Nucleic Acids Research
|
|
|
- Sydney Waters
- 6 years ago
- Views:
Transcription
1 Volume 15 Number Nucleic Acids Research Precise and imprecise somatic excision of the transposon Tcl in the nematode C.elegans Ke-San Ruan* and Scott W.Emmons Department of Molecular Biology, Division of Biological Sciences, Albert Einstein College of Medicine, Bronx, NY 10461, USA Received June 8, 1987; Revised and Accepted August 4, 1987 ABSTRACT Eleven chromosomal products of somatic excision of Tcl transposable elements have been cloned and sequenced. The cloning method did not involve genetic reversion; therefore the products analyzed should be representative. Six empty religated target sites were from excision of one Tcl element inserted near actin genes on linkage group V; five were from a second Tcl element inserted elsewhere on the same linkage group. All six products from the first element were identical in sequence to an empty target site from a second strain, indicating excision had been precise. Two of the products from the second element were also precise, whereas the other three contained four extra nucleotides at the point of excision, indicating an imprecise excision. The four nucleotides are the same in all cases and could represent two terminal nucleotides of the transposon plus a two-nucleotide target site duplication. The difference in the ratio of precise to imprecise excision at the two insertion sites suggests a possible chromosomal position effect on the pathway of Tcl somatic excision. INTRODUCTION Certain eukaryotic transposable elements undergo excision from the chromosomal DNA at significant frequencies. For such elements excision may be the first step of a conservative transposition pathway. Excision can be followed by ligation of the chromosomal target site. The nucleotide sequence of the religated site has been determined for the plant transposable elements Taml of Antirrhinum majus, and Ac/Ds and Spm-I8 of Zea mays (1), and for the FB and P elements of Drosophila (2,3). Religated sites have generally been recovered from revertants of transposon-induced mutations. Nevertheless, excision has usually been found to have been imprecise, and to have left behind or deleted a small and variable number of nucleotides. Religated sites have been obtained without genetic selection for the Spm-I8 element, which undergoes frequent excision in somatic cells. Their structure was found to be similar to products isolated from genetic revertants (1,4). Tcl transposable elements are highly-conserved DNA segments of 1610 nucleotides, bounded by perfect 54 nucleotide inverted repeats (5). They are I R L Press Limited, Oxford, England. 6875
2 found in all C. elegans strains examined so far. Tcl elements undergo excision at low frequency in the germline (6,7), and at much higher frequency in somatic cells (8). Extrachromosomal linear and circular monomer copies of the transposon can be detected that are presumably products of somatic excision (9,10). Religated target sites arising from somatic excision can be detected in preparations of genomic DNA by Southern hybridization using sequences flanking Tcl elements as probes. By this method, somatic excision has been demonstrated to occur at each of several genomic sites where Tcl elements have been examined (6-8,11). We report here that excision of Tcl elements in somatic cells of C. elegans can be both precise and imprecise. The ratio of precise to imprecise events appears to be different for two elements at different chromosomal locations, suggesting that flanking sequences at the insertion site may influence the excision pathway. MATERIALS AND METHODS Isolation of Chromosomal Empty Sites Isolation of the 3.1 kb EcoRI/KpnI fragment containing the excision site of Tcl.l was accomplished as follows. Twenty micrograms of Bergerac DNA double digested with EcoRI and KpnI was applied across the top of a 0.7% agarose gel and fractionated together with standards as shown in Figure 1. The standards were cut from the gel and stained with ethidium bromide for photographing. The appropriate region of the gel containing unstained Bergerac DNA was cut out and the DNA isolated as follows: electroelution into IX Tris-borate buffer in a dialysis bag at 10 V/cm for 1-2 hr (12), purification and concentration on a NACS Bio-60 column (Bethesda Research laboratories) or Elutip D (Schleicher & Schuell), ethanol precipitation, and resuspension in 40 microliters of TE buffer (1 mm Tris-HCl, ph 7.5; 1 mm EDTA). The isolated Bergerac restriction fragments were ligated to the sequencing vector puc18 (13) which was linearized by double digestion with EcoRI and RpnI. The ligation reaction was performed in a solution containing 300 ng of nematode DNA, 50 ng of vector DNA, 50 mm Tris-HCl, ph 7.5, 20 mm MgC12, 1 mm ATP, 1 mm dithiothreitol, and 1-5 units of T4 ligase (International Biotechnologies, Inc.). The reaction was allowed to proceed for 2-12 hr at room temperature. E. coli strain JM109 (14), made competent by the standard CaC12 method (12), was transformed with the ligation mixture (50 ng DNA/1.5 x 108 competent cells). Transformed cells were selected by plating on agar containing ampicillin at 50 ng/ml, and colonies containing religated target sites were identified by colony hybridization (15), using 6876
3 as probe the 3.5 kb EcoRI fragment from Bristol. The probe was gel-purified to eliminate vector sequences, and labeled by nick-translation. An identical procedure was used to isolate the 155 bp SstI fragment from the Tcl.2 religated site, except that a 1.5% agarose gel was used to fractionate the genomic DNA, and the puc18 vector was linearized by digestion with SstI. Transformed colonies were screened with gel-purified 7.0 kb BamHI fragment of pcelool cloned from Bristol. DNA Sequencing Nucleotide sequences of cloned religated target sites were determined by the dideoxy chain termination method (16), using primers homologous to the pucl8 vector. In every case the DNA sequence was determined at least twice; in some cases the sequence of both strands was determined. RESULTS Cloning of Chromosomal Empty Sites We have directly isolated without genetic selection religated target sites from two genomic locations of Tcl elements in the Bergerac strain, and determined their nucleotide sequence. The two Tcl elements involved are denoted Tcl.1 and Tcl.2 (11). Both elements are located on linkage group V in the Bergerac strain, and were isolated as DNA polymorphisms between Bergerac and Bristol, which lacks elements at these locations (17,18). The sequence of the Tcl.1 element and its target site, and the sequence of the Tcl.2 target site have been reported (5,19). The sequence of the ends of the Tcl.2 element has been determined starting from 17 nucleotides within one end and 21 nucleotides within the other end of the element (this work), and is identical to that of Tcl.l. Otherwise Tc1.2 has not been sequenced, but it is the same length as Tcl.1 and does not differ from Tcl.1 with respect to any of a number of restriction sites that have been examined. Somatic excision of Tcl.1 and Tcl.2 is demonstrated by the faint bands on the lanes of Bergerac DNA in Figure 1A and B, where Southern hybridizations of genomic DNA probed with sequences flanking Tcl.1 and Tcl.2 are shown. The flanking probes reveal that the DNA is polymorphic, consisting of a mixture of filled and empty sites. This is due to spontaneous excision of the Tcl element, which occurs generally in somatic tissues at a frequency of about 5Z per generation, and throughout the life cycle (8,11). Restriction fragments containing empty sites were cloned by cutting out the appropriate region of an agarose gel, electroeluting the DNA, ligating the eluted DNA to a sequencing vector, puc18 (13), and screening the 6877
4 t=z,:7.7,:>z2'.s>a~ Nucleic Acids Research A g>~ B,,>,c 84,~~~~~~~~4 6 K:1) 3-r! f i :._.,, r -8,;a.e.5 anb 01-:'. ->BR 322 CUT OUTC 5 GEL. - - kb EL.FC:TROEL. ITIC' PROBE: penbri PROBE- i)ce t 4 PURIIAl(hTI1ON B Y NACStOLUM N Li+l-- _ ~ ~~~~~~~~ TRANSF(9PMAeT:ON L. IGAT)QN303ng DNA NSERTrS WITH 50n VESC70R touc 18) ins1 $TU FDi;ERI WitZAh vnf' - CELD WITle k-:i ' F,M PT N'.F 5:5 a T rn Figure 1. Method of cloning empty target sites. (A) Somatic excision of Tcl.1. Plasmid pce(bri)tl containing a 3.5 kb EcoRI fragment cloned from the Bristol strain (17) was used as a probe in a Southern hybridization of EcoRI-digested genomic DNA from Bristol and Bergerac. Genomic DNA was fractionated on a 0.7% agarose gel and transferred to nitrocellulose for hybridization to the nick-translated probe (12). The 3.5 kb probe hybridizes primarily to a fragment of 5.1 kb in Bergerac, due to the presence of a 1.6 kb Tcl element. A small amount of 3.5 kb fragment is also observable in Bergerac, due to somatic excision of the element. A restriction map of the 3.5 kb EcoRI fragment is shown. The fragment isolated for sequencing was a 3.1 kb KpnI/EcoRI fragment spanning the target site. (B) Somatic excision of Tc1.2. A similar Southern hybridization is shown for Tcl.2, using as a probe plasmid pcelool containing a 7.0 kb BamHI Bristol fragment (18). The 155 bp SstI fragment spanning the insertion site and shown on the restriction map was isolated from Bergerac for sequencing. (C) Outline of the strategy for cloning chromosomal empty sites described under MATERIALS AND METHODS. partial genomic library with the flanking sequence probe (Figure 1C). DNA Sequence Comparison of Empty Sites with Corresponding Filled Sites The nucleotide sequence of 11 religated excision sites isolated in this way is shown in Figure 2. Each site was isolated from DNA of a separate population of Bergerac worms grown from a single self-fertilizing hermaphrodite, to ensure that independent excision events were analyzed. In view of the low frequency of germline excision in this strain (10-3 to 10-5) (6,7), the overwhelming likelihood is that these DNA fragments arose from 6878
5 A. TC1.1 Nucleic Acids Research FILLED SITE BERGERAC AAATTTTTCAAA TGTGCTACA EMPTY SITE BERGERAC CLONE 1 AAATTT1TCAAATA TATGTGCTACA 2 AAATTTTTCAAATA -----TATGTGCTACA 3 AAAITTTTCAAATA TATGTGCTACA 4 AAATT1TTCAAATA TATGTGCTACA 5 AAAITTTTCAAATA TATGTGCTACA 6 AAATIITICAAATA TATGTGCTACA CORRESPONDING SITE BRISTOL AAATT1TTCAAATA TATGTGCTACA B. Tcl.2 FILLED SITE BERGERAC ATAGTACATATTTA 3 GCACT-ATGTACTTACAG EMPTY SITE BERGERAC CLONE 1 ATAGTACATATTT TGTATGTACTTACAG 2 ATAGTACATATTTA TGTACTTACAG 3 ATAGTACATATTA TGTACTTACAG 4 ATAGTACATATTA TGTATGTACTTACAG 5 ATAGTACATATTT TGITGTACTTACAG CORRESPONDING SITE BRISTOL ATAGTACATATTTA TGTACTTACAG Figure 2. DNA sequence comparisons. (A) Comparison of religated target sites from Bergerac, formed by excision of Tcl.1, to the corresponding filled site from Bergerac and empty site from Bristol (5). Transposon sequences are boxed; the putative target site duplication is double underlined. The alignment of the single TA dinucleotide in the empty sites with the left-hand TA of the filled site is arbitrary. (B) Similar comparison for religated target sites of Tcl.2. Extra nucleotides in three of these sequences are aligned to show their identity to the two terminal nucleotides of the 3' end of the transposon (TG) and the two bases of the target site duplication (TA). somatic excision events. The sequences of the empty sites from Bergerac are compared to the sequence of the corresponding filled site from Bergerac and the empty site from Bristol (5,19). Six empty sites produced by excision of Tcl.1 in Bergerac are identical in sequence to the corresponding empty site from Bristol, and therefore resulted from precise excision. One copy of a pair of TA dinucleotides flanking the Tcl element in the filled site is absent from the excision products. The origin of these TA dinucleotides is ambiguous. All Tcl elements so far examined at the nucleotide level have inserted at a TA dinucleotide and are flanked by a pair of TA dinucleotides. One interpretation of the flanking TA dinucleotides is that they represent a duplication of the target site TA. A second interpretation is that one T and A are part of the element, and Tcl duplicates no target site sequences. The ambiguity can 6879
6 only be resolved if a Tcl element inserted at a sequence other than TA is found. Two empty sites from Tcl.2 excision also have the same sequence as the corresponding empty site from Bristol, and are therefore the products of precise excision. The remaining three, however, have four extra nucleotides left at the site of excision, indicating an imprecise excision. The four extra nucleotides, TGTA, are identical in each case and are the same as the last two nucleotides of the 3' end of the Tcl element (TG) plus the putative target duplication (TA). DISCUSSION Both precise and imprecise excision products have also been isolated for P elements of Drosophila (3). In this case as well, the structure of the products of imprecise excision events could be accounted for by assuming that nucleotides from the target site duplication (8 nucleotides in length) and from the ends of the transposon are variably left behind at the excision site. A somewhat different interpretation was required for the results obtained with plant transposable elements, where extra nucleotides found in religated target sites could only be accounted for by assuming that sequences in the target site had been duplicated upon excision of the element (1). The ratio of precise to imprecise excision of Tcl appears to be different at the two genomic locations we have examined. Sequence analysis of additional excision products is required to confirm this conclusion. Possibly the nucleotides at the insertion site influence the detailed pathway of excision and the resolution or repair of its products. Apparent position effects on the behavior of transposable elements in maize have long been known from genetic observations (20). Muller-Neumann et al. (21) have proposed that the differences in the timing and frequency of excision of two Ac elements in maize is caused by differences in flanking sequences. Tcl-induced mutations in the unc-54 and unc-22 loci of the same strain of C. elegans undergo germline reversion at very different frequencies (6,22). Although the frequencies of somatic excision of Tcl at five examined genomic sites showed no significant differences (11,23), a position effect on the pattern of excision products is suggested here. ACKNOWLEDGEMENTS We thank M. Malone and S. Smith for careful preparation of media. This work was supported by grants from the NSF (DCB ), and the American Cancer Society (FRA-227) to SE. Some nematode strains used in this work were 6880
7 provided by the Caenorhabditis Genetics Center, which is funded by a grant from the NIH to the University of Missouri. *Permanent address: Institute of Biophysics, Academy of Sciences of China, Beijing, China REFERENCES 1. Saedler, H. and Nevers, P. (1985) EMB0 J. 4, Collins, M. and Rubin, G. M. (1983) Nature (London) 303, Rio, P. C., Laski, F. A. and Rubin, G. M. (1986) Cell 44, Schwarz-Sommer, Z., Gierl, A., Cuypers, H., Peterson, P. A. and Saedler, H. (1985) EMBO J. 4, Rosenzweig, B., Liao, L. W. & Hirsh, D. (1983) Nucl. Acids Res. 11, Eide, D. and Anderson, P. (1985) Proc. Natl. Acad. Sci., USA 82, Moerman, D., Benian, G. and Waterston, R. (1986) Proc. Natl. Acad. Sci., USA 83, Emmons, S. W. and Yesner, L. (1984) Cell 36, Ruan, K.-S. and Emmons, S. W. (1984) Proc. Natl. Acad. Sci., USA 81, Rose, A. M. and Snutch, T. P. (1984) Nature 31, Emmons, S. W., Roberts, S. and Ruan, K.-S. (1986) Mol. Gen. Genet. 202, Maniatis, T., Fritsch, E. F. and Sambrook, J. (1983) Molecular Clonin A Laboratory Manual. Cold Spring Harbor, New York (1982). 13. Norrander, J., Kempe, T. and Messing, J. (1983) Gene 26, Messing, J. (1983) Meth. Enzymol. 101, Davis, R. W., Botstein, D. and Roth, J. R. (1980) A Manual For Genetics Engineering. Cold Spring Harbor, New York. 16. Sanger, F., Nicklen, S. and Coulson, A. R. (1977) Proc. Natl. Acad. Sci., USA 74, Liao, L. W., Rosenzweig, B. and Hirsh, D. (1983) Proc. Natl. Acad. Sci., USA 80, Emmons, S. W., Yesner, L., Ruan, K.-S. and Katzenberg, D. (1983) Cell 32, Rosenzweig, B., Liao, L. W. and Hirsh, D. (1983) Nucl. Acids Res. 11, Peterson, P. A. (1977) In Bukhari, A. I. (ed.), DNA Insertion Elements, Plasmids and Episomes Cold Spring Harbor, N. Y., pp Muller-Neumann, M., Yoder, J. I. and Starlinger, P. (1984) Mol. Gen. Genet. 198, (1984). 22. Moerman, D. and Waterston, R. (1984) Genetics 108, Emmons, S. W., Ruan, K. S., Levitt, A. and Yesner, L. (1985) Cold Spring Harbor Symp. Quant. Biol. 50, (1985). 6881
Somatic Excision of Transposable Element Tcl from the Bristol Genome of Caenorhabditis elegans
 MOLECULAR AND CELLULAR BIOLOGY, May 1986, p. 1782-1786 Vol. 6, No. 5 0270-7306/86/051782-05$02.00/0 Copyright 1986, American Society for Microbiology Somatic Excision of Transposable Element Tcl from the
MOLECULAR AND CELLULAR BIOLOGY, May 1986, p. 1782-1786 Vol. 6, No. 5 0270-7306/86/051782-05$02.00/0 Copyright 1986, American Society for Microbiology Somatic Excision of Transposable Element Tcl from the
An estimate of the physical distance between two linked markers in Haemophilus influenzae
 J. Biosci., Vol. 13, No. 3, September 1988, pp. 223 228. Printed in India. An estimate of the physical distance between two linked markers in Haemophilus influenzae Ε. Β. SAMIWALA, VASUDHA P. JOSHI and
J. Biosci., Vol. 13, No. 3, September 1988, pp. 223 228. Printed in India. An estimate of the physical distance between two linked markers in Haemophilus influenzae Ε. Β. SAMIWALA, VASUDHA P. JOSHI and
BIO 202 Midterm Exam Winter 2007
 BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
Computational Biology I LSM5191
 Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Computational Biology I LSM5191 Lecture 5 Notes: Genetic manipulation & Molecular Biology techniques Broad Overview of: Enzymatic tools in Molecular Biology Gel electrophoresis Restriction mapping DNA
Mos1 insertion. MosTIC protocol-11/2006. repair template - Mos1 transposase expression - Mos1 excision - DSB formation. homolog arm.
 MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
MosTIC (Mos1 excision induced Transgene Instructed gene Conversion) Valérie Robert (vrobert@biologie.ens.fr) and Jean-Louis Bessereau (jlbesse@biologie.ens.fr) (November 2006) Introduction: MosTIC (Robert
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Biology 4100 Minor Assignment 1 January 19, 2007
 Biology 4100 Minor Assignment 1 January 19, 2007 This assignment is due in class on February 6, 2007. It is worth 7.5% of your final mark for this course. Your assignment must be typed double-spaced on
Biology 4100 Minor Assignment 1 January 19, 2007 This assignment is due in class on February 6, 2007. It is worth 7.5% of your final mark for this course. Your assignment must be typed double-spaced on
Genomic Sequencing. Genomic Sequencing. Maj Gen (R) Suhaib Ahmed, HI (M)
 Maj Gen (R) Suhaib Ahmed, HI (M) The process of determining the sequence of an unknown DNA is called sequencing. There are many approaches for DNA sequencing. In the last couple of decades automated Sanger
Maj Gen (R) Suhaib Ahmed, HI (M) The process of determining the sequence of an unknown DNA is called sequencing. There are many approaches for DNA sequencing. In the last couple of decades automated Sanger
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Genetic Engineering & Recombinant DNA
 Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
BIO 304 Fall 2000 Exam II Name: ID #: 1. Fill in the blank with the best answer from the provided word bank. (2 pts each)
 1. Fill in the blank with the best answer from the provided word bank. (2 pts each) incomplete dominance conditional mutation penetrance expressivity pleiotropy Southern blotting hybridization epistasis
1. Fill in the blank with the best answer from the provided word bank. (2 pts each) incomplete dominance conditional mutation penetrance expressivity pleiotropy Southern blotting hybridization epistasis
Data Sheet Quick PCR Cloning Kit
 Data Sheet Quick PCR Cloning Kit 6044 Cornerstone Ct. West, Ste. E DESCRIPTION: The Quick PCR Cloning Kit is a simple and highly efficient method to insert any gene or DNA fragment into a vector, without
Data Sheet Quick PCR Cloning Kit 6044 Cornerstone Ct. West, Ste. E DESCRIPTION: The Quick PCR Cloning Kit is a simple and highly efficient method to insert any gene or DNA fragment into a vector, without
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Answer sheet. Student number:
 Page 1 of 9 MIDTERM EXAM OF BIO/BPS3151 2016 Answer sheet Name: Student number: Part II: Calculations 1 128g 2 58.5g 3 NaCl: 1L Water: 0.2L 4 2.5 g/l 5 0.4 6 1:4:2 7 900 ml 8 Plasmid A: 3.75 µl Plasmid
Page 1 of 9 MIDTERM EXAM OF BIO/BPS3151 2016 Answer sheet Name: Student number: Part II: Calculations 1 128g 2 58.5g 3 NaCl: 1L Water: 0.2L 4 2.5 g/l 5 0.4 6 1:4:2 7 900 ml 8 Plasmid A: 3.75 µl Plasmid
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
B. Incorrect! Ligation is also a necessary step for cloning.
 Genetics - Problem Drill 15: The Techniques in Molecular Genetics No. 1 of 10 1. Which of the following is not part of the normal process of cloning recombinant DNA in bacteria? (A) Restriction endonuclease
Genetics - Problem Drill 15: The Techniques in Molecular Genetics No. 1 of 10 1. Which of the following is not part of the normal process of cloning recombinant DNA in bacteria? (A) Restriction endonuclease
Ligation Independent Cloning (LIC) Procedure
 Ligation Independent Cloning (LIC) Procedure Ligation Independent Cloning (LIC) LIC cloning allows insertion of DNA fragments without using restriction enzymes into specific vectors containing engineered
Ligation Independent Cloning (LIC) Procedure Ligation Independent Cloning (LIC) LIC cloning allows insertion of DNA fragments without using restriction enzymes into specific vectors containing engineered
BIOTECHNOLOGY. Sticky & blunt ends. Restriction endonucleases. Gene cloning an overview. DNA isolation & restriction
 BIOTECHNOLOGY RECOMBINANT DNA TECHNOLOGY Recombinant DNA technology involves sticking together bits of DNA from different sources. Made possible because DNA & the genetic code are universal. 2004 Biology
BIOTECHNOLOGY RECOMBINANT DNA TECHNOLOGY Recombinant DNA technology involves sticking together bits of DNA from different sources. Made possible because DNA & the genetic code are universal. 2004 Biology
Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe:
 1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
This article reprinted from: Dooley, M. M Restriction endonuclease digestion of a plasmid.
 This article reprinted from: Dooley, M. M. 2008. Restriction endonuclease digestion of a plasmid. Pages 389-392, in Tested Studies for Laboratory Teaching, Volume 29 (K.L. Clase, Editor). Proceedings of
This article reprinted from: Dooley, M. M. 2008. Restriction endonuclease digestion of a plasmid. Pages 389-392, in Tested Studies for Laboratory Teaching, Volume 29 (K.L. Clase, Editor). Proceedings of
Tcl(Hin): a form of the transposable element Tcl in Caenorhabditis elegans
 ; the Tcl(Hin): a form of the transposable element Tcl in Caenorhabditis elegans A. M. ROSE,' L. J. HARRIS, N. R. MAWJI, AND W. J. MORRIS Department of Medical Genetics, The University of British Columbia,
; the Tcl(Hin): a form of the transposable element Tcl in Caenorhabditis elegans A. M. ROSE,' L. J. HARRIS, N. R. MAWJI, AND W. J. MORRIS Department of Medical Genetics, The University of British Columbia,
ITS Sequencing in Millepora. 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector
 Page 1 of 5 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector 1. Digest 1 µg of pbluescript with Eco RI 2. Following digestion, add 0.1 volumes of 3M sodium acetate (ph
Page 1 of 5 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector 1. Digest 1 µg of pbluescript with Eco RI 2. Following digestion, add 0.1 volumes of 3M sodium acetate (ph
Chapter 20 DNA Technology & Genomics. If we can, should we?
 Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
Synthetic Biology for
 Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Synthetic Biology for Plasmids and DNA Digestion Plasmids Plasmids are small DNA molecules that are separate from chromosomal DNA They are most commonly found as double stranded, circular DNA Typical plasmids
Biotechnology. Chapter 20. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Chapter 20 Biotechnology PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp Copyright
Molecular Genetics Techniques. BIT 220 Chapter 20
 Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
(i) A trp1 mutant cell took up a plasmid containing the wild type TRP1 gene, which allowed that cell to multiply and form a colony
 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
DNA Ligation Kit Ver. 1 Manual
 Table of content Description... 2 Procedures and Examples A. Insertion of DNA into plasmid vectors... 3 B. Insertion of DNA into λ phage vectors... 4 C. Self-circulization of linear DNA... 4 D. Linker
Table of content Description... 2 Procedures and Examples A. Insertion of DNA into plasmid vectors... 3 B. Insertion of DNA into λ phage vectors... 4 C. Self-circulization of linear DNA... 4 D. Linker
BSCI410-Liu/Spring 06 Exam #1 Feb. 23, 06
 Your Name: Your UID# 1. (20 points) Match following mutations with corresponding mutagens (X-RAY, Ds transposon excision, UV, EMS, Proflavin) a) Thymidine dimmers b) Breakage of DNA backbone c) Frameshift
Your Name: Your UID# 1. (20 points) Match following mutations with corresponding mutagens (X-RAY, Ds transposon excision, UV, EMS, Proflavin) a) Thymidine dimmers b) Breakage of DNA backbone c) Frameshift
BS 50 Genetics and Genomics Week of Nov 29
 BS 50 Genetics and Genomics Week of Nov 29 Additional Practice Problems for Section Problem 1. A linear piece of DNA is digested with restriction enzymes EcoRI and HinDIII, and the products are separated
BS 50 Genetics and Genomics Week of Nov 29 Additional Practice Problems for Section Problem 1. A linear piece of DNA is digested with restriction enzymes EcoRI and HinDIII, and the products are separated
Bi 8 Lecture 4. Ellen Rothenberg 14 January Reading: from Alberts Ch. 8
 Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Bi 8 Lecture 4 DNA approaches: How we know what we know Ellen Rothenberg 14 January 2016 Reading: from Alberts Ch. 8 Central concept: DNA or RNA polymer length as an identifying feature RNA has intrinsically
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Supplementary Methods pcfd5 cloning protocol
 Supplementary Methods cloning protocol vermilion trna grna trna grna U6:3 Terminator AmpR attb is a vector for expressing one or multiple trna-flanked Cas9 grnas under the control of the strong, ubiquitous
Supplementary Methods cloning protocol vermilion trna grna trna grna U6:3 Terminator AmpR attb is a vector for expressing one or multiple trna-flanked Cas9 grnas under the control of the strong, ubiquitous
The nucleotide sequences and expression of genes for the beta and epsilon subunits of ATP synthase from rice (Oryza sativa L.)
 Jpn. J. Genet. (1989) 64, pp. 223-229 The nucleotide sequences and expression of genes for the beta and epsilon subunits of ATP synthase from rice (Oryza sativa L.) Yoko NIsxtzAwA1 and Atsushi HIRAI* Graduate
Jpn. J. Genet. (1989) 64, pp. 223-229 The nucleotide sequences and expression of genes for the beta and epsilon subunits of ATP synthase from rice (Oryza sativa L.) Yoko NIsxtzAwA1 and Atsushi HIRAI* Graduate
Hetero-Stagger PCR Cloning Kit
 Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
DNA miniprep by Alkaline Lysis (activity)
 DNA miniprep by Alkaline Lysis (activity) Contents 1 Alkaline Lysis 2 Exercise 1: Plasmid DNA Mini-Prep by Alkaline Lysis 3 Identification of Plasmid DNA 4 Exercise 2: Restriction Digestion Identification
DNA miniprep by Alkaline Lysis (activity) Contents 1 Alkaline Lysis 2 Exercise 1: Plasmid DNA Mini-Prep by Alkaline Lysis 3 Identification of Plasmid DNA 4 Exercise 2: Restriction Digestion Identification
PLNT2530 (2018) Unit 6b Sequence Libraries
 PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
Bootcamp: Molecular Biology Techniques and Interpretation
 Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Bootcamp: Molecular Biology Techniques and Interpretation Bi8 Winter 2016 Today s outline Detecting and quantifying nucleic acids and proteins: Basic nucleic acid properties Hybridization PCR and Designing
Table of Contents. i. Insertion of DNA fragments into plasmid vector...4. ii. Self-circularization of linear blunt-ended DNA...4
 Table of Contents I. Description...2 II. Kit Components...2 III. Storage...2 IIV. Notes...2 V. Reference...3 VI. PROCEDURES A. Dephosphorylation of vector DNA...3 B. Blunting reaction...3 C. Ligation reaction
Table of Contents I. Description...2 II. Kit Components...2 III. Storage...2 IIV. Notes...2 V. Reference...3 VI. PROCEDURES A. Dephosphorylation of vector DNA...3 B. Blunting reaction...3 C. Ligation reaction
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
The Effect of Size of Chromosomal DNA from Escherichia coli VC10 on Transformation of Escherichia coli HB101 by the Plasmid p328.5
 The Effect of Size of Chromosomal DNA from Escherichia coli VC10 on Transformation of Escherichia coli HB101 by the Plasmid p328.5 CHARLENE CHU, CHRISTINA HAN, HIROMI SHIMIZU, AND BONNIE WONG Department
The Effect of Size of Chromosomal DNA from Escherichia coli VC10 on Transformation of Escherichia coli HB101 by the Plasmid p328.5 CHARLENE CHU, CHRISTINA HAN, HIROMI SHIMIZU, AND BONNIE WONG Department
1a. What is the ratio of feathered to unfeathered shanks in the offspring of the above cross?
 1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive genes at two loci; the
1. Whether or not the shanks of chickens contains feathers is due to two independently assorting genes. Individuals have unfeathered shanks when they are homozygous for recessive genes at two loci; the
Chapter 10 (Part II) Gene Isolation and Manipulation
 Biology 234 J. G. Doheny Chapter 10 (Part II) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. What does PCR stand for? 2. What does the
Biology 234 J. G. Doheny Chapter 10 (Part II) Gene Isolation and Manipulation Practice Questions: Answer the following questions with one or two sentences. 1. What does PCR stand for? 2. What does the
Supplemental Materials and Methods
 Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
Supplemental Materials and Methods Proteins and reagents Proteins were purified as described previously: RecA, RecQ, and SSB proteins (Harmon and Kowalczykowski 1998); RecF protein (Morimatsu and Kowalczykowski
Journal of Experimental Microbiology and Immunology (JEMI) Vol. 6:20-25 Copyright December 2004, M&I UBC
 Preparing Plasmid Constructs to Investigate the Characteristics of Thiol Reductase and Flavin Reductase With Regard to Solubilizing Insoluble Proteinase Inhibitor 2 in Bacterial Protein Overexpression
Preparing Plasmid Constructs to Investigate the Characteristics of Thiol Reductase and Flavin Reductase With Regard to Solubilizing Insoluble Proteinase Inhibitor 2 in Bacterial Protein Overexpression
Fun with DNA polymerase
 Fun with DNA polymerase Why would we want to be able to make copies of DNA? Can you think of a situation where you have only a small amount and would like more? Enzymatic DNA synthesis To use DNA polymerase
Fun with DNA polymerase Why would we want to be able to make copies of DNA? Can you think of a situation where you have only a small amount and would like more? Enzymatic DNA synthesis To use DNA polymerase
Fast-Link DNA Ligation Kits
 Fast-Link DNA Ligation Kits Cat. Nos. LK0750H and LK6201H Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA086E Fast-Link DNA Ligation Kits 6/2017 1 1. Introduction The Fast-Link
Fast-Link DNA Ligation Kits Cat. Nos. LK0750H and LK6201H Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA086E Fast-Link DNA Ligation Kits 6/2017 1 1. Introduction The Fast-Link
Microsatellite markers
 Microsatellite markers Review of repetitive sequences 25% 45% 8% 21% 13% 3% Mobile genetic elements: = dispersed repeat included: transposition: moving in the form of DNA by element coding for transposases.
Microsatellite markers Review of repetitive sequences 25% 45% 8% 21% 13% 3% Mobile genetic elements: = dispersed repeat included: transposition: moving in the form of DNA by element coding for transposases.
Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for :
 Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Genome research in eukaryotes
 Functional Genomics Genome and EST sequencing can tell us how many POTENTIAL genes are present in the genome Proteomics can tell us about proteins and their interactions The goal of functional genomics
Functional Genomics Genome and EST sequencing can tell us how many POTENTIAL genes are present in the genome Proteomics can tell us about proteins and their interactions The goal of functional genomics
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
2054, Chap. 13, page 1
 2054, Chap. 13, page 1 I. Microbial Recombination and Plasmids (Chapter 13) A. recombination = process of combining genetic material from 2 organisms to produce a genotype different from either parent
2054, Chap. 13, page 1 I. Microbial Recombination and Plasmids (Chapter 13) A. recombination = process of combining genetic material from 2 organisms to produce a genotype different from either parent
Quantitation of ssdna using OliGreen Fluorescent Stain
 Quantitation of ssdna using OliGreen Fluorescent Stain Several different techniques require the use of short synthetic oligonucleotide molecules, often referred to as primers. In each case, the use of
Quantitation of ssdna using OliGreen Fluorescent Stain Several different techniques require the use of short synthetic oligonucleotide molecules, often referred to as primers. In each case, the use of
A. Incorrect! This statement is true. Transposable elements can cause chromosome rearrangements.
 Genetics - Problem Drill 17: Transposable Genetic Elements No. 1 of 10 1. Which of the following statements is NOT true? (A) Transposable elements can cause chromosome rearrangements. (B) Transposons can
Genetics - Problem Drill 17: Transposable Genetic Elements No. 1 of 10 1. Which of the following statements is NOT true? (A) Transposable elements can cause chromosome rearrangements. (B) Transposons can
A) (5 points) As the starting step isolate genomic DNA from
 GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
GS Final Exam Spring 00 NAME. bub ts is a recessive temperature sensitive mutation in yeast. At º C bub ts cells grow normally, but at º C they die. Use the information below to clone the wild-type BUB
Characteristics of bacterial Plasmid : Size : Conformation : Replication origin of replication : Replication Protein :
 Characteristics of bacterial Plasmid : Size : Conformation : Replication origin of replication : Replication Protein : Definition of Plasmid Plasmids are extrachromosomal circular, double stranded DNA
Characteristics of bacterial Plasmid : Size : Conformation : Replication origin of replication : Replication Protein : Definition of Plasmid Plasmids are extrachromosomal circular, double stranded DNA
DNA Cloning with Cloning Vectors
 Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Biotechnology. Biotechnology is difficult to define but in general it s the use of biological systems to solve problems.
 MITE 2 S Biology Biotechnology Summer 2004 Austin Che Biotechnology is difficult to define but in general it s the use of biological systems to solve problems. Recombinant DNA consists of DNA assembled
MITE 2 S Biology Biotechnology Summer 2004 Austin Che Biotechnology is difficult to define but in general it s the use of biological systems to solve problems. Recombinant DNA consists of DNA assembled
2. Outline the levels of DNA packing in the eukaryotic nucleus below next to the diagram provided.
 AP Biology Reading Packet 6- Molecular Genetics Part 2 Name Chapter 19: Eukaryotic Genomes 1. Define the following terms: a. Euchromatin b. Heterochromatin c. Nucleosome 2. Outline the levels of DNA packing
AP Biology Reading Packet 6- Molecular Genetics Part 2 Name Chapter 19: Eukaryotic Genomes 1. Define the following terms: a. Euchromatin b. Heterochromatin c. Nucleosome 2. Outline the levels of DNA packing
BSCI410-Liu/Spring 09/Feb 26 Exam #1 Your name:
 1. (20 points) Give the name of a mutagen that could cause the following damages to DNA: a) Thymidine dimers UV b) Breakage of DNA backbone X-Ray c) 2 bp insertion (frameshift mutation) proflavin, acridine
1. (20 points) Give the name of a mutagen that could cause the following damages to DNA: a) Thymidine dimers UV b) Breakage of DNA backbone X-Ray c) 2 bp insertion (frameshift mutation) proflavin, acridine
Escherichia coli plasmids (Si nuclease/gel electrophoresis/electron microscope)
 Proc. Natl. Acad. Sci. USA Vol. 73, No. 7, pp. 2316-2320, July 1976 Biochemistry Isolation of inverted repeat sequences, including ISi, IS2, and IS3, in Escherichia coli plasmids (Si nuclease/gel electrophoresis/electron
Proc. Natl. Acad. Sci. USA Vol. 73, No. 7, pp. 2316-2320, July 1976 Biochemistry Isolation of inverted repeat sequences, including ISi, IS2, and IS3, in Escherichia coli plasmids (Si nuclease/gel electrophoresis/electron
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
BIOTECHNOLOGY : PRINCIPLES AND PROCESSES
 CHAPTER 11 BIOTECHNOLOGY : PRINCIPLES AND PROCESSES POINTS TO REMEMBER Bacteriophage : A virus that infects bacteria. Bioreactor : A large vessel in which raw materials are biologically converted into
CHAPTER 11 BIOTECHNOLOGY : PRINCIPLES AND PROCESSES POINTS TO REMEMBER Bacteriophage : A virus that infects bacteria. Bioreactor : A large vessel in which raw materials are biologically converted into
Chapter 4. Recombinant DNA Technology
 Chapter 4 Recombinant DNA Technology 5. Plasmid Cloning Vectors Plasmid Plasmids Self replicating Double-stranded Mostly circular DNA ( 500 kb) Linear : Streptomyces, Borrelia burgdorferi Replicon
Chapter 4 Recombinant DNA Technology 5. Plasmid Cloning Vectors Plasmid Plasmids Self replicating Double-stranded Mostly circular DNA ( 500 kb) Linear : Streptomyces, Borrelia burgdorferi Replicon
XXII DNA cloning and sequencing. Outline
 XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
XXII DNA cloning and sequencing 1) Deriving DNA for cloning Outline 2) Vectors; forming recombinant DNA; cloning DNA; and screening for clones containing recombinant DNA [replica plating and autoradiography;
PCR Cloning II fusion domains for purification His 6 GST chitin binding protein MBP
 PCR cloning : why secondary amplification? May 26, 2009 secondary amplification amplification of the coding sequence alone or of defined fragments ligation into expression vector insertion of fusion domains
PCR cloning : why secondary amplification? May 26, 2009 secondary amplification amplification of the coding sequence alone or of defined fragments ligation into expression vector insertion of fusion domains
SELECTED TECHNIQUES AND APPLICATIONS IN MOLECULAR GENETICS
 SELECTED TECHNIQUES APPLICATIONS IN MOLECULAR GENETICS Restriction Enzymes 15.1.1 The Discovery of Restriction Endonucleases p. 420 2 2, 3, 4, 6, 7, 8 Assigned Reading in Snustad 6th ed. 14.1.1 The Discovery
SELECTED TECHNIQUES APPLICATIONS IN MOLECULAR GENETICS Restriction Enzymes 15.1.1 The Discovery of Restriction Endonucleases p. 420 2 2, 3, 4, 6, 7, 8 Assigned Reading in Snustad 6th ed. 14.1.1 The Discovery
_ DNA absorbs light at 260 wave length and it s a UV range so we cant see DNA, we can see DNA only by staining it.
 * GEL ELECTROPHORESIS : its a technique aim to separate DNA in agel based on size, in this technique we add a sample of DNA in a wells in the gel, then we turn on the electricity, the DNA will travel in
* GEL ELECTROPHORESIS : its a technique aim to separate DNA in agel based on size, in this technique we add a sample of DNA in a wells in the gel, then we turn on the electricity, the DNA will travel in
Biol/Chem 475 Spring 2007
 Biol/Chem 475 Spring 2007 Goal of lab: For most of the quarter, we will be exploring a gene family that was first discovered in fruitlfies and then found to be present in humans and worms and fish and
Biol/Chem 475 Spring 2007 Goal of lab: For most of the quarter, we will be exploring a gene family that was first discovered in fruitlfies and then found to be present in humans and worms and fish and
FROM EXPERIMENTS IN BACTERIAL GENETICS AND GENE TECHNIQUE
 Uppsala 2001-04-01 REPORT FROM EXPERIMENTS IN BACTERIAL GENETICS AND GENE TECHNIQUE Laboratory assistants: Maria Jönsson Amera Gibreel Students: Contents ASSIGNMENT:... 3 INTRODUCTION:... 3 MATERIAL AND
Uppsala 2001-04-01 REPORT FROM EXPERIMENTS IN BACTERIAL GENETICS AND GENE TECHNIQUE Laboratory assistants: Maria Jönsson Amera Gibreel Students: Contents ASSIGNMENT:... 3 INTRODUCTION:... 3 MATERIAL AND
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
QUICK-Clone TM User Manual. cdna
 QUICK-Clone TM User Manual cdna PT1150-1 (PR752268) Published 25 May 2007 Table of Contents I. Introduction 3 II. Applications Discussion 4 A. Primer Design 4 B. Setting up the PCR Reaction 4 C. Example
QUICK-Clone TM User Manual cdna PT1150-1 (PR752268) Published 25 May 2007 Table of Contents I. Introduction 3 II. Applications Discussion 4 A. Primer Design 4 B. Setting up the PCR Reaction 4 C. Example
Applicazioni biotecnologiche
 Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Applicazioni biotecnologiche Analisi forense Sintesi di proteine ricombinanti Restriction Fragment Length Polymorphism (RFLP) Polymorphism (more fully genetic polymorphism) refers to the simultaneous occurrence
Cold Fusion Cloning Kit. Cat. #s MC100A-1, MC101A-1. User Manual
 Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability
 Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
Antisense RNA Targeting the First Periplasmic Domain of YidC in Escherichia coli Appears to Induce Filamentation but Does Not Affect Cell Viability Riaaz Lalani, Nathaniel Susilo, Elisa Xiao, Andrea Xu
Recitation CHAPTER 9 DNA Technologies
 Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks
 A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
A Modified Digestion-Circularization PCR (DC-PCR) Approach to Detect Hypermutation- Associated DNA Double-Strand Breaks SARAH K. DICKERSON AND F. NINA PAPAVASILIOU Laboratory of Lymphocyte Biology, The
Chapter 20. Gene creatures, partii: jumping genes and junk DNA. Prepared by Woojoo Choi
 Chapter 20. Gene creatures, partii: jumping genes and junk DNA Prepared by Woojoo Choi 1) Transposable elements (transposons): segment of DNA that can move as a unit from one location to another, but which
Chapter 20. Gene creatures, partii: jumping genes and junk DNA Prepared by Woojoo Choi 1) Transposable elements (transposons): segment of DNA that can move as a unit from one location to another, but which
FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE.
 Instruction manual Blunting high 0810 Blunting high F0990K BLK-101 20 reactions Store at -20 C. Contents [1] Introduction [2] Components [3] Protocol 1. Blunting 2. Ligation [4] Troubleshooting [5] References
Instruction manual Blunting high 0810 Blunting high F0990K BLK-101 20 reactions Store at -20 C. Contents [1] Introduction [2] Components [3] Protocol 1. Blunting 2. Ligation [4] Troubleshooting [5] References
Analysis of a transposable element in Caenorhabditis elegans (nematode genome organization/repetitive DNA/inverted repeat/strain variation)
 Proc. Natl. cad. Sci. US Vol. 80, pp. 3585-3589, June 1983 Biochemistry nalysis of a transposable element in Caenorhabditis elegans (nematode genome organization/repetitive DN/inverted repeat/strain variation)
Proc. Natl. cad. Sci. US Vol. 80, pp. 3585-3589, June 1983 Biochemistry nalysis of a transposable element in Caenorhabditis elegans (nematode genome organization/repetitive DN/inverted repeat/strain variation)
Please sign below if you wish to have your grades posted by the last five digits of your SSN
 BIO 226R EXAM III (Sample) PRINT YOUR NAME Please sign below if you wish to have your grades posted by the last five digits of your Signature BIO 226R Exam III has 8 pages, and 26 questions. There are
BIO 226R EXAM III (Sample) PRINT YOUR NAME Please sign below if you wish to have your grades posted by the last five digits of your Signature BIO 226R Exam III has 8 pages, and 26 questions. There are
Use of Drosophila Melanogaster as a Model System in the Study of Human Sodium- Dependent Multivitamin Transporter. Michael Brinton BIOL 230W.
 Use of Drosophila Melanogaster as a Model System in the Study of Human Sodium- Dependent Multivitamin Transporter Michael Brinton BIOL 230W.001 28 October 2013 TA: Sashi Gollapudi Introduction Many human
Use of Drosophila Melanogaster as a Model System in the Study of Human Sodium- Dependent Multivitamin Transporter Michael Brinton BIOL 230W.001 28 October 2013 TA: Sashi Gollapudi Introduction Many human
Supporting Information
 Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
Supporting Information Kilian et al. 10.1073/pnas.1105861108 SI Materials and Methods Determination of the Electric Field Strength Required for Successful Electroporation. The transformation construct
1. A brief overview of sequencing biochemistry
 Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending)
 peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
Nucleic acid-free silica-matrix: Regeneration of DNA binding columns
 MAXXBOND ready-to-use - Kit for the regeneration of DNA binding columns with pure silica matrices Product No. MB007 Nucleic acid-free silica-matrix: Regeneration of DNA binding columns efficient and easy
MAXXBOND ready-to-use - Kit for the regeneration of DNA binding columns with pure silica matrices Product No. MB007 Nucleic acid-free silica-matrix: Regeneration of DNA binding columns efficient and easy
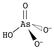 ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I) We have made considerable progress in our analysis of the gene for
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I) We have made considerable progress in our analysis of the gene for
Ready_to_use Fast Seamless Cloning Kit. User Manual
 For general laboratory use. Not for use in diagnostic procedures. FOR IN VITRO USE ONLY. Ready_to_use Fast Seamless Cloning Kit User Manual 1 / 6 Tel: 021-58975266 Fax: 021-50800270 Email:tech@dogene.com
For general laboratory use. Not for use in diagnostic procedures. FOR IN VITRO USE ONLY. Ready_to_use Fast Seamless Cloning Kit User Manual 1 / 6 Tel: 021-58975266 Fax: 021-50800270 Email:tech@dogene.com
Chapter 15 Recombinant DNA and Genetic Engineering. Restriction Enzymes Function as Nature s Pinking Shears
 Chapter 15 Recombinant DNA and Genetic Engineering In this chapter you will learn How restriction enzyme work and why they are essential to DNA technology. About various procedures such as cloning and
Chapter 15 Recombinant DNA and Genetic Engineering In this chapter you will learn How restriction enzyme work and why they are essential to DNA technology. About various procedures such as cloning and
Cat # Box1 Box2. DH5a Competent E. coli cells CCK-20 (20 rxns) 40 µl 40 µl 50 µl x 20 tubes. Choo-Choo Cloning TM Enzyme Mix
 Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
GENOME 371, Problem Set 6
 GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
Chapter 6 - Molecular Genetic Techniques
 Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Chapter 6 - Molecular Genetic Techniques Two objects of molecular & genetic technologies For analysis For generation Molecular genetic technologies! For analysis DNA gel electrophoresis Southern blotting
Bi 8 Lecture 5. Ellen Rothenberg 19 January 2016
 Bi 8 Lecture 5 MORE ON HOW WE KNOW WHAT WE KNOW and intro to the protein code Ellen Rothenberg 19 January 2016 SIZE AND PURIFICATION BY SYNTHESIS: BASIS OF EARLY SEQUENCING complex mixture of aborted DNA
Bi 8 Lecture 5 MORE ON HOW WE KNOW WHAT WE KNOW and intro to the protein code Ellen Rothenberg 19 January 2016 SIZE AND PURIFICATION BY SYNTHESIS: BASIS OF EARLY SEQUENCING complex mixture of aborted DNA
d. reading a DNA strand and making a complementary messenger RNA
 Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
