2D difference gel electrophoresis (2D-DIGE) Spot picking and in-gel digestion
|
|
|
- Judith Bennett
- 6 years ago
- Views:
Transcription
1 2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the 7 biological replicates, one analytical 2D-DIGE gel was prepared for ph and one for ph , each containing a total of 150 µg protein (50 µg each of Cy5 labeled proteins of control platelets, Cy3 labeled proteins of GPVI-activated platelets and Cy2 labeled internal pooled standard proteins). The process of detection of differentially abundant spots was carried out in several consecutive steps including minimal labeling of the samples, 2D-gel electrophoresis consisting of isoelectric focusing (IEF) and sodium dodecylsulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), acquisition of the gel images and DeCyder analysis. The whole strategy consisted of 14 protein gels and thus a total of 42 readout images for the three different labeling dyes. Changes in protein abundance were considered biologically relevant, if (I) the spot was present in all 7 gels, (II) the average ratio exceeded ±2.0 for ph 4 7 gels and ±1.5 for ph 6 9 gels and (III) the p-value was below Isoelectric focusing (IEF) was performed using the Multiphor II IEF system (GE Healthcare Biosciences). Samples were applied onto 18-cm immobilized ph gradient (IPG) strips ph 4 7 and ph 6 9 (ImmobilineTM DryStrips, GE Healthcare Biosciences). Rehydration of DryStrips using rehydration buffer (7M urea, 2M thiourea, 4% (w/v) CHAPS, 1% (v/v) PharmalyteTM ph 3 10, 13 mm DTE, 0,8% (v/v) bromphenol blue; for ph gradient ph 6 9 DryStrips DTE replaced by 0.012% (v/v) DeStreakTM reagent) was conducted over night at room temperature. Anodic sample cup loading was used for all analytical and ph 6 9 preparative samples and in-gel rehydration was used for ph 4 7 preparative samples. Analytical and preparative samples on ph 4 7 gradients were focused for a total of 44 kvh, analytical samples on ph 6 9 gradients were focused for a total of 34.5 kvh and preparative samples on ph 6 9 gradients were focused for a total of 55 kvh. Focused IPG strips were stored at 80 C. Prior to SDS-electrophoresis IPG strips were equilibrated in DTEequilibration buffer (6 M urea, 30% (v/v) glycerin, 2% (w/v) SDS, 10% (v/v) 0.5 M Tris-HCl ph 6.8, 1% (w/v) DTE) and subsequently in iodoacetamide-equilibration buffer (6 M urea, 30% (v/v) glycerin, 2% (w/v) SDS, 10% (v/v) 0.5 M Tris-HCl ph 6.8, 2% (w/v) iodoacetamide) for 10 minutes each. SDS-PAGE (gel size 16 cm 18 cm 1 mm) was performed in a Protean II xi device (Bio-Rad) using a 2 cm 4% polyacrylamide stacking gel (0.5 M Tris-HCl ph 6.8, 4% acrylamide/bisacrylamide (30%), 0.1% SDS, 0.05% APS, 0.1% TEMED) and a 12% polyacrylamide separation gel (1.5 M Tris-HCl ph 8.8, 12% acrylamide/bisacrylamide (30%), 0.1% SDS, 0.05% APS, 0.05% TEMED). The IPG strips were cut to a length of 14 cm and loaded onto the SDS-polyacrylamide-gels and fixed with 0.5% (w/v) agarose in running buffer (25 mm TRIS, 192 mm glycin, 3.4 mm SDS). Gels were run at 9 C, 30 ma/gel for 30 minutes and 40 ma/gel for 3 hours in running buffer. For preparative gels, 500 µg of unlabeled platelet protein were applied to each gel. The gels were post-stained with Roti -Blue colloidal coomassie stain for evaluation of total protein content. All spots of interest were manually picked from the gels (suppl. fig. 1). From the location of the spots in the gel, the approximate protein size and the isoelectric point (pi) were estimated for each excised spot. All spots were digested with trypsin and subjected to mass spectrometry. Spot picking and in-gel digestion Spots of preparative 2-D gels were stained with Roti -Blue colloidal coomassie stain (Roth) as noted in the manufacturer s instructions. The gel spots were excised manually from preparative 2-D gels containing 500 µg of sample proteins. Excised gel pieces were washed in ddh 2 O twice and 50 mm NH 4 HCO 3 -solution on a shaker for 15 min each. The gel pieces
2 were dehydrated with 100 µl of 100% acetonitrile. 10µL of 50 mm NH 4 HCO 3 -solution with 70 ng of sequencing grade modified porcine trypsin (Promega) in resuspension buffer were added to each gel piece and proteins were digested overnight at 37 C. The supernatant was stored on ice and the gel pieces were washed with 50 mm NH 4 HCO 3 and 70% acetonitrile on a shaker for 15 minutes each. The peptide solution was dried in a vacuum concentrator. Peptides for subsequent MALDI-TOF/TOF MS were reconstituted in 10 µl 0.5% trifluoroacetic acid and sonicated for 3 minutes. Reversed-Phase ZipTip µ-c18 Pipette Tips (Millipore GmbH) were used for desalting and concentration of the peptide solution. Bound peptides were eluted with 2 µl of matrix solution (α-cyano-4-hydroxy-cinnamic acid, Bruker Daltonics) at a matrix concentration of 5 mg/ml and immediately spotted onto an Opti- TOF MALDI target (Applied Biosystems). Mass spectrometry and database search MS and MS/MS spectra were acquired with 1600 laser shots and 500 laser shots, respectively (Nd:YAG laser, 355 nm, air collision in the MS/MS mode) on a 4800 MALDI-TOF-TOF mass spectrometer (Applied Biosystems). A minimum S/N of 10 was applied for peak list generation. Database search of all MS and MS/MS spectra using Mascot v2.1 (Matrix Science) was carried out using the GPS Explorer v3.6 Software (Applied Biosystems) searching the human subset of the Swiss-Prot Database (Release 51.0). The following parameters were used for the combined MS and MS/MS data search: I) fixed modifications: carbamidomethyl (C), II) variable modifications: oxidation (M), III) cleavage enzyme: trypsin, IV) maximum number of missed cleavages: 1, V) precursor tolerance: 200 ppm and VI) MS/MS fragment tolerance: 0.5 Da. For the LC-MS/MS analysis the dried samples were dissolved in 40 µl 0.1% formic acid and injected in a 1D-nano-LC-MS/MS setup, consisting of an online-coupled chromatography system (Ettan MDLC, GE Healtcare, Germany) to a linear ion trap mass spectrometer (Thermo LTQ, Thermo Electron). For peptide desalting and concentration a C18 trap column (C18 PepMap100, Particle size: 5 µm, 100Å, Column size: 300 µm 5 mm, LC Packings Dionex, Sunnyvale, CA, USA) was used at a flow rate of 10 µl/min using 0.1% formic acid. The peptides of the mixture were separated by RP-chromatography (column: C18 PepMap 100, 3µm, 75µm i.d. 15cm, LC Packings). As mobile phase A water and as mobile phase B 84% ACN was used both containing 0.1% formic acid. The LC method was composed of a 30 min ramp from 0% mobile phase A to 30% mobile phase B, a subsequent 20 min ramp to 60% B and finally held for 10 min at 100% B. ESI ionization was done using distal coated SilicaTips (FS D-20, Woburn, MA, USA) at a needle voltage of 1.4 kv. The MS method consisted of a cycle of one full MS scan with three subsequent data dependent MS/MS events (35% collision energy). The dynamic exclusion was set to 30 s. For peak list generation Bioworks 3.2 (Thermo Electron) was used according to the manufacturer s recommendations for this type of instrument (absolute threshold: 1000; relative threshold: 5.00; precursor ion tolerance: 1.4; minimum ion count 10). The generated MS/MS spectra were searched against the IPI Human v3.2 with the MASCOT v2.1 algorithm using the following parameters: (I) enzyme: trypsin (II) fixed modifications: carbamidomethyl (C), (III) variable modifications: oxidation (M), (IV) peptide tolerance: 2Da, (V) MS/MS tolerance: 0.8Da, (VI) peptide charge: 1+, 2+ and 3+, (VII) instrument: ESI-TRAP, (VIII) allowing up to 2 missed cleavages. The settings were adjusted to specify the search and to reflect modifications like alkylations made on the protein fragments during gel electrophoresis and accompanied processes. For MALDI TOF/TOF as well as LC-ESI-MS/MS analysis only results with a Mascot Protein Score > 100 were accepted as positively identified proteins.
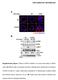
3 Figure S1. Preparative gels showing all picked spots Gels contained 500 µg protein and were stained with Roti -Blue. Left lanes contain Dalton Mark VII-L standard mixture for SDS-PAGE. (A) Preparative gel ph (B) Preparative gel ph Figure S2 (A) Platelet activation with the mab HGP4C9 is specific for GPVI. P-selectin (CD62P) surface expression was determined on isolated human platelets after stimulation with mab HGP4C9 (1 µg/ml), or control IgG1 (1µg/ml) for 10 minutes in the absence and presence of soluble human GPVI-Fc (0.5µg/ml or 2µg/ml hgpvi-fc). Results are given as mean fluorescence intensity (arbitrary units, AU) (n=3, mean ± SEM, *P < 0.01 for 4C9 versus 2µg/ml hgpvi-fc). (B) Effect of the HGP4C9 Fab antibody on platelet activation. P-selectin (CD62P) and activated integrin αiibβ3 (PAC-1) surface expression was determined on isolated human platelets after stimulation with HGP4C9 Fab antibody (10 µg/ml; n=4). Results are given as mean fluorescence intensity (arbitrary units, AU) (n=3 4, mean ± SEM, P=n.s.). Figure S3. Variability within all seven biological replicates Depicted are the 3D views of differentially abundant protein spots (A (spots no. 1 4), B (spots no. 5 8), C (spots no. 9 13)), their corresponding graph views and p-values. Figure S4. Detection of platelet proteins by Western blot analysis Cellular lysates from platelets treated with control IgG (RmC7H8) or GPVI-antibody HGP4C9 (1 μg/ml) for 10 min, were obtained, and protein expression was determined by Western blot analysis. Detection of (A) charged multivesicular body protein 3 (25-kDa protein), (B) src substrate cortactin (61-kDa protein), and (C) pleckstrin (40-kDa protein).
4 Figure S1 A B
5 Figure S2 A * 4C B
6 Figure S3 A 1) Ig gamma-1 chain C region (rat) p=1.3e-05 2) Ig gamma-1 chain C region (rat) p= 4.2e-11 3) Ig gamma-1 chain C region (rat) / ERp57 p= 3.8e-07 4) Src substrate cortactin p= 1.2e-08
7 Figure S3 B 5) Pleckstrin p=7.4e-03 6) Pleckstrin p=9.8e-03 7) Pleckstrin/beta-centractin p= 1.2e-02 8) Charged multivesicular body protein 3 p= 8.5e-12
8 Figure S3 C 9) Pyruvate kinase isozymes (M1/M2)/N(2),N(2)- Dimethylguanosine trna methyltransferase p=3.2e-04 10) Pleckstrin p= 2.7e-02 11) Pleckstrin p= 4.6e-02 12) Pleckstrin p= 3.8e-02 13) Aldose reductase p=1.1e-02
9 Figure S4 A CHMP3 Tubulin B Cortactin Tubulin C Pleckstrin Tubulin
Automated platform of µlc-ms/ms using SAX trap column for
 Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis
 Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Advanced Protein Electrophoresis. Advanced Protein Electrophoresis. Learning Objective. Proteomics ; Molecular & cell Biology
 Electrophoresis is a powerful technique for finer protein separation and visualization of separated proteins. It is based on the principle of migration of charged proteins in an electric field. Electrophoretic
Electrophoresis is a powerful technique for finer protein separation and visualization of separated proteins. It is based on the principle of migration of charged proteins in an electric field. Electrophoretic
Quantification of Isotope Encoded Proteins in 2D Gels
 Quantification of Isotope Encoded Proteins in 2D Gels Using Surface Enhanced Resonance Raman Giselle M. Knudsen 1, Brandon M. Davis 2, Shirshendu K. Deb 1, Yvette Loethen 2, Ravindra Gudihal 1, Pradeep
Quantification of Isotope Encoded Proteins in 2D Gels Using Surface Enhanced Resonance Raman Giselle M. Knudsen 1, Brandon M. Davis 2, Shirshendu K. Deb 1, Yvette Loethen 2, Ravindra Gudihal 1, Pradeep
IEF-2DE Analysis and Protein Identification Xia Wu 1, Steven J. Clough 2, Steven C. Huber 3 and Man-Ho Oh 4*
 IEF-2DE Analysis and Protein Identification Xia Wu 1, Steven J. Clough 2, Steven C. Huber 3 and Man-Ho Oh 4* 1 Genome Sciences, University of Washington, Seattle, USA; 2 Crop Sciences, USDA-ARS and the
IEF-2DE Analysis and Protein Identification Xia Wu 1, Steven J. Clough 2, Steven C. Huber 3 and Man-Ho Oh 4* 1 Genome Sciences, University of Washington, Seattle, USA; 2 Crop Sciences, USDA-ARS and the
Proteomics. Proteomics is the study of all proteins within organism. Challenges
 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SOP Title: Extraction and quantitative analysis of proteins from FFPE tissue
 SOP Title: Extraction and quantitative analysis of proteins from FFPE tissue The STORE processing methods were shown to be fit-for purpose for DNA, RNA and protein extraction from FFPE material. The STORE
SOP Title: Extraction and quantitative analysis of proteins from FFPE tissue The STORE processing methods were shown to be fit-for purpose for DNA, RNA and protein extraction from FFPE material. The STORE
Separation of Native Monoclonal Antibodies and Identification of Charge Variants:
 Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham. Bioscience) were washed four times with 1 mm HCl and twice with coupling buffer
 Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
Supporting information
 Supporting information Simple and Integrated Spintip-based Technology Applied for Deep Proteome Profiling Wendong Chen,, Shuai Wang, Subash Adhikari, Zuhui Deng, Lingjue Wang, Lan Chen, Mi Ke, Pengyuan
Supporting information Simple and Integrated Spintip-based Technology Applied for Deep Proteome Profiling Wendong Chen,, Shuai Wang, Subash Adhikari, Zuhui Deng, Lingjue Wang, Lan Chen, Mi Ke, Pengyuan
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Conventional isoelectric focusing with the Agilent 3100 OFFGEL Fractionator
 Conventional isoelectric focusing with the Agilent 3100 OFFGEL Fractionator Technical Overview Introduction This Technical Overview demonstrates the ability of the Agilent 3100 OFFGEL Fractionator to fractionate
Conventional isoelectric focusing with the Agilent 3100 OFFGEL Fractionator Technical Overview Introduction This Technical Overview demonstrates the ability of the Agilent 3100 OFFGEL Fractionator to fractionate
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System
 Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Objective. Introduction. IP assisted LC/MS/MS making study protein complexes easy. Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2
 IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection.
 Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection Application Note Christian Wenz Andreas Rüfer Abstract Agilent Equipment Agilent 3100
Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection Application Note Christian Wenz Andreas Rüfer Abstract Agilent Equipment Agilent 3100
Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium
 Supplementary data itraq-based Proteomics Analyses Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium supplemented with 5g L -1 glucose for 48 hours at 37 C with
Supplementary data itraq-based Proteomics Analyses Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium supplemented with 5g L -1 glucose for 48 hours at 37 C with
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore, India
 Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
Disulfide Linkage Analysis of IgG1 using an Agilent 1260 Infinity Bio inert LC System with an Agilent ZORBAX RRHD Diphenyl sub 2 µm Column
 Disulfide Linkage Analysis of IgG1 using an Agilent 126 Infinity Bio inert LC System with an Agilent ZORBAX RRHD Diphenyl sub 2 µm Column Application Note Biotherapeutics & Biosimilars Author M. Sundaram
Disulfide Linkage Analysis of IgG1 using an Agilent 126 Infinity Bio inert LC System with an Agilent ZORBAX RRHD Diphenyl sub 2 µm Column Application Note Biotherapeutics & Biosimilars Author M. Sundaram
Improving Productivity with Applied Biosystems GPS Explorer
 Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
2-Dimensional Electrophoresis
 2-Dimensional Electrophoresis Proteomics Pathway Proteomics Pathway History of 2-DE 1956 -paper & starch gel 1975 -Coupling of IEF & SDS-PAGE IPG-IEF 2DE-Techniques Immobilized ph Gradient IsoElectric
2-Dimensional Electrophoresis Proteomics Pathway Proteomics Pathway History of 2-DE 1956 -paper & starch gel 1975 -Coupling of IEF & SDS-PAGE IPG-IEF 2DE-Techniques Immobilized ph Gradient IsoElectric
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B
 Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi: 1.138/nnano.21.25 Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Zhou J. Deng, Mingtao Liang, Michael Monteiro, Istvan
SUPPLEMENTARY INFORMATION doi: 1.138/nnano.21.25 Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Zhou J. Deng, Mingtao Liang, Michael Monteiro, Istvan
electrophoresis tech Versatile Separation Capabilities of the PROTEAN i12 IEF System
 electrophoresis tech note 6139 Versatile Separation Capabilities of the PROTEAN i12 IEF System ChengJun Sun, Adriana Harbers, Tom Berkelman, and Katy McGirr, Life Science Group, Bio-Rad Laboratories Inc.,
electrophoresis tech note 6139 Versatile Separation Capabilities of the PROTEAN i12 IEF System ChengJun Sun, Adriana Harbers, Tom Berkelman, and Katy McGirr, Life Science Group, Bio-Rad Laboratories Inc.,
Identification of human serum proteins detectable after Albumin removal with Vivapure Anti-HSA Kit
 Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
1-DE 2-DE??? PCPPAU 04/2011
 1-DE 2-DE??? PCPPAU 04/2011 THEORY Any charged ion or group of ions will migrate when placed in an electric field. ph -----IEP (pi) Migration is dependent on charge density Charge/Mass + - PCPPAU 04/2011
1-DE 2-DE??? PCPPAU 04/2011 THEORY Any charged ion or group of ions will migrate when placed in an electric field. ph -----IEP (pi) Migration is dependent on charge density Charge/Mass + - PCPPAU 04/2011
Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics
 Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics INTRODUCTION Peptide sequencing by nano LC-ESI-MS/MS is a widespread technique used for protein identification in proteomics.
Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics INTRODUCTION Peptide sequencing by nano LC-ESI-MS/MS is a widespread technique used for protein identification in proteomics.
Application Note. Abstract. Author. Biopharmaceutical
 Innovator and Biosimilar Monoclonal Antibody-Peptide Map Comparison Using the Agilent 70 Capillary Electrophoresis System and Agilent MatchCompare Software Application Note Biopharmaceutical Author Arunkumar
Innovator and Biosimilar Monoclonal Antibody-Peptide Map Comparison Using the Agilent 70 Capillary Electrophoresis System and Agilent MatchCompare Software Application Note Biopharmaceutical Author Arunkumar
Supplementary Material. The MIAPE-GE glossary (MIAPE-GE version 1.4).
 Supplementary Material. The MIAPE-GE glossary (MIAPE-GE version 1.4). Classification Definition 1. General features 1.1.1 Date stamp The date on which the work described was initiated; given in the standard
Supplementary Material. The MIAPE-GE glossary (MIAPE-GE version 1.4). Classification Definition 1. General features 1.1.1 Date stamp The date on which the work described was initiated; given in the standard
Agilent 3100 OFFGEL Fractionator Kit
 Agilent 3100 OFFGEL Fractionator Kit Quick Start Guide Before you begin 2 Reagent Kit Checklist 2 Required Equipment and Reagents 2 Sample and Stock Solution Preparation Notes 3 Procedures 5 To prepare
Agilent 3100 OFFGEL Fractionator Kit Quick Start Guide Before you begin 2 Reagent Kit Checklist 2 Required Equipment and Reagents 2 Sample and Stock Solution Preparation Notes 3 Procedures 5 To prepare
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements
 Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Peptide enrichment and fractionation
 Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Proteomics and some of its Mass Spectrometric Applications
 Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Mass Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System
 GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
Lecture 5: 8/31. CHAPTER 5 Techniques in Protein Biochemistry
 Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
OMX-S Device for In-Gel Digestion
 INSTRUCTION MANUAL OMX-S Device for In-Gel Digestion (Cat. No. 39240.01, 39240.02) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 Heidelberg Phone +49-6221-138400, Fax +49-6221-1384010 e-mail: info@serva.de
INSTRUCTION MANUAL OMX-S Device for In-Gel Digestion (Cat. No. 39240.01, 39240.02) SERVA Electrophoresis GmbH Carl-Benz-Str. 7 D-69115 Heidelberg Phone +49-6221-138400, Fax +49-6221-1384010 e-mail: info@serva.de
Supporting Information I
 Single-shot top-down proteomics with capillary zone electrophoresiselectrospray ionization-tandem mass spectrometry for identification of nearly 600 Escherichia coli proteoforms Rachele A. Lubeckyj 1,,
Single-shot top-down proteomics with capillary zone electrophoresiselectrospray ionization-tandem mass spectrometry for identification of nearly 600 Escherichia coli proteoforms Rachele A. Lubeckyj 1,,
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins
 Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Proteomics and Vaccine Potency Testing
 Proteomics and Vaccine Potency Testing Louisa B. Tabatabai, PhD Respiratory Diseases of Livestock Research Unit National Animal Disease Center, ARS, USDA, Ames, Iowa What is proteomics Methodologies of
Proteomics and Vaccine Potency Testing Louisa B. Tabatabai, PhD Respiratory Diseases of Livestock Research Unit National Animal Disease Center, ARS, USDA, Ames, Iowa What is proteomics Methodologies of
Biotherapeutic Non-Reduced Peptide Mapping
 Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
SOP: PP021.6 Modified: 2/23/2017 by MCL Preparation of Purified Ag85 Individual Components (a, b, c)
 SOP: PP021.6 Modified: 2/23/2017 by MCL Preparation of Purified Ag85 Individual Components (a, b, c) Materials and Reagents: 1. Culture filtrate proteins (CFP) from M. tuberculosis, 300-600 mg 2. Ammonium
SOP: PP021.6 Modified: 2/23/2017 by MCL Preparation of Purified Ag85 Individual Components (a, b, c) Materials and Reagents: 1. Culture filtrate proteins (CFP) from M. tuberculosis, 300-600 mg 2. Ammonium
Characterize Fab and Fc Fragments by Cation-Exchange Chromatography
 Characterize Fab and Fc Fragments by Cation-Exchange Chromatography Application Note Biologics and Biosimilars Authors Isabel Vandenheede, Emmie Dumont, Pat Sandra, and Koen Sandra Research Institute for
Characterize Fab and Fc Fragments by Cation-Exchange Chromatography Application Note Biologics and Biosimilars Authors Isabel Vandenheede, Emmie Dumont, Pat Sandra, and Koen Sandra Research Institute for
Developing a natural partnership for rapid, sensitive identification of binding partners
 GE Healthcare Technology Note 24 Biacore systems Developing a natural partnership for rapid, sensitive identification of binding partners Biacore 3000 and mass spectrometry Summary The coupling of label-free
GE Healthcare Technology Note 24 Biacore systems Developing a natural partnership for rapid, sensitive identification of binding partners Biacore 3000 and mass spectrometry Summary The coupling of label-free
Amersham CyDye DIGE Fluor Labeling Kit for Scarce Samples
 GE Healthcare Amersham CyDye DIGE Fluor Labeling Kit for Scarce Samples Reagents for labeling protein with CyDye DIGE Fluor Cy3 and Cy5 saturation dyes, before 2-dimensional electrophoresis Product Booklet
GE Healthcare Amersham CyDye DIGE Fluor Labeling Kit for Scarce Samples Reagents for labeling protein with CyDye DIGE Fluor Cy3 and Cy5 saturation dyes, before 2-dimensional electrophoresis Product Booklet
CyDye DIGE Fluor Labelling Kit for Scarce Samples
 instructions product code 25-8009-83 25-8009-84 CyDye DIGE Fluor Labelling Kit for Scarce Samples Reagents for labelling protein with CyDye DIGE Fluor Cy 3 and Cy5 saturation dyes, before 2-dimensional
instructions product code 25-8009-83 25-8009-84 CyDye DIGE Fluor Labelling Kit for Scarce Samples Reagents for labelling protein with CyDye DIGE Fluor Cy 3 and Cy5 saturation dyes, before 2-dimensional
Fluorescent Difference Gel Electrophoresis
 1 of 5 8/21/2008 10:50 AM Please cite as: CSH Protocols; 2006; doi:10.1101/pdb.prot4233 Protocol Fluorescent Difference Gel Electrophoresis Angelika Görg, Oliver Drews, and Walter Weiss This protocol was
1 of 5 8/21/2008 10:50 AM Please cite as: CSH Protocols; 2006; doi:10.1101/pdb.prot4233 Protocol Fluorescent Difference Gel Electrophoresis Angelika Görg, Oliver Drews, and Walter Weiss This protocol was
Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer
 Supporting Information Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer Alexander S Hebert 1,2, Satendra Prasad 3, Michael W Belford 3, Derek J Bailey 3, Graeme C
Supporting Information Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer Alexander S Hebert 1,2, Satendra Prasad 3, Michael W Belford 3, Derek J Bailey 3, Graeme C
Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs
 Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Automation of MALDI-TOF Analysis for Proteomics
 Application Note #MT-50 BIFLEX III / REFLEX III Automation of MALDI-TOF Analysis for Proteomics D. Suckau, C. Köster, P.Hufnagel, K.-O. Kräuter and U. Rapp, Bruker Daltonik GmbH, Bremen, Germany Introduction
Application Note #MT-50 BIFLEX III / REFLEX III Automation of MALDI-TOF Analysis for Proteomics D. Suckau, C. Köster, P.Hufnagel, K.-O. Kräuter and U. Rapp, Bruker Daltonik GmbH, Bremen, Germany Introduction
Multiple SDS-PAGE on Vertical Electrophoresis Units
 Please cite as: CSH Protocols; 2006; doi:10.1101/pdb.prot4232 Protocol Multiple SDS-PAGE on Vertical Electrophoresis Units Angelika Görg, Oliver Drews, and Walter Weiss This protocol was adapted from "Separation
Please cite as: CSH Protocols; 2006; doi:10.1101/pdb.prot4232 Protocol Multiple SDS-PAGE on Vertical Electrophoresis Units Angelika Görg, Oliver Drews, and Walter Weiss This protocol was adapted from "Separation
Complete kit for the systematic analysis of protein kinases from a proteome such as cell lysate, tissue or microorganisms.
 Protein Kinase Connect (Small Format PEP) Catalog # AB000404 Complete kit for the systematic analysis of protein kinases from a proteome such as cell lysate, tissue or microorganisms. Please read this
Protein Kinase Connect (Small Format PEP) Catalog # AB000404 Complete kit for the systematic analysis of protein kinases from a proteome such as cell lysate, tissue or microorganisms. Please read this
Peptide-mass fingerprinting (PMF) for the characterization of histone variants
 BioScience Trends. 26; (5):357-364. Supporting Information Supplementary Materials and Methods Peptide-mass fingerprinting (PMF) for the characterization of histone variants For the digestion, each fraction
BioScience Trends. 26; (5):357-364. Supporting Information Supplementary Materials and Methods Peptide-mass fingerprinting (PMF) for the characterization of histone variants For the digestion, each fraction
Put the PRO in Protein Characterization
 Put the PRO in Protein Characterization Aaron Boice LC/Q-TOF Product Manager 5-June-2017 Agenda The Right Tools for the Job Tackling Intact Proteins Break it Down Peptides and PTMs Sweet Success Released
Put the PRO in Protein Characterization Aaron Boice LC/Q-TOF Product Manager 5-June-2017 Agenda The Right Tools for the Job Tackling Intact Proteins Break it Down Peptides and PTMs Sweet Success Released
Fast and Efficient Peptide Mapping of a Monoclonal Antibody (mab): UHPLC Performance with Superficially Porous Particles
 Fast and Efficient Peptide Mapping of a Monoclonal Antibody (mab): UHPLC Performance with Superficially Porous Particles Application Note Biotherapeutics and Biosimilars Authors James Martosella, Alex
Fast and Efficient Peptide Mapping of a Monoclonal Antibody (mab): UHPLC Performance with Superficially Porous Particles Application Note Biotherapeutics and Biosimilars Authors James Martosella, Alex
Advances in analytical biochemistry and systems biology: Proteomics
 Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research
 PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
The best proteomics: combination of upfront reduction of sample complexity, reproducible resolution, and effective follow up
 The best proteomics: combination of upfront reduction of sample complexity, reproducible resolution, and effective follow up Helen Kim, Ph.D. Dept of Pharmacology & Toxicology University of Alabama at
The best proteomics: combination of upfront reduction of sample complexity, reproducible resolution, and effective follow up Helen Kim, Ph.D. Dept of Pharmacology & Toxicology University of Alabama at
Two-Dimensional LC Protein Separation on Monolithic Columns in a Fully Automated Workflow
 Technical Note Two-Dimensional LC Protein Separation on Monolithic Columns in a Fully Automated Workflow Introduction The complex nature of proteomics samples requires high-resolution separation methods
Technical Note Two-Dimensional LC Protein Separation on Monolithic Columns in a Fully Automated Workflow Introduction The complex nature of proteomics samples requires high-resolution separation methods
Identification of Microprotein-Protein Interactions via APEX Tagging
 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
BACTERIAL PRODUCTION EXPRESSION METHOD OVERVIEW: PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT kda (full-length) 34.
 BACTERIAL PRODUCTION PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT 2015-XXXX XXXX pet-32a 50.9 kda (full-length) 34.0 kda (cleaved) EXPRESSION METHOD OVERVIEW: Plasmid DNA was transformed into BL21
BACTERIAL PRODUCTION PEF # GENE NAME EXPRESSION VECTOR MOLECULAR WEIGHT 2015-XXXX XXXX pet-32a 50.9 kda (full-length) 34.0 kda (cleaved) EXPRESSION METHOD OVERVIEW: Plasmid DNA was transformed into BL21
High-resolution Analysis of Charge Heterogeneity in Monoclonal Antibodies Using ph-gradient Cation Exchange Chromatography
 High-resolution Analysis of Charge Heterogeneity in Monoclonal Antibodies Using ph-gradient Cation Exchange Chromatography Agilent 1260 Infinity Bio-inert Quaternary LC System with Agilent Bio Columns
High-resolution Analysis of Charge Heterogeneity in Monoclonal Antibodies Using ph-gradient Cation Exchange Chromatography Agilent 1260 Infinity Bio-inert Quaternary LC System with Agilent Bio Columns
Introduction. Benefits of the SWATH Acquisition Workflow for Metabolomics Applications
 SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
The CebE/MsiK Transporter is a Doorway to the Cellooligosaccharide-mediated. Pathogenicity
 The CebE/MsiK Transporter is a Doorway to the Cellooligosaccharide-mediated Induction of Streptomyces scabies Pathogenicity Samuel Jourdan, Isolde Maria Francis, Min Jung Kim, Joren Jeico C. Salazar, Sören
The CebE/MsiK Transporter is a Doorway to the Cellooligosaccharide-mediated Induction of Streptomyces scabies Pathogenicity Samuel Jourdan, Isolde Maria Francis, Min Jung Kim, Joren Jeico C. Salazar, Sören
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins*
 Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
Appendix. Table of contents
 Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
CaptiveSpray nanobooster
 CaptiveSpray nanobooster The Revolution in Proteomics Ionization Innovation with Integrity LC-MS Source The Key to Performance and Reliability for nano flow MS Bruker CaptiveSpray principle: Stable and
CaptiveSpray nanobooster The Revolution in Proteomics Ionization Innovation with Integrity LC-MS Source The Key to Performance and Reliability for nano flow MS Bruker CaptiveSpray principle: Stable and
Universal Solution for Monoclonal Antibody Quantification in Biological Fluids Using Trap-Elute MicroLC-MS Method
 Universal Solution for Monoclonal Antibody Quantification in Biological Fluids Using Trap-Elute MicroLC-MS Method Featuring the SCIEX QTRAP 6500+ LC-MS/MS System with OptiFlow Turbo V source and M5 MicroLC
Universal Solution for Monoclonal Antibody Quantification in Biological Fluids Using Trap-Elute MicroLC-MS Method Featuring the SCIEX QTRAP 6500+ LC-MS/MS System with OptiFlow Turbo V source and M5 MicroLC
Click Chemistry Capture Kit
 http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
Rapid UHPLC Analysis of Reduced Monoclonal Antibodies using an Agilent ZORBAX Rapid Resolution High Definition (RRHD) 300SB-C8 Column
 Rapid UHP Analysis of Reduced Monoclonal Antibodies using an Agilent ZORBAX Rapid Resolution High Definition (RRHD) 3SB-C8 Column Application Note BioPharma Authors James Martosella, Phu Duong, Susanne
Rapid UHP Analysis of Reduced Monoclonal Antibodies using an Agilent ZORBAX Rapid Resolution High Definition (RRHD) 3SB-C8 Column Application Note BioPharma Authors James Martosella, Phu Duong, Susanne
ProteoExtract Protein Precipitation Kit Cat. No
 User Protocol 539180 Rev. 12-October-06 JSW ProteoExtract Protein Precipitation Kit Cat. No. 539180 Note that this user protocol is not lot-specific and is representative of the current specifications
User Protocol 539180 Rev. 12-October-06 JSW ProteoExtract Protein Precipitation Kit Cat. No. 539180 Note that this user protocol is not lot-specific and is representative of the current specifications
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein. Application
 Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
Comparison of different methods for purification analysis of a green fluorescent Strep-tag fusion protein Application Petra Sebastian Meike Kuschel Stefan Schmidt Abstract This Application Note describes
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH)
 User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
The effect of simulated microgravity on the Brassica napus seedling proteome
 10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
Biotherapeutic Peptide Mapping Information Dependent Acquisition (IDA) Method
 Biotherapeutic Peptide Mapping Information Dependent Acquisition (IDA) Method Routine biotherapeutic accurate mass peptide mapping analysis on the X500B QTOF System Method details for the routine peptide
Biotherapeutic Peptide Mapping Information Dependent Acquisition (IDA) Method Routine biotherapeutic accurate mass peptide mapping analysis on the X500B QTOF System Method details for the routine peptide
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note TOF/MS
 Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
mabs and ADCs analysis by RP
 mabs and ADCs analysis by RP Shanhua Lin, Ph.D. The world leader in serving science Protein and mab Separation by HPLC Size difference? YES Size Exclusion Chromatography (SEC) MAbPac SEC-1 NO NO Charge
mabs and ADCs analysis by RP Shanhua Lin, Ph.D. The world leader in serving science Protein and mab Separation by HPLC Size difference? YES Size Exclusion Chromatography (SEC) MAbPac SEC-1 NO NO Charge
timstof Pro powered by PASEF and the Evosep One for high speed and sensitive shotgun proteomics
 timstof Pro powered by PASEF and the Evosep One for high speed and sensitive shotgun proteomics The Parallel Accumulation Serial Fragmentation (PASEF) method for trapped ion mobility spectrometry (TIMS)
timstof Pro powered by PASEF and the Evosep One for high speed and sensitive shotgun proteomics The Parallel Accumulation Serial Fragmentation (PASEF) method for trapped ion mobility spectrometry (TIMS)
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS
 Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest and Lei Xiong SCIEX, Redwood City,
Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest and Lei Xiong SCIEX, Redwood City,
Computing with large data sets
 Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
pt7ht vector and over-expressed in E. coli as inclusion bodies. Cells were lysed in 6 M
 Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Supplementary Methods MIG6 production, purification, inhibition, and kinase assays MIG6 segment 1 (30mer, residues 334 364) peptide was synthesized using standard solid-phase peptide synthesis as described
Complete kit for the systematic analysis of protein kinases from a proteome such as cell lysate, tissue or microorganisms.
 Protein Kinase Landscape (Large Format PEP) Catalog # AB000504 Complete kit for the systematic analysis of protein kinases from a proteome such as cell lysate, tissue or microorganisms. Please read this
Protein Kinase Landscape (Large Format PEP) Catalog # AB000504 Complete kit for the systematic analysis of protein kinases from a proteome such as cell lysate, tissue or microorganisms. Please read this
FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE.
 User Protocol 122643 Rev. 12 May 2005 JSW Page 1 of 7 ProteoExtract Albumin/IgG Removal Kit, Maxi Cat. No. 122643 1. Introduction One of the major challenges in functional proteomics is the handling of
User Protocol 122643 Rev. 12 May 2005 JSW Page 1 of 7 ProteoExtract Albumin/IgG Removal Kit, Maxi Cat. No. 122643 1. Introduction One of the major challenges in functional proteomics is the handling of
2-D Fractionation Kit
 GE Healthcare 2-D Fractionation Kit Ettan sample preparation kits and reagents. Product Booklet Code: 80-6501-04 Page finder 1. Legal 3 2. Handling 4 2.1. Safety warnings and precautions 4 2.2. Storage
GE Healthcare 2-D Fractionation Kit Ettan sample preparation kits and reagents. Product Booklet Code: 80-6501-04 Page finder 1. Legal 3 2. Handling 4 2.1. Safety warnings and precautions 4 2.2. Storage
Bioanalytical LC-MS of monoclonal antibody therapeutics
 Bioanalytical LC-MS of monoclonal antibody therapeutics William D van Dongen, product manager pharma bioanalysis small molecules immunogenicity 40 years experience in bioanalysis Bioanalytical Expertise
Bioanalytical LC-MS of monoclonal antibody therapeutics William D van Dongen, product manager pharma bioanalysis small molecules immunogenicity 40 years experience in bioanalysis Bioanalytical Expertise
The best proteomics: combination of good gels, addressing quality control, and mining the data
 Mar 23,2010 UAB BMG744 The best proteomics: combination of good gels, addressing quality control, and mining the data Helen Kim, Ph.D. Dept of Pharmacology & Toxicology University of Alabama at Birmingham
Mar 23,2010 UAB BMG744 The best proteomics: combination of good gels, addressing quality control, and mining the data Helen Kim, Ph.D. Dept of Pharmacology & Toxicology University of Alabama at Birmingham
MassPREP On-Line Desalting Cartridge
 CONTENTS I. INTRODUCTION II. INSTALLING THE MASSPREP ON-LINE DESALTING CARTRIDGE INTO THE SENTRY.1 X 1 MM GUARD COLUMN HOLDER III. RECOMMENDED LC/MS SYSTEM CONFIGURATION TO MINIMIZE MS SOURCE CONTAMINATION
CONTENTS I. INTRODUCTION II. INSTALLING THE MASSPREP ON-LINE DESALTING CARTRIDGE INTO THE SENTRY.1 X 1 MM GUARD COLUMN HOLDER III. RECOMMENDED LC/MS SYSTEM CONFIGURATION TO MINIMIZE MS SOURCE CONTAMINATION
Improving Sensitivity for an Immunocapture LC-MS Assay of Infliximab in Rat Plasma Using Trap-and-Elute MicroLC-MS
 Improving Sensitivity for an Immunocapture LC-MS Assay of in Rat Plasma Using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest
Improving Sensitivity for an Immunocapture LC-MS Assay of in Rat Plasma Using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest
ADVANCING ATTRIBUTE CONTROL OF ANTIBODIES AND ITS DERIVATIVES USING HIGH RESOLUTION ANALYTICS
 ADVANCING ATTRIBUTE CONTROL OF ANTIBODIES AND ITS DERIVATIVES USING HIGH RESOLUTION ANALYTICS Henry Shion 1, Jing Fang 1, William Alley 1, Barbara Sullivan 2, Nick Tomczyk 3, Liuxi Chen 1, Ying-Qing Yu
ADVANCING ATTRIBUTE CONTROL OF ANTIBODIES AND ITS DERIVATIVES USING HIGH RESOLUTION ANALYTICS Henry Shion 1, Jing Fang 1, William Alley 1, Barbara Sullivan 2, Nick Tomczyk 3, Liuxi Chen 1, Ying-Qing Yu
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
Western Blot Tool Box
 Western Blot Tool Box BOX12/BOX12-03 V1.1 Store at 2-8 C For research use only Introduction The Western Blot Tool Box is designed to conveniently provide reagents/buffers needed for Western blotting, from
Western Blot Tool Box BOX12/BOX12-03 V1.1 Store at 2-8 C For research use only Introduction The Western Blot Tool Box is designed to conveniently provide reagents/buffers needed for Western blotting, from
LECTURE-3. Protein Chemistry to proteomics HANDOUT. Proteins are the most dynamic and versatile macromolecules in a living cell, which
 LECTURE-3 Protein Chemistry to proteomics HANDOUT PREAMBLE Proteins are the most dynamic and versatile macromolecules in a living cell, which regulates essential activities of the cell. The classical protein
LECTURE-3 Protein Chemistry to proteomics HANDOUT PREAMBLE Proteins are the most dynamic and versatile macromolecules in a living cell, which regulates essential activities of the cell. The classical protein
PLRP-S Polymeric Reversed-Phase Column for LC/MS Separation of mabs and ADC
 PLRP-S Polymeric Reversed-Phase Column for LC/MS Separation of mabs and ADC Analysis of Intact and Fragmented mabs and ADC Application Note Biotherapeutics and Biologics Author Suresh Babu C.V. Agilent
PLRP-S Polymeric Reversed-Phase Column for LC/MS Separation of mabs and ADC Analysis of Intact and Fragmented mabs and ADC Application Note Biotherapeutics and Biologics Author Suresh Babu C.V. Agilent
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
