SOP Title: Extraction and quantitative analysis of proteins from FFPE tissue
|
|
|
- Toby Benson
- 6 years ago
- Views:
Transcription
1 SOP Title: Extraction and quantitative analysis of proteins from FFPE tissue The STORE processing methods were shown to be fit-for purpose for DNA, RNA and protein extraction from FFPE material. The STORE characterisation methods were shown to be fit for-purpose for Quality Control of the extracted DNA, RNA and proteins.
2 Helmholtz Zentrum München German Research Center for Environmental Health Institute of Radiation Biology, Proteomics Group Standard Operating Procedure for Extraction and quantitative analysis of proteins from FFPE tissue Created Date: By: Dr. Omid Azimzadeh
3 Summary The following protocol describes the procedure of FFPE label-free quantitative proteomics analysis. The quality of the FFPE tissue samples is examined by conventional histology and and immunoblotting analysis. This is followed by protein extraction, using detergent containing buffer and accurate protein concentration measurement. The extracted proteins are subjected to be resolved on 1D SDS-PAGE before in-gel digestion and LC/MS-MS analysis. The obtained peptide spectra are quantified using the Progenesis software (Nonlinear). Safety considerations Gloves and protective clothing should be worn throughout. Reagents and Equipments Reagents needed: SDS and ammonium bicarbonate (Sigma, St. Luis, MO); RapiGest from (Waters, USA); acetone, acetonitrile, formic acid, trifluoroacetic acid (TFA) (Roth, Karlsuhe, Germany); dithiothreitol (DTT), iodoacetamide, urea, thiourea, tris-(hydroxymethyl) aminomethane (Tris), CHAPS and IPG buffer 3-10 (GE Healthcare, Freiburg, Germany). Sequencing grade trypsin (Promega, Madison, WI); cyano-4- hydroxycinnamic acid (Bruker Daltonik, Bremen, Germany). All solutions are prepared using HPLC grade water (Roth, Karlsuhe, Germany). Equipment needed: Staining dishes for deparaffinization of FFPE sections, microcentrifuge (with rotor for 1.5 ml tubes), vortexer, water bath or heating block capable of reaching 100 C, thermomixer (e.g. Eppendorf, Hamburg, Germany). Histology To examine the quality of the FFPE samples, tissue morphology and the tissue structure of the samples are controlled by histology. 3 µm paraffin sections are cut from the paraffin embedded tissue blocks for HE staining. Deparaffinisation of the microscope slides is performed according to the following protocol: xylene (4 changes), 96 % (v/v) ethanol (3 changes), 70 % (v/v) 2
4 ethanol (3 changes) followed by 2 washes by de-ionized water. Each step is performed for 3 minutes. For HE staining, slides are placed in Mayer s acid heamalum for 3 min, rinsed by running tap water 10 min, followed by incubating the slides in Eosin for 2 min. Slides are dehydrated in increasing ethanol series and mounted with Eukitt (Sigma- Aldrich, Germany) and cover slips from Menzel-Glas (Menzel, Germany). Images are taken with the Olympus Dotslide2.0 Slide scanning system. Immunoblotting Analysis To examine the quality of the extracted proteins from FFPE samples, the samples are controlled by western blot and antibody detection as follow: Solubilise 20 µg samples of the protein extract from FFPE tissue in SDS- PAGE sample buffer, and separate by 12% SDS-PAGE under reducing conditions. 2 Transfer proteins to nitrocellulose membranes (GE Healthcare) using a TE 77 semidry blotting system (GE Healthcare) at 1 ma/cm for 1h. Block the membranes using 3 % BSA in PBS, ph 7.4, for 1 h at room temperature wash three times in 10 mm Tris-HCl, ph 7.4, 150 mm NaCl for 5 min Incubate overnight at 4 C with antibodies against candidate proteins using dilutions recommended by the manufacturer. Wash the membrane 3 times as described above. Incubate the blots with secondary antibody and developed using the ECL system (GE Healthcare) following standard procedures. 3
5 Workflow of FFPE tissue label free quantitative proteomics Protein extraction The protein extraction and digestion would be performed as described before. 1 Place FFPE tissue sections (20 µm thick, 80mm 2 ) on microscope slides. Deparaffinise the slides by incubating twice with xylene for 10 min at room temperature before rehydration in a graded series of ethanol (100%, 95% and 70%) for 10 min each. Scrap the tissue sections from the slides and collect in 1.5 ml tubes. Wash the scrapped tissue with 0.5% beta-octylglucoside at RT for 10 min. Centrifuge the tubes at for 5 min at 14,000 g at 4 C. Resuspend the tissue in a extraction buffer containing 20 mm Tris-HCl, ph 8.8, 2% SDS, 1% beta-octylglucoside, 200 mm DTT, 200 mm glycine and mixture of protease inhibitor cocktail (Complete, Roche Diagnostics). Incubate samples in the buffer at 100 C for 20 min, and at 80 C for 2 h with shaking. Centrifuge the extract for 30 min at 14,000 g at 4 C. 4
6 Precipitate protein extract with the 2D clean up kit (GE Healthcare) following the manufacturer s instructions. Quantify the protein amount from the pellet resuspended in Tris buffer in triplicate using both Bradford method and 2D Quant Kit (GE Healthcare) following the manufacturer s instructions. Protein separation 1D SDS PAGE Solubilise 60 µg samples of the protein extract from FFPE tissue in SDS- PAGE sample buffer, and separate by 12% SDS-PAGE under reducing conditions. 2 Stain the gel with Brilliant blue G In-gel Digestion Cut 1D stained gel slices from polyacrylamide gel and transfer into an Eppendorf tube. Wash 1x 10 minutes with 200 μl 60 % acetonitrile in 50 mm NH 4 HCO 3. Wash 1x 10 minutes with 200 μl H 2 O. Add 100 μl 5 mm DTT, incubate 15 minutes 60 C. Remove DTT solution from gel pieces. Add 100 μl freshly prepared 25 mm iodacetamide (0.046 g / 10 ml), incubate 15 minutes at room temperature in the dark. Remove iodacetamid from gel pieces. Wash 1 x 5 minutes with 100 μl H 2 O. Wash 1 x 10 minutes with 100 μl 100 % acetonitrile. Wash 1 x 10 minutes with 100 μl 50 mm ammoniumbicarbonate. Wash 1 x 10 minutes with 100 μl 60 % acetonitrile. Wash 1 x 10 minutes with 100 μl 100 % acetonitrile. Prepare the trypsin stock with 1 mm HCl as described in the manual. Trypsin digestion: Overlay the gel pieces with 0,01 μg / μl trypsin diluted in 50 mm ammoniumbicarbonat, and digest the samples overnight at 37 C. 5
7 Peptide elution: Add 1-2 μl 0.5 % TFA, shake 15 minutes to stop the trypsin activity. Transfer supernatant to a tube and combine it with the solutions from the next two elution steps. Add enough volume of 60 % acetonitrile / 0.1 %TFA in order to cover the gel pieces completely and shake for 15 minutes. Add 100 % acetonitrile / 0.1 % TFA and shake again for 15 minutes and combine with the other fractions. As a result the gel pieces should shrink and become white. Dry samples in the speed vac to complete dryness. Re-dissolve in 2 % ACN / 0.5 % TFA. Mass Spectrometry LC-MSMS analysis is performed on an Ultimate3000 nano HPLC system (Dionex, Sunnyvale, CA) online coupled to a LTQ OrbitrapXL mass spectrometer (Thermo Fisher Scientific) by a nano spray ion source. Peptides generated from in-gel digests of FFPE are acidified with TFA and automatically loaded to the HPLC system equipped with a nano trap column (100 μm i.d. 2 cm, packed with Acclaim PepMap100 C18, 5 μm, 100 Å, Dionex) at a flow rate of 30 μl/minute in 5% buffer B (80% acetonitrile, 0.1% formic acid (FA) in HPLCgrade water) and 95% buffer A (5% acetonitrile, 0.1% FA in HPLC-grade water). After 5 min, the peptides are eluted and separated on the analytical column (75 μm i.d. 15 cm, Acclaim PepMap100 C18, 3 μm, 100Å, Dionex) by a gradient from 5% to 50% of buffer B at 300 nl/minute flow over 140 min. Remaining peptides are eluted by a short gradient from 50% to 100% buffer B in 5 min. The eluting peptides are analysed by the LTQ OrbitrapXL. From the high resolution MS pre-scan, the five most intense peptide ions are selected for fragment analysis in the linear ion trap if they exceed an intensity of at least 200 counts and if they are at least doubly charged. The normalised collision energy for CID is set to a value of 35 and the resulting fragments are detected with normal resolution in the linear ion trap. The lock mass option is activated, and the background signal with the mass of is used as lock mass. Every ion selected for fragmentation is excluded for 30s by dynamic exclusion. 6
8 Label-Free Peptide Quantification The label-free quantitative approach is performed using Progenesis software (version 2.5, Nonlinear). 4 Profile data of the MS scans are transformed to peak lists with Progenesis LC-MS using a proprietary algorithm. This method uses wavelet-based filtering to smooth the peak envelopes and identify noisy areas, i.e. areas deemed to contain no ion peaks. Peaks are then modelled in non-noisy areas to record their peak m/z value, intensity, abundance (area under the peak) and m/z width. MS/MS spectra are transformed similarly and then stored in peak lists comprising m/z and abundance. After selecting one sample as a reference, the retention times of all other samples within the experiment are aligned by manually creating three to five landmarks followed by automatic alignment of all retention times to maximal overlay of the 2D feature maps. Features with only one charge or more than seven charges are masked at this point and excluded from further analyses. After alignment and feature exclusion, samples are divided into the appropriate groups (healthy and ERU), and raw abundances of all features are normalized. Normalization corrects for factors resulting from experimental variation and is automatically calculated over all features in all samples. It results in a unique factor for each sample that corrects all features in the sample in a similar way for experimental variation. After normalization, statistical analysis is performed using transformed ( log-like arcsinh(.)function) normalized abundances for one-way analysis of variance (ANOVA) calculations of all detected features. No minimal thresholds are set neither for the method of peak picking nor selection of data to use for quantification. Co-detection across all runs ensures that abundance data used for relative quantification is obtained for every peptide ion in every run resulting in data variance that is representative of the full dataset. For quantification, all peptides (with Mascot score 30 and p < 0.01) of an identified protein are included and the total cumulative abundance is calculated by summing the abundances of all peptides allocated to the respective protein. Calculations of the protein p value (one-way ANOVA) are then performed on the sum of the normalised abundances across all runs. ANOVA values of p 0.05 and additionally regulation of 1.5-fold or 0.7-fold are regarded as significant for all further results. After quantification of peptides, those features not having an MS/MS scan from the initial samples run are exported to Excel (Microsoft) and used as an inclusion list for a replicate run of samples on the Orbitrap. Resulting raw data files are aligned to the 7
9 experiments in Progenesis and additional MS/MS scans resulting from these measurements are added to the previous ones; however, the original quantifications and statistics are not changed. Database Search and Protein Identification All MS/MS spectra are exported from the Progenesis software as Mascot generic file (mgf) and used for peptide identification with Mascot (version 2.2) in the Ensembl database for Mus musculus. Search parameters used are: 10 ppm peptide mass tolerance and 1-Da fragment mass tolerance, one missed cleavage allowed, carbamidomethylation is set as fixed modification and methionine oxidation, Phosphorylation of tyrosine, serine, and threonine are allowed as variable modifications. A Mascot-integrated decoy database search calculates a false discovery rate. Searching is performed on the concatenated mgf files with an ion score cut-off of 30 and a significance threshold of p Only peptides with ion scores of 30 and above and only proteins with at least one unique peptide ranked as top candidate (bold red in Mascot) are considered and re-imported into Progenesis software. REFERENCES 1. Azimzadeh, O.; Barjaktarovic, Z.; Aubele, M.; Calzada-Wack, J.; Sarioglu, H.; Atkinson, M. J.; Tapio, S., Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J Proteome Res 9, (9), Laemmli, U. K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, (5259), Heukeshoven, J.; Dernick, R., Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. ELECTROPHORESIS 1985, 6, (3), Hauck, S. M.; Dietter, J.; Kramer, R. L.; Hofmaier, F.; Zipplies, J. K.; Amann, B.; Feuchtinger, A.; Deeg, C. A.; Ueffing, M., Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol Cell Proteomics 9, (10),
Supporting information
 Supporting information Simple and Integrated Spintip-based Technology Applied for Deep Proteome Profiling Wendong Chen,, Shuai Wang, Subash Adhikari, Zuhui Deng, Lingjue Wang, Lan Chen, Mi Ke, Pengyuan
Supporting information Simple and Integrated Spintip-based Technology Applied for Deep Proteome Profiling Wendong Chen,, Shuai Wang, Subash Adhikari, Zuhui Deng, Lingjue Wang, Lan Chen, Mi Ke, Pengyuan
Automated platform of µlc-ms/ms using SAX trap column for
 Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
2D difference gel electrophoresis (2D-DIGE) Spot picking and in-gel digestion
 2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the
2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins*
 Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
Click-&-Go TM Protein Enrichment Kit *for capture alkyne-modified proteins* Product No. 1039 Introduction The Click-&-Go TM Protein Enrichment Kit is an efficient, biotin-streptavidin free tool for capture
Click Chemistry Capture Kit
 http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
http://www.clickchemistrytools.com tel: 480 584 3340 fax: 866 717 2037 Click Chemistry Capture Kit Product No. 1065 Introduction The Click Chemistry Capture Kit provides all the necessary reagents* to
Appendix. Table of contents
 Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
Objective. Introduction. IP assisted LC/MS/MS making study protein complexes easy. Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2
 IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
IP assisted LC/MS/MS making study protein complexes easy Jon Hao 1, Yi Liu 1, Xiaozhi Ren 2, and King-Wai Yau 2 1 Poochon Scientific, Frederick, Maryland 21701 2 Department of Neuroscience, Johns Hopkins
FFPE DNA Extraction Protocol
 FFPE DNA Extraction Protocol Introduction The number of archival formalin-fixed paraffin embedded (FFPE) samples is in the millions, providing an invaluable repository of information for genetic analysis.
FFPE DNA Extraction Protocol Introduction The number of archival formalin-fixed paraffin embedded (FFPE) samples is in the millions, providing an invaluable repository of information for genetic analysis.
Cell Signaling Technology
 Cell Signaling Technology PTMScan Direct: Multipathway v2.0 Proteomics Service Group January 14, 2013 PhosphoScan Deliverables Project Overview Methods PTMScan Direct: Multipathway V2.0 (Tables 1,2) Qualitative
Cell Signaling Technology PTMScan Direct: Multipathway v2.0 Proteomics Service Group January 14, 2013 PhosphoScan Deliverables Project Overview Methods PTMScan Direct: Multipathway V2.0 (Tables 1,2) Qualitative
E.Z.N.A. FFPE RNA Kit. R preps R preps R preps
 E.Z.N.A. FFPE RNA Kit R6954-00 5 preps R6954-01 50 preps R6954-02 200 preps January 2015 E.Z.N.A. FFPE RNA Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing
E.Z.N.A. FFPE RNA Kit R6954-00 5 preps R6954-01 50 preps R6954-02 200 preps January 2015 E.Z.N.A. FFPE RNA Kit Table of Contents Introduction and Overview...2 Kit Contents/Storage and Stability...3 Preparing
Peptide enrichment and fractionation
 Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Protein sample preparation Successful analysis of low-abundance proteins and/or the identification of posttranslationally modified peptides often require several steps: enrichment, fractionation, and/or
Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection.
 Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection Application Note Christian Wenz Andreas Rüfer Abstract Agilent Equipment Agilent 3100
Alternative to 2D gel electrophoresis OFFGEL electrophoresis combined with high-sensitivity on-chip protein detection Application Note Christian Wenz Andreas Rüfer Abstract Agilent Equipment Agilent 3100
GenElute FFPE DNA Purification Kit. Catalog number DNB400 Storage temperature -20 ºC TECHNICAL BULLETIN
 GenElute FFPE DNA Purification Kit Catalog number DNB400 Storage temperature -20 ºC TECHNICAL BULLETIN Product Description GenElute FFPE DNA Purification Kit provides a rapid method for the isolation and
GenElute FFPE DNA Purification Kit Catalog number DNB400 Storage temperature -20 ºC TECHNICAL BULLETIN Product Description GenElute FFPE DNA Purification Kit provides a rapid method for the isolation and
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research
 PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
SOP: PP021.6 Modified: 2/23/2017 by MCL Preparation of Purified Ag85 Individual Components (a, b, c)
 SOP: PP021.6 Modified: 2/23/2017 by MCL Preparation of Purified Ag85 Individual Components (a, b, c) Materials and Reagents: 1. Culture filtrate proteins (CFP) from M. tuberculosis, 300-600 mg 2. Ammonium
SOP: PP021.6 Modified: 2/23/2017 by MCL Preparation of Purified Ag85 Individual Components (a, b, c) Materials and Reagents: 1. Culture filtrate proteins (CFP) from M. tuberculosis, 300-600 mg 2. Ammonium
PreAnalytiX Supplementary Protocol
 PreAnalytiX Supplementary Protocol Purification of full-length proteins from sections of PAXgene Tissue fixed, paraffin-embedded (PFPE) tissue This protocol describes using the Qproteome FFPE Tissue kit
PreAnalytiX Supplementary Protocol Purification of full-length proteins from sections of PAXgene Tissue fixed, paraffin-embedded (PFPE) tissue This protocol describes using the Qproteome FFPE Tissue kit
Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham. Bioscience) were washed four times with 1 mm HCl and twice with coupling buffer
 Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
Supporting Information Materials and Methods Affinity chromatography. Three-hundred microliters of N-hydroxysuccinimideactivated Separose 4 Fast Flow beads stored in 100% isopropanol (Amersham Bioscience)
FFPE DNA Extraction kit
 FFPE DNA Extraction kit Instruction Manual FFPE DNA Extraction kit Cat. No. C20000030, Format 50 rxns Version 1 / 14.08.13 Content Introduction 4 Overview of FFPE DNA extraction workflow 4 Kit contents
FFPE DNA Extraction kit Instruction Manual FFPE DNA Extraction kit Cat. No. C20000030, Format 50 rxns Version 1 / 14.08.13 Content Introduction 4 Overview of FFPE DNA extraction workflow 4 Kit contents
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS
 Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-DNA FFPE Kit Catalog Nos. D3067 Highlights Streamlined purification of high-quality FFPE tissue DNA that is ideal for PCR, Next-Gen library prep, enzymatic manipulations, etc.
INSTRUCTION MANUAL Quick-DNA FFPE Kit Catalog Nos. D3067 Highlights Streamlined purification of high-quality FFPE tissue DNA that is ideal for PCR, Next-Gen library prep, enzymatic manipulations, etc.
The effect of simulated microgravity on the Brassica napus seedling proteome
 10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu *
 RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu * School of Biomedical Sciences, Thomas Jefferson University, Philadelphia, USA *For correspondence:
RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu * School of Biomedical Sciences, Thomas Jefferson University, Philadelphia, USA *For correspondence:
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
ProteoExtract Protein Precipitation Kit Cat. No
 User Protocol 539180 Rev. 12-October-06 JSW ProteoExtract Protein Precipitation Kit Cat. No. 539180 Note that this user protocol is not lot-specific and is representative of the current specifications
User Protocol 539180 Rev. 12-October-06 JSW ProteoExtract Protein Precipitation Kit Cat. No. 539180 Note that this user protocol is not lot-specific and is representative of the current specifications
Protocol(Research use only)
 Immunohistochemistry (without pretreatment) p2 Immunohistochemistry (Microwave pretreatment) p3 Immunohistochemistry (Autoclave pretreatment) p4 Immunohistochemistry (Trypsin pretreatment) p5 Immunohistochemistry
Immunohistochemistry (without pretreatment) p2 Immunohistochemistry (Microwave pretreatment) p3 Immunohistochemistry (Autoclave pretreatment) p4 Immunohistochemistry (Trypsin pretreatment) p5 Immunohistochemistry
Supporting Information I
 Single-shot top-down proteomics with capillary zone electrophoresiselectrospray ionization-tandem mass spectrometry for identification of nearly 600 Escherichia coli proteoforms Rachele A. Lubeckyj 1,,
Single-shot top-down proteomics with capillary zone electrophoresiselectrospray ionization-tandem mass spectrometry for identification of nearly 600 Escherichia coli proteoforms Rachele A. Lubeckyj 1,,
Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium
 Supplementary data itraq-based Proteomics Analyses Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium supplemented with 5g L -1 glucose for 48 hours at 37 C with
Supplementary data itraq-based Proteomics Analyses Protein preparation and digestion. P. aeruginosa strains were grown in in AB minimal medium supplemented with 5g L -1 glucose for 48 hours at 37 C with
GeneMATRIX Tissue DNA Purification Kit
 GeneMATRIX Tissue DNA Purification Kit Kit for isolation of total DNA from human and animal tissues Cat. no. E3550 Version 5.1 April, 2008 Distributor: Roboklon GmbH Robert-Rössle-Str.10 B55 13125 Berlin
GeneMATRIX Tissue DNA Purification Kit Kit for isolation of total DNA from human and animal tissues Cat. no. E3550 Version 5.1 April, 2008 Distributor: Roboklon GmbH Robert-Rössle-Str.10 B55 13125 Berlin
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements
 Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
SUPPLEMENTARY INFORMATION
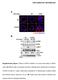 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
IEF-2DE Analysis and Protein Identification Xia Wu 1, Steven J. Clough 2, Steven C. Huber 3 and Man-Ho Oh 4*
 IEF-2DE Analysis and Protein Identification Xia Wu 1, Steven J. Clough 2, Steven C. Huber 3 and Man-Ho Oh 4* 1 Genome Sciences, University of Washington, Seattle, USA; 2 Crop Sciences, USDA-ARS and the
IEF-2DE Analysis and Protein Identification Xia Wu 1, Steven J. Clough 2, Steven C. Huber 3 and Man-Ho Oh 4* 1 Genome Sciences, University of Washington, Seattle, USA; 2 Crop Sciences, USDA-ARS and the
The preparation of native chromatin from cultured human cells.
 Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Component. Buffer RL
 GenElute FFPE RNA Purification Catalog number RNB400 Product Description Sigma s GenElute FFPE RNA Purification Kit provides a rapid method for the isolation and purification of total RNA (including microrna)
GenElute FFPE RNA Purification Catalog number RNB400 Product Description Sigma s GenElute FFPE RNA Purification Kit provides a rapid method for the isolation and purification of total RNA (including microrna)
The CebE/MsiK Transporter is a Doorway to the Cellooligosaccharide-mediated. Pathogenicity
 The CebE/MsiK Transporter is a Doorway to the Cellooligosaccharide-mediated Induction of Streptomyces scabies Pathogenicity Samuel Jourdan, Isolde Maria Francis, Min Jung Kim, Joren Jeico C. Salazar, Sören
The CebE/MsiK Transporter is a Doorway to the Cellooligosaccharide-mediated Induction of Streptomyces scabies Pathogenicity Samuel Jourdan, Isolde Maria Francis, Min Jung Kim, Joren Jeico C. Salazar, Sören
Protocol for in vitro transcription
 Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
Protocol for in vitro transcription Assemble the reaction at room temperature in the following order: Component 10xTranscription Buffer rntp T7 RNA Polymerase Mix grna PCR DEPC H 2 O volume 2μl 2μl 2μl
(Spin Column) For purification of DNA and RNA from formalin-fixed, paraffin-embedded. tissue sections. Instruction for Use. For Research Use Only
 AmoyDx FFPE DNA/RNA Kit (Spin Column) For purification of DNA and RNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use For Research Use Only Instruction Version: B1.0 Revision
AmoyDx FFPE DNA/RNA Kit (Spin Column) For purification of DNA and RNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use For Research Use Only Instruction Version: B1.0 Revision
Western Blot Tool Box
 Western Blot Tool Box BOX12/BOX12-03 V1.1 Store at 2-8 C For research use only Introduction The Western Blot Tool Box is designed to conveniently provide reagents/buffers needed for Western blotting, from
Western Blot Tool Box BOX12/BOX12-03 V1.1 Store at 2-8 C For research use only Introduction The Western Blot Tool Box is designed to conveniently provide reagents/buffers needed for Western blotting, from
Proteomics. Proteomics is the study of all proteins within organism. Challenges
 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B
 Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
ReliaPrep FFPE gdna Miniprep System
 TECHNICAL MANUAL ReliaPrep FFPE gdna Miniprep System Instructions for Use of Products A2351 and A2352 Revised 12/15 TM352 ReliaPrep FFPE gdna Miniprep System All technical literature is available at: www.promega.com/protocols/
TECHNICAL MANUAL ReliaPrep FFPE gdna Miniprep System Instructions for Use of Products A2351 and A2352 Revised 12/15 TM352 ReliaPrep FFPE gdna Miniprep System All technical literature is available at: www.promega.com/protocols/
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore, India
 Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
(Spin Column) For purification of DNA and RNA from formalin-fixed, paraffin-embedded tissue sections. Instruction for Use
 AmoyDx FFPE DNA/RNA Kit (Spin Column) For purification of DNA and RNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use Instruction Version: B1.3 Revision Date: September 2015
AmoyDx FFPE DNA/RNA Kit (Spin Column) For purification of DNA and RNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use Instruction Version: B1.3 Revision Date: September 2015
Supplementary Fig. 1. S-1. Supplementary Fig. 2. S-2. Supplementary Fig. 3. S-3. Supplementary Fig. 4. S-4. Supplementary Fig. 5.
 Supplementary Information - An IonStar experimental strategy for MS1 ion current-based quantification using ultra-high-field Orbitrap: reproducible, in-depth and accurate protein measurement in larger
Supplementary Information - An IonStar experimental strategy for MS1 ion current-based quantification using ultra-high-field Orbitrap: reproducible, in-depth and accurate protein measurement in larger
DNA isolation from tissue DNA isolation from eukaryotic cells (max. 5 x 106 cells) DNA isolation from paraffin embedded tissue
 INDEX KIT COMPONENTS 3 STORAGE AND STABILITY 3 BINDING CAPACITY 3 INTRODUCTION 3 IMPORTANT NOTES 4 EUROGOLD TISSUE DNA MINI KIT PROTOCOLS 5 A. DNA isolation from tissue 5 B. DNA isolation from eukaryotic
INDEX KIT COMPONENTS 3 STORAGE AND STABILITY 3 BINDING CAPACITY 3 INTRODUCTION 3 IMPORTANT NOTES 4 EUROGOLD TISSUE DNA MINI KIT PROTOCOLS 5 A. DNA isolation from tissue 5 B. DNA isolation from eukaryotic
Biotherapeutic Non-Reduced Peptide Mapping
 Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
Biotherapeutic Non-Reduced Peptide Mapping Routine non-reduced peptide mapping of biotherapeutics on the X500B QTOF System Method details for the routine non-reduced peptide mapping of a biotherapeutic
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Supplementary Figures Supplementary Figure 1. FANCD2 Western blot. Lane 1 marker, lane 2 LN9SV, lane 3 LN9SV treated with hydroxyurea (HU), lane 4 GM6914, lane 5 GM6914 treated with HU, lane 6 VU697L,
Method Optimisation in Bottom-Up Analysis of Proteins
 Method Optimisation in Bottom-Up Analysis of Proteins M. Styles, 1 D. Smith, 2 J. Griffiths, 2 L. Pereira, 3 T. Edge 3 1 University of Manchester, UK; 2 Patterson Institute, Manchester, UK; 3 Thermo Fisher
Method Optimisation in Bottom-Up Analysis of Proteins M. Styles, 1 D. Smith, 2 J. Griffiths, 2 L. Pereira, 3 T. Edge 3 1 University of Manchester, UK; 2 Patterson Institute, Manchester, UK; 3 Thermo Fisher
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis
 Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Investigating Biological Variation of Liver Enzymes in Human Hepatocytes
 Investigating Biological Variation of Liver Enzymes in Human Hepatocytes SWATH Acquisition on the TripleTOF Systems Xu Wang 1, Hui Zhang 2, Christie Hunter 1 1 SCIEX, USA, 2 Pfizer, USA MS/MS ALL with
Investigating Biological Variation of Liver Enzymes in Human Hepatocytes SWATH Acquisition on the TripleTOF Systems Xu Wang 1, Hui Zhang 2, Christie Hunter 1 1 SCIEX, USA, 2 Pfizer, USA MS/MS ALL with
Liver Mitochondria Proteomics Employing High-Resolution MS Technology
 Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Instructions SILAC Protein Quantitation Kits
 Instructions SILAC Protein Quantitation Kits CIL Catalog No. Description DMEM LYS C SILAC Protein Quantitation Kit DMEM (Dulbecco s Modified Eagle s Medium) Kit contains: SILAC DMEM Media, 2 x 500 ml Dialyzed
Instructions SILAC Protein Quantitation Kits CIL Catalog No. Description DMEM LYS C SILAC Protein Quantitation Kit DMEM (Dulbecco s Modified Eagle s Medium) Kit contains: SILAC DMEM Media, 2 x 500 ml Dialyzed
Identification of human serum proteins detectable after Albumin removal with Vivapure Anti-HSA Kit
 Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Characterization of a fusion protein under native and denaturing conditions with maxis II
 Characterization of a fusion protein under native and denaturing conditions with maxis II Abstract The maxis II UHR-QTOF was used to characterize key attributes of a fusion protein in its native conformation
Characterization of a fusion protein under native and denaturing conditions with maxis II Abstract The maxis II UHR-QTOF was used to characterize key attributes of a fusion protein in its native conformation
Spectral Counting Approaches and PEAKS
 Spectral Counting Approaches and PEAKS INBRE Proteomics Workshop, April 5, 2017 Boris Zybailov Department of Biochemistry and Molecular Biology University of Arkansas for Medical Sciences 1. Introduction
Spectral Counting Approaches and PEAKS INBRE Proteomics Workshop, April 5, 2017 Boris Zybailov Department of Biochemistry and Molecular Biology University of Arkansas for Medical Sciences 1. Introduction
AmoyDx FFPE DNA Kit. (Spin Column) For purification of DNA from formalin-fixed, paraffin-embedded tissue sections. Instruction for Use
 AmoyDx FFPE DNA Kit (Spin Column) For purification of DNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use 8.0223501X036G 36 tests Amoy Diagnostics Co., Ltd. 39 Dingshan Road,
AmoyDx FFPE DNA Kit (Spin Column) For purification of DNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use 8.0223501X036G 36 tests Amoy Diagnostics Co., Ltd. 39 Dingshan Road,
SILAC Protein Quantitation Kits
 INSTRUCTIONS SILAC Protein Quantitation Kits A33969 A33970 A33971 A33972 A33973 MAN0016245 Rev C.0 Pub. Part No. 2161996.8 Number A33969 Description SILAC Protein Quantitation Kit (LysC) DMEM DMEM for
INSTRUCTIONS SILAC Protein Quantitation Kits A33969 A33970 A33971 A33972 A33973 MAN0016245 Rev C.0 Pub. Part No. 2161996.8 Number A33969 Description SILAC Protein Quantitation Kit (LysC) DMEM DMEM for
EZ-10 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK
 EZ-0 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK (Bacteria, Plant, Animal, Blood) Version 8 Rev 05/0/03 EZ-0 Genomic DNA Kit Handbook Table of Contents Introduction Limitations of Use Features Applications
EZ-0 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK (Bacteria, Plant, Animal, Blood) Version 8 Rev 05/0/03 EZ-0 Genomic DNA Kit Handbook Table of Contents Introduction Limitations of Use Features Applications
1. QUANTITY OF LYSATE 2. LYSIS BUFFER
 SAMPLE PREPARATION 1. QUANTITY OF LYSATE The amount of protein requested for the Kinex KAM-880 Antibody Microarray service is 100 µg per sample at an approximate concentration of 2 mg/ml. If your samples
SAMPLE PREPARATION 1. QUANTITY OF LYSATE The amount of protein requested for the Kinex KAM-880 Antibody Microarray service is 100 µg per sample at an approximate concentration of 2 mg/ml. If your samples
Table of contents. NucleoSpin mirna Separation of small and large RNA
 Table of contents 1 Protocols 2 1.1 RNA purification from animal tissue and cultured cells with separation in small and large RNA 2 1.2 RNA purification from plant tissue 7 1.3 Fractionation of pre-purified
Table of contents 1 Protocols 2 1.1 RNA purification from animal tissue and cultured cells with separation in small and large RNA 2 1.2 RNA purification from plant tissue 7 1.3 Fractionation of pre-purified
Separation of Native Monoclonal Antibodies and Identification of Charge Variants:
 Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-DNA/RNA FFPE Kit Catalog No. R1009 Highlights High performance sample prep technology for total nucleic acid (DNA & RNA) recovery from the same FFPE tissue sample/section. High
INSTRUCTION MANUAL Quick-DNA/RNA FFPE Kit Catalog No. R1009 Highlights High performance sample prep technology for total nucleic acid (DNA & RNA) recovery from the same FFPE tissue sample/section. High
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD
 Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Technical Note Detection of post-immunoprecipitation proteins by Western blot using the Quick Western Kit IRDye 680RD Developed for: Aerius, Odyssey Classic, Odyssey CLx and Odyssey Sa Imaging Systems
Cat. # MK600. For Research Use. ApopLadder Ex. Product Manual. v201608da
 Cat. # MK600 For Research Use ApopLadder Ex Product Manual Table of Contents I. Description... 3 II. Principle... 3 III. Features... 3 IV. Components... 3 V. Storage... 3 VI. Materials Required but not
Cat. # MK600 For Research Use ApopLadder Ex Product Manual Table of Contents I. Description... 3 II. Principle... 3 III. Features... 3 IV. Components... 3 V. Storage... 3 VI. Materials Required but not
relative fitness score (12 M SeMet)
 4.00 relative fitness score (12 M SeMet) 2.00 0.00-2.00-4.00-6.00-6.00-4.00-2.00 0.00 2.00 4.00 relative fitness score (20 M SeMet) Supplementary Figure 1. Relative fitness scores of the deletion strains
4.00 relative fitness score (12 M SeMet) 2.00 0.00-2.00-4.00-6.00-6.00-4.00-2.00 0.00 2.00 4.00 relative fitness score (20 M SeMet) Supplementary Figure 1. Relative fitness scores of the deletion strains
ProteinPilot Software Overview
 ProteinPilot Software Overview High Quality, In-Depth Protein Identification and Protein Expression Analysis Sean L. Seymour and Christie L. Hunter SCIEX, USA As mass spectrometers for quantitative proteomics
ProteinPilot Software Overview High Quality, In-Depth Protein Identification and Protein Expression Analysis Sean L. Seymour and Christie L. Hunter SCIEX, USA As mass spectrometers for quantitative proteomics
LavaDigest Protease Monitoring Kit
 LavaDigest Protease Monitoring Kit Ordering LP-031020 LavaDigest protease monitoring kit for up to 2000 assays Order from http://www.fluorotechnics.com.au L.avaDigest Protocol March 2013 Content 2 LavaDigest
LavaDigest Protease Monitoring Kit Ordering LP-031020 LavaDigest protease monitoring kit for up to 2000 assays Order from http://www.fluorotechnics.com.au L.avaDigest Protocol March 2013 Content 2 LavaDigest
For Research Use Only. Not for use in diagnostic procedures.
 Printed December 13, 2011 Version 1.0 For Research Use Only. Not for use in diagnostic procedures. DDDDK-tagged Protein PURIFICATION GEL with Elution Peptide (MoAb. clone FLA-1) CODE No. 3326 / 3327 PURIFICATION
Printed December 13, 2011 Version 1.0 For Research Use Only. Not for use in diagnostic procedures. DDDDK-tagged Protein PURIFICATION GEL with Elution Peptide (MoAb. clone FLA-1) CODE No. 3326 / 3327 PURIFICATION
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins
 Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
EPIGENTEK. EpiQuik Tissue Chromatin Immunoprecipitation Kit. Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
OPPF-UK Standard Protocols: Mammalian Expression
 OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
ProteinPilot Report for ProteinPilot Software
 ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
Quantification of Isotope Encoded Proteins in 2D Gels
 Quantification of Isotope Encoded Proteins in 2D Gels Using Surface Enhanced Resonance Raman Giselle M. Knudsen 1, Brandon M. Davis 2, Shirshendu K. Deb 1, Yvette Loethen 2, Ravindra Gudihal 1, Pradeep
Quantification of Isotope Encoded Proteins in 2D Gels Using Surface Enhanced Resonance Raman Giselle M. Knudsen 1, Brandon M. Davis 2, Shirshendu K. Deb 1, Yvette Loethen 2, Ravindra Gudihal 1, Pradeep
Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde
 Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Supplementary text Supplementary materials and methods Histopathological examination Segments of the obstructed intestinal loops were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin wax with
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System
 Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
Isotopic Resolution of Chromatographically Separated IdeS Subunits Using the X500B QTOF System Fan Zhang 2, Sean McCarthy 1 1 SCIEX, MA, USA, 2 SCIEX, CA USA Analysis of protein subunits using high resolution
ProMass HR Applications!
 ProMass HR Applications! ProMass HR Features Ø ProMass HR includes features for high resolution data processing. Ø ProMass HR includes the standard ProMass deconvolution algorithm as well as the full Positive
ProMass HR Applications! ProMass HR Features Ø ProMass HR includes features for high resolution data processing. Ø ProMass HR includes the standard ProMass deconvolution algorithm as well as the full Positive
Product # Specifications. Kit Specifications Maximum Column Binding Capacity (gdna) Maximum Column Loading Volume.
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com FFPE DNA Purification Kit Product # 47400 Product Insert Norgen
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com FFPE DNA Purification Kit Product # 47400 Product Insert Norgen
Analysis of Monoclonal Antibodies and Their Fragments by Size Exclusion Chromatography Coupled with an Orbitrap Mass Spectrometer
 Analysis of Monoclonal Antibodies and Their Fragments by Size Exclusion Chromatography Coupled with an Orbitrap Mass Spectrometer Shanhua Lin, 1 Hongxia Wang, 2 Zhiqi Hao, 2 Patrick Bennett, 2 and Xiaodong
Analysis of Monoclonal Antibodies and Their Fragments by Size Exclusion Chromatography Coupled with an Orbitrap Mass Spectrometer Shanhua Lin, 1 Hongxia Wang, 2 Zhiqi Hao, 2 Patrick Bennett, 2 and Xiaodong
Ren Lab ENCODE in situ HiC Protocol for Tissue
 Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows
 Supporting Information Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows Ekaterina Stepanova 1, Steven P. Gygi 1, *, Joao A. Paulo 1, * 1 Department of
Supporting Information Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows Ekaterina Stepanova 1, Steven P. Gygi 1, *, Joao A. Paulo 1, * 1 Department of
Tissue Acetyl-Histone H4 ChIP Kit
 Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
ab Ran Activation Assay Kit
 ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
ab173247 Ran Activation Assay Kit Instructions for Use For the simple and fast measurement of Ran activation. This product is for research use only and is not intended for diagnostic use. Version 1 Last
Tissue & Cell Genomic DNA Purification Kit. Cat. #:DP021/ DP Size:50/150 reactions Store at RT For research use only
 Tissue & Cell Genomic DNA Purification Kit Cat. #:DP021/ DP021-150 Size:50/150 reactions Store at RT For research use only 1 Description: The Tissue & Cell Genomic DNA Purification Kit provides a rapid,
Tissue & Cell Genomic DNA Purification Kit Cat. #:DP021/ DP021-150 Size:50/150 reactions Store at RT For research use only 1 Description: The Tissue & Cell Genomic DNA Purification Kit provides a rapid,
Cdc42 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
Jan 25, 05 His Bind Kit (Novagen)
 Jan 25, 05 His Bind Kit (Novagen) (1) Prepare 5ml of 1X Charge buffer (stock is 8X= 400mM NiSO4): 0.625ml of the stock + 4.375ml DH2O. (2) Prepare 13ml of 1X Binding buffer (stock is 8X = 40mM imidazole,
Jan 25, 05 His Bind Kit (Novagen) (1) Prepare 5ml of 1X Charge buffer (stock is 8X= 400mM NiSO4): 0.625ml of the stock + 4.375ml DH2O. (2) Prepare 13ml of 1X Binding buffer (stock is 8X = 40mM imidazole,
Genomic DNA Extraction Kit (Tissue)2.0
 Genomic DNA Extraction Kit (Tissue)2.0 Ver. Q0125 No part of these protocols may be reproduced in any form or by any mean, transmitted, or translated into a machine language without the permission of YEASTERN
Genomic DNA Extraction Kit (Tissue)2.0 Ver. Q0125 No part of these protocols may be reproduced in any form or by any mean, transmitted, or translated into a machine language without the permission of YEASTERN
An Antibody-free Approach for the Global Analysis of. Protein Methylation
 Supplemental materials to An Antibody-free Approach for the Global Analysis of Protein Methylation Keyun Wang,, Mingming Dong,, Jiawei Mao,, Yan Wang,, Yan Jin,, Mingliang Ye,, *,,, Hanfa Zou CAS Key Lab
Supplemental materials to An Antibody-free Approach for the Global Analysis of Protein Methylation Keyun Wang,, Mingming Dong,, Jiawei Mao,, Yan Wang,, Yan Jin,, Mingliang Ye,, *,,, Hanfa Zou CAS Key Lab
AmpliScribe T7-Flash Biotin-RNA Transcription Kit
 AmpliScribe T7-Flash Biotin-RNA Transcription Kit Cat. No. ASB71110 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA276E AmpliScribe T7-Flash Biotin-RNA Transcription Kit 12/2016
AmpliScribe T7-Flash Biotin-RNA Transcription Kit Cat. No. ASB71110 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA276E AmpliScribe T7-Flash Biotin-RNA Transcription Kit 12/2016
AmoyDx FFPE RNA Kit. (Spin Column) For purification of total RNA from formalin-fixed, paraffin-embedded tissue sections. Instruction for Use
 AmoyDx FFPE RNA Kit (Spin Column) For purification of total RNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use 8.02.24101X036G 36 tests Amoy Diagnostics Co., Ltd. 39 Dingshan
AmoyDx FFPE RNA Kit (Spin Column) For purification of total RNA from formalin-fixed, paraffin-embedded tissue sections Instruction for Use 8.02.24101X036G 36 tests Amoy Diagnostics Co., Ltd. 39 Dingshan
Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005
 INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005 Highlights Fast protocol to purify Strep-tagged proteins from cell-free extracts Screen your recombinant colonies directly for
INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004, P2005 Highlights Fast protocol to purify Strep-tagged proteins from cell-free extracts Screen your recombinant colonies directly for
For Research Use Only. Not for use in diagnostic procedures. Anti-NRF2 mab
 Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-NRF2 mab CODE No. M200-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 1F2 Mouse IgG1 100 L,
Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-NRF2 mab CODE No. M200-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 1F2 Mouse IgG1 100 L,
Gel/PCR Extraction Kit
 Gel/PCR Extraction Kit Item No: EX-GP200 (200rxns) Content Content Binding Buffer BD Wash Buffer PE Elution Buffer (10 mm Tris-HCl, ph 8.5) Spin Columns EX-GP200 80 ml 20 mlx3 10 ml 200 each Description
Gel/PCR Extraction Kit Item No: EX-GP200 (200rxns) Content Content Binding Buffer BD Wash Buffer PE Elution Buffer (10 mm Tris-HCl, ph 8.5) Spin Columns EX-GP200 80 ml 20 mlx3 10 ml 200 each Description
Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Quantitation techniques Label-free TMT SILAC Peptide identification and FDR 194 Lecture
Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Quantitation techniques Label-free TMT SILAC Peptide identification and FDR 194 Lecture
For Research Use Only. Not for use in diagnostic procedures. Anti-NRF2 mab
 Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-NRF2 mab CODE No. M200-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 1F2 Mouse IgG1 κ 100 µl,
Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-NRF2 mab CODE No. M200-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal 1F2 Mouse IgG1 κ 100 µl,
EZ-10 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK
 EZ-0 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK (Bacteria, Plant, Animal, Blood) Version 5.0 Rev 03/25/205 Table of Contents Introduction 2 Limitations of Use 2 Features 2 Applications 2 Storage 2
EZ-0 SPIN COLUMN GENOMIC DNA MINIPREPS KIT HANDBOOK (Bacteria, Plant, Animal, Blood) Version 5.0 Rev 03/25/205 Table of Contents Introduction 2 Limitations of Use 2 Features 2 Applications 2 Storage 2
Strep-Spin Protein Miniprep Kit Catalog No. P2004 & P2005
 INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004 & P2005 Highlights Fast & Simple: Purify Strep-tagged proteins from cell-free extracts using a spin-column in 7 minutes High-Quality:
INSTRUCTION MANUAL Strep-Spin Protein Miniprep Kit Catalog No. P2004 & P2005 Highlights Fast & Simple: Purify Strep-tagged proteins from cell-free extracts using a spin-column in 7 minutes High-Quality:
Gα 13 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
igem2016 Microbiology BMB SDU
 igem2016 Microbiology BMB SDU Project type: Plastic Project title: characterizing and optimizing PHB productin in E. coli. Sub project: 1. Creation date: 2016.09.26 Written by: Joel Performed by: Joel
igem2016 Microbiology BMB SDU Project type: Plastic Project title: characterizing and optimizing PHB productin in E. coli. Sub project: 1. Creation date: 2016.09.26 Written by: Joel Performed by: Joel
