostained with the
Supplementary Materials and Methods
 Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
Supplementary Materials and Methods sirna sequences used in this study The sequences of Stealth Select RNAi for ALK and FLOT-1 were as follows: ALK sense no.1 (ALK): 5 -AAUACUGACAGCCACAGGCAAUGUC-3 ; ALK
EPIGENTEK. EpiQuik In Situ DNA Damage Assay Kit. Base Catalog # P-6001 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik In Situ DNA Damage Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik In Situ DNA Damage Assay Kit is suitable for specifically measuring DNA damage in situ through
EpiQuik In Situ DNA Damage Assay Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik In Situ DNA Damage Assay Kit is suitable for specifically measuring DNA damage in situ through
TITLE: Chromatin shearing with SDS detergent buffers
 TITLE: Chromatin shearing with SDS detergent buffers Summary of Operating Conditions Target Base Pair (Range) 300-700 Duty Cycle 20% Intensity 8 Cycles per Burst 200 Processing Time We suggest an initial
TITLE: Chromatin shearing with SDS detergent buffers Summary of Operating Conditions Target Base Pair (Range) 300-700 Duty Cycle 20% Intensity 8 Cycles per Burst 200 Processing Time We suggest an initial
Cat. # MK600. For Research Use. ApopLadder Ex. Product Manual. v201608da
 Cat. # MK600 For Research Use ApopLadder Ex Product Manual Table of Contents I. Description... 3 II. Principle... 3 III. Features... 3 IV. Components... 3 V. Storage... 3 VI. Materials Required but not
Cat. # MK600 For Research Use ApopLadder Ex Product Manual Table of Contents I. Description... 3 II. Principle... 3 III. Features... 3 IV. Components... 3 V. Storage... 3 VI. Materials Required but not
ab Ran Activation Assay Kit
 ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
EPIGENTEK. EpiQuik Chromatin Immunoprecipitation Kit. Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Chromatin Immunoprecipitation Kit Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Chromatin Immunoprecipitation Kit is suitable for combining the specificity of
EpiQuik Chromatin Immunoprecipitation Kit Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Chromatin Immunoprecipitation Kit is suitable for combining the specificity of
Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript;
 Supplemental Methods Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript; Bio-Rad, Hercules, CA, USA) and standard RT-PCR experiments were carried out using the 2X GoTaq
Supplemental Methods Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript; Bio-Rad, Hercules, CA, USA) and standard RT-PCR experiments were carried out using the 2X GoTaq
Supplementary Information: Materials and Methods. GST and GST-p53 were purified according to standard protocol after
 Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
Supplementary Information: Materials and Methods Recombinant protein expression and in vitro kinase assay. GST and GST-p53 were purified according to standard protocol after induction with.5mm IPTG for
longitudinally and placed on a clean glass slide with luminal surface facing up. Epithelial cells
 SUPPLEMENTARY MATERIALS AND METHODS: Protein extraction and Western blot Colons collected from mice were flushed with PBS. Each colon was then cut open longitudinally and placed on a clean glass slide
SUPPLEMENTARY MATERIALS AND METHODS: Protein extraction and Western blot Colons collected from mice were flushed with PBS. Each colon was then cut open longitudinally and placed on a clean glass slide
Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry
 Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
ChIP-seq Protocol for RNA-Binding Proteins
 ChIP-seq: Cells were grown according to the approved ENCODE cell culture protocols. Cells were fixed in 1% formaldehyde and resuspended in lysis buffer. Chromatin was sheared to 100-500 bp using a Branson
ChIP-seq: Cells were grown according to the approved ENCODE cell culture protocols. Cells were fixed in 1% formaldehyde and resuspended in lysis buffer. Chromatin was sheared to 100-500 bp using a Branson
MEFs were treated with the indicated concentrations of LLOMe for three hours, washed
 Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
Gα 13 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
In Situ Hybridization
 In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
A Supersandwich Fluorescence in Situ Hybridization (SFISH) Strategy. for Highly Sensitive and Selective mrna Imaging in Tumor Cells
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) A Supersandwich Fluorescence in Situ Hybridization (SFISH)
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2015 Electronic Supplementary Information (ESI) A Supersandwich Fluorescence in Situ Hybridization (SFISH)
ab Hypoxic Response Human Flow Cytometry Kit
 ab126585 Hypoxic Response Human Flow Cytometry Kit Instructions for Use For measuring protein levels by flow cytometry: hypoxia-inducible factor 1-alpha (HIF1A) and BCL2/adenovirus E1B 19 kda proteininteracting
ab126585 Hypoxic Response Human Flow Cytometry Kit Instructions for Use For measuring protein levels by flow cytometry: hypoxia-inducible factor 1-alpha (HIF1A) and BCL2/adenovirus E1B 19 kda proteininteracting
EPIGENTEK. EpiQuik Tissue Chromatin Immunoprecipitation Kit. Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
For in vitro killing assays with lysed cells, neutrophils were sonicated using a 550 Sonic
 Supplemental Information Cell Host & Microbe, Volume 8 Statins Enhance Formation of Phagocyte Extracellular Traps Ohn A. Chow, Maren von Köckritz-Blickwede, A. Taylor Bright, Mary E. Hensler, Annelies
Supplemental Information Cell Host & Microbe, Volume 8 Statins Enhance Formation of Phagocyte Extracellular Traps Ohn A. Chow, Maren von Köckritz-Blickwede, A. Taylor Bright, Mary E. Hensler, Annelies
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
TUNEL Apoptosis Detection Kit (Green Fluorescence)
 TUNEL Apoptosis Detection Kit (Green Fluorescence) Item NO. KTA2010 Product Name TUNEL Apoptosis Detection Kit (Green Fluorescence) ATTENTION For laboratory research use only. Not for clinical or diagnostic
TUNEL Apoptosis Detection Kit (Green Fluorescence) Item NO. KTA2010 Product Name TUNEL Apoptosis Detection Kit (Green Fluorescence) ATTENTION For laboratory research use only. Not for clinical or diagnostic
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION SUPPLEMENTARY MATERIALS AND METHODS Antibodies and sirna The antibodies against BAF170, BAF155, BAF60a, BAF57 and BAF53 were purchased from Santa Cruz (TX), and the antibodies
SUPPLEMENTARY INFORMATION SUPPLEMENTARY MATERIALS AND METHODS Antibodies and sirna The antibodies against BAF170, BAF155, BAF60a, BAF57 and BAF53 were purchased from Santa Cruz (TX), and the antibodies
Global Histone H4 Acetylation Assay Kit
 Global Histone H4 Acetylation Assay Kit Catalog Number KA0634 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Global Histone H4 Acetylation Assay Kit Catalog Number KA0634 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
rev. 05/31/12 Box 2 contains the following buffers/reagents:
 Store at RT, 4 C, and 20 C #9002 SimpleChIP Enzymatic Chromatin IP Kit (Agarose Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 05/31/12 Orders
Store at RT, 4 C, and 20 C #9002 SimpleChIP Enzymatic Chromatin IP Kit (Agarose Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 05/31/12 Orders
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
Supplemental Experimental Procedures
 Supplemental Experimental Procedures Generation of BLM protein segments and mutants. For immunoprecipitation experiments with topoisomerase IIα, BLM N-terminal segments were generated by PCR amplification
Supplemental Experimental Procedures Generation of BLM protein segments and mutants. For immunoprecipitation experiments with topoisomerase IIα, BLM N-terminal segments were generated by PCR amplification
Supplemental Data. Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes
 Cell, Volume 135 Supplemental Data Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes Andrzej T. Wierzbicki, Jeremy R. Haag, and
Cell, Volume 135 Supplemental Data Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes Andrzej T. Wierzbicki, Jeremy R. Haag, and
Northern Blotting (acc. to Maniatis)
 1 TMM,4-06 Northern Blotting (acc. to Maniatis) For work with RNA: Be aware of RNases! Use gloves, wash gloved hands with water, dry. Always use freshly autoclaved not yet opened pipet tips / Eppis! Treat
1 TMM,4-06 Northern Blotting (acc. to Maniatis) For work with RNA: Be aware of RNases! Use gloves, wash gloved hands with water, dry. Always use freshly autoclaved not yet opened pipet tips / Eppis! Treat
SANTA CRUZ BIOTECHNOLOGY, INC.
 TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
TECHNICAL SERVICE GUIDE: Western Blotting 2. What size bands were expected and what size bands were detected? 3. Was the blot blank or was a dark background or non-specific bands seen? 4. Did this same
Supplemental Data. Ohnishi et al. Plant Cell. (2010) /tpc anti-lci1 NH 4+ NO 3- DAPI IF DAPI IF
 Supplemental Data. Ohnishi et al. Plant Cell. (2010). 10.1105/tpc.109.071811 A DAPI HC IF DAPI LC IF antilci1 B DAPI IF DAPI IF C2 E4 Supplemental Figure 1. Indirect immunofluorescence analyses of LCI1
Supplemental Data. Ohnishi et al. Plant Cell. (2010). 10.1105/tpc.109.071811 A DAPI HC IF DAPI LC IF antilci1 B DAPI IF DAPI IF C2 E4 Supplemental Figure 1. Indirect immunofluorescence analyses of LCI1
Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg
![Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg](/thumbs/88/115416970.jpg) Product Datasheet Histone H4 [Dimethyl Lys20] Antibody NB21-2089SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related
Product Datasheet Histone H4 [Dimethyl Lys20] Antibody NB21-2089SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related
CytoChip Oligo Summary Protocol
 FOR RESEARCH USE ONLY ILLUMINA PROPRIETARY October 2014 Page 1 of 15 Sample Preparation Sample Preparation Materials Table 1 Starting materials for sample preparation Starting materials Amount DNA (unsheared,
FOR RESEARCH USE ONLY ILLUMINA PROPRIETARY October 2014 Page 1 of 15 Sample Preparation Sample Preparation Materials Table 1 Starting materials for sample preparation Starting materials Amount DNA (unsheared,
Chromatin Immunoprecipitation (ChIP) Assay (PROT11)
 Chromatin Immunoprecipitation (ChIP) Assay (PROT11) Antigone Kouskouti & Irene Kyrmizi Institute of Molecular Biology and Biotechnology FORTH 1527 Vassilika Vouton 711 10, Heraklion, Crete, Greece Email
Chromatin Immunoprecipitation (ChIP) Assay (PROT11) Antigone Kouskouti & Irene Kyrmizi Institute of Molecular Biology and Biotechnology FORTH 1527 Vassilika Vouton 711 10, Heraklion, Crete, Greece Email
DNA Laddering Assay Kit
 DNA Laddering Assay Kit Item No. 660990 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Precautions 4 If You
DNA Laddering Assay Kit Item No. 660990 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Precautions 4 If You
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
Supplemental Data. Regulating Gene Expression. through RNA Nuclear Retention
 Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Supplemental Data Regulating Gene Expression through RNA Nuclear Retention Kannanganattu V. Prasanth, Supriya G. Prasanth, Zhenyu Xuan, Stephen Hearn, Susan M. Freier, C. Frank Bennett, Michael Q. Zhang,
Gα i Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα i Activation Assay Kit Catalog Number 80301 20 assays NewEast Biosciences, Inc 1 Table of Content Product
Tissue Acetyl-Histone H4 ChIP Kit
 Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
EZ-10 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK
 EZ-0 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK (Bacteria, Plant, Animal, Blood) Version 8 Rev 05/0/03 EZ-0 Genomic DNA Kit Handbook Table of Contents Introduction Limitations of Use Features Applications
EZ-0 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK (Bacteria, Plant, Animal, Blood) Version 8 Rev 05/0/03 EZ-0 Genomic DNA Kit Handbook Table of Contents Introduction Limitations of Use Features Applications
0.5% Triton X-100 for 5 min at room temperature. Fixed and permeabilized cells were
 1 Supplementary Methods Immunohistochemistry EBC-1 cells were fixed in 4% paraformaldehyde for 15 min at room temperature, followed by 0.5% Triton X-100 for 5 min at room temperature. Fixed and permeabilized
1 Supplementary Methods Immunohistochemistry EBC-1 cells were fixed in 4% paraformaldehyde for 15 min at room temperature, followed by 0.5% Triton X-100 for 5 min at room temperature. Fixed and permeabilized
Establishing sirna assays in primary human peripheral blood lymphocytes
 page 1 of 7 Establishing sirna assays in primary human peripheral blood lymphocytes This protocol is designed to establish sirna assays in primary human peripheral blood lymphocytes using the Nucleofector
page 1 of 7 Establishing sirna assays in primary human peripheral blood lymphocytes This protocol is designed to establish sirna assays in primary human peripheral blood lymphocytes using the Nucleofector
ChIPmentation CeMM v1.14 (September 2016)
 ChIPmentation CeMM v1.14 (September 2016) This protocol works well for efficient histone modification and transcription factor antibodies. For less efficient antibodies sonication different sonication-,
ChIPmentation CeMM v1.14 (September 2016) This protocol works well for efficient histone modification and transcription factor antibodies. For less efficient antibodies sonication different sonication-,
Zebrafish ChIP-array protocol: version 1 August 5 th 2005
 Zebrafish ChIP-array protocol: version 1 August 5 th 2005 PROCEDURE I. Fixation of embryos 1. Dechorionate embryos (Pronase treatment) 2. Fix embryos, 15 mins in 1.85% formaldehyde in Embryo medium, RT,
Zebrafish ChIP-array protocol: version 1 August 5 th 2005 PROCEDURE I. Fixation of embryos 1. Dechorionate embryos (Pronase treatment) 2. Fix embryos, 15 mins in 1.85% formaldehyde in Embryo medium, RT,
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex
 Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
Immunofluorescence Confocal Microscopy of 3D Cultures Grown on Alvetex 1.0. Introduction Immunofluorescence uses the recognition of cellular targets by fluorescent dyes or antigen-specific antibodies coupled
TUNEL Universal Apoptosis Detection Kit (FITC-labeled)
 TUNEL Universal Apoptosis Detection Kit (FITC-labeled) Cat. No. L00427 Technical Manual No. TM0623 Version 03102011 I Description. 1 II Key Features.... 1 III Kit Contents.. 1 IV Storage.. 2 V Protocol..
TUNEL Universal Apoptosis Detection Kit (FITC-labeled) Cat. No. L00427 Technical Manual No. TM0623 Version 03102011 I Description. 1 II Key Features.... 1 III Kit Contents.. 1 IV Storage.. 2 V Protocol..
EpiSeeker ChIP Kit Histone H4 (acetyl) Tissue
 ab117151 EpiSeeker ChIP Kit Histone H4 (acetyl) Tissue Instructions for Use For successful chromatin immunoprecipitation for acetyl-histone H4 from mammalian tissues This product is for research use only
ab117151 EpiSeeker ChIP Kit Histone H4 (acetyl) Tissue Instructions for Use For successful chromatin immunoprecipitation for acetyl-histone H4 from mammalian tissues This product is for research use only
Cell culture. HeLa cells were cultured as monolayers in Dulbecco s Minimal Essential
 Supporting Online Material Materials and methods Cell culture. HeLa cells were cultured as monolayers in Dulbecco s Minimal Essential Medium (Gibco BRL, Invitrogen Corporation, Carlsbad, CA, USA), supplemented
Supporting Online Material Materials and methods Cell culture. HeLa cells were cultured as monolayers in Dulbecco s Minimal Essential Medium (Gibco BRL, Invitrogen Corporation, Carlsbad, CA, USA), supplemented
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD
 Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin
 ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin Article (Unspecified) Goodarzi, A.A., Noon, A.T., Deckbar, D., Ziv, Y., Shiloh, Y., Lobrich, M. and Jeggo, P.A.
ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin Article (Unspecified) Goodarzi, A.A., Noon, A.T., Deckbar, D., Ziv, Y., Shiloh, Y., Lobrich, M. and Jeggo, P.A.
HOOK 6X His Protein Spin Purification (Bacteria)
 222PR 03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Spin Purification (Bacteria) For the Purification of His Tagged
222PR 03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Spin Purification (Bacteria) For the Purification of His Tagged
SOP for Quantitation of the Hepatitis C Virus (HCV) Genome by RT-PCR
 Virus Bank SOP-HCV-001 1. Scope SOP for Quantitation of the Hepatitis C Virus (HCV) Genome by RT-PCR 1.1 This procedure describes a method for quantitation of the HCV genome in HCV-infected cells by RT-PCR.
Virus Bank SOP-HCV-001 1. Scope SOP for Quantitation of the Hepatitis C Virus (HCV) Genome by RT-PCR 1.1 This procedure describes a method for quantitation of the HCV genome in HCV-infected cells by RT-PCR.
HOOK 6X His Protein Purification (Bacteria)
 182PR-02 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Purification (Bacteria) For The Purification Of His-Tagged Proteins
182PR-02 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name HOOK 6X His Protein Purification (Bacteria) For The Purification Of His-Tagged Proteins
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Plasmid DNA transfection of SW480 human colorectal cancer cells with the Biontex K2 Transfection System
 Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Plasmid DNA transfection of human colorectal cancer cells with the Biontex K2 Transfection System Stephanie Hehlgans and Franz Rödel, Department of Radiotherapy and Oncology, Goethe- University Frankfurt,
Protocol for in vitro transcription
 Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
