Staby TM Codon T7 kit Manual for GST fusion
|
|
|
- Felix Daniels
- 5 years ago
- Views:
Transcription
1 Staby TM Codon T7 kit Manual for GST fusion (v2.0) for GE-SCGST-. kits
2 . Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o Overview of Staby Express TM 5 o Expression of heterologous genes in E. coli 6 o Benefits of the Staby TM Codon system 7 o Experimental Outline: easy 4 steps procedure 8 Step 1. Cloning Insert 8 Step 2. Transformation into the CYS22 strain 10 Step 3. Transformation into the expression host 12 Step 4. Gene expression 14 About protein detection and purification using GST 16 Troubleshooting 17 References 18 Related Staby TM products and services 19 Staby TM products ordering information 20 Assurance letter about the use of the T7 expression system 21 Worldwide ordering 22 Copyright 2013 by Delphi Genetics SA. All rights reserved.
3 Content and storage : The Staby TM Codon T7 expression kit is shipped on dry ice. Storage: -80 C Two different types of Staby TM Codon T7 kits are available: one containing electrocompetent cells (in non self-standing tubes), and the other type containing chemically-competent cells (in self standing tubes). Each kit contains one box with the following items: Name pscodongst1.2 DNA Grey cap CYS22 strain (for cloning) Pink cap SE1 strain (for expression) Blue cap Staby reverse primer Red cap Staby forward primer Red cap Regeneration medium White cap Concentration/remarks Amount (SCGST-0505 and SCGST-0707) Amount (SCGST-1010 and SCGST-1212) 0.1µg/µl 1 tube of 50µl 1 tube of 50µl Competent cells 5 tubes 10 tubes Competent cells 5 tubes 10 tubes 0.1µg/µl in water 5 -CCA ACT CAG CTT CCT TTC G-3 0.1µg/µl in water 5 -GCG TCC GGC GTA GAG GAT C-3 2% Tryptone 0.5% Yeast extract 0.05% NaCl 2.5mM KCl 10mM MgCl2 1 tube of 20µl 1 tube of 20µl 1 tube of 20µl 1 tube of 20µl 5 tubes of 1.5 ml 10 tubes of 1.5 ml Manual 1 1 The genotype of the CYS22 strain is: F - enda1 glnv44 thi-1 reca1 rela1 gyra96 dcm - nupg Φ80dlacZ M15 (laczya-argf)u169 hsdr17(r K - m K+ ), λ -, ccdb +. The genotype of the SE1 strain is: derivates from E. coli B strain, F -, Cm R, ompt, lon, hsds B (restriction -, modification - ), gal, dcm, DE3 (laci, T7polymerase under the control of the PlacUV5 promoter), ccdb +. Copyright 2013 by Delphi Genetics SA. All rights reserved. 2
4 Material Safety Data Sheet: Product and company identification: Delphi Genetics SA Tel: Rue Antoine de St-Exupéry, 5 Fax: B-6041 Charleroi, Belgium delphigenetics@delphigenetics.com Hazards identification No specific hazard concerning the products of the StabyCodon T7 kit. First aid measures Inhalation: If one of the products of the StabyCodon T7 kit is inhaled, remove to fresh air. If not breathing, give artificial respiration. If breathing is difficult, give oxygen. Get medical attention. Ingestion: Do not induce vomiting unless directed to do so by medical personnel. Never give anything by mouth to an unconscious person. If large quantities of the products of the StabyCodon T7 kit are swallowed, call a physician immediately. Skin contact: In case of contact, immediately flush skin with plenty of water. Remove contaminated clothing and shoes. Wash clothing before reuse. Thoroughly clean shoes before reuse. Get medical attention. Eye contact: In case of contact with one of the products of the StabyCodon T7 kit, immediately flush eyes with plenty of water for at least 15 minutes. Get medical attention. Fire-fighting measures Use foam or all purpose dry chemicals to extinguish. Fire fighters should wear positive self-contained breathing apparatus and full turnout gear. Accidental release measures Immediately contact emergency personnel. Use suitable protective equipment (see below exposure controls and personal protection). For small spills add absorbent, scoop up material and place in a sealed, liquid-proof container for disposal. For large spills dike spilled material or otherwise contain material to ensure runoff does not reach a waterway. Place spilled material in an appropriate container for disposal. Minimize contact of spilled material with soils to prevent runoff to surface waterways. Handling and storing Keep the container tightly closed, in a cool and well-ventilated area. Personal protection The occupational exposure limits were not determined. Protect your skin and body using uniform or laboratory coat, chemical resistant, impervious gloves. Use safety glasses, face shield or other fullface protection if potential exists for direct exposure to aerosols or splashes. Disposal consideration Waste must be disposed of in accordance with federal, state and local environmental control regulations. N.B.: Final determination of suitability of any material is the sole responsibility of the user. All materials may present unknown hazards and should be used with caution. To the best of our knowledge, the information contained herein is accurate. However, neither Delphi Genetics SA nor any of its subsidiaries assumes any liability whatsoever for the accuracy or completeness of the information contained herein. Copyright 2013 by Delphi Genetics SA. All rights reserved. 3
5 Licenses The Staby TM Codon T7 expression kit is covered by worldwide patents (US , US , US and other patents pending). The kit is sold under a license from the Université Libre de Bruxelles (Belgium). The kit is sold for research purpose only. A license from Delphi Genetics SA is required for any commercial use. (Please, contact Delphi Genetics at delphigenetics@delphigenetics.com) T7 expression kit is based on technology developed at Brookhaven National Laboratory under contract with the U.S. Department of Energy and is the subject of patent applications assigned to Brookhaven Science Associate (BSA) in the United States of America (see assurance letter below). User Guide The Staby TM Codon T7 kit combines several technologies: T7 expression, plasmid stabilization and efficient supply of rare trnas to obtain a high yield of heterologous-protein expression even when the protein contains rare codons. Overview of the T7 expression system The T7 expression system is based on the use of the T7 bacteriophage promoter and RNA polymerase. The T7 RNA polymerase is useful for synthesizing large amounts of RNA selectively: the T7 RNA polymerase only recognizes the T7 promoter and not the E. coli promoters. Conversely, the E. coli RNA polymerase does not recognize the T7 promoter (see below). The T7 RNA polymerase is able to transcribe genes five times faster than the E. coli RNA polymerase (Chamberlin and Ring, 1973; Golomb and Chamberlin, 1974). The gene encoding the T7 RNA polymerase is inserted into the chromosome of the expression bacteria (SE1, figure 1). The expression of this gene is under the control of the lacuv5 promoter and therefore is basically controlled by the same mechanisms as the lac operon. Thus, the expression of the T7 RNA polymerase is repressed by the binding of the lac repressor (encoded by the laci gene) to the laco operator sequence. The gene encoding the repressor is present in the bacterial chromosome and also in the pscodongst1.2 plasmid to ensure high amount of repressor molecules. Consequently, in normal conditions, the T7 RNA polymerase is not or very weakly expressed. An additional repression of the lac promoter can be obtained using medium containing glucose. The presence of glucose in the medium (especially in the stationary phase) induces the metabolic repression: the bacteria will first use glucose as a carbon source and will reduce the concentration of cyclic AMP, ensuring a better repression of the lac promoter (cyclic AMP stimulates the lac and lacuv5 promoters). Moreover, Studier et al. (1990) have shown that a better regulation of the expression of the gene of interest is obtained by adding the laco operator sequence between the T7 promoter and the beginning of the gene of interest. This sequence is present in the pscodongst1.2 vector. Consequently, the lac repressor will also repress the expression of the gene of interest. Copyright 2013 by Delphi Genetics SA. All rights reserved. 4
6 Figure 1: The T7 expression system used in the SE1 strain Plasmid laci gene lac repressor lacuv5 promoter lac operator T7 promoter T7 RNA polymerase gene T7 RNA polymerase Gene of interest Product of interest Plasmid IPTG induction, expression Adding isopropyl-β-d-thiogalactoside (IPTG) to the medium will induce the expression of (i) the T7 RNA polymerase and of (ii) the gene of interest by removing the lac repressor bound to the laco sequence (figure 1). A powerful feature of the T7 expression system is the ability to clone the gene of interest under conditions of extremely low or no transcriptional activity, that is, in the absence of the T7 RNA polymerase (as the CYS22 genetic background). The expression of the gene of interest is minimal in the absence of the T7 RNA polymerase because this gene is under the control of the T7 promoter which is only recognized by the T7 RNA polymerase and not by the E. coli RNA polymerase. If the target gene is cloned directly into the expression strain, even a low basal expression of the T7 RNA polymerase can interfere with growth and selection of the right construct. After the cloning step into a cloning strain lacking the T7 RNA polymerase (CYS22), the plasmid construct is transferred into the expression strain encoding the T7 RNA polymerase (SE1) to produce the protein of interest. As the the Staby TM Codon T7 system is based on commonly used T7 promoter, you can easily change between your existing expression system and Staby Codon. Overview of the stabilization system: Higher plasmid stability= More proteins Principle: The stabilization system is based on the use of bacterial antidote/poison ccda/ccdb genes naturally found in the F plasmid of Escherichia coli (for more information about this system, see Bernard et al., 1992 and 1994 or Gabant et al and 2002). In the Staby TM Codon system, the antidote gene (ccda, 218bp) is introduced in the plasmid DNA under the control of a constitutive promoter. On the other hand, the bacterial toxic gene (ccdb, 305bp) is introduced in the chromosome of the bacteria (cf. fig. below). Expression of the poison gene is under the control of a promoter strongly repressed in the presence of the plasmid. When the plasmid is lost, the antidote protein is degraded and the production of the toxin is induced, causing cell death. Copyright 2013 by Delphi Genetics SA. All rights reserved. 5
7 Figure 2: Principle of the stabilization system Practically this means that when bacteria are growing during the pre-induction phase, 100% of these bacteria will carry the vector. If they lose the vector, they will not obtain a growth advantage, but will die. Upon induction 100% of the bacteria will start producing the recombinant protein leading to higher yields of the target protein and less background caused by unwanted proteins. For manufacturers of recombinant proteins this system offers a great benefit because it is an antibiotic free expression system. Therefore the manufactured protein will also be free of traces of antibiotics. Expression of heterologous genes in E. coli: In all organisms, most amino acids are encoded by more than one codon: 61 codons are available for 20 amino acids. But each organism is characterized by a specific codon bias (see table below), i.e. it preferentially uses some codons over others. In practice, when a heterologous gene is expressed in E. coli, this gene might exhibit some codons that are common in the original host but are rarely used in E. coli. Whereas, the presence of only a small number of rare codons might not severely depress target protein synthesis, the presence of clusters of and/or numerous rare codons generates a demand for one or more rare trnas. In turn, the rarity of some trnas leads to very low expression of the target protein due to premature translation termination, translation frameshifting, amino acid misincorporation, growth inhibition and plasmid instability. Six rare codons can cause problems in E. coli B (e.g.; BL21(DE3) or SE1): AGG and AGA (both encoding arginine using the argu trna), AUA (isoleucine, ilex trna), CUA (leucine, leuw trna), GGA (glycine, glyt trna), and CCC (proline, prol trna). An analysis of your gene-of-interest can be performed using Staby TM Soft. Copyright 2013 by Delphi Genetics SA. All rights reserved. 6
8 Amino acid Codon Frequency in E. Frequency in Frequency in Frequency in coli B (SE1)(%) Homo sapiens Arabidopsis Saccharomyces (%) thaliana (%) cerevisiae (%) Arginine CGT CGC CGA* 5* CGG AGA AGG Glycine GGT GGC GGA GGG Isoleucine ATT ATC ATA Leucine TTA TTG CTT CTC CTA CTG Proline CCT CCC CCA CCG *: CGA codon does not cause problem because large amounts of the corresponding trna are present In the Staby TM Codon T7 kit, we solve the problem by the use of the pscodongst1.2 expression plasmid encoding the trna genes of the six rare codons. Hence, this plasmid contains the T7 promoter for a strong expression, the ccda gene for plasmid stabilization and supplies the rare trnas. Benefits of the Staby TM Codon system: High yield of heterologous-protein expression even when the protein contains rare codons; Not necessary to mutate each rare codon; Recombinant plasmid perfectly stabilized even without the use of antibiotics; Reduced background of parasite proteins ; No additional plasmid in bacteria; The promoters typically used in protein production remain available; System is usable in any culture medium. Copyright 2013 by Delphi Genetics SA. All rights reserved. 7
9 Experimental outline: Easy 4 steps procedure. 1. Cloning of your gene of interest in the pscodongst1.2 vector 2. Transformation into the CYS22 E. coli cells and selection of the desired construction 3. Transformation of your plasmid DNA into the SE1 E. coli competent cells 4. Expression of your gene of interest without antibiotic Step 1: Cloning of your gene of interest in the pscodongst1.2 vector: Many strategies can be used for cloning your gene of interest (goi) into the pscodongst1.2 vector. The most convenient strategy is to use restriction enzymes: the single-cutter enzymes from the multiple cloning sites are indicated on the map (see below: from NdeI to XhoI). Use the buffer and incubation conditions provided by the restriction enzyme manufacturer. After restriction, the DNA fragment encoding the gene of interest is inserted by ligation using compatible ends. XhoI PspXI EagI NotI HindIII SalI SacI EcoRI BamHI NheI NdeI SphI SphI XbaI Stabyfwd ApaI PciI T7 ccda EcoRV pscodongst1.2 laci GST Stabyrev 6513 bps NcoI His NruI Amp glyt prol argu ilex leuw rep PvuI PciI Figure 3: Restriction map of the pscodongst1.2 vector Features: -Staby forward primer: T7 promoter: GST: His: Staby reverse primer: (C) The complete sequence of the vector is available on our website ( Copyright 2013 by Delphi Genetics SA. All rights reserved. 8
10 Remarks: Take into account the reading frame of the GST gene (glutathione-s-transferase) to fuse your sequence to the GST tag at the N-terminal end of your protein (cf. figure below). This tag facilitates detection and purification of the target protein. Note that an Enterokinase site is present in the pscodongst1.2 vector at the C- terminal end of the GST tag to cleave the fused protein (removal of the GST tag). The cleavage position is indicated in the figure below (after the recognition site DDDDK). The pscodongst1.2 vector allows also the fusion of 6 histidine residues at the C- terminal end of the protein. If needed, the C-terminal fusion can be skipped by including a stop codon (TAA or TGA) at the end of your gene of interest. Please, note that the 6 histidine residues are not in the same frame than GST to avoid any fortuitous fusion (the 6 histidine codons are indicated in blue in the figure below). Consequently, you must take into account the reading frame of this His-tag if you want to use it. Fig. 4. Sequence of the cloning region of pscodongst1.2: (unique restriction sites are indicated in bold or italized) Start of your gene GST C-terminal end NdeI NheI ttt ggt ggt ggc gac cat cct cca aaa gat gat gac gat aag cat atg gct agc aaa cca cca ccg ctg gta gga ggt ttt cta cta ctg cta ttc gta tac cga tcg F G G G D H P P K D D D D K H M A S Enterokinase site Ecl136II BamHI EcoRI SacI SalI atg act ggt gga cag caa atg ggt cgc gga tcc gaa ttc gag ctc cgt cga caa tac tga cca cct gtc gtt tac cca gcg cct agg ctt aag ctc gag gca gct gtt M T G G Q Q M G R G S E F E L R R Q AvaI HindIII NotI XhoI his-tag Stop gct tgc ggc cgc act cga g cac cac cac cac cac cac tgagatccggctgctaa cga acg ccg gcg tga gct c gtg gtg gtg gtg gtg gtg actctaggccgacgatt A C G R T R H H H H H H - Important: Delphi Genetics can help you with a software-based optimization of the nucleotide sequence of your gene-of-interest for a best protein production (please contact us at stabysoft@delphigenetics.com). Copyright 2013 by Delphi Genetics SA. All rights reserved. 9
11 Step 2. Transformation into the CYS22 strain and selection of the desired construction: Selection of the desired construction is performed in CYS22 E. coli cells lacking the T7 RNA polymerase gene (*). These cells contain the ccdb gene in their chromosome. This enables: (i) High efficiency of transformation (the transformation efficiency of SE1 - derivative of BL21 is lower than that of CYS22), (ii) (iii) Stabilization of plasmids for high DNA production, Selection of the desired construction without expression of the gene of interest (goi). (*)Remark: It is not recommended to clone directly the goi into the expression host containing the T7 RNA polymerase gene: the T7 gene basal expression, and the resulting goi basal expression, would reduce the efficiency of recovery of the desired construction. Protocol: Two different types of Staby TM Codon T7 kits are available: one containing electrocompetent cells, and the other type containing chemically-competent cells. a) Transformation by electroporation: 1) Prepare LB plates containing 100µg/ml Ampicillin. Let the plates dry and then warm them at 37 C. 2) Set up your electroporator for bacterial transformation. Use the manufacturer s instructions. Classically, electroporation conditions are: 2,5 kv, 25 µf, and 200 Ohms. 3) For each cloning reaction, place one vial of the CYS22 electrocompetent cells (pink cap) and one electroporation cuvette on ice. Allow the cells to thaw on ice for 5-10 minutes. 4) Add 1 or 2 µl of the ligation to the vial of the CYS22 electrocompetent cells (pink cap). Stir gently to mix. Do not mix by pipetting up and down. If you wish to use more than 2µl of the ligation mix, it is recommended to dialyze it against sterile water using a 0.025µm filter. Add the sterile water in a Petri dish and carefully place the filter on the water surface. Delicately, put the ligation mix on the filter. Wait 10min, pipet back the ligation mix and add the dialyzed solution to the electrocompetent cells. 5) Transfer all the content of the tube (cells+dna) to the pre-chilled electroporation cuvette. 6) Electroporate the cells according to the manufacturer s instructions. 7) Quickly add 500µl of the regeneration medium (white cap) at room temperature and mix well. 8) Spread immediately 20 to 150µl on the pre-warmed plates containing ampicillin. Copyright 2013 by Delphi Genetics SA. All rights reserved. 10
12 9) Incubate the plates overnight at 37 C. 10) Pick about 10 colonies and culture them overnight in 10ml of LB medium with or without ampicillin (100µg/ml). Note: The stabilization is now effective; the ccdb gene is activated. Consequently, the plasmid is stabilized in the CYS22 strain and no antibiotic is needed to select bacteria containing the plasmid. However, the ampicilline resistance is still available. The stabilization system will insure high yield of plasmid DNA. 11) Extract plasmid DNA and analyze the constructions using your method of choice (restriction, sequencing, ). Sequencing primers (Staby reverse and Staby forward primers) are included in the kit (0.1 µg/µl). The complete sequence of the pscodongst1.2 vector is available on our website: 12) Choose one of the clones containing the desired construct. Mix well 800µl of the culture with 800µl of sterile glycerol and transfer to a cryo-vial. Store at -80 C. b) Transformation using chemically competent cells: 1. Prepare LB plates containing 100 µg/ml Ampicillin. Let the plates dry and then warm them up at 37 C. 2. Set a water bath or a heating-bloc to 42 C 3. Thaw (bring to room temperature) one vial of regeneration medium (white cap) per cloning reaction. 4. For each cloning reaction, place one vial of the CYS22 chemically-competent cells (self-standing tube with pink cap) on ice. Allow the cells to thaw on ice for 5-10 minutes. 5. Add 5 µl of the ligation product to one vial of the CYS22 chemically competent cells (self-standing tube with pink cap). Stir gently to mix. Do not mix by pipetting up and down. 6. Incubate on ice for 30 minutes. 7. Heat-shock the bacteria by placing the vial at 42 C for 30 seconds without shaking. 8. Immediately transfer the tubes to ice. 9. Add 250µl of room-temperature regeneration medium (white cap) and mix well. 10. Spread immediately 10, 20 and 100µl of the product (from step 9) on different pre-warmed plates. If you wish to have more clones, incubate the product (from step 9) at 37 C for one hour for regeneration of the bacteria before spreading of 10, 20 and 100µl on different pre-warmed plates. 11. Incubate the plates overnight at 37 C. 12. Pick about 10 colonies and culture them overnight in 10ml of LB medium with or without ampicillin (100µg/ml). Note: The stabilization is now effective; the ccdb gene is activated. Consequently, the plasmid is stabilized in the CYS22 strain and no antibiotic is needed to select bacteria containing the plasmid. However, the ampicilline resistance is still available. The stabilization system will insure high yield of plasmid DNA. 13. Extract plasmid DNA and analyze the constructions using your method of choice (restriction, sequencing, ). Sequencing primers (Staby reverse and Copyright 2013 by Delphi Genetics SA. All rights reserved. 11
13 Staby forward primers) are included in the kit (0.1 µg/µl). The complete sequence of the pscodongst1.2 vector is available on our website: Choose one of the clones containing the desired construct. Mix well 800µl of the culture with 800µl of sterile glycerol and transfer to a cryo-vial. Store at -80 C. Step 3. Transformation in the expression host: Positive control: The pscodongst1.2 vector can be used without gene of interest as a positive control of your experiment. To do so, transform it according to the protocol below. a) Transformation by electroporation: 1) Prepare LB plates containing 100µg/ml Ampicillin. Let the plates dry and then warm them at 37 C. Note: Addition of 1% glucose (from a sterile filtered 20% stock solution) in the plates can be useful to better repress the promoter and to avoid basal expression. 2) Set up your electroporator for bacterial transformation. Use the manufacturer s instructions. Classically, electroporation conditions are: 2,5 kv, 25 µf, and 200 Ohms. 3) For each transformation, place one vial of the SE1 electrocompetent cells (blue cap) and one electroporation cuvette on ice. Allow the cells to thaw on ice for 5-10 minutes. Note: to use the pscodongst1.2 vector without gene of interest as a control, do not forget to add an additional vial of SE1 and an additional cuvette. 4) Add 1µl of the selected plasmid DNA (steps 11 and 12 above) to the SE1 cells and mix gently. 5) Transfer all the content of the tube (cells+dna) to the pre-chilled electroporation cuvette. 6) Electroporate the cells according to the manufacturer s instructions. 7) Quickly add 500µl of the regeneration medium (white cap) at room temperature and mix well. 8) Spread immediately 20 to 150µl on the pre-warmed LB plates. 9) Incubate the plates overnight at 37 C. 10) Optional: Pick about 5 colonies and culture them overnight in 10ml of LB medium. Note: The plasmid is now stabilized in the SE1 strain using the Staby TM Codon system, no antibiotic is needed to select bacteria containing the plasmid. However, the ampicilline resistance gene is still available. Addition of 1% glucose (from a sterile filtered 20% stock solution) in the medium can be useful to better repress the promoter and to avoid basal expression. 11) Optional: Extract plasmid DNA and analyze the constructions using your method of choice (restriction, sequencing, ). Sequencing primers are included in the kit (0.1 µg/µl). The complete sequence of the pscodongst1.2 vector is available on our website: Copyright 2013 by Delphi Genetics SA. All rights reserved. 12
14 12) Optional: Select one of the clones containing the desired construction. Mix well 800µl of the culture with 800µl of sterile glycerol and transfer to a cryovial. Store at -80 C. b) Transformation using chemically competent cells: 1. Prepare LB plates containing 100 µg/ml Ampicillin. Let the plates dry and then warm them up at 37 C. Note: Addition of 1% glucose (from a sterile filtered 20% stock solution) in the plates can be useful to better repress the promoter and to avoid undesirable expression. 2. Set a water bath or a heating-bloc to 42 C 3. Thaw (bring to room temperature) one vial of regeneration medium (white cap) per cloning reaction. 4. For each transformation, place one vial of the SE1 chemically-competent cells (self-standing tube with blue cap) on ice. Allow the cells to thaw on ice for 5-10 minutes. Note: to use the pscodongst1.2 vector without gene of interest as a positive control, do not forget to thaw an additional vial of SE1. 5. Add 1µl or 2µl of the selected plasmid DNA (steps 11 and 12 above) to one vial of the SE1 chemically competent cells (self-standing tube with blue cap). Stir gently to mix. Do not mix by pipetting up and down. 6. Incubate on ice for 30 minutes. 7. Heat-shock the bacteria by placing the vial at 42 C for 30 seconds without shaking. 8. Immediately transfer the tubes to ice. 9. Add 250µl of room-temperature regeneration medium (white cap) and mix well. 10. Spread immediately 10, 20 and 100µl of the product (from step 9) on different pre-warmed plates. 11. Incubate the plates overnight at 37 C. 12. Optional: Pick about 5 colonies and culture them overnight in 10ml of LB medium. Note: The plasmid is now stabilized in the SE1 strain using the Staby TM Codon system, no antibiotic is needed to select bacteria containing the plasmid. However, the ampicilline resistance gene is still available. Addition of 1% glucose (from a sterile filtered 20% stock solution) in the medium can be useful to better repress the promoter and to avoid basal expression. 13. Optional: Extract plasmid DNA and analyze the constructions using your method of choice (restriction, sequencing, ). Sequencing primers are included in the kit (0.1 µg/µl). The complete sequence of the pscodongst1.2 vector is available on our website: Optional: Select one of the clones containing the desired construction. Mix well 800µl of the culture with 800µl of sterile glycerol and transfer to a cryovial. Store at -80 C. Copyright 2013 by Delphi Genetics SA. All rights reserved. 13
15 Step 4. Expression of your gene of interest: The T7 RNA polymerase is under the control of the PlacUV5 promoter (Studier and Moffat, 1986; Studier et al., 1990). Both the SE1 strain and the pscodongst1.2 vector carry the laci gene. LacI represses both the expression of the T7 RNA polymerase and the transcription of the gene of interest. Consequently, the expression of the T7 RNA polymerase is inducible by isopropyl-β-d-thiogalactoside (IPTG): addition of IPTG to the culture of the SE1 strain containing the pscodongst1.2 plasmid will induce the expression of the T7 RNA polymerase which, in turn, will transcribe the gene of interest. Expression can also be performed using Staby TM Switch medium (auto-inducible medium without IPTG). Perform the small-scale expression following the manual instructions. The small-scale protocols below will allow you to verify that the target protein is produced upon induction and to verify for the presence of detection tags in the target protein. Experiment control: The SE1 bacteria containing the pscodongst1.2 vector alone will allow you to test the expression of the GST protein alone (without his tag, 262 amino acids, molecular weight: 30.3kDa, presents as homodimer : ~60kDa). Protocol for a small-scale expression using Staby TM Switch autoinducible medium: 1. Inoculate two containers containing the desired volume of pre-warmed Staby TM Switch medium with a few microliters (1 or 2 µl / 10ml culture) from a glycerol stock of the SE1 strain containing your construction in the pscodongst1.2 vector. Alternatively, inoculate containers with a single colony from a plate streaked with this strain. For a 10ml culture volume, the use of 50ml tubes with conical bottom (28mm x 114mm) is ideal. The tubes can be maintained closed during all the whole expression experiment. For bigger culture volumes, use Erlenmeyer flasks with a capacity of 5 times the culture volume. For 96 well plates, use 1 single colony or volume of a glycerol stock per well. Antibiotics are not required but can be used. 2. Add 1% sterile glucose (from a sterile-filtered 20% stock solution) to one of the two containers. This culture will be used as a non-induced control and/or to prepare a glycerol stock. 3. Incubate the containers at 37 C for approximately 24 hours with shaking (200rpm max, rotary shaker, 2.54cm orbit). Note: (1) If your protein is unstable, add 1% lactose (from a sterile-filtered 20% stock solution) 2 hours before the end of the culture. (2) It is essential to grow the bacteria to stationary phase for full induction. If you want to incubate your cultures at lower temperature (<37 C), it is necessary to adapt the incubation time. Continue incubation for several hours (8 to 10 hours) after saturation. The first time, it is recommended to take a sample every hour and to check the protein expression (red colour or SDS-PAGE analysis). Copyright 2013 by Delphi Genetics SA. All rights reserved. 14
16 4. After incubation, measure the Optical Density at 600nm for each culture. Transfer 1ml sample of each flask in a microcentrifuge tube. Centrigufe at maximum speed (13000 g) for 10 min (if possible at 4 C) and continue with protein extraction (see below). Protocol for a small-scale expression using IPTG: 1) Inoculate two Erlenmeyer flasks containing 10ml of LB medium with a few microliters from a glycerol stock. Alternatively, pick two single colonies from a plate streaked with the SE1 bacteria containing your construction; inoculate two flasks containing 10ml of LB medium. 2) Incubate with shaking at 37 C until OD600 reaches (the best range is between 0.6 and 0.8). 3) In one of the two flasks, add IPTG (100µl of a fresh 100mM stock solution) to reach a final concentration of 1mM. The other flask is used as a non-induced control. Continue incubation of both flasks for 2-3 hours. 4) Measure the Optical Density at 600nm for each culture. Transfer 1ml sample of each flask in a microcentrifuge tube. Centrigufe at maximum speed (13000 g) for 10 min (if possible at 4 C) and continue with protein extraction (see below). Protein extraction under denaturing conditions 1) Transfer 1ml sample of each flask in a microcentrifuge tube. Centrigufe at maximum speed (13000 g) for 10 min (if possible at 4 C). Discard the supernatant, add 1ml H2O and resuspend the bacteria. Add 50µl of cold 100% Trichloroacetic acid (TCA) (w/v) to each sample and vortex for a few seconds. Note: The TCA precipitation allows the analysis of the total protein content of the cells. Other methods can be used to specifically analyze different fractions (soluble, insoluble, periplasm, ) in order to identify the cellular localization of the target protein. For more information, please, check specialized literature or protocols (e.g., Sambrook et al., Ausubel et al.). 2) Place on ice for 10 min. 3) Centrifuge at maximum speed (13000 g) for 10 min (if possible at 4 C). 4) Remove carefully and discard the supernatant. 5) Wash the pellet with cold acetone (+4 C): add 500µl of acetone, vortex, and centrifuge for 5 min at maximum speed (if possible at 4 C). 6) Repeat steps 7 and 8 7) Remove carefully the supernatant. Air dry the final pellet: leave the tube opened on the bench or use vacuum drying. 8) Add (OD600 x 200)µl of 1X sample buffer (2X sample buffer= 100mM DTT, 2% SDS, 80mM Tris-HCl, ph 6.8, 0.006% bromophenol blue, 15% glycerol). Vortex vigorously to resuspend the pellet. Note: Taking into account the OD 600 allows comparison of Coomassie-stained band intensities between samples. Copyright 2013 by Delphi Genetics SA. All rights reserved. 15
17 9) Heat the samples at 70 C-100 C (10min.) to resuspend and denature the proteins. The samples can be used directly for SDS-PAGE analysis or stored at - 20 C. 10) Load 4 to 10 µl of each sample in a SDS-PAGE gel containing the appropriate concentration of polyacrylamide (according to the size of the overproduced protein). Add a molecular size marker. Note: The sample volume that needs to be loaded will depend on the gel size, the expression level, and the extraction efficiency. 11) After migration, visualize the proteins with Coomassie-blue staining or continue the analysis with western blot. Note: Western blot analysis is a more specific and sensitive method but needs proteinspecific antibodies or fusion tag-specific antibodies. For more information, please, check specialized literature or protocols (e.g., Sambrook et al, Ausubel et al.). About protein detection and purification using GST: GST (Glutathione-S-Transferase) is commonly used for protein detection and purification. This protein is expressed in E. coli with full enzymatic activity. GST is a protein of 26 kda with an isolelectric point pi = 5.0. The protein is soluble and presents as a homodimer in E. coli cytoplasm (molecular weight of the dimer: 58.5 kda). In denaturing conditions, the dimer is resolved and a single band at ~26kDa is visible. However, if proteins were not completely denatured before loading on gel, the dimer is observed (band at ~58.5 kda, even when using SDS-PAGE gels). It is thus necessary to take into account the presence of GST dimer when you analyse your gels when denaturing conditions were not used. GST detection can be performed using SDS-PAGE analysis, enzymatic detection (using GST substrate 1-cloro-2,4 dinitrobenzene, CDNB) or Western blot. The Western blot is often the most powerful method. For more information about the use of anti-gst antibody, please, consult documentation of the manufacturer but do not forget to run controls (protein samples of E. coli that do and do not contain parental pscodongst1.2 vector). For protein purification using GST, several products exist and can be used with your fusion protein. One of the most useful is the glutathione-sepharose column: the glutathione is attached to sepharose and is complementary to the binding site of the GST binding site. One of the most important parameters affecting the binding of GST fusion proteins to glutathione-sepharose column is the flow rate. Since the binding kinetics between glutathione and GST are relatively slow, it is necessary to keep the flow rate constant and low during sample application to achieve maximum binding capacity. Another point to consider is the protein solubility. GST is soluble and can help to solubilise your protein. However, if your fusion protein is insoluble, it is often easier to purify it using the His-tag of the vector: using (His)6 fusion protein enables the use of denaturing agents during purification (and refolding after purification) or on-column refolding procedure. For more information about these procedures, please, consult documentation of the column manufacturer. In most cases, functional tests can be performed using the fusion protein but if removal is necessary, the pscodongst1.2 vector contains an enterokinase cleavage site at the C-terminal end of GST. This site is the specific cleavage site of the enterokinase protease. It allows the removal of GST after purification or detection Copyright 2013 by Delphi Genetics SA. All rights reserved. 16
18 without leaving extra amino acids. Take into account that after cleavage, the protease should be removed. Columns with specific affinity to enterokinase exist and could be combined to the purification column. For more information, please, contact the manufacturer. Troubleshooting: Please note that problems with cloning or expression efficiencies can result from the following parameters. Most of these problems can be fixed as explained below. However, due to intrinsic and specific properties of your gene or protein, the cloning or expression efficiencies may vary. Problem Only a few or no colonies are observed after transformation (ligation mix into the CYS22 bacteria or plasmid construct into the SE1 bacteria). No expression Solution - Check the DNA concentration of your insert and the ligation conditions. - Check the quality of your insert (one single band must be visible after agarose gel electrophoresis of the purified DNA fragment). - Be sure that the DNA transformation was optimal. When using electrocompetent bacteria, check the electroporation conditions (see above). When using chemically competent bacteria: check the temperature of the water bath, incubate the transformation product during one hour at 37 C to allow regeneration of the bacteria before spreading. - Check your plates with another strain which is resistant to the ampicillin antibiotic. If no growth is observed, check your antibiotic solution. - Your cloned fragment could be toxic for the bacteria. Check the literature (if data are available). Add 1% glucose (from a sterile 20% stock solution) to the plates to better repress the promoter. -Test the transformation efficiency with the pscodongst1.2 vector without gene of interest - Check the gene sequence for mutations. - Check your expression conditions. - Check expression starting with a single colony or a glycerol stock (do not use preculture) - When using Staby TM Swicth medium, pre-warm the medium at room temperature or 37 C before inoculation. - Check expression using the expression control. - Your protein might be unstable, try different induction conditions (lower temperature, Staby TM Switch medium, longer induction time, ). - Use Cherry TM Express or Cherry TM Codon kits to follow your protein with your eyes (for more information, please consult our website). Copyright 2013 by Delphi Genetics SA. All rights reserved. 17
19 References: Ausubel F., Brent R., Kingston R., Moore D., Seidman J.G., Smith J.A., Struhl K Current Protocols in Molecular Biology. John Wiley and Sons edition. USA. Bernard P. and Couturier M Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226: Bernard P., Gabant P., Bahassi El M., Couturier M Positive-selection vectors using the F plasmid ccdb killer gene. Gene 148: Chamberlin M., and Ring J Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J. Biol. Chem. 248: Gabant P., Szpirer C.Y., Couturier M., Faelen M Direct selection cloning vectors adapted to the genetic analysis of Gram-negative bacteria and their plasmids. Gene 207: Gabant P., C.Y. Szpirer, Van Melderen L Plasmid poison-antidote systems: functions and technological applications. In Recent advances in plasmid science ed.: S.G. Pandalai (Transworld research network edition). Vol1, pg: Golomb M. and Chamberlin M Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J. Biol. Chem. 249: Sambrook J., Fritsch E., Maniatis T in Molecular Cloning: a laboratory manual, Second edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. Studier W. and Moffat B.A Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189: Studier W., Rosenberg A., Dunn J., Dubendorff J Use of T7 RNA polymerase to direct expression of cloned genes. Methods in Enzymology vol. 185: Copyright 2013 by Delphi Genetics SA. All rights reserved. 18
20 Related Staby TM products and services: The StabyCloning TM kit is designed for the rapid, precise and efficient DNA cloning of PCR products. The complete cloning procedure is performed in one hour (including plating), the background is basically nil (the bacteria containing vectors without insert are killed), the PCR product is oriented, the plasmid is stabilized, and the export of the insert to another vector is easily selected. The StabyExpress TM T7 kit contains all the key elements for cloning of a gene-of-interest and its expression in Escherichia coli. The kit combines two technologies (T7 expression and plasmid stabilization) that allow high-yield protein expression and standardization of the production-protocol. Vectors with his-tag and/or GST-tag are available. The GetStaby TM kit allows easy addition of Delphi-Genetics stabilization technology into your favourite vector. The technology is compatible with any expression system. Using this technology, your vectors are perfectly stabilized even without antibiotics. The Staby TM Codon T7 kit combines three technologies to ensure high-yield and standardized expression of eukaryote proteins in Escherichia coli. These technologies are (i) T7-controled expression, (ii) plasmid stabilization, and (iii) codon-usage adaptation of E. coli for the efficient expression of proteins that contain rare codons. The Cherry TM Express kit allows direct visualization (by eye!) of your protein of interest during protein production in E. coli and protein purification. Special requirements or reagents are not needed. It is also possible to quantify the protein concentration at any step by spectral measurement. The Cherry TM Express kit combines multiple advantages: protein visualization, T7 expression, plasmid stabilization and codon-usage adaptation. The Staby TM Switch medium is an auto-inducible medium (ready-touse) designed for high-level protein expression using Staby TM products or any other IPTG-inducible bacterial expression system. Using Staby TM Switch medium, protein expression is automatically induced when high cell density is reached. Thus, it is neither necessary to add IPTG nor to monitor optical density during bacterial growth. Staby TM Soft was specifically designed by Delphi Genetics to support the users of the Staby TM Operating System. This software package can perform customized gene-of-interest analysis to choose the most adapted kit and to optimize protein production. For more information, please, consult Copyright 2013 by Delphi Genetics SA. All rights reserved. 19
21 Staby TM products ordering information: StabyExpress TM GE-SET pstaby1.2 expression kit, electro-competent 5 reactions GE-SET pstaby1.2 expression kit, chemically-competent 5 reactions GE-SET pstaby1.2 expression kit, electro-competent 10 reactions GE-SET pstaby1.2 expression kit, chemically-competent 10 reactions GE-SET pstaby1.2 expression kit, electro-competent 20 reactions GE-SET pstaby1.2 expression kit, chemically-competent 20 reactions GE-SEGST-0505 pstabygst1.2 expression kit, electro-competent 5 reactions GE-SEGST-0707 pstabygst1.2 expression kit, chemically-competent 5 reactions GE-SEGST-1010 pstabygst1.2 expression kit, electro-competent 10 reactions GE-SEGST-1212 pstabygst1.2 expression kit, chemically-competent 10 reactions GE-SET x 50µl CYS22 and 10x50µl SE1 bacteria, electro-competent 10 reactions GE-SET x 100µl CYS22 and 10x100µl SE1 bacteria, chimio-competent 10 reactions GE-SET Set of 20 expression bacteria (SE1), electro, 50µl/tube 20 reactions GE-SET Set of 20 expression bacteria (SE1), chimio, 100µl/tube 20 reactions GetStaby TM GE-GSA1-10 GetStaby kit, electro-competent cells 10 reactions GE-GSA1-12 GetStaby kit, chemically-competent cells 10 reactions StabyCloning TM GE-STC1-10 StabyCloning kit, electro-competent cells 10 reactions GE-STC1-12 StabyCloning kit, chemically-competent cells 10 reactions GE-STC1-20 StabyCloning kit, electro-competent cells 20 reactions GE-STC1-22 StabyCloning kit, chemically-competent cells 20 reactions GE-STCB-20 Set of 20 cloning bacteria (CYS22) electro (50µl/tube) 20 reactions GE-STCB-22 Set of 20 cloning bacteria (CYS22) chimio (100µl/tube) 20 reactions Staby TM Codon GE-SCT pscodon1.3 expression kit, electro-competent 5 reactions GE-SCT pscodon1.3 expression kit, chimio-competent 5 reactions GE-SCT pscodon1.3 expression kit, electro-competent 10 reactions GE-SCT pscodon1.3 expression kit, chimio-competent 10 reactions GE-SCGST-0505 pscodongst1.2 expression kit, electro-competent 5 reactions GE-SCGST-0707 pscodongst1.2 expression kit, chimio-competent 5 reactions GE-SCGST-1010 pscodongst1.2 expression kit, electro-competent 10 reactions GE-SCGST-1212 pscodongst1.2 expression kit, chimio-competent 10 reactions Staby TM Switch GE-AIME-04 Auto-induction medium 2L Cherry TM Express GE-CET7-05 CherryExpress T7 expression kit, N-terminal tag, electro 5 reactions GE-CET7-06 CherryExpress T7 expression kit, C-terminal tag, electro 5 reactions GE-CET7-07 CherryExpress T7 expression kit, N-terminal tag, chimio 5 reactions GE-CET7-08 CherryExpress T7 expression kit, C-terminal tag, chimio 5 reactions GE-CET7-10 CherryExpress T7 expression kit, N-terminal tag, electro 10 reactions GE-CET7-11 CherryExpress T7 expression kit, C-terminal tag, electro 10 reactions GE-CET7-12 CherryExpress T7 expression kit, N-terminal tag, chimio 10 reactions GE-CET7-13 CherryExpress T7 expression kit, C-terminal tag, chimio 10 reactions Cherry TM Codon GE-CCT7-05 CherryCodon T7 expression kit, N-terminal tag, electro 5 reactions GE-CCT7-06 CherryCodon T7 expression kit, C-terminal tag, electro 5 reactions GE-CCT7-07 CherryCodon T7 expression kit, N-terminal tag, chimio 5 reactions GE-CCT7-08 CherryCodon T7 expression kit, C-terminal tag, chimio 5 reactions GE-CCT7-10 CherryCodon T7 expression kit, N-terminal tag, electro 10 reactions GE-CCT7-11 CherryCodon T7 expression kit, C-terminal tag, electro 10 reactions GE-CCT7-12 CherryCodon T7 expression kit, N-terminal tag, chimio 10 reactions GE-CCT7-13 CherryCodon T7 expression kit, C-terminal tag, chimio 10 reactions Copyright 2013 by Delphi Genetics SA. All rights reserved. 20
22 ACADEMIC AND NON-PROFIT LABORATORY ASSURANCE LETTER REGARDING THE USE OF THE T7 EXPRESSION SYSTEM The T7 expression system is based on technology developed at Brookhaven National Laboratory under contract with the U. S. Department of Energy and is the subject of patent applications assigned to Brookhaven Science Associates, LLC. (BSA). BSA will grant a non-exclusive license for use of this technology, including the enclosed materials, based upon the following assurances: 1. These materials are to be used for noncommercial research purposes only. A separate license is required for any commercial use, including the use of these materials for research purposes or production purposes by any commercial entity. Information about commercial licenses may be obtained from the Office of Intellectual Property & Sponsored Research, Brookhaven National Laboratory, Bldg. 475D, P. O. Box 5000, Upton, New York , telephone (631) No materials that contain the cloned copy of T7 gene 1, the gene for T7 RNA polymerase, may be distributed further to third parties outside of your laboratory, unless the recipient receives a copy of this license and agrees to be bound by its terms. This limitation applies to strains BL21(DE3), BL21(DE3)pLysS, BL21(DE3)pLysE and SE1 and any derivatives you may make of them. You may refuse this license by returning the enclosed materials unused. By keeping or using the enclosed materials, you agree to be bound by the terms of this license. Copyright 2013 by Delphi Genetics SA. All rights reserved. 21
23 Worldwide ordering: You can order our products directly from Delphi Genetics worldwide (with the exception of Asia) using our online ordering platform ( Should you prefer to work with a local dealer, you can find a list of distributors on our website: Copyright 2013 by Delphi Genetics SA. All rights reserved. 22
24 Contact us: Delphi Genetics SA Rue Antoine de St-Exupéry, 5 B-6041 Charleroi Belgium Tel: Fax: delphigenetics@delphigenetics.com Copyright 2013 by Delphi Genetics SA. All rights reserved.
StabyExpress TM kit Manual for GST fusion
 StabyExpress TM kit Manual for GST fusion (v2.0) for GE-SEGST-. kits www.delphigenetics.com Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of
StabyExpress TM kit Manual for GST fusion (v2.0) for GE-SEGST-. kits www.delphigenetics.com Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of
BL21(DE3) Competent cells
 BL21(DE3) Competent cells Chemically-competent E. coli cells supplied in 96-tube tray Instruction manual (v2.1) Table of contents Content and storage 1 Material Safety and Data Sheet 2 User Guide 3 o Introduction
BL21(DE3) Competent cells Chemically-competent E. coli cells supplied in 96-tube tray Instruction manual (v2.1) Table of contents Content and storage 1 Material Safety and Data Sheet 2 User Guide 3 o Introduction
Staby TM Codon T7 kit Manual (v2.0)
 Staby TM Codon T7 kit Manual (v2.0) Staby TM CodonT7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o
Staby TM Codon T7 kit Manual (v2.0) Staby TM CodonT7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o
StabyExpress TM T7 kit Manual (v1.7)
 StabyExpress TM T7 kit Manual (v1.7) Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o Overview of Staby Express
StabyExpress TM T7 kit Manual (v1.7) Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o Overview of Staby Express
StabyExpress TM T7 kit Manual (v1.6)
 StabyExpress TM T7 kit Manual (v1.6) Staby Express TM T7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system
StabyExpress TM T7 kit Manual (v1.6) Staby Express TM T7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system
Cherry TM Express kit
 Cherry TM Express kit Manual (v3.0) www.delphigenetics.com Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the Cherry TM tag system 4 o Overview
Cherry TM Express kit Manual (v3.0) www.delphigenetics.com Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the Cherry TM tag system 4 o Overview
StabyExpress TM T7 kit Manual (v1.5)
 StabyExpress TM T7 kit Manual (v1.5) Staby Express TM T7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system
StabyExpress TM T7 kit Manual (v1.5) Staby Express TM T7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system
Staby TM Codon T7 kit Manual (v1.5)
 Staby TM Codon T7 kit Manual (v1.5) Staby TM CodonT7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o
Staby TM Codon T7 kit Manual (v1.5) Staby TM CodonT7 manual. Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the T7 expression system 4 o
Manual (v1.4) Manual (v1.3)
 Cherry TM Codon StabyCloning kit TM kit Manual (v1.4) Manual (v1.3) . Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the Cherry TM tag system
Cherry TM Codon StabyCloning kit TM kit Manual (v1.4) Manual (v1.3) . Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the Cherry TM tag system
Cat. # Product Size DS130 DynaExpress TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1
 Product Name: Kit Component TA PCR Cloning Kit (ptakn-2) Cat. # Product Size DS130 TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1 2 Ligation Buffer
Product Name: Kit Component TA PCR Cloning Kit (ptakn-2) Cat. # Product Size DS130 TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1 2 Ligation Buffer
GetStaby TM kit Manual (v1.4)
 GetStaby TM kit Manual (v1.4) Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the stabilization principle 4 o Benefits of the Staby TM system
GetStaby TM kit Manual (v1.4) Table of contents Content and storage 2 Material Safety and Data Sheet 3 Licenses 4 User Guide 4 o Overview of the stabilization principle 4 o Benefits of the Staby TM system
PRODUCT INFORMATION. Composition of SOC medium supplied :
 Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
BL21(DE3) expression competent cell pack DS265. Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube)
 Product Name : Code No. : Kit Component : BL21(DE3) expression competent cell pack DS265 Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube) transfromation efficiency: 5 10
Product Name : Code No. : Kit Component : BL21(DE3) expression competent cell pack DS265 Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube) transfromation efficiency: 5 10
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
MacBlunt PCR Cloning Kit Manual
 MacBlunt PCR Cloning Kit Manual Shipping and Storage MacBlunt PCR Cloning Kits are shipped on dry ice. Each kit contains a box with cloning reagents and an attached bag with Eco-Blue Competent Cells (optional).
MacBlunt PCR Cloning Kit Manual Shipping and Storage MacBlunt PCR Cloning Kits are shipped on dry ice. Each kit contains a box with cloning reagents and an attached bag with Eco-Blue Competent Cells (optional).
Genlantis A division of Gene Therapy Systems, Inc Telesis Court San Diego, CA USA Telephone: or (US toll free)
 TurboCells BL21(DE3) TurboCells BL21(DE3)pLysS Chemically Competent E. coli Instruction Manual Catalog Numbers C302020 C303020 A division of Gene Therapy Systems, Inc. 10190 Telesis Court San Diego, CA
TurboCells BL21(DE3) TurboCells BL21(DE3)pLysS Chemically Competent E. coli Instruction Manual Catalog Numbers C302020 C303020 A division of Gene Therapy Systems, Inc. 10190 Telesis Court San Diego, CA
Materials Protein synthesis kit. This kit consists of 24 amino acids, 24 transfer RNAs, four messenger RNAs and one ribosome (see below).
 Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Expression of Recombinant Proteins
 Expression of Recombinant Proteins Uses of Cloned Genes sequencing reagents (eg, probes) protein production insufficient natural quantities modify/mutagenesis library screening Expression Vector Features
Expression of Recombinant Proteins Uses of Cloned Genes sequencing reagents (eg, probes) protein production insufficient natural quantities modify/mutagenesis library screening Expression Vector Features
HI-Control BL21(DE3) & HI-Control 10G Chemically Competent Cells
 HI-Control BL21(DE3) & HI-Control 10G Chemically Competent Cells FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE MA156 Rev.31OCT2016 Table of Contents Components & Storage Conditions... 3 HI-Control
HI-Control BL21(DE3) & HI-Control 10G Chemically Competent Cells FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE MA156 Rev.31OCT2016 Table of Contents Components & Storage Conditions... 3 HI-Control
E. cloni EXPRESS Electrocompetent Cells
 E. cloni EXPRESS Electrocompetent Cells FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608) 831-9011 FAX:
E. cloni EXPRESS Electrocompetent Cells FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608) 831-9011 FAX:
Table S1. Bacterial strains (Related to Results and Experimental Procedures)
 Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR
 Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR 1 The problem We wish to clone a yet unknown gene from a known
Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR 1 The problem We wish to clone a yet unknown gene from a known
Supplementary. Table 1: Oligonucleotides and Plasmids. complementary to positions from 77 of the SRα '- GCT CTA GAG AAC TTG AAG TAC AGA CTG C
 Supplementary Table 1: Oligonucleotides and Plasmids 913954 5'- GCT CTA GAG AAC TTG AAG TAC AGA CTG C 913955 5'- CCC AAG CTT ACA GTG TGG CCA TTC TGC TG 223396 5'- CGA CGC GTA CAG TGT GGC CAT TCT GCT G
Supplementary Table 1: Oligonucleotides and Plasmids 913954 5'- GCT CTA GAG AAC TTG AAG TAC AGA CTG C 913955 5'- CCC AAG CTT ACA GTG TGG CCA TTC TGC TG 223396 5'- CGA CGC GTA CAG TGT GGC CAT TCT GCT G
Genomics and Gene Recognition Genes and Blue Genes
 Genomics and Gene Recognition Genes and Blue Genes November 1, 2004 Prokaryotic Gene Structure prokaryotes are simplest free-living organisms studying prokaryotes can give us a sense what is the minimum
Genomics and Gene Recognition Genes and Blue Genes November 1, 2004 Prokaryotic Gene Structure prokaryotes are simplest free-living organisms studying prokaryotes can give us a sense what is the minimum
Supporting Information. Copyright Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 2006
 Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Supporting Information for Expanding the Genetic
Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Supporting Information for Expanding the Genetic
Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost
 Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen University,
Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen University,
Overnight Express TM Autoinduction
 About the System Overnight Express Autoinduction System 1 71300-3 71300-4 Overnight Express Autoinduction System 2 71366-3 71366-4 Description The Overnight Express Autoinduction 1 and 2 are designed for
About the System Overnight Express Autoinduction System 1 71300-3 71300-4 Overnight Express Autoinduction System 2 71366-3 71366-4 Description The Overnight Express Autoinduction 1 and 2 are designed for
Supporting Information
 Supporting Information Transfection of DNA Cages into Mammalian Cells Email: a.turberfield@physics.ox.ac.uk Table of Contents Supporting Figure 1 DNA tetrahedra used in transfection experiments 2 Supporting
Supporting Information Transfection of DNA Cages into Mammalian Cells Email: a.turberfield@physics.ox.ac.uk Table of Contents Supporting Figure 1 DNA tetrahedra used in transfection experiments 2 Supporting
ORFs and genes. Please sit in row K or forward
 ORFs and genes Please sit in row K or forward https://www.flickr.com/photos/teseum/3231682806/in/photostream/ Question: why do some strains of Vibrio cause cholera and others don t? Methods Mechanisms
ORFs and genes Please sit in row K or forward https://www.flickr.com/photos/teseum/3231682806/in/photostream/ Question: why do some strains of Vibrio cause cholera and others don t? Methods Mechanisms
Quantitative reverse-transcription PCR. Transcript levels of flgs, flgr, flia and flha were
 1 Supplemental methods 2 3 4 5 6 7 8 9 1 11 12 13 14 15 16 17 18 19 21 22 23 Quantitative reverse-transcription PCR. Transcript levels of flgs, flgr, flia and flha were monitored by quantitative reverse-transcription
1 Supplemental methods 2 3 4 5 6 7 8 9 1 11 12 13 14 15 16 17 18 19 21 22 23 Quantitative reverse-transcription PCR. Transcript levels of flgs, flgr, flia and flha were monitored by quantitative reverse-transcription
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
Supplementary Information
 Supplementary Information A general solution for opening double-stranded DNA for isothermal amplification Gangyi Chen, Juan Dong, Yi Yuan, Na Li, Xin Huang, Xin Cui* and Zhuo Tang* Supplementary Materials
Supplementary Information A general solution for opening double-stranded DNA for isothermal amplification Gangyi Chen, Juan Dong, Yi Yuan, Na Li, Xin Huang, Xin Cui* and Zhuo Tang* Supplementary Materials
An engineered tryptophan zipper-type peptide as a molecular recognition scaffold
 SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
Engineering D66N mutant using quick change site directed mutagenesis. Harkewal Singh 09/01/2010
 Engineering D66N mutant using quick change site directed mutagenesis Harkewal Singh 09/01/2010 1 1- What is quick change site directed mutagenesis? 2- An overview of the kit contents. 3- A brief information
Engineering D66N mutant using quick change site directed mutagenesis Harkewal Singh 09/01/2010 1 1- What is quick change site directed mutagenesis? 2- An overview of the kit contents. 3- A brief information
Supplemental Data Supplemental Figure 1.
 Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Disease and selection in the human genome 3
 Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Hetero-Stagger PCR Cloning Kit
 Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases For research use only Cat. # : GVT202 Size : 20 Reactions
 pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases Cat. # : GVT202 Size : 20 Reactions Store at -20 For research use only 1 pgm-t Cloning Kit Cat. No.: GVT202 Kit Contents
pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases Cat. # : GVT202 Size : 20 Reactions Store at -20 For research use only 1 pgm-t Cloning Kit Cat. No.: GVT202 Kit Contents
Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH).
 Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20
 pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending)
 peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
Y-chromosomal haplogroup typing Using SBE reaction
 Schematic of multiplex PCR followed by SBE reaction Multiplex PCR Exo SAP purification SBE reaction 5 A 3 ddatp ddgtp 3 T 5 A G 3 T 5 3 5 G C 5 3 3 C 5 ddttp ddctp 5 T 3 T C 3 A 5 3 A 5 5 C 3 3 G 5 3 G
Schematic of multiplex PCR followed by SBE reaction Multiplex PCR Exo SAP purification SBE reaction 5 A 3 ddatp ddgtp 3 T 5 A G 3 T 5 3 5 G C 5 3 3 C 5 ddttp ddctp 5 T 3 T C 3 A 5 3 A 5 5 C 3 3 G 5 3 G
Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC
 Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Supplemental material
 Supplemental material Diversity of O-antigen repeat-unit structures can account for the substantial sequence variation of Wzx translocases Yaoqin Hong and Peter R. Reeves School of Molecular Bioscience,
Supplemental material Diversity of O-antigen repeat-unit structures can account for the substantial sequence variation of Wzx translocases Yaoqin Hong and Peter R. Reeves School of Molecular Bioscience,
strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular
![strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular](/thumbs/72/66361212.jpg) Additional file 2 Identification of AOX1 in P. pastoris GS115 with a Mut s phenotype Results and Discussion The HBsAg producing strain was originally identified as a Mut s (methanol utilization slow) strain
Additional file 2 Identification of AOX1 in P. pastoris GS115 with a Mut s phenotype Results and Discussion The HBsAg producing strain was originally identified as a Mut s (methanol utilization slow) strain
Det matematisk-naturvitenskapelige fakultet
 UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam: Friday
UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam: Friday
GeNei TM Transformation Teaching Kit Manual
 Teaching Kit Manual Cat No. New Cat No. KT07 107385 KT07A 106220 Revision No.: 00060505 CONTENTS Page No. Objective 3 Principle 3 Kit Description 6 Materials Provided 7 Procedure 9 Observation & Interpretation
Teaching Kit Manual Cat No. New Cat No. KT07 107385 KT07A 106220 Revision No.: 00060505 CONTENTS Page No. Objective 3 Principle 3 Kit Description 6 Materials Provided 7 Procedure 9 Observation & Interpretation
Molecular Techniques Third-year Biology
 PLANNING Genetics Lab practices Molecular Techniques. Genetics Lab practices protocol. 2015-16 PCR-DIRECTED MUTAGENESIS, MOLECULAR CLONING AND RESTRICTION ANALYSIS Sessions 1 & 2 (2x3 hours): PCR-directed
PLANNING Genetics Lab practices Molecular Techniques. Genetics Lab practices protocol. 2015-16 PCR-DIRECTED MUTAGENESIS, MOLECULAR CLONING AND RESTRICTION ANALYSIS Sessions 1 & 2 (2x3 hours): PCR-directed
GST Fusion Protein Purification Kit
 Glutathione Resin GST Fusion Protein Purification Kit Cat. No. L00206 Cat. No. L00207 Technical Manual No. TM0185 Version 01042012 Index 1. Product Description 2. Related Products 3. Purification Procedure
Glutathione Resin GST Fusion Protein Purification Kit Cat. No. L00206 Cat. No. L00207 Technical Manual No. TM0185 Version 01042012 Index 1. Product Description 2. Related Products 3. Purification Procedure
CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution
 CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution Cat. Nos. C400EL10, C400CH10, and CIS40025 Available exclusively thru Lucigen. lucigen.com/epibio
CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution Cat. Nos. C400EL10, C400CH10, and CIS40025 Available exclusively thru Lucigen. lucigen.com/epibio
Lecture 19A. DNA computing
 Lecture 19A. DNA computing What exactly is DNA (deoxyribonucleic acid)? DNA is the material that contains codes for the many physical characteristics of every living creature. Your cells use different
Lecture 19A. DNA computing What exactly is DNA (deoxyribonucleic acid)? DNA is the material that contains codes for the many physical characteristics of every living creature. Your cells use different
TaKaRa MiniBEST Plasmid Purification Kit Ver.4.0
 Cat. # 9760 For Research Use TaKaRa MiniBEST Plasmid Purification Kit Ver.4.0 Product Manual Table of Contents I. Description... 3 II. Kit Components... 3 III. Shipping and Storage... 4 IV. Preparation
Cat. # 9760 For Research Use TaKaRa MiniBEST Plasmid Purification Kit Ver.4.0 Product Manual Table of Contents I. Description... 3 II. Kit Components... 3 III. Shipping and Storage... 4 IV. Preparation
RPA-AB RPA-C Supplemental Figure S1: SDS-PAGE stained with Coomassie Blue after protein purification.
 RPA-AB RPA-C (a) (b) (c) (d) (e) (f) Supplemental Figure S: SDS-PAGE stained with Coomassie Blue after protein purification. (a) RPA; (b) RPA-AB; (c) RPA-CDE; (d) RPA-CDE core; (e) RPA-DE; and (f) RPA-C
RPA-AB RPA-C (a) (b) (c) (d) (e) (f) Supplemental Figure S: SDS-PAGE stained with Coomassie Blue after protein purification. (a) RPA; (b) RPA-AB; (c) RPA-CDE; (d) RPA-CDE core; (e) RPA-DE; and (f) RPA-C
Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis
 1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
PCR analysis was performed to show the presence and the integrity of the var1csa and var-
 Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Figure 1. Map of cloning vector pgem T-Easy (bacterial plasmid DNA)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Arabidopsis actin depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB
 Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
NZYGene Synthesis kit
 Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Hurricane Miniprep Kit PROTOCOL
 Hurricane Miniprep Kit PROTOCOL Description: The Hurricane Miniprep Kit is designed for purification of up to 25 ug of high purity plasmid DNA from a starting volume of 2-5 ml of bacterial culture. The
Hurricane Miniprep Kit PROTOCOL Description: The Hurricane Miniprep Kit is designed for purification of up to 25 ug of high purity plasmid DNA from a starting volume of 2-5 ml of bacterial culture. The
Supplemental Data. mir156-regulated SPL Transcription. Factors Define an Endogenous Flowering. Pathway in Arabidopsis thaliana
 Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli
 TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300102, EC300110, EC300150, and C300C105 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300102, EC300110, EC300150, and C300C105 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com
SUPPLEMENTAL TABLE S1. Additional descriptions of plasmid constructions and the oligonucleotides used Plasmid or Oligonucleotide
 SUPPLEMENTAL TABLE S1. Additional descriptions of plasmid constructions and the oligonucleotides used Plasmid or Oligonucleotide former/ working Description a designation Plasmids pes213a b pes213-tn5
SUPPLEMENTAL TABLE S1. Additional descriptions of plasmid constructions and the oligonucleotides used Plasmid or Oligonucleotide former/ working Description a designation Plasmids pes213a b pes213-tn5
SAY IT WITH DNA: Protein Synthesis Activity by Larry Flammer
 TEACHER S GUIDE SAY IT WITH DNA: Protein Synthesis Activity by Larry Flammer SYNOPSIS This activity uses the metaphor of decoding a secret message for the Protein Synthesis process. Students teach themselves
TEACHER S GUIDE SAY IT WITH DNA: Protein Synthesis Activity by Larry Flammer SYNOPSIS This activity uses the metaphor of decoding a secret message for the Protein Synthesis process. Students teach themselves
High-throughput cloning and expression in recalcitrant bacteria
 High-throughput cloning and expression in recalcitrant bacteria Eric R Geertsma & Bert Poolman Supplementary text and figures: Supplementary Figure 1 Frequency of SfiI sites yielding identical 3 extensions
High-throughput cloning and expression in recalcitrant bacteria Eric R Geertsma & Bert Poolman Supplementary text and figures: Supplementary Figure 1 Frequency of SfiI sites yielding identical 3 extensions
PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells
 Supplementary Information for: PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells Ju Hye Jang 1, Hyun Kim 2, Mi Jung Jang 2, Ju Hyun Cho 1,2,* 1 Research Institute
Supplementary Information for: PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells Ju Hye Jang 1, Hyun Kim 2, Mi Jung Jang 2, Ju Hyun Cho 1,2,* 1 Research Institute
NAME:... MODEL ANSWER... STUDENT NUMBER:... Maximum marks: 50. Internal Examiner: Hugh Murrell, Computer Science, UKZN
 COMP710, Bioinformatics with Julia, Test One, Thursday the 20 th of April, 2017, 09h30-11h30 1 NAME:...... MODEL ANSWER... STUDENT NUMBER:...... Maximum marks: 50 Internal Examiner: Hugh Murrell, Computer
COMP710, Bioinformatics with Julia, Test One, Thursday the 20 th of April, 2017, 09h30-11h30 1 NAME:...... MODEL ANSWER... STUDENT NUMBER:...... Maximum marks: 50 Internal Examiner: Hugh Murrell, Computer
Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR
 Supplemental Dataset Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. DNA sequence Amino acid sequence WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR Allele 1 CCTGTC------------------GATAGC
Supplemental Dataset Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. DNA sequence Amino acid sequence WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR Allele 1 CCTGTC------------------GATAGC
Gene synthesis by circular assembly amplification
 Gene synthesis by circular assembly amplification Duhee Bang & George M Church Supplementary figures and text: Supplementary Figure 1. Dpo4 gene (1.05kb) construction by various methods. Supplementary
Gene synthesis by circular assembly amplification Duhee Bang & George M Church Supplementary figures and text: Supplementary Figure 1. Dpo4 gene (1.05kb) construction by various methods. Supplementary
Hes6. PPARα. PPARγ HNF4 CD36
 SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
Phage T1-Resistant TransforMax EPI300 -T1 R Electrocompetent E. coli TransforMax EPI300 -T1 R Chemically Competent E. coli
 Phage T1-Resistant TransforMax EPI300 -T1 R Electrocompetent E. coli TransforMax EPI300 -T1 R Chemically Competent E. coli Cat. Nos. EC02T15, EC02T110, and CT1C0210 Connect with Epicentre on our blog (epicentral.blogspot.com),
Phage T1-Resistant TransforMax EPI300 -T1 R Electrocompetent E. coli TransforMax EPI300 -T1 R Chemically Competent E. coli Cat. Nos. EC02T15, EC02T110, and CT1C0210 Connect with Epicentre on our blog (epicentral.blogspot.com),
Endura Chemically Competent and Electrocompetent Cells
 Endura Chemically Competent and Electrocompetent Cells FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE. Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608)
Endura Chemically Competent and Electrocompetent Cells FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE. Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608)
I-Blue Midi Plasmid Kit. I-Blue Midi Plasmid Kit. (Endotoxin Free) IBI SCIENTIFIC. Instruction Manual Ver For Research Use Only.
 Instruction Manual Ver. 05.11.17 For Research Use Only I-Blue Midi Plasmid Kit & I-Blue Midi Plasmid Kit (Endotoxin Free) IB47180, IB47190 (2 Preparation Sample Kit) IB47181, IB47191 (25 Preparation Kit)
Instruction Manual Ver. 05.11.17 For Research Use Only I-Blue Midi Plasmid Kit & I-Blue Midi Plasmid Kit (Endotoxin Free) IB47180, IB47190 (2 Preparation Sample Kit) IB47181, IB47191 (25 Preparation Kit)
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli
 TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300105, EC300110, EC300150, and C300C105 Connect with Epicentre on our blog (epicentral.blogspot.com),
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300105, EC300110, EC300150, and C300C105 Connect with Epicentre on our blog (epicentral.blogspot.com),
Epicurian Coli BL21(DE3) Competent Cells, Epicurian Coli BL21(DE3)pLysS Competent Cells, and Epicurian Coli BL21 Competent Cells
 Epicurian Coli BL21(DE3) Competent Cells, Epicurian Coli BL21(DE3)pLysS Competent Cells, and Epicurian Coli BL21 Competent Cells INSTRUCTION MANUAL Catalog #200131 (Epicurian Coli BL21(DE3) Competent Cells,
Epicurian Coli BL21(DE3) Competent Cells, Epicurian Coli BL21(DE3)pLysS Competent Cells, and Epicurian Coli BL21 Competent Cells INSTRUCTION MANUAL Catalog #200131 (Epicurian Coli BL21(DE3) Competent Cells,
Certificate of Analysis
 Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Certificate of Analysis pet6xhn Expression Vector Set Contents Product Information... 1 pet6xhn-n, pet6xhn-c, and pet6xhn-gfpuv Vector Information... 2 Location of Features... 4 Additional Information...
Converting rabbit hybridoma into recombinant antibodies with effective transient production in an optimized human expression system
 Converting rabbit hybridoma into recombinant antibodies with effective transient production in an optimized human expression system Dr. Tim Welsink Molecular Biology Transient Gene Expression OUTLINE Short
Converting rabbit hybridoma into recombinant antibodies with effective transient production in an optimized human expression system Dr. Tim Welsink Molecular Biology Transient Gene Expression OUTLINE Short
Supplementary Information. Construction of Lasso Peptide Fusion Proteins
 Supplementary Information Construction of Lasso Peptide Fusion Proteins Chuhan Zong 1, Mikhail O. Maksimov 2, A. James Link 2,3 * Departments of 1 Chemistry, 2 Chemical and Biological Engineering, and
Supplementary Information Construction of Lasso Peptide Fusion Proteins Chuhan Zong 1, Mikhail O. Maksimov 2, A. James Link 2,3 * Departments of 1 Chemistry, 2 Chemical and Biological Engineering, and
ITS Sequencing in Millepora. 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector
 Page 1 of 5 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector 1. Digest 1 µg of pbluescript with Eco RI 2. Following digestion, add 0.1 volumes of 3M sodium acetate (ph
Page 1 of 5 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector 1. Digest 1 µg of pbluescript with Eco RI 2. Following digestion, add 0.1 volumes of 3M sodium acetate (ph
Supporting Information
 Supporting Information Barderas et al. 10.1073/pnas.0801221105 SI Text: Docking of gastrin to Constructed scfv Models Interactive predocking of the 4-WL-5 motif into the central pocket observed in the
Supporting Information Barderas et al. 10.1073/pnas.0801221105 SI Text: Docking of gastrin to Constructed scfv Models Interactive predocking of the 4-WL-5 motif into the central pocket observed in the
TECHNICAL BULLETIN. In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits
 In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
In Vitro Bacterial Split Fluorescent Protein Fold n Glow Solubility Assay Kits Catalog Numbers APPA001 In Vitro Bacterial Split GFP "Fold 'n' Glow" Solubility Assay Kit (Green) APPA008 In Vitro Bacterial
SUPPLEMENTARY MATERIALS AND METHODS. E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1)
 SUPPLEMENTARY MATERIALS AND METHODS E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1) dinb::kan (lab stock) derivative was used as wild-type. MG1655 alka tag dinb (2) is
SUPPLEMENTARY MATERIALS AND METHODS E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1) dinb::kan (lab stock) derivative was used as wild-type. MG1655 alka tag dinb (2) is
Geneaid Maxi Plasmid Kit & Geneaid Maxi Plasmid Kit (Endotoxin Free)
 Geneaid Maxi Plasmid Kit & Geneaid Maxi Plasmid Kit (Endotoxin Free) PM002, PME02 (2 Preparation Sample Kit) PM010, PME10 (10 Preparation Kit) PM025, PME25 (25 Preparation Kit) Instruction Manual Ver.
Geneaid Maxi Plasmid Kit & Geneaid Maxi Plasmid Kit (Endotoxin Free) PM002, PME02 (2 Preparation Sample Kit) PM010, PME10 (10 Preparation Kit) PM025, PME25 (25 Preparation Kit) Instruction Manual Ver.
Project 07/111 Final Report October 31, Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines
 Project 07/111 Final Report October 31, 2007. Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines Project Leader: Dr Douglas C. Hodgins (519-824-4120 Ex 54758, fax 519-824-5930)
Project 07/111 Final Report October 31, 2007. Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines Project Leader: Dr Douglas C. Hodgins (519-824-4120 Ex 54758, fax 519-824-5930)
Supporting information for Biochemistry, 1995, 34(34), , DOI: /bi00034a013
 Supporting information for Biochemistry, 1995, 34(34), 10807 10815, DOI: 10.1021/bi00034a013 LESNIK 10807-1081 Terms & Conditions Electronic Supporting Information files are available without a subscription
Supporting information for Biochemistry, 1995, 34(34), 10807 10815, DOI: 10.1021/bi00034a013 LESNIK 10807-1081 Terms & Conditions Electronic Supporting Information files are available without a subscription
Bacterial Hosts and Protein Studies. Brian & Phuong
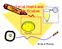 Bacterial Hosts and Protein Studies Brian & Phuong Expression Studies: Purpose To find cell strain that gives best expression of protein(s) you are studying at. Bacterial Hosts Commonly Used for Expression
Bacterial Hosts and Protein Studies Brian & Phuong Expression Studies: Purpose To find cell strain that gives best expression of protein(s) you are studying at. Bacterial Hosts Commonly Used for Expression
Supplemental Information. Human Senataxin Resolves RNA/DNA Hybrids. Formed at Transcriptional Pause Sites. to Promote Xrn2-Dependent Termination
 Supplemental Information Molecular Cell, Volume 42 Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination Konstantina Skourti-Stathaki, Nicholas
Supplemental Information Molecular Cell, Volume 42 Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination Konstantina Skourti-Stathaki, Nicholas
Lambda CE6 Induction Kit
 Lambda CE6 Induction Kit INSTRUCTION MANUAL Catalog #235200 Revision A For In Vitro Use Only 235200-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other
Lambda CE6 Induction Kit INSTRUCTION MANUAL Catalog #235200 Revision A For In Vitro Use Only 235200-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Investigation of the Biosynthesis of the Lasso Peptide Chaxapeptin Using an E. coli-based Production System Helena Martin-Gómez, Uwe Linne, Fernando Albericio, Judit Tulla-Puche,*
SUPPORTING INFORMATION Investigation of the Biosynthesis of the Lasso Peptide Chaxapeptin Using an E. coli-based Production System Helena Martin-Gómez, Uwe Linne, Fernando Albericio, Judit Tulla-Puche,*
Human Cell-Free Protein Expression System
 Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Cat. # 3281 For Research Use Human Cell-Free Protein Expression System Product Manual Table of Contents I. Description... 3 II. Components... 3 III. Materials Required but not Provided... 4 IV. Storage...
Cat # Box1 Box2. DH5a Competent E. coli cells CCK-20 (20 rxns) 40 µl 40 µl 50 µl x 20 tubes. Choo-Choo Cloning TM Enzyme Mix
 Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
Supporting Online Information
 Supporting Online Information Isolation of Human Genomic DNA Sequences with Expanded Nucleobase Selectivity Preeti Rathi, Sara Maurer, Grzegorz Kubik and Daniel Summerer* Department of Chemistry and Chemical
Supporting Online Information Isolation of Human Genomic DNA Sequences with Expanded Nucleobase Selectivity Preeti Rathi, Sara Maurer, Grzegorz Kubik and Daniel Summerer* Department of Chemistry and Chemical
CopyCutter EPI400 Electrocompetent E. coli. CopyCutter EPI400 Chemically Competent E. coli
 Cat. Nos. C400EL10, C400CH10, and CIS40025 CopyCutter EPI400 Electrocompetent and Chemically Competent E. coli* cells were developed to significantly lower the copy number of a wide variety of common vectors
Cat. Nos. C400EL10, C400CH10, and CIS40025 CopyCutter EPI400 Electrocompetent and Chemically Competent E. coli* cells were developed to significantly lower the copy number of a wide variety of common vectors
Supplementary Figure 1A A404 Cells +/- Retinoic Acid
 Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
ENDEXT Technology. Instruction manual for protein synthesis. with wheat germ cell-free system
 ENDEXT Technology Instruction manual for protein synthesis with wheat germ cell-free system 1 Protocol Overview Plasmid DNA construction (see Section 3.1) Preparation of plasmid DNA for transcription (see
ENDEXT Technology Instruction manual for protein synthesis with wheat germ cell-free system 1 Protocol Overview Plasmid DNA construction (see Section 3.1) Preparation of plasmid DNA for transcription (see
2.5. Equipment and materials supplied by user Template preparation by cloning into plexsy_invitro-2 vector 4. 6.
 Manual 15 Reactions LEXSY in vitro Translation Cell-free protein expression kit based on Leishmania tarentolae contains the vector plexsy_invitro-2 Cat. No. EGE-2002-15 FOR RESEARCH USE ONLY. NOT INTENDED
Manual 15 Reactions LEXSY in vitro Translation Cell-free protein expression kit based on Leishmania tarentolae contains the vector plexsy_invitro-2 Cat. No. EGE-2002-15 FOR RESEARCH USE ONLY. NOT INTENDED
Legends for supplementary figures 1-3
 High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology Sidsel Nag 1,2 *, Marlene D. Dalgaard 3, Poul-Erik
High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology Sidsel Nag 1,2 *, Marlene D. Dalgaard 3, Poul-Erik
Plasmid Midiprep Purification Kit
 Plasmid Midiprep Purification Kit Cat. # : DP01MD/ DP01MD-100 Size : 20/100 Reactions Store at RT For research use only 1 Description: The Plasmid Midiprep Purification Kit provides simple rapid protocol
Plasmid Midiprep Purification Kit Cat. # : DP01MD/ DP01MD-100 Size : 20/100 Reactions Store at RT For research use only 1 Description: The Plasmid Midiprep Purification Kit provides simple rapid protocol
Anti-Pim-1 (Cat#3247), anti-met (Cat#3127), anti-ron (Cat#2654), Anti-EGFR
 Supplementary Methods Antibodies Anti-Pim-1 (Cat#3247), anti-met (Cat#3127), anti-ron (Cat#2654), Anti-EGFR (Cat#2646), anti-igf1r (Cat#3018), anti-insr (Cat#3020), anti-akt (pan, Cat#4691), anti-phospho-akt
Supplementary Methods Antibodies Anti-Pim-1 (Cat#3247), anti-met (Cat#3127), anti-ron (Cat#2654), Anti-EGFR (Cat#2646), anti-igf1r (Cat#3018), anti-insr (Cat#3020), anti-akt (pan, Cat#4691), anti-phospho-akt
EasyPrep TM Plasmid Maxiprep Manual
 EasyPrep TM Plasmid Maxiprep Manual Catalog#: PD05-01, PD05-02 PD05-11, PD05-12 Bioland Scientific LLC 14925 Paramount Blvd., Suite C Paramount, CA 90723 USA Tel: (877) 603-8882 Fax: (562) 733-6008 Email:
EasyPrep TM Plasmid Maxiprep Manual Catalog#: PD05-01, PD05-02 PD05-11, PD05-12 Bioland Scientific LLC 14925 Paramount Blvd., Suite C Paramount, CA 90723 USA Tel: (877) 603-8882 Fax: (562) 733-6008 Email:
