Molecular cytogenetic characterization of Thinopyrum intermedium-derived wheat germplasm specifying resistance to wheat streak mosaic virus
|
|
|
- Erik Marsh
- 6 years ago
- Views:
Transcription
1 Theor Appl Genet (1998) 96 : 1 7 Springer-Verlag 1998 Q. Chen B. Friebe R. L. Conner A. Laroche J. B. Thomas B. S. Gill Molecular cytogenetic characterization of Thinopyrum intermedium-derived wheat germplasm specifying resistance to wheat streak mosaic virus Received: 2 May 1997 / Accepted: 20 May 1997 Abstract ¹hinopyrum intermedium is a promising source of resistance to wheat streak mosaic virus (WSMV), a devastating disease of wheat. Three wheat germplasm lines possessing resistance to WSMV, derived from ¹riticum aestivum ¹h. intermedium crosses, are analyzed by C-banding and genomic in situ hybridization (GISH) to determine the amount and location of alien chromatin in the transfer lines. Line CI15092 was confirmed as a disomic substitution line in which wheat chromosome 4A was replaced by ¹h. intermedium chromosome 4Ai 2. The other two lines, CI17766 and A , carry an identical Robertsonian translocation chromosome in which the complete short arm of chromosome 4Ai 2 was transferred to the long arm of wheat chromosome 4A. Fluorescence in situ hybridization (FISH) using ABD genomic DNA from wheat as a probe and S genomic DNA from Pseudoroegneria stipifolia as the blocker, and vice versa, revealed that the entire short arm of the translocation was derived from the short arm of chromosome 4Ai 2 and the breakpoint was located at the centromere. Chromosomal arm ratios (L/S) of 2.12 in CI17766 and 2.15 in A showed that the translocated chromosome is submetacentric. This Lethbridge Research Centre, Canada, contribution No , and Contribution no J of the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, KS, USA Communicated by G. Wenzel Q. Chen ( ) R. L. Conner A. Laroche J. B. Thomas Research Centre, Agriculture and Agri-Food Canada, P.O. Box 3000, Lethbridge, T1J 4B1, AB Canada Fax:#1 (403) chenqi@em.agr.ca B. Friebe B. S. Gill Department of Plant Pathology, Wheat Genetic Resource Centre, Throckmorton Hall, Kansas State University, Manhattan, KS , USA translocated chromosome is designated as T4AL 4Ai 2S as suggested by Friebe et al. (1991). Key words Fluorescence in situ hybridization Translocation WSMV resistance ¹hinopyrum intermedium ¹riticum aestivum Introduction Wheat streak mosaic virus (WSMV) is a damaging viral disease of wheat found in Canada, the United States and many other countries of the world. There is no resistance in wheat but several resistant germplasms from wheat ¹hinopyrum intermedium hybrids have been developed (Friebe et al. 1991). The wheat germplasm line CI15092, derived from a cross of wheat with ¹h. intermedium (Wells et al. 1973), was identified as a substitution line with immunity to WSMV. The resistance to WSMV exhibited by this line is controlled by a gene(s) located on the short arm of a ¹h. intermedium chromosome, 4Ai 2 (Wang and Liang 1977; Friebe et al. 1991). A translocation line also carrying this resistance was released as CI17766 (Liang et al. 1979). Based on C-banding and in situ hybridization, Friebe et al. (1991) concluded that the translocation involved the short arm of chromosome 4Ai 2 and the long arm of chromosome 4A, which had broken and rejoined at the centromere (i.e., a Robertsonian translocation). These observations contradicted Wang et al. (1980) who, based on chromosome pairing, had concluded that only a part of the short arm of 4Ai 2 had replaced a distal segment of the short arm of chromosome 4A (formerly 4B) in CI Recently, using fluorescence in situ hybridization (FISH) analysis, Wang and Zhang (1996) again reiterate that the translocated chromosome consists of the complete long arm of 4A, the proximal 45% of the short arm of 4A, and the entire short arm of the ¹h. intermedium chromosome 4Ai 2.
2 2 However, 4AS is genetically homoeologous to the 4AL and 4DL arms, and its recombination with 4Ai 2S seems highly improbable. To resolve this apparent conflict, genomic in situ hybridization (GISH), C-banding, and phase-contrast microscopy analyses were used in the present study to unequivocally map the physical location of the translocation breakpoint and to estimate the size of the transferred ¹h. intermedium chromosome segment. Material and methods Three WSMV-resistant wheat-¹h. intermedium derivatives were analysed in the present study. Germplasm CI15092 was produced by Wells et al. (1973) and was identified as a 4Ai 2(4D) chromosome substitution line (Friebe et al. 1991). Lines CI17766 and A are wheat-¹h. intermedium translocation stocks that were both derived from CI Line CI17766 was derived from the cross of Chinese Spring monosomic for chromosome 5B CI The resulting F progeny were crossed with the Chinese Spring ph1 mutant and line CI17766 was selected from the resulting progeny of this second cross and is resistant to all known strains of WSMV (Liang et al. 1979). Line A is a re-selection from CI17766 and exhibits resistance to WSMV at or below 25 C (Wang and Zhang 1996). To determine the breakpoint and size of the transferred ¹h. intermedium segments and their genomic origin, GISH analysis was used with total genomic DNA probes from the following species: ¹riticum aestivum L. cv Chinese Spring (CS) (2n"6x"42, genomes ABD); ¹hinopyrum elongatum (Host) Beauv. (2n"2x"14, genome E); ¹hinopyrum bessarabicum (Savul. and Rayss) A. Love (2n"2x"14, genome J); and Pseudoroegneria stipifolia (Czern ex Nevski) A. Love (2n"2x"14, genome S). C-banding was performed using the technique described by Gill et al. (1991). The GISH protocol followed the procedure described by Chen et al. (1995). Chromosome preparations were carried out according to Chen et al. (1996). To clarify the position of the primary constriction, mitotic chromosomes in root-tip cells were photographed in phase contrast before GISH analysis. The length of alien or translocated chromosomes were measured after GISH and the mean length of the short and long arms and the total length of the translocated chromosomes in these lines were compared using Student s t-test (Steel and Torrie 1980). The rationale for the notational designations of ¹h. intermedium chromosomes was discussed by Friebe et al. (1991, 1996). Thus, for chromosome 4Ai 2 the Arabic number 4 identifies that it belongs to homoeologous group 4, the letters Ai denote its origin from Agropyron intermedium (now called ¹h. intermedium) and 2 identifies it as the second group-4 ¹h. intermedium chromosome transferred to wheat (Friebe et al. 1991). For T4AL 4Ai 2S, the letter T identifies a translocation chromosome, 4AL"long arm of chromosome 4A of wheat, the symbol z indicates a breakpoint at the centromere, and 4Ai 2S"short arm of 4Ai 2 of¹h.intermedium (Raupp et al. 1995). Results Germplasm line CI15092 GISH analysis of CI15092, using genomic DNA of Chinese Spring as a probe and Ps. stipifolia S genomic DNA as the blocker, labelled 20 out of the 21 chromosome pairs over their entire lengths (yellow) but one chromosome pair remained unlabelled (appears red because of counterstaining with propidine iodide) (Fig. 1 a). The reverse analysis using S genomic DNA as a probe and wheat genomic DNA as the blocker, labelled one chromosome pair and left the other 20 pairs unlabelled (Fig. 1b). Under the reverse conditions, the alien pair showed strongest labelling around the telomeres and centromeres with a weakly labelled segment at the center of each arm (Fig. 1 b). Measurement of 20 ¹h. intermedium 4Ai 2 chromosomes in ten GISH-labelled cells of CI15092 gave an average length of 6.26 μm (S"2.88, L"3.38) and an arm ratio (L/S) of 1.18 (Table 1). The C-banding pattern of 4Ai 2 in CI15092 is distinctive, with a small subtelomeric C- band in the short arm and a proximal C-band and prominent terminal C-band in the long arm (Fig. 2), substituting a chromosome pair 4A of wheat, as previously reported by Friebe et al. (1991). Germplasm lines CI17766 and A GISH analysis of CI17766 and its re-selection, A , using a wheat genomic DNA probe and an S genome DNA blocker (Figs. 1 c, g), together with the reverse GISH analysis (S-genome DNA as a probe and wheat genomic DNA as the blocker) (Figs. 1 d, h), all demonstrated that these lines carry a translocated Fig. 1a h GISH on somatic metaphase cells from root tips of substitution line CI15092 and translocation lines CI17766 and A using genomic DNA from Chinese Spring (ABD genomes), Pseudoroegneria stipifolia (S genome) and ¹h. bessarabicum (J genome) as probes. Sites of probe hybridization fluoresce yellow or greenish yellow with FITC, while non-hybridized sites fluoresce red with propidium-iodide counterstain. a Line CI15092 shows two red submetacentric ¹h. intermedium 4Ai 2 chromosomes and 40 yellow wheat chromosomes labelled with wheat genomic DNA probe in the presence of an S genomic DNA blocker. b The two alien chromosomes 4Ai 2 in line CI15092 strongly hybridized at terminal and centromeric regions when detected with the S genomic DNA probe and the wheat genomic DNA blocker. c One pair of submetacentric translocated chromosomes in line CI17766 was detected by a wheat genomic DNA probe and an S genomic DNA blocker. The translocation breakpoint is at the centromere. d Line CI17766 displays the two yellow 4Ai 2 chromosome arms carrying a GISH fingerprint detected by an S genomic DNA probe attached onto two red wheat chromosome arms with breakpoints at the centromere. e Detection of one or two artificial tertiaries constrictions occurring on the long arm of wheat chromosomes in line CI17766 after GISH with wheat genomic DNA as a probe and S genomic DNA as a blocker. f Detection of one pair of submetacentric translocated chromosomes with a breakpoint at the centromere by the J genomic DNA probe in line CI Fourteen smaller wheat chromosomes were also stained light yellow by the S genomic DNA. g Detection of one pair of a submetacentric translocated chromosomes in line A using wheat genomic DNA as a probe. h A cell of A showing strong hybridization signals near the centromere area on short arms of the two translocated chromosomes when S genomic DNA was used as a probe and wheat genomic DNA was used as a blocker
3 3
4 4 Table 1 Length (μm) of different chromosomes in CS, CI15092, CI17766 and A a, withincolumns, means followed by the same letter are not significantly different from each other according to Student s t-test at P"0.01 Chromosome Short Long Total Arm arm arm ratio 4A (CS) 3.68$0.43a 6.01$0.70a 9.69$1.08a 1.64b 4Ai 2 (CI15092) 2.88$0.34b 3.38$0.31b 6.26$0.55c 1.18c 4AL 4Ai 2S (CI17766) 2.76$0.33b 5.86$0.79a 8.62$1.07b 2.12a 4AL 4Ai 2S (A ) 2.95$0.54b 6.33$1.13a 9.27$1.64ab 2.15a Fig. 2 C-banding patterns of critical chromosomes present in the WSMV-resistant germplasm chromosome in which a short segment from ¹h. intermedium was joined to a long wheat segment with the breakpoint apparently located at the centromere. This coincidence of the breakpoint and centromere has been the subject of conflicting reports in the literature (Friebe et al. 1991; Wang and Zhang 1996). Unequivocal identification of primary and secondary constrictions is possible if mitotic metaphases are first analysed in phase contrast followed by chromosome banding or GISH analyses (Endo and Gill 1984). Under negative phase contrast, only primary constrictions (centromeres) and secondary constrictions (nucleolar organisers) are clearly visible (Fig. 3 a and c). However, following GISH-staining tertiary constrictions sometimes became evident, especially in those chromosomes that were labelled by the wheat genomic-dna labelled probe (Figs. 1 e and 3 b). Based on observations of 30 GISH-labelled cells in CI17766 using a wheat genomic DNA probe, 47% of the translocated chromosomes showed a narrow band with no hybridisation signal in the middle of the long arm, thus creating the appearance of a double centromeric chromosome (Fig. 1 e). For this reason, a comparison of chromosomes in phase contrast with GISH (CI1776)- and reverse GISH (A )-labelled chromosomes of the same cell was used to clearly demonstrate that the breakpoint in the translocated chromosome pair coincided with the centromere (Fig. 3). GISH images of CI17766 were also produced using genomic DNA from ¹h. bessarabicum (genome J) and ¹h. elongatum (genome E) as probes in the presence of wheat genomic DNA as a blocker. As with the S genome probe, the J genome probe also detected a pair of translocated chromosomes with a breakpoint at the centromere. The entire short arm was labelled and the long arm was unlabelled (Fig. 1f ). In addition to the signal on the short arm of the translocated chromosome, 14 other chromosomes were also weakly labelled light yellow by J genomic DNA (Fig. 1f ). Because these chromosomes were smaller than average, they probably belong to the D genome of wheat. Genomic DNA from ¹h. elongatum gave similar results to the S or J genomic DNA probes except that its hybridization signal on the translocated chromosome was weaker. These differences in signal intensities may be a reflection of genomic repetitive DNA sequence homology. C-banding analysis of CI17766 and A showed identical results with respect to the translocated chromosome (Fig. 2). In each case, the short arm of the translocated pair was identical to the short arm of the alien pair in CI15092 (4Ai 2), while the long arm was identical to the long arm of chromosome 4A of Chinese Spring (Fig. 2). Measurements of chromosome length were also made on comparable preparations of Chinese Spring, CI15092, CI17766 and A (Table 1). The average length of the translocated chromosome in CI17766 is 8.62 μm (S"2.76; L"5.86) with an arm ratio (L/S) of The average length of the translocated chromosome in line A is 9.27 μm (S"2.95 μm; L"6.33 μm) with a arm ratio (L/S) of The total length of the translocated chromosome in line A is a little longer than that found in CI17766, but the estimated value of
5 5 Fig. 3a d Comparison of metaphase chromosomes under phase contrast and GISH in the same cell of the translocation lines CI17766 (a and b) and A (c and d). Centromeres are marked by arrows, while arrow heads indicated the tertiary constriction that occurred after GISH when wheat genomic DNA is used as the probe. a Phase-contrast image of a root-tip cell of CI17766 showing the position of the centromere on the translocated chromosome. b Tertiary constrictions (arrow heads) that became evident along the long arm of same wheat chromosomes shown in after GISH using a wheat genomic DNA probe and an S genomic DNA blocker. c Phase-contrast image of a root-tip cell of A showing the position of the centromere on the translocated chromosome. d GISH using an S genomic DNA probe and a wheat DNA blocker for the same cell as in c t"!1.81 was not significant (P"0.01; df 58). No statistically significant difference was found among the lines CI15092, CI17766, and A for the mean length of the short arm of 4Ai 2. Similarly, no significant differences were found between the long arms of 4A of Chinese Spring and those of the T4AL 4Ai 2S translocated chromosomes of CI17766 (t"!0.7; df 48). The results for the size and arm ratio of the translocated chromosome in both CI17766 and A strongly support the GISH and C-banding results that the translocated chromosome resulted through breakage at the centromere and re-union in a Robertsonian translocation between the short arm of 4Ai 2 and the long arm of 4A. Discussion C-banding and GISH analyses revealed the presence of an identical Robertsonian translocation in lines
6 6 CI17766 and A Comparisons of the C-banding pattern and chromosome lengths of the original 4A, 4Ai 2, and the translocated chromosomes T4A 4Ai 2 showed that the translocation consisted of the entire short arm of 4Ai 2 of¹h.intermedium and the entire long arm of chromosome 4A of wheat joined at the centromere. Furthermore comparison of chromosomes under phase contrast and after GISH revealed that the breakpoints in both CI17766 and A were located at the centromere. The translocated chromosomes in CI17766 and A were both submetacentric with similar arm ratios (L/S) of 2.12 and 2.15, respectively. Wang and Zhang (1996) reported that the translocated chromosome in A was metacentric with a non-centromeric breakpoint located about the middle of the short arm, whereas previous studies based on C-banding analysis identified the translocated chromosome as sub-metacentric and suggested a centromeric breakpoint with the complete arms of 4AL and 4Ai 2S being present (Friebe et al. 1991). The present data are in agreement with the latter study and identified the translocated chromosome as T4AL 4Ai 2S. It should be noted that in about half of the GISH-labelled cells analysed, the T4AL 4Ai 2S translocated chromosome appeared to have, in addition to the centromere, another constriction-like structure located in the middle of the long arm (Fig. 1 e). This artefact was caused by the lack of hybridization signal in this region, most likely resulting from twisting of the chromosome arms, and was probably the reason for the mis-identification of the translocated chromosome in the study by Wang and Zhang (1996). Lack of a hybridization signal in some chromosome regions is common in GISH experiments (Xu et al. 1994) and was also observed for the long arms of other wheat chromosomes in the present study (Fig. 3 b). When examined under phase contrast, each chromosome possessed only one primary constriction or centromere. In the few cases where more than one constriction occurred, they were nucleolar organisers (secondary constrictions) on chromosomes 1B or 6B of wheat (Endo and Gill 1984). Although no other constriction-like structures were noted under phase contrast, obvious examples of tertiary constrictions appeared in the same chromosomes following GISH with wheat genomic DNA as the probe. Consequently, determining the location of the centromere from GISHstained preparations alone may become somewhat subjective. While GISH staining provides unambiguous evidence of translocation breakpoints, prior phasecontrast photography of cells provided a useful adjunct to this method in order to demonstrate convincingly the relationship between the primary constriction or centromere and the translocation breakpoint. The wheat-¹h. intermedium translocated chromosome in CI17766 and A was assumed to be derived from induced homoeologous recombination (Liang et al. 1979). However, the wheat chromosome 4A involved in this translocation is structurally rearranged (Naranjo et al. 1988). Chromosome 4A was involved in a large paracentric inversion and a cyclical translocation with chromosomes 5A and 7B. As a result, the physically shorter arm is genetically related to other group-4 long arms and the physically longer arm shows homoeology to group-4 short arms, with the distal segment being homoeologous to group-7 short arms (Mickelson-Young et al. 1995). Thus, the 4Ai 2 short arm can not compensate for the loss of the short arm of chromosome 4A which is homoeologous to the group-4 long arms. Because of the rearranged structure of chromosome 4A, the 4Ai 2S arm probably had no chance for homoeologous pairing and recombination with either arm of chromosome 4A. In the F hybrid of CS mono 5B CI15092 three univalents were present (Liang et al. 1979), involving chromosomes 5B, 4A, and 4Ai 2. Univalents have a tendency to break at the centromere, followed by fusion of the broken chromosome arms (Sears 1952). Depending on which arms are involved in the fusion, different whole-arm translocations can be produced (Lukaszewski and Gustafson 1983). The centromeric breakpoint in T4AL 4Ai 2S and the blurred homoeology of chromosome 4A suggests that this translocation was most likely produced by centromeric breakage and re-fusion. In the present GISH study, the 4Ai 2 chromosome showed special GISH patterns with different genomic DNA probes. The S genomic DNA probe hybridized strongly at centromeric and distal regions, but not at the middle of the 4Ai 2 arm. This indicates that chromosome 4Ai 2 is genetically related to the S genome but not directly derived from this genome. The GISH results, using J or E genomic DNA probes, showed that these probes strongly hybridized to the entire length of the 4Ai 2 arms. These results suggest that chromosome 4Ai 2 was most likely derived from the J or E genomes. However, it is interesting to note that the S genomic probe provided better discrimination of the ¹h. intermedium chromatin than did J or E genomic DNA. This is most likely due to the fact that the S genome is less closely related to the ABD genomes than are the J or E genomes. This would explain why the J and E genomic probes hybridized with about 14 small wheat chromosomes which were possibly those of the D genome of wheat. Other molecular and cytogenetic studies have also confirmed these points (Dvorak 1980; Zhang et al. 1996). References Chen Q, Conner RL, Laroche A (1995) Identification of the parental chromosomes of the wheat-alien amphiploid Agrotana by genomic in situ hybridization. Genome 38 :
7 7 Chen Q, Conner RL, Laroche A (1996) Molecular characterization of Haynaldia villosa chromatin in wheat lines carrying resistance to wheat curl mite colonization. Theor Appl Genet 93 : Dvorak J (1980) Homoeology between Agropyron elongatum chromosomes and ¹riticum aestivum chromosomes. Can J Genet Cytol 22 : Endo TR, Gill BS (1984) The heterochromatin distribution and genome evolution in diploid species of Elymus and Agropyron. Can J Genet Cytol 26 : Friebe B, Mukai Y, Dhaliwal HS, Martin TJ, Gill BS (1991) Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germ plasm by C-banding and in situ hybridization. Theor Appl Genet 81 : Friebe B, Jiang JM, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to disease and pests: current status. Euphytica 91 : Gill BS, Friebe B, Endo TR (1991) Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (¹riticum aestivum L.). Genome 34 : Liang GH, Wang RRC, Niblett CL, Heyne ER (1979) Registration of B wheat germ plasm. Crop Sci 19 : 427 Lukaszewski AS, Gustafson JP (1983) Translocation and modifications of chromosomes in ¹riticale wheat hybrids. Theor Appl Genet 64 : Mickelson-Young L, Endo TR, Gill BS (1995) A cytogenetic ladder map of wheat homoeologous group-4 chromosomes. Theor Appl Genet 90 : Naranjo T, Roca A, Goicoechea PG, Giraldez R (1988) Chromosome structure of common wheat: genome reassignment of chromosomes 4A and 4B. In: Miller TE, Koebner RMD (eds) Proc 7th Int Wheat Genet Symp Cambridge, UK, pp Raupp WJ, Friebe B, Gill BS (1995) Suggested guidelines for the nomenclature and abbreviation of the genetic stocks of wheat, ¹riticum aestivum L., and its relatives. Wheat Inf Serv 81 : Sears ER (1952) Misdivision of univalents in common wheat. Chromosoma 4 : Steel RGD, Torrie JH (1980) Principles and procedures of statistics: a biometrical approach. McGraw-Hill, New York Wang RRC, Liang GH (1977) Cytogenetic location of genes for resistance to wheat streak mosaic in an Agropyron substitution line. J Hered 68 : Wang RRC, Zhang ZY (1996) Characterization of the translocated chromosome using fluorescence in situ hybridization and random amplified polymorphic DNA on two ¹riticum aestivum- ¹hinopyrum intermedium translocation lines resistant to wheat streak mosaic or barley yellow dwarf virus. Chromosome Res 4 : Wang RRC, Barnes EE, Cook LL (1980) Transfer of wheat streak mosaic virus resistance from Agropyron to the homoeologous chromosome of wheat. Cereal Res Commun 8 : Wells DG, Wong RSC, Lay CL, Gardner WAS, Buchenau GW (1973) Registration of C.I and C.I wheat germplasm. Crop Sci 13 : 776 Xu J, Conner RL, Laroche A (1994) C-banding and fluorescence in situ hybridization studies of the wheat-alien hybrid Agrotana. Genome 37 : Zhang XY, Koul A, Petroski R, Ouellet T, Fedak G, Dong YS, Wang RRC (1996) Molecular verification and characterization of BYDY-resistant germ plasms derived from hybrids of wheat with ¹hinopyrum ponticum and ¹h. intermedium. Theor Appl Genet 93 :
Study on the Translocation of Wheat- E. elongatum by Fluorescence in Situ Hybridization
 Available online at www.sciencedirect.com Procedia Environmental Sciences 12 (2012 ) 1220 1224 2011 International Conference on Environmental Science and Engineering (ICESE 2011) Study on the Translocation
Available online at www.sciencedirect.com Procedia Environmental Sciences 12 (2012 ) 1220 1224 2011 International Conference on Environmental Science and Engineering (ICESE 2011) Study on the Translocation
A NEW SPECIES OF WHEAT THAT CONTINUES TO GROW AFTER HARVEST
 P E R E N N I A L C R O P S F O R F O O D S E C U R I T Y P R O C E E D I N G S O F T H E F A O E X P E R T W O R K S H O P A G R O - S Y S T E M S, E C O L O G Y A N D N U T R I T I O N 21 A NEW SPECIES
P E R E N N I A L C R O P S F O R F O O D S E C U R I T Y P R O C E E D I N G S O F T H E F A O E X P E R T W O R K S H O P A G R O - S Y S T E M S, E C O L O G Y A N D N U T R I T I O N 21 A NEW SPECIES
Wheat Chromosome Engineering and Breeding Jianli Chen
 Wheat Chromosome Engineering and Breeding Jianli Chen Chromosome Engineering A process to transfer favorable alleles through inter-specific hybridization and interchange of chromatin using aneupolids Aneuploids?
Wheat Chromosome Engineering and Breeding Jianli Chen Chromosome Engineering A process to transfer favorable alleles through inter-specific hybridization and interchange of chromatin using aneupolids Aneuploids?
Wide Hybridization in Plant Breeding
 Wide Hybridization in Plant Breeding Wide hybridization - a cross of two individuals belonging to different species (also called interspecific hybridization) - Such a cross can be (rarely) realized in
Wide Hybridization in Plant Breeding Wide hybridization - a cross of two individuals belonging to different species (also called interspecific hybridization) - Such a cross can be (rarely) realized in
Genomic differentiation of Hordeum chilense from H. vulgare as revealed by repetitive and EST sequences
 Genes Genet. Syst. (2005) 80, p. 147 159 Genomic differentiation of Hordeum chilense from H. vulgare as revealed by repetitive and EST sequences Adel Abdel-Aziz Hagras 1, Masahiro Kishii 1, Hiroyuki Tanaka
Genes Genet. Syst. (2005) 80, p. 147 159 Genomic differentiation of Hordeum chilense from H. vulgare as revealed by repetitive and EST sequences Adel Abdel-Aziz Hagras 1, Masahiro Kishii 1, Hiroyuki Tanaka
Molecular Characterization of a New Wheat-Thinopyrum intermedium Translocation Line with Resistance to Powdery Mildew and Stripe Rust
 Int. J. Mol. Sci. 2015, 16, 2162-2173; doi:10.3390/ijms16012162 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Molecular Characterization of a
Int. J. Mol. Sci. 2015, 16, 2162-2173; doi:10.3390/ijms16012162 Article OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Molecular Characterization of a
I 1967, 1972) we have shown that the introduction of a particular heterochromatic
 FORMATION OF MEGACHROMOSOMES FROM HETEROCHROMATIC BLOCKS OF NZCOTZANA TOMENTOSZFORMIS1 J. A. BURNS AND D. U. GERSTEL Department of Crop Science, North Carolina State University at Raleigh, North Carolina
FORMATION OF MEGACHROMOSOMES FROM HETEROCHROMATIC BLOCKS OF NZCOTZANA TOMENTOSZFORMIS1 J. A. BURNS AND D. U. GERSTEL Department of Crop Science, North Carolina State University at Raleigh, North Carolina
Molecular cytogenetic characterization of a new leaf rolling triticale
 Molecular cytogenetic characterization of a new leaf rolling triticale E.N. Yang 1,2, Z.J. Yang 3, J.F. Zhang 2, Y.C. Zou 2 and Z.L. Ren 1 1 State Key Laboratory for Plant Genetics and Breeding, Sichuan
Molecular cytogenetic characterization of a new leaf rolling triticale E.N. Yang 1,2, Z.J. Yang 3, J.F. Zhang 2, Y.C. Zou 2 and Z.L. Ren 1 1 State Key Laboratory for Plant Genetics and Breeding, Sichuan
Phenotypic response conferred by the Lr22a leaf rust resistance gene against ten Swiss P. triticina isolates.
 Supplementary Figure 1 Phenotypic response conferred by the leaf rust resistance gene against ten Swiss P. triticina isolates. The third leaf of Thatcher (left) and RL6044 (right) is shown ten days after
Supplementary Figure 1 Phenotypic response conferred by the leaf rust resistance gene against ten Swiss P. triticina isolates. The third leaf of Thatcher (left) and RL6044 (right) is shown ten days after
Liu et al. Genetics 2013 (in press)
 Liu et al. Genetics 2013 (in press) For over 40 years, the hybrid sunflower seed industry has largely relied on CMS PET-1and its corresponding Rf 1 gene Alternative CMS/Rf systems could expand the diversity
Liu et al. Genetics 2013 (in press) For over 40 years, the hybrid sunflower seed industry has largely relied on CMS PET-1and its corresponding Rf 1 gene Alternative CMS/Rf systems could expand the diversity
Genetics - Problem Drill 05: Genetic Mapping: Linkage and Recombination
 Genetics - Problem Drill 05: Genetic Mapping: Linkage and Recombination No. 1 of 10 1. A corn geneticist crossed a crinkly dwarf (cr) and male sterile (ms) plant; The F1 are male fertile with normal height.
Genetics - Problem Drill 05: Genetic Mapping: Linkage and Recombination No. 1 of 10 1. A corn geneticist crossed a crinkly dwarf (cr) and male sterile (ms) plant; The F1 are male fertile with normal height.
Recombination. The kinetochore ("spindle attachment ) always separates reductionally at anaphase I and equationally at anaphase II.
 Recombination Chromosome Separations At Anaphase I And II Mather (1935 pp. 53-62). Reductional vs. equational separations Reductional Division: Sister chromatids go to same pole at anaphase I Equational
Recombination Chromosome Separations At Anaphase I And II Mather (1935 pp. 53-62). Reductional vs. equational separations Reductional Division: Sister chromatids go to same pole at anaphase I Equational
Chapter 5 DNA and Chromosomes
 Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
Chapter 5 DNA and Chromosomes DNA as the genetic material Heat-killed bacteria can transform living cells S Smooth R Rough Fred Griffith, 1920 DNA is the genetic material Oswald Avery Colin MacLeod Maclyn
American Society of Cytopathology Core Curriculum in Molecular Biology
 American Society of Cytopathology Core Curriculum in Molecular Biology American Society of Cytopathology Core Curriculum in Molecular Biology Chapter 3 Molecular Techniques Separation and Detection, Part
American Society of Cytopathology Core Curriculum in Molecular Biology American Society of Cytopathology Core Curriculum in Molecular Biology Chapter 3 Molecular Techniques Separation and Detection, Part
C. Incorrect! Second Law: Law of Independent Assortment - Genes for different traits sort independently of one another in the formation of gametes.
 OAT Biology - Problem Drill 20: Chromosomes and Genetic Technology Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as needed, (3) Pick
OAT Biology - Problem Drill 20: Chromosomes and Genetic Technology Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as needed, (3) Pick
Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance
 Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance Niaz Ali 1, 2, JS (Pat) Heslop-Harrison 1, Habib Ahmad 2, Robert A Graybosch
Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance Niaz Ali 1, 2, JS (Pat) Heslop-Harrison 1, Habib Ahmad 2, Robert A Graybosch
Detection of alien genetic introgressions in bread wheat using dot-blot genomic hybridisation
 Mol Breeding (2017) 37: 32 DOI 10.1007/s11032-017-0629-5 Detection of alien genetic introgressions in bread wheat using dot-blot genomic hybridisation María -Dolores Rey & Pilar Prieto Received: 4 October
Mol Breeding (2017) 37: 32 DOI 10.1007/s11032-017-0629-5 Detection of alien genetic introgressions in bread wheat using dot-blot genomic hybridisation María -Dolores Rey & Pilar Prieto Received: 4 October
A strategy for detecting chromosome-specific rearrangements in rye
 A strategy for detecting chromosome-specific rearrangements in rye I E. Alvarez, C. Alonso-Blanco, R. Garcia Suarez, J.J. Ferreira, A. Roca, and R. Giraldez Abstract: To obtain translocations involving
A strategy for detecting chromosome-specific rearrangements in rye I E. Alvarez, C. Alonso-Blanco, R. Garcia Suarez, J.J. Ferreira, A. Roca, and R. Giraldez Abstract: To obtain translocations involving
Problem Set 2B Name and Lab Section:
 Problem Set 2B 9-26-06 Name and Lab Section: 1. Define each of the following rearrangements (mutations) (use one phrase or sentence for each). Then describe what kind of chromosomal structure you might
Problem Set 2B 9-26-06 Name and Lab Section: 1. Define each of the following rearrangements (mutations) (use one phrase or sentence for each). Then describe what kind of chromosomal structure you might
Structural variation. Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona
 Structural variation Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Genetic variation How much genetic variation is there between individuals? What type of variants
Structural variation Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Genetic variation How much genetic variation is there between individuals? What type of variants
Fluorescent in-situ Hybridization
 Fluorescent in-situ Hybridization Presented for: Presented by: Date: 2 Definition In situ hybridization is the method of localizing/ detecting specific nucleotide sequences in morphologically preserved
Fluorescent in-situ Hybridization Presented for: Presented by: Date: 2 Definition In situ hybridization is the method of localizing/ detecting specific nucleotide sequences in morphologically preserved
Molecular cytogenetics, cytology and genomic comparisons in the Triticeae
 Hereditas 116: 93-99 (1992) Molecular cytogenetics, cytology and genomic comparisons in the Triticeae J. S. HESLOP-HARRISON Karyobiology Group, Cell Biology Department, JI Centre for Plant Science Research,
Hereditas 116: 93-99 (1992) Molecular cytogenetics, cytology and genomic comparisons in the Triticeae J. S. HESLOP-HARRISON Karyobiology Group, Cell Biology Department, JI Centre for Plant Science Research,
Annamária SCHNEIDER and Márta MOLNÁR-LÁNG
 Detection of Various U and M Chromosomes in Wheat-Aegilops biuncialis Hybrids and Derivatives Using Fluorescence in situ Hybridisation and Molecular Markers Annamária SCHNEIDER and Márta MOLNÁR-LÁNG Agricultural
Detection of Various U and M Chromosomes in Wheat-Aegilops biuncialis Hybrids and Derivatives Using Fluorescence in situ Hybridisation and Molecular Markers Annamária SCHNEIDER and Márta MOLNÁR-LÁNG Agricultural
If today s lecture is perplexing, there is a useful film at
 1 Roadmap Chromosome evolution: Inversion Transposition Fission and fusion If today s lecture is perplexing, there is a useful film at http://www.youtube.com/watch?v=zcnymmhlkaw 2 One-minute responses
1 Roadmap Chromosome evolution: Inversion Transposition Fission and fusion If today s lecture is perplexing, there is a useful film at http://www.youtube.com/watch?v=zcnymmhlkaw 2 One-minute responses
DO NOT OPEN UNTIL TOLD TO START
 DO NOT OPEN UNTIL TOLD TO START BIO 312, Section 1, Spring 2011 February 21, 2011 Exam 1 Name (print neatly) Instructor 7 digit student ID INSTRUCTIONS: 1. There are 11 pages to the exam. Make sure you
DO NOT OPEN UNTIL TOLD TO START BIO 312, Section 1, Spring 2011 February 21, 2011 Exam 1 Name (print neatly) Instructor 7 digit student ID INSTRUCTIONS: 1. There are 11 pages to the exam. Make sure you
7.17: Writing Up Results and Creating Illustrations
 7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
7.17: Writing Up Results and Creating Illustrations A Results Exercise: Kansas and Pancakes Write a 5-sentence paragraph describing the results illustrated in this figure: - Describe the figure: highlights?
ZytoLight SPEC ABL2 Dual Color Break Apart Probe
 ZytoLight SPEC ABL2 Dual Color Break Apart Probe Z-2200-200 20 (0.2 ml) For the detection of translocations involving the ABL2 gene at 1q25.2 by fluorescence in situ hybridization (FISH).... In vitro diagnostic
ZytoLight SPEC ABL2 Dual Color Break Apart Probe Z-2200-200 20 (0.2 ml) For the detection of translocations involving the ABL2 gene at 1q25.2 by fluorescence in situ hybridization (FISH).... In vitro diagnostic
E/70C-64C/56B-51E/87B-91A/15F-cm. of X
 8 GENETICS: B. P. KA UFMA NN A COMPLEX INDUCED REARRANGEMENT OF DROSOPHILA CHROMOSOMES AND ITS BEARING ON THE PROBLEM OF CHROMOSOME RECOMBINA TION DEPARTMENT OF GENETICS, By.B. P. KAUFMANN CARNEGIE INSTITUTION
8 GENETICS: B. P. KA UFMA NN A COMPLEX INDUCED REARRANGEMENT OF DROSOPHILA CHROMOSOMES AND ITS BEARING ON THE PROBLEM OF CHROMOSOME RECOMBINA TION DEPARTMENT OF GENETICS, By.B. P. KAUFMANN CARNEGIE INSTITUTION
BENG 183 Trey Ideker. Genome Assembly and Physical Mapping
 BENG 183 Trey Ideker Genome Assembly and Physical Mapping Reasons for sequencing Complete genome sequencing!!! Resequencing (Confirmatory) E.g., short regions containing single nucleotide polymorphisms
BENG 183 Trey Ideker Genome Assembly and Physical Mapping Reasons for sequencing Complete genome sequencing!!! Resequencing (Confirmatory) E.g., short regions containing single nucleotide polymorphisms
ZytoLight SPEC FGFR3 Dual Color Break Apart Probe
 ZytoLight SPEC FGFR3 Dual Color Break Apart Probe Z-2170-200 20 (0.2 ml) For the detection of translocations involving the FGFR3 gene at 4p16.3 by fluorescence in situ hybridization (FISH).... In vitro
ZytoLight SPEC FGFR3 Dual Color Break Apart Probe Z-2170-200 20 (0.2 ml) For the detection of translocations involving the FGFR3 gene at 4p16.3 by fluorescence in situ hybridization (FISH).... In vitro
Regents Biology REVIEW 5: GENETICS
 Period Date REVIEW 5: GENETICS 1. Chromosomes: a. Humans have chromosomes, or homologous pairs. Homologous: b. Chromosome pairs carry genes for the same traits. Most organisms have two copies of the gene
Period Date REVIEW 5: GENETICS 1. Chromosomes: a. Humans have chromosomes, or homologous pairs. Homologous: b. Chromosome pairs carry genes for the same traits. Most organisms have two copies of the gene
Fluorescence In Situ Hybridization Based Karyotyping of Soybean Translocation Lines
 INVESTIGATION Fluorescence In Situ Hybridization Based Karyotyping of Soybean Translocation Lines Seth D. Findley,* Allison L. Pappas, Yaya Cui,* James A. Birchler, Reid G. Palmer, and Gary Stacey*,1 *National
INVESTIGATION Fluorescence In Situ Hybridization Based Karyotyping of Soybean Translocation Lines Seth D. Findley,* Allison L. Pappas, Yaya Cui,* James A. Birchler, Reid G. Palmer, and Gary Stacey*,1 *National
Analysis of the expression of homoeologous genes in polyploid cereals using fluorescence cdna-sscp
 Analysis of the expression of homoeologous genes in polyploid cereals using fluorescence cdna-sscp De Bustos A., Perez R., Jouve N. in Molina-Cano J.L. (ed.), Christou P. (ed.), Graner A. (ed.), Hammer
Analysis of the expression of homoeologous genes in polyploid cereals using fluorescence cdna-sscp De Bustos A., Perez R., Jouve N. in Molina-Cano J.L. (ed.), Christou P. (ed.), Graner A. (ed.), Hammer
Population Cytology of the Genus Phaulacridium V. * Ph. marginale - The Omarama Population
 Aust. J. BioI. Sci., 1977, 30, 319-28 Population Cytology of the Genus Phaulacridium V. * Ph. marginale - The Omarama Population M. Westerman Department of Genetics and Human Variation, La Trobe University,
Aust. J. BioI. Sci., 1977, 30, 319-28 Population Cytology of the Genus Phaulacridium V. * Ph. marginale - The Omarama Population M. Westerman Department of Genetics and Human Variation, La Trobe University,
Comparison of bread wheat lines selected by doubled haploid, single-seed descent and pedigree selection methods
 Theor Appl Genet (1998) 97: 550 556 Springer-Verlag 1998 M. N. Inagaki G. Varughese S. Rajaram M. van Ginkel A. Mujeeb-Kazi Comparison of bread wheat lines selected by doubled haploid, single-seed descent
Theor Appl Genet (1998) 97: 550 556 Springer-Verlag 1998 M. N. Inagaki G. Varughese S. Rajaram M. van Ginkel A. Mujeeb-Kazi Comparison of bread wheat lines selected by doubled haploid, single-seed descent
NBS-LRR sequence family is associated with leaf and stripe rust resistance on the end of homoeologous chromosome group 1S of wheat
 Theor Appl Genet (2000) 101:1139 1144 Springer-Verlag 2000 ORIGINAL ARTICLE W. Spielmeyer L. Huang H. Bariana A. Laroche B.S. Gill E.S. Lagudah NBS-LRR sequence family is associated with leaf and stripe
Theor Appl Genet (2000) 101:1139 1144 Springer-Verlag 2000 ORIGINAL ARTICLE W. Spielmeyer L. Huang H. Bariana A. Laroche B.S. Gill E.S. Lagudah NBS-LRR sequence family is associated with leaf and stripe
Inheritance Biology. Unit Map. Unit
 Unit 8 Unit Map 8.A Mendelian principles 482 8.B Concept of gene 483 8.C Extension of Mendelian principles 485 8.D Gene mapping methods 495 8.E Extra chromosomal inheritance 501 8.F Microbial genetics
Unit 8 Unit Map 8.A Mendelian principles 482 8.B Concept of gene 483 8.C Extension of Mendelian principles 485 8.D Gene mapping methods 495 8.E Extra chromosomal inheritance 501 8.F Microbial genetics
Development and application of CGHPRO, a novel software package for retrieving, handling and analysing array CGH data
 Aus dem Max Planck Institut für molekulare Genetik DISSERTATION Development and application of CGHPRO, a novel software package for retrieving, handling and analysing array CGH data Zur Erlangung des akademischen
Aus dem Max Planck Institut für molekulare Genetik DISSERTATION Development and application of CGHPRO, a novel software package for retrieving, handling and analysing array CGH data Zur Erlangung des akademischen
Overview of Human Genetics
 Overview of Human Genetics 1 Structure and function of nucleic acids. 2 Structure and composition of the human genome. 3 Mendelian genetics. Lander et al. (Nature, 2001) MAT 394 (ASU) Human Genetics Spring
Overview of Human Genetics 1 Structure and function of nucleic acids. 2 Structure and composition of the human genome. 3 Mendelian genetics. Lander et al. (Nature, 2001) MAT 394 (ASU) Human Genetics Spring
Chapter 4 Gene Linkage and Genetic Mapping
 Chapter 4 Gene Linkage and Genetic Mapping 1 Important Definitions Locus = physical location of a gene on a chromosome Homologous pairs of chromosomes often contain alternative forms of a given gene =
Chapter 4 Gene Linkage and Genetic Mapping 1 Important Definitions Locus = physical location of a gene on a chromosome Homologous pairs of chromosomes often contain alternative forms of a given gene =
Inheritance (IGCSE Biology Syllabus )
 Inheritance (IGCSE Biology Syllabus 2016-2018) Key definitions Chromosome Allele Gene Haploid nucleus Diploid nucleus Genotype Phenotype Homozygous Heterozygous Dominant Recessive A thread of DNA, made
Inheritance (IGCSE Biology Syllabus 2016-2018) Key definitions Chromosome Allele Gene Haploid nucleus Diploid nucleus Genotype Phenotype Homozygous Heterozygous Dominant Recessive A thread of DNA, made
How to use genetic methods for detecting linkage. David D. Perkins. Background
 How to use genetic methods for detecting linkage. David D. Perkins Background It took 75 years using classical genetic methods to assign the first 1000 loci in N. crassa to the seven linkage groups (Perkins
How to use genetic methods for detecting linkage. David D. Perkins Background It took 75 years using classical genetic methods to assign the first 1000 loci in N. crassa to the seven linkage groups (Perkins
Chromosomes. Ms. Gunjan M. Chaudhari
 Chromosomes Ms. Gunjan M. Chaudhari Chromsomes Chromosome structure Chromatin structure Chromosome variations The new cytogenetics Prokaryotic chromosomes Circular double helix Complexed with protein in
Chromosomes Ms. Gunjan M. Chaudhari Chromsomes Chromosome structure Chromatin structure Chromosome variations The new cytogenetics Prokaryotic chromosomes Circular double helix Complexed with protein in
Dr. Bikram S. Gill University Distinguished Professor of Plant Pathology Wheat Genetics Resource Center Plant Pathology Department
 Address: Department of Plant Pathology Kansas State University Manhattan, KS 66506-5502 Telephone: 913-532-6176 Fax: 913-532-5692 Dr. Bikram S. Gill University Distinguished Professor of Plant Pathology
Address: Department of Plant Pathology Kansas State University Manhattan, KS 66506-5502 Telephone: 913-532-6176 Fax: 913-532-5692 Dr. Bikram S. Gill University Distinguished Professor of Plant Pathology
CHROMOSOMA. Identification by Giemsa Technique of the Translocations Separating Cultivated Rye from Three Wild Species of Secale
 Chromosoma (Berl.) 59, 217-225 (1977) CHROMOSOMA 9 by Springer-Verlag 1977 Identification by Giemsa Technique of the Translocations Separating Cultivated Rye from Three Wild Species of Secale R.J. Singh
Chromosoma (Berl.) 59, 217-225 (1977) CHROMOSOMA 9 by Springer-Verlag 1977 Identification by Giemsa Technique of the Translocations Separating Cultivated Rye from Three Wild Species of Secale R.J. Singh
Expressed sequence tag-pcr markers for identification of alien barley chromosome 2H in wheat
 Expressed sequence tag-pcr markers for identification of alien barley chromosome 2H in wheat M.J. Wang 1 *, H.D. Zou 1 *, Z.S. Lin 2, Y. Wu 1, X. Chen 2 and Y.P. Yuan 1 1 College of Plant Science, Jilin
Expressed sequence tag-pcr markers for identification of alien barley chromosome 2H in wheat M.J. Wang 1 *, H.D. Zou 1 *, Z.S. Lin 2, Y. Wu 1, X. Chen 2 and Y.P. Yuan 1 1 College of Plant Science, Jilin
What is his karyotype?
 Methods of chromosome analysis What is his karyotype? He is 4 years old (looks like 2 y.o.) Knows few words Inadequate reaction Abnormal face Short hands Heart abnormalities, kidney abnormalities Plurimapformative
Methods of chromosome analysis What is his karyotype? He is 4 years old (looks like 2 y.o.) Knows few words Inadequate reaction Abnormal face Short hands Heart abnormalities, kidney abnormalities Plurimapformative
Chapter 5: Overview. Overview. Introduction. Genetic linkage and. Genes located on the same chromosome. linkage. recombinant progeny with genotypes
 Chapter 5: Genetic linkage and chromosome mapping. Overview Introduction Linkage and recombination of genes in a chromosome Principles of genetic mapping Building linkage maps Chromosome and chromatid
Chapter 5: Genetic linkage and chromosome mapping. Overview Introduction Linkage and recombination of genes in a chromosome Principles of genetic mapping Building linkage maps Chromosome and chromatid
3I03 - Eukaryotic Genetics Repetitive DNA
 Repetitive DNA Satellite DNA Minisatellite DNA Microsatellite DNA Transposable elements LINES, SINES and other retrosequences High copy number genes (e.g. ribosomal genes, histone genes) Multifamily member
Repetitive DNA Satellite DNA Minisatellite DNA Microsatellite DNA Transposable elements LINES, SINES and other retrosequences High copy number genes (e.g. ribosomal genes, histone genes) Multifamily member
PDGFRB FISH PRODUCT DATASHEET. Proprietary Name: PDGFRB FISH for Gleevec Eligibility in Myelodysplastic Syndrome/Myeloproliferative Disease (MDS/MPD)
 PDGFRB FISH PRODUCT DATASHEET Proprietary Name: PDGFRB FISH for Gleevec Eligibility in Myelodysplastic Syndrome/Myeloproliferative Disease (MDS/MPD) Established Name: PDGFRB FISH for Gleevec in MDS/MPD
PDGFRB FISH PRODUCT DATASHEET Proprietary Name: PDGFRB FISH for Gleevec Eligibility in Myelodysplastic Syndrome/Myeloproliferative Disease (MDS/MPD) Established Name: PDGFRB FISH for Gleevec in MDS/MPD
Name Class Date. Practice Test
 Name Class Date 12 DNA Practice Test Multiple Choice Write the letter that best answers the question or completes the statement on the line provided. 1. What do bacteriophages infect? a. mice. c. viruses.
Name Class Date 12 DNA Practice Test Multiple Choice Write the letter that best answers the question or completes the statement on the line provided. 1. What do bacteriophages infect? a. mice. c. viruses.
ZytoLight Dual Color Dual Fusion Probe
 ZytoLight SPEC ETV6/RUNX1 Dual Color Dual Fusion Probe Z-2157-200 20 (0.2 ml) For the detection of the translocation t(12;21)(p13;q22) by fluorescence in situ hybridization (FISH).... In vitro diagnostic
ZytoLight SPEC ETV6/RUNX1 Dual Color Dual Fusion Probe Z-2157-200 20 (0.2 ml) For the detection of the translocation t(12;21)(p13;q22) by fluorescence in situ hybridization (FISH).... In vitro diagnostic
Linkage & Genetic Mapping in Eukaryotes. Ch. 6
 Linkage & Genetic Mapping in Eukaryotes Ch. 6 1 LINKAGE AND CROSSING OVER! In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even
Linkage & Genetic Mapping in Eukaryotes Ch. 6 1 LINKAGE AND CROSSING OVER! In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even
Gene Linkage and Genetic. Mapping. Key Concepts. Key Terms. Concepts in Action
 Gene Linkage and Genetic 4 Mapping Key Concepts Genes that are located in the same chromosome and that do not show independent assortment are said to be linked. The alleles of linked genes present together
Gene Linkage and Genetic 4 Mapping Key Concepts Genes that are located in the same chromosome and that do not show independent assortment are said to be linked. The alleles of linked genes present together
Further Reading - DNA
 Further Reading - DNA DNA BACKGROUND What is DNA? DNA (short for deoxyribonucleic acid ) is a complex molecule found in the cells of all living things. The blueprint for life, DNA contains all the information
Further Reading - DNA DNA BACKGROUND What is DNA? DNA (short for deoxyribonucleic acid ) is a complex molecule found in the cells of all living things. The blueprint for life, DNA contains all the information
Chapter 6 Linkage and Chromosome Mapping in Eukaryotes
 Chapter 6 Linkage and Chromosome Mapping in Eukaryotes Early Observations By 1903 Sutton pointed out likelihood that there were many more unit factors than chromosomes in most species Shortly, observations
Chapter 6 Linkage and Chromosome Mapping in Eukaryotes Early Observations By 1903 Sutton pointed out likelihood that there were many more unit factors than chromosomes in most species Shortly, observations
1. A brief overview of sequencing biochemistry
 Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Supplementary reading materials on Genome sequencing (optional) The materials are from Mark Blaxter s lecture notes on Sequencing strategies and Primary Analysis 1. A brief overview of sequencing biochemistry
Sept 2. Structure and Organization of Genomes. Today: Genetic and Physical Mapping. Sept 9. Forward and Reverse Genetics. Genetic and Physical Mapping
 Sept 2. Structure and Organization of Genomes Today: Genetic and Physical Mapping Assignments: Gibson & Muse, pp.4-10 Brown, pp. 126-160 Olson et al., Science 245: 1434 New homework:due, before class,
Sept 2. Structure and Organization of Genomes Today: Genetic and Physical Mapping Assignments: Gibson & Muse, pp.4-10 Brown, pp. 126-160 Olson et al., Science 245: 1434 New homework:due, before class,
Biology 201 (Genetics) Exam #3 120 points 20 November Read the question carefully before answering. Think before you write.
 Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Name KEY Section Biology 201 (Genetics) Exam #3 120 points 20 November 2006 Read the question carefully before answering. Think before you write. You will have up to 50 minutes to take this exam. After
Q1. Figure 1 shows a pair of chromosomes at the start of meiosis. The letters represent alleles. Figure (1)
 Q1. Figure 1 shows a pair of chromosomes at the start of meiosis. The letters represent alleles. Figure 1 (a) What is an allele? (b) Explain the appearance of one of the chromosomes in Figure 1. (c) The
Q1. Figure 1 shows a pair of chromosomes at the start of meiosis. The letters represent alleles. Figure 1 (a) What is an allele? (b) Explain the appearance of one of the chromosomes in Figure 1. (c) The
REVIEWS. Structural variation in the human genome
 REVIEWS Structural variation in the human genome Lars Feuk, Andrew R. Carson and Stephen W. Scherer Abstract The first wave of information from the analysis of the human genome revealed SNPs to be the
REVIEWS Structural variation in the human genome Lars Feuk, Andrew R. Carson and Stephen W. Scherer Abstract The first wave of information from the analysis of the human genome revealed SNPs to be the
MICROSATELLITE MARKER AND ITS UTILITY
 Your full article ( between 500 to 5000 words) - - Do check for grammatical errors or spelling mistakes MICROSATELLITE MARKER AND ITS UTILITY 1 Prasenjit, D., 2 Anirudha, S. K. and 3 Mallar, N.K. 1,2 M.Sc.(Agri.),
Your full article ( between 500 to 5000 words) - - Do check for grammatical errors or spelling mistakes MICROSATELLITE MARKER AND ITS UTILITY 1 Prasenjit, D., 2 Anirudha, S. K. and 3 Mallar, N.K. 1,2 M.Sc.(Agri.),
CHROMOSOME EXCHANGES IN HUMAN LEUKOCYTES INDUCED BY MITOMYCIN C
 CHROMOSOME EXCHANGES IN HUMAN LEUKOCYTES INDUCED BY MITOMYCIN C MARGERY W. SHAW AND MAIMON M. COHEN Department of Human Genztics, Universily of Michigan Medical School, Ann Arbor Received July 3. 964 MITOMYCIN
CHROMOSOME EXCHANGES IN HUMAN LEUKOCYTES INDUCED BY MITOMYCIN C MARGERY W. SHAW AND MAIMON M. COHEN Department of Human Genztics, Universily of Michigan Medical School, Ann Arbor Received July 3. 964 MITOMYCIN
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
GENE ACTION STUDIES OF DIFFERENT QUANTITATIVE TRAITS IN MAIZE
 Pak. J. Bot., 42(2): 1021-1030, 2010. GENE ACTION STUDIES OF DIFFERENT QUANTITATIVE TRAITS IN MAIZE MUHAMMAD IRSHAD-UL-HAQ 1, SAIF ULLAH AJMAL 2*, MUHAMMAD MUNIR 2 AND MUHAMMAD GULFARAZ 3 1 Millets Research
Pak. J. Bot., 42(2): 1021-1030, 2010. GENE ACTION STUDIES OF DIFFERENT QUANTITATIVE TRAITS IN MAIZE MUHAMMAD IRSHAD-UL-HAQ 1, SAIF ULLAH AJMAL 2*, MUHAMMAD MUNIR 2 AND MUHAMMAD GULFARAZ 3 1 Millets Research
Effect of 5-azacytidine and trichostatin A on somatic centromere association in wheat
 399 Effect of 5-azacytidine and trichostatin A on somatic centromere association in wheat Maria Vorontsova, Peter Shaw, Steve Reader, and Graham Moore Abstract: Both homologous and non-homologous chromosomes
399 Effect of 5-azacytidine and trichostatin A on somatic centromere association in wheat Maria Vorontsova, Peter Shaw, Steve Reader, and Graham Moore Abstract: Both homologous and non-homologous chromosomes
IQFISH on Dako Omnis. Panel for Lung Cancer. Dako FAST RESULTS. ALK, ROS1, RET and MET IQFISH. Dako Omnis. Agilent Pathology Solutions
 PRODUCT INFORMATION Dako Omnis ALK, ROS1, RET and MET IQFISH Dako Agilent Pathology Solutions IQFISH on Dako Omnis Panel for Lung Cancer FAST RESULTS Fast, high-quality FISH Integrated into your IHC workflow
PRODUCT INFORMATION Dako Omnis ALK, ROS1, RET and MET IQFISH Dako Agilent Pathology Solutions IQFISH on Dako Omnis Panel for Lung Cancer FAST RESULTS Fast, high-quality FISH Integrated into your IHC workflow
Genetics. Genetics- is the study of all manifestation of inheritance from the distributions of traits to the molecules of the gene itself
 What is Genetics? Genetics Mapping of genes Basis of life Inheritable traits Abnormalities Disease Development DNA RNA Proteins Central dogma - Watson & Crick Genes- segments of DNA that code for proteins
What is Genetics? Genetics Mapping of genes Basis of life Inheritable traits Abnormalities Disease Development DNA RNA Proteins Central dogma - Watson & Crick Genes- segments of DNA that code for proteins
DNA and DNA Replication
 Name Period PreAP Biology QCA 2 Review Your EOS exam is approximately 70 MC questions. This review, coupled with your QCA 1 review you received in October should lead you back through the important concepts
Name Period PreAP Biology QCA 2 Review Your EOS exam is approximately 70 MC questions. This review, coupled with your QCA 1 review you received in October should lead you back through the important concepts
Genomic in Situ hybridization in intergeneric hybrids between Raphanus sativus and Brassica oleracea
 296 GENETICS AND BREEDING: Genetics and Germplasm Genomic in Situ hybridization in intergeneric hybrids between Raphanus sativus and Brassica oleracea CHENG Yugui 1, 2, WU Jiangsheng 1, ZHANG Minghai 2,
296 GENETICS AND BREEDING: Genetics and Germplasm Genomic in Situ hybridization in intergeneric hybrids between Raphanus sativus and Brassica oleracea CHENG Yugui 1, 2, WU Jiangsheng 1, ZHANG Minghai 2,
Concepts: What are RFLPs and how do they act like genetic marker loci?
 Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
Restriction Fragment Length Polymorphisms (RFLPs) -1 Readings: Griffiths et al: 7th Edition: Ch. 12 pp. 384-386; Ch.13 pp404-407 8th Edition: pp. 364-366 Assigned Problems: 8th Ch. 11: 32, 34, 38-39 7th
CHAPTER 20 DNA TECHNOLOGY AND GENOMICS. Section A: DNA Cloning
 Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Answers to additional linkage problems.
 Spring 2013 Biology 321 Answers to Assignment Set 8 Chapter 4 http://fire.biol.wwu.edu/trent/trent/iga_10e_sm_chapter_04.pdf Answers to additional linkage problems. Problem -1 In this cell, there two copies
Spring 2013 Biology 321 Answers to Assignment Set 8 Chapter 4 http://fire.biol.wwu.edu/trent/trent/iga_10e_sm_chapter_04.pdf Answers to additional linkage problems. Problem -1 In this cell, there two copies
3. human genomics clone genes associated with genetic disorders. 4. many projects generate ordered clones that cover genome
 Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Fusion Gene Analysis. ncounter Elements. Molecules That Count WHITE PAPER. v1.0 OCTOBER 2014 NanoString Technologies, Inc.
 Fusion Gene Analysis NanoString Technologies, Inc. Fusion Gene Analysis ncounter Elements v1.0 OCTOBER 2014 NanoString Technologies, Inc., Seattle WA 98109 FOR RESEARCH USE ONLY. Not for use in diagnostic
Fusion Gene Analysis NanoString Technologies, Inc. Fusion Gene Analysis ncounter Elements v1.0 OCTOBER 2014 NanoString Technologies, Inc., Seattle WA 98109 FOR RESEARCH USE ONLY. Not for use in diagnostic
Los Alamos NATIONAL LABORATORY. h situ Hybridization
 Title: Applications of Strand-Specific h situ Hybridization Author(s): E. H. Goodwin, LS-4 J. Meyne, LS-3 S. M. Bailey, LS-4 D. Quigley, University of Oregon L. Smith, University of New Mexico R. Tennyson,
Title: Applications of Strand-Specific h situ Hybridization Author(s): E. H. Goodwin, LS-4 J. Meyne, LS-3 S. M. Bailey, LS-4 D. Quigley, University of Oregon L. Smith, University of New Mexico R. Tennyson,
Pilar Rubio, Lourdes Daza, Nicolás Jouve. To cite this version: HAL Id: hal https://hal.archives-ouvertes.fr/hal
 Meiotic behaviour, chromosome stability and genetic analysis of the preferential transmission of 1B-1R, 1A-1R and 1R(1D) chromosomes in intervarietal hybrids of wheat Pilar Rubio, Lourdes Daza, Nicolás
Meiotic behaviour, chromosome stability and genetic analysis of the preferential transmission of 1B-1R, 1A-1R and 1R(1D) chromosomes in intervarietal hybrids of wheat Pilar Rubio, Lourdes Daza, Nicolás
Candidate-gene cloning and targeted marker enrichment of wheat chromosomal regions using RNA fingerprinting differential display
 Candidate-gene cloning and targeted marker enrichment of wheat chromosomal regions using RNA fingerprinting differential display Kulvinder S. Gill and Devinder Sandhu 633 Abstract: The usefulness of the
Candidate-gene cloning and targeted marker enrichment of wheat chromosomal regions using RNA fingerprinting differential display Kulvinder S. Gill and Devinder Sandhu 633 Abstract: The usefulness of the
melanogaster has acquired He-T DNA sequences at
 Proc. Nati. Acad. Sci. USA Vol. 85, pp. 8116-8120, November 1988 Genetics A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres (heterochromatin/polytene
Proc. Nati. Acad. Sci. USA Vol. 85, pp. 8116-8120, November 1988 Genetics A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres (heterochromatin/polytene
Identification and Chromosomal Location of Tandemly Repeated DNA Sequences in Allium cepa
 Genes Genet. Syst. (2001) 76, p. 53 60 Identification and Chromosomal Location of Tandemly Repeated DNA Sequences in Allium cepa Geum Sook Do 1, Bong Bo Seo 1 *, Maki Yamamoto 2, Go Suzuki 3 and Yasuhiko
Genes Genet. Syst. (2001) 76, p. 53 60 Identification and Chromosomal Location of Tandemly Repeated DNA Sequences in Allium cepa Geum Sook Do 1, Bong Bo Seo 1 *, Maki Yamamoto 2, Go Suzuki 3 and Yasuhiko
grassgenomics: davidkopecký
 grassgenomics: howto portiona mammoth? davidkopecký laboratory of molecular cytogenetics and cytometry centre of the region haná for biotechnological and agricultural research olomouc, czech republic Olomouc
grassgenomics: howto portiona mammoth? davidkopecký laboratory of molecular cytogenetics and cytometry centre of the region haná for biotechnological and agricultural research olomouc, czech republic Olomouc
Minor comments 1. Figture 2b The ordinate scale is not appropriate. 2. Please supplement the pollen sterility picture of different ms1 mutant.
 Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper identified a nuclear encoded recessive male sterile gene, Tams1, encoding a glycosylphosphatidylinositol anchored lipid transfer protein
Reviewers' comments: Reviewer #1 (Remarks to the Author): This paper identified a nuclear encoded recessive male sterile gene, Tams1, encoding a glycosylphosphatidylinositol anchored lipid transfer protein
7 Gene Isolation and Analysis of Multiple
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Chapter 17. PCR the polymerase chain reaction and its many uses. Prepared by Woojoo Choi
 Chapter 17. PCR the polymerase chain reaction and its many uses Prepared by Woojoo Choi Polymerase chain reaction 1) Polymerase chain reaction (PCR): artificial amplification of a DNA sequence by repeated
Chapter 17. PCR the polymerase chain reaction and its many uses Prepared by Woojoo Choi Polymerase chain reaction 1) Polymerase chain reaction (PCR): artificial amplification of a DNA sequence by repeated
Characterizing Phenotypes of Bacteria by Staining Method
 Experiment 3 Laboratory to Biology III Diversity of Microorganisms / Wintersemester / page 1 Experiment Characterizing Phenotypes of Bacteria by Staining Method Advisor Reading NN Chapters 3.1, 3.7, 3.8,
Experiment 3 Laboratory to Biology III Diversity of Microorganisms / Wintersemester / page 1 Experiment Characterizing Phenotypes of Bacteria by Staining Method Advisor Reading NN Chapters 3.1, 3.7, 3.8,
Cell Nucleus. Chen Li. Department of Cellular and Genetic Medicine
 Cell Nucleus Chen Li Department of Cellular and Genetic Medicine 13 223 chenli2008@fudan.edu.cn Outline A. Historical background B. Structure of the nucleus: nuclear pore complex (NPC), lamina, nucleolus,
Cell Nucleus Chen Li Department of Cellular and Genetic Medicine 13 223 chenli2008@fudan.edu.cn Outline A. Historical background B. Structure of the nucleus: nuclear pore complex (NPC), lamina, nucleolus,
ESTIMATING GENETIC VARIABILITY WITH RESTRICTION ENDONUCLEASES RICHARD R. HUDSON1
 ESTIMATING GENETIC VARIABILITY WITH RESTRICTION ENDONUCLEASES RICHARD R. HUDSON1 Department of Biology, University of Pennsylvania, Philadelphia, Pennsylvania 19104 Manuscript received September 8, 1981
ESTIMATING GENETIC VARIABILITY WITH RESTRICTION ENDONUCLEASES RICHARD R. HUDSON1 Department of Biology, University of Pennsylvania, Philadelphia, Pennsylvania 19104 Manuscript received September 8, 1981
The process of new DNA to another organism. The goal is to add one or more that are not already found in that organism.
 Genetic Engineering Notes The process of new DNA to another organism. The goal is to add one or more that are not already found in that organism. Selective Breeding Carefully choosing which plants and
Genetic Engineering Notes The process of new DNA to another organism. The goal is to add one or more that are not already found in that organism. Selective Breeding Carefully choosing which plants and
Lesson 3 Gel Electrophoresis of Amplified PCR Samples and Staining of Agarose Gels
 Lesson 3 Gel Electrophoresis of Amplified PCR Samples and Staining of Agarose Gels What Are You Looking At? Before you analyze your PCR products, let s take a look at the target sequence being explored.
Lesson 3 Gel Electrophoresis of Amplified PCR Samples and Staining of Agarose Gels What Are You Looking At? Before you analyze your PCR products, let s take a look at the target sequence being explored.
Segregation Ratios. Single genes in diploids: backcross will yield a 1:1; intercross 3:1 if dominant, 1:2:1 if codominant
 Segregation Ratios Reference: Mather (1957) Single genes and no linkage. SEGREGATION IN DIPLOIDS Analysis of single gene segregations determine what type of genetic control exists for a single trait i.e.,
Segregation Ratios Reference: Mather (1957) Single genes and no linkage. SEGREGATION IN DIPLOIDS Analysis of single gene segregations determine what type of genetic control exists for a single trait i.e.,
CHROMOSOMAL HOMOLOGY AND DIVERGENCE BETWEEN SIBLING SPECIES OF DEER MICE: PEROMYSCUS MANICULA TUS AND
 Evolution, 32(2), 1978, pp. 334-341 CHROMOSOMAL HOMOLOGY AND DIVERGENCE BETWEEN SIBLING SPECIES OF DEER MICE: PEROMYSCUS MANICULA TUS AND P. MELANOTIS (RODENTIA, CRICETIDAE) IRAF. GREENBAUD?,ROBERTJ. BAKER'AND
Evolution, 32(2), 1978, pp. 334-341 CHROMOSOMAL HOMOLOGY AND DIVERGENCE BETWEEN SIBLING SPECIES OF DEER MICE: PEROMYSCUS MANICULA TUS AND P. MELANOTIS (RODENTIA, CRICETIDAE) IRAF. GREENBAUD?,ROBERTJ. BAKER'AND
Existing potato markers and marker conversions. Walter De Jong PAA Workshop August 2009
 Existing potato markers and marker conversions Walter De Jong PAA Workshop August 2009 1 What makes for a good marker? diagnostic for trait of interest robust works even with DNA of poor quality or low
Existing potato markers and marker conversions Walter De Jong PAA Workshop August 2009 1 What makes for a good marker? diagnostic for trait of interest robust works even with DNA of poor quality or low
Mutations, Genetic Testing and Engineering
 Mutations, Genetic Testing and Engineering Objectives Describe how techniques such as DNA fingerprinting, genetic modifications, and chromosomal analysis are used to study the genomes of organisms (TEKS
Mutations, Genetic Testing and Engineering Objectives Describe how techniques such as DNA fingerprinting, genetic modifications, and chromosomal analysis are used to study the genomes of organisms (TEKS
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
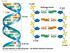 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Recombinant DNA Technology. The Role of Recombinant DNA Technology in Biotechnology. yeast. Biotechnology. Recombinant DNA technology.
 PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University C H A P T E R 8 Recombinant DNA Technology The Role of Recombinant DNA Technology in Biotechnology Biotechnology?
2. Outline the levels of DNA packing in the eukaryotic nucleus below next to the diagram provided.
 AP Biology Reading Packet 6- Molecular Genetics Part 2 Name Chapter 19: Eukaryotic Genomes 1. Define the following terms: a. Euchromatin b. Heterochromatin c. Nucleosome 2. Outline the levels of DNA packing
AP Biology Reading Packet 6- Molecular Genetics Part 2 Name Chapter 19: Eukaryotic Genomes 1. Define the following terms: a. Euchromatin b. Heterochromatin c. Nucleosome 2. Outline the levels of DNA packing
From DNA to Protein: Genotype to Phenotype
 12 From DNA to Protein: Genotype to Phenotype 12.1 What Is the Evidence that Genes Code for Proteins? The gene-enzyme relationship is one-gene, one-polypeptide relationship. Example: In hemoglobin, each
12 From DNA to Protein: Genotype to Phenotype 12.1 What Is the Evidence that Genes Code for Proteins? The gene-enzyme relationship is one-gene, one-polypeptide relationship. Example: In hemoglobin, each
Dros. Inf. Serv. 99 (2016) Teaching Notes 97. CRISPR/Cas9 induced mutations of the white gene of haplo-x and diplo-x Drosophila melanogaster.
 Dros. Inf. Serv. 99 () Teaching Notes 9 CRISPR/Cas9 induced mutations of the white gene of haplo-x and diplo-x Drosophila melanogaster. 33. Everett, Ashley M., Kelsey Allyse Buettner, Marissa Rachelle
Dros. Inf. Serv. 99 () Teaching Notes 9 CRISPR/Cas9 induced mutations of the white gene of haplo-x and diplo-x Drosophila melanogaster. 33. Everett, Ashley M., Kelsey Allyse Buettner, Marissa Rachelle
