Regular Paper. Introduction
|
|
|
- Randolph Arnold
- 6 years ago
- Views:
Transcription
1 Evidence that Proliferation of Golgi Apparatus Depends on Both De Novo Generation from the Endoplasmic Reticulum and Formation from Pre-Existing Stacks During the Growth of Tobacco BY-2 Cells Moses Olabiyi Abiodun 1 and Ken Matsuoka 1,2,3, * 1 Laboratory of Plant Nutrition, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University, Hakozaki, Higashi-ku, Fukuoka, Japan 2 Laboratory of Plant Nutrition, Faculty of Agriculture, Kyushu University, Hakozaki, Higashi-ku, Fukuoka, Japan 3 Biotron Application Centre, Kyushu University, Hakozaki, Higashi-ku, Fukuoka, Japan *Corresponding author: , kenmat@agr.kyushu-u.ac.jp; Fax, (Received December 18, 2012; Accepted January 16, 2013) Regular Paper In higher plants, the numbers of cytoplasmic-distributed Golgi stacks differ based on function, age and cell type. It has not been clarified how the numbers are controlled, whether all the Golgi apparatus in a cell function equally and whether the increase in Golgi number is a result of the de novo formation from the endoplasmic reticulum (ER) or fission of pre-existing stacks. A tobacco prolyl 4-hydroxylase (NtP4H1.1), which is a cis-golgi-localizing type II membrane protein, was tagged with a photoconvertible fluorescent protein, mkikgr (monomeric Kikume green red), and expressed in tobacco bright yellow 2 (BY-2) cells. Transformed cells were exposed to purple light to convert the fluorescence from green to red. A time-course analysis after the conversion revealed a progressive increase in green puncta and a decrease in the red puncta. From 3 to 6 h, we observed red, yellow and green fluorescent puncta corresponding to pre-existing Golgi; Golgi containing both pre-existing and newly synthesized protein; and newly synthesized Golgi. Analysis of the number and fluorescence of Golgi at different phases of the cell cycle suggested that an increase in Golgi number with both division and de novo synthesis occurred concomitantly with DNA replication. Investigation with different inhibitors suggested that the formation of new Golgi and the generation of Golgi containing both pre-existing and newly synthesized protein are mediated by different machineries. These results and modeling based on quantified results indicate that the Golgi apparatuses in tobacco BY-2 cells are not uniform and suggest that both de novo synthesis from the ER and Golgi division contribute almost equally to the increase in proliferating cells. Keywords: Cell cycle Golgi apparatus Photoconvertible fluorescence protein Tobacco BY-2 cells. Abbreviations: BFA, brefeldin A; BY-2, bright yellow 2; CHX, cycloheximide; DAPI, 4 0,6-diamidino-2-phenylindole; DMSO, dimethylsulfoxide; ER, endoplasmic reticulum; GFP, green fluorescent protein; 3MA, 3-methyl adenine; mkikgr, monomeric Kikume green red; mrfp, monomeric red fluorescent protein; TIBA, 2,3,5-triiodobenzoic acid. Introduction The Golgi apparatus is an important organelle concerned primarily with synthesis and trafficking of complex carbohydrates for the plant cell wall, and transport of proteins to vacuoles (Matheson et al. 2006). Unlike those in animal cells, Golgi stacks in plants remain structurally and functionally intact throughout the cell cycle. This may be linked to the need for plant cells to maintain active secretion for the formation of the cell plate which defines the plane of plant cell division (Faso et al. 2009). In Arabidopsis shoot meristem cells, the number of Golgi stacks has been shown to double from approximately 34 to approximately 66 during the G 2 stage of the cell cycle, i.e. just prior to mitosis (Segui-Simarro and Staehelin 2006). Cells with high secretory activity such as pollen tubes and root hairs seem to produce large numbers of new Golgi stacks depending on their task and growth status (Samaj et al. 2006). Although it is known that active and increased secretion is required from the Golgi apparatus during plant cell division, yet the question of how and when the Golgi proliferate remains a matter of controversy. Endoplasmic reticulum (ER)-derived de novo biogenesis and subsequent fission have been implicated in this process based on a study using brefeldin A (BFA)-treated tobacco bright yellow 2 (BY-2) cells as a model (Langhans et al. 2007). Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014, available online at The Author Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists. All rights reserved. For permissions, please journals.permissions@oup.com Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author
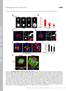
2 M. O. Abiodun and K. Matsuoka However, since this de novo regeneration of the Golgi apparatus during recovery from BFA treatment is obviously an artificial situation (Langhans et al, 2007), it has not been demonstrated in plants that intact cells operate the same process during the proliferation of cells. These same models have been proposed in mammalian cells (Ho et al. 2006), but the actual mechanism is still under debate (Hawes et al. 2010). Despite these revelations, the actual level of contribution of each mechanism to the increase in Golgi number has never been elucidated. Garcia-Herdugo et al. (1988) posited that Golgi stacks double in number during metaphase in onion root meristems, but Hirose and Komamine (1989) claimed that the duplication occurs during cytokinesis in synchronized cultures of Catharanthus roseus. However, in BY-2 cells, Nebenfuhr et al. (2000) could not allocate a distinctive increase in Golgi stacks to any particular phase in the cell cycle. Prior to and during cell division, increased synthesis/transportation of certain proteins and cell wall materials are necessary, and a subset of the Golgi apparatus in tobacco BY-2 cells accumulates in the phragmoplast region (Nebenfuhr et al. 2000). In regard to the increase in Golgi apparatus during these cellular events, a question arises as to whether the pre-existing populations of Golgi carry out this function or new ones are synthesized to assist the pre-existing ones. To be able to visualize the dynamics of the Golgi stacks in vivo physically, Hawes et al. (2010) suggested utilization of photoconvertible fluorescent proteins e.g. Kaede tagged with Golgi-resident proteins. When irradiated with UV or violet light, the fluorescence emitted by populations of Golgi stacks was converted from green to red. On incubation of the tissue, the new Golgi marker protein emitted green fluorescence, and when transported to existing Golgi stacks, the protein gave a mix of red, green and yellow signals (Brown et al. 2010). However, the use of a microscope for photoconversion has limitations in application as the condition is usually stressful to cells. In addition, the Kaede protein is not easy to use as it is a tetrameric protein that might affect localization and turnover; and the intensity of fluorescence before and after conversion is quite different. We thus chose the monomeric derivative of Kikume green red protein mkikgr as a photoconvertible reporter protein because this protein shows similar intensities of fluorescence before and after conversion (Habuchi et al. 2008). We used a cis-golgi protein type 1 prolyl 4-hydroxylase, NtP4H1.1 (formerly called tobacco PH; Yuasa et al. 2005) as a Golgi marker because the cis-golgi is not highly acidic and the acid dissociation constant (pka) of green mkikgr is slightly acidic, which allowed sufficiently high fluorescence of mkikgr at the cis-golgi. Using this fusion protein, we addressed the question as to whether and to what extent do de novo formation from the ER and fission of pre-existing stacks contribute to the increase in Golgi number in proliferating plant cells using tobacco BY-2 cells as a model. Results Subcellular localization of NtP4H1.1 mkikgr The previous characterization of NtP4H1.1 revealed that it is a type 1 P4H of tobacco predominantly localizing in the cis-side of the Golgi apparatus. Protein fusion with both green fluorescent protein (GFP) and monomeric red fluorescent protein (mrfp) targeted these proteins to the cis-golgi in tobacco BY-2 cells (Yuasa et al. 2005, Toyooka et al. 2009, Munemasa et al. 2012). Therefore, we expected that a fusion of NtP4H1.1 and another fluorescent protein, mkikgr (Habuchi et al. 2008), would also be targeted to the cis-golgi. As expected, the fusion protein (NtP4H1.1 mkikgr) expressed in tobacco BY-2 cells showed punctate fluorescent signals that are indistinguishable from the pattern of NtP4H1.1 GFP and NtP4H1.1 mrfp (Fig. 1). Thus, we concluded that the NtP4H1.1nmKikGR is targeted to the cis-golgi in tobacco BY-2 cells. Differences in the population of Golgi apparatus in growing tobacco BY-2 cells To analyze the different populations of Golgi apparatus existing in BY-2 cells at different time points of cell growth, a suspension culture of transformed BY-2 cells expressing NtP4H1.1 mkikgr at the logarithmic phase of growth in a regular Erlenmeyer flask was exposed to purple light for 3 h to convert the fluorescence of NtP4H1.1 mkikgr from green to red. After illumination, most of the fluorescent puncta in the cells emitted red fluorescence (Fig. 2A After exposure ; Supplementary Fig. S2), an indication that the conversion of the fluorochrome of NtP4H1.1 mkikgr from green to red fluorescence was efficient under this exposure condition. In addition, we did not observe differences in the growth of cells Fig. 1 Localization of fusion protein, NtP4H1.1 mkikgr. Confocal images of log phase BY-2 cells showing the localization of NtP4H1.1 fused with different fluorescent proteins. The pattern of fluorescence in NtP4H1.1 mkikgr is compared with previously reported NtP4H1.1 GFP and NtP4H1.1 mrfp. Images of non-transformed cells were also analyzed for background fluorescence. Scale bar = 10 mm. 542 Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author 2013.
3 Proliferation of Golgi apparatus in BY-2 cells Fig. 2 Definition of the intensity of the three observed populations of Golgi in tobacco BY-2 cells. (A) Confocal images of logarithmic cells transformed with NtP4H1.1 mkikgr showing the fluorescence puncta of different Golgi populations at different time points. Scale bar = 10 mm. (B) Both green and red fluorescent punctate dots from time-course confocal images of NtP4H1.1 mkikgr-expressing cells were separately subjected to intensity analysis using Image J software, and 40 dots were analyzed for intensities per time point and for the control. Logarithms of the ratio of green to red values are represented graphically. Value ranges of green, yellow and red Golgi were defined. Before conversion is a confocal image of a non-photoconverted transformed log phase cell; After conversion is a confocal image of a cell immediately after photoconversion; 6 h and 12 h are confocal images of cultured photoconverted cells at 6 and 12 h, respectively. after exposure, an indication that this exposure caused little stress, at least on the criterion of cell growth. Thus, we chose this exposure condition for the subsequent analyses. After purple light exposure, cells were subcultured in fresh medium and cultured in the dark. Samples were collected from the culture and viewed under a microscope, and confocal images of cells at the non-cell division cycle were collected using both green and red fluorescence ranges. Optical sections of cells containing the middle of the cells and parallel to the long axis of the cells were chosen for analyses. The intensities of both the green and red signals of each dot in the cells were quantified. For controls, transformed cells without exposure to purple light were used. We first compared the intensities of green fluorescence before conversion and red fluorescence after conversion to determine whether some normalization of the values of the intensities is required for quantification. As shown in Supplementary Fig. S1, the distribution of green and red signals before and the red after photoconversion were similar under the data collection condition used, and most of the intensities of signals were not saturated. The red signals before and the green after photoconversion were also similar, except for the appearance of some green signals after photoconversion (Supplementary Fig. S1). Thus, we chose this fluorescence recording condition for the subsequent analyses. We analyzed the pattern of both green and red fluorescence in the cells before color conversion, just after color conversion and 6 and 12 h after color conversion. During the incubation, the green fluorescence increased in the cells but the pattern was distinct from what was seen in the control. Cells at 6 and 12 h contained more fluorescence at the perinuclear region and just below the plasma membrane, and these signals were less in the control. This suggests that a significant proportion of newly synthesized NtP4H1.1 mkikgr was still not targeted to the Golgi and possibly was retained in the ER. We then quantified the intensities of both green and red fluorescence from Golgi puncta. For each condition or time point indicated, the ratio of the intensities of green to red signals was calculated using a total of 40 punctate dots from five randomly chosen cells. A graphical plot of the logarithm of the intensity ratio (Fig. 2B) revealed that almost all of the dots in non-converted cells showed values above +1.30, which were defined as the green Golgi population, which contain <5% of the purple-converted (red) NtP4H1.1 mkikgr signal relative to the non-converted (green) NtP4H1.1 mkikgr signal. Similarly, the red Golgi populations were defined as those in which the puncta had values between 2.40 and 1.30, containing >95% of purple-converted (red) NtP4H1.1 mkikgr signal relative to green NtP4H1.1 mkikgr signal. Populations of Golgi that yielded values between 1.30 and were grouped as the yellow Golgi population, and contained both red and green NtP4H1.1 mkikgr proteins in significant amounts. Most of the puncta in cells after conversion were classified as either red or yellow, and the percentage of the green Golgi was always 7.5%. In all cases tested, 40 80% of the population was classified as red but the percentage varied among the experiments. When illuminated cells were further grown for 6 or 12 h, the red population decreased and the green population increased over time (Fig. 2B). This suggests that pre-existing Golgi acquired newly synthesized protein and that some of the Golgi were formed de novo during the incubation. To gain more insight into the turnover of NtP4H1.1 mkikgr and the Golgi apparatus, a detailed time-course Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author
4 M. O. Abiodun and K. Matsuoka analysis was carried out up to 48 h after color conversion. The green fluorescence appeared just after conversion and increased progressively over time. At the early time points, relatively high non-punctate fluorescence at the perinuclear region was observed which turned weaker at later time points, and green dots that correspond to the Golgi stacks became prominent (Fig. 3A). Quantification of the number of Golgi with different colors indicated that the green Golgi increased progressively from 0 h (immediately after photoconversion) up to 48 h (Fig. 3A, B). Conversely, the red (old pre-existing) Golgi decreased progressively in the cell immediately after conversion to absolutely zero per cell after 42 h (Fig. 3B). The yellow population also showed a decrease after approximately 10 h of lag time and disappeared after 45 h of culturing following the disappearance of the red population. Since two out of the three observed populations of the Golgi were reduced, we determined the trend of the total number of Golgi apparatus during the experiment. An increase in the total number of Golgi stacks was observed until 12 h, after which a reduction was recorded at around 24 h, followed by a plateau. The numbers of Golgi per cell did not change more than 2-fold during the culture period (Supplementary Fig. S3). To correlate this change with the growth of cells, the volume of photoconverted cells and the size of the cells were measured every 12 h for 48 h. Using this information, we calculated the relative increase of the Golgi numbers per volume of cell culture and per cell during the incubation periods. The Golgi number per volume of cell culture increased steadily up to approximately 18-fold within 48 h and this increase correlated well with the growth of cells (Fig. 3C). The relative increase of Golgi number per cell behaved differently (Fig. 3C) but similarly to the total number of Golgi per cell (Supplementary Fig. S3). We next analyzed the rate of increase of green fluorescence in the cells as not all the NtP4H1.1 mkikgr seemed localized in the Golgi, especially at earlier times after biosynthesis. We found that the relative increase of the green fluorescence per cell increased about 2-fold within the first 12 h and remained constant until the end of the incubation period (Fig, 3D). Based on these data and the data of cell growth, we estimated the increase in green NtP4H1.1 mkikgr during the growth. The total intensity of green fluorescence per culture increased up to approximately 30-fold during the 48 h of incubation. We also quantified the intensity of red fluorescence in the cell during the incubation period and the change in the intensity per culture was calculated using the cellular fluorescence value and the rate of cell growth. We observed a rapid decrease in the red fluorescence in the cell. A slower but significant decrease in the red fluorescence per culture was observed (Fig. 3E). This observation means that NtP4H1.1 mkikgr is degraded slowly, having a half-life of about 24 h with an initial lag time of approximately 8 h for our experimental conditions (Table 1). We then analyzed whether the increase in green Golgi during the culture could be explained by the fission and subsequent division of pre-existing Golgi after receiving the newly synthesized NtP4H1. mkikgr. As shown in Fig. 3G, this model cannot explain the rapid and significant increase of green Golgi. We also tested whether the increase in Golgi number was solely the result of de novo formation of Golgi from newly synthesized NtP4H1.1 mkikgr (Fig. 3H; Supplementary Fig. S4c). This model also did not match the actual change in the populations of the Golgi. A mixed model of de novo formation from the ER and Golgi fission was then constructed considering the turnover rate of the red-converted NtP4H1.1 mkikgr (Fig. 3I; Supplementary Fig. S4d) to determine whether the change in Golgi color can be explained using a combination of these two models. The mixed model revealed a similar pattern of change to that observed in the actual data (Fig. 3F; and Supplementary Fig. S4a). The model allowed us to estimate the contribution of the two pathways to the Golgi increase, and the contribution seemed almost the same during the growth of the BY-2 cells (Table 1). Effect of BFA on the population change It has been reported that BFA inhibits protein trafficking through inhibition of vesicle formation at the Golgi apparatus and other membranes (Nebenfuhr et al. 2002). BFA treatment reverses at least the morphology of the Golgi apparatus as the washout of BFA clearly regenerates Golgi stacks in tobacco BY-2 cells (Langhans et al. 2007). We reported previously (Yuasa et al. 2005) that treatment of BY-2 cells with 14.8 mm BFA changed the distribution of NtP4H1.1 GFP to the ER. To examine the effect of BFA on the distribution of the differently colored Golgi apparatus, 12-h-old photoconverted BY-2 cells expressing the NtP4H1.1 mkikgr fusion protein were treated with 14.8 mm BFA for 60 min and then fluorescence images were collected. Thereafter, cells were washed with fresh medium without BFA and cultured in fresh medium for 2 h, and additional images were collected. It was discovered that BFA altered the fluorescence pattern emitted by the Golgi puncta with green, yellow and red colors to a strong ER pattern of yellow fluorescence (Fig. 4A, middle planes). After 2 h of cultivation in fresh medium, the punctate pattern of fluorescence was restored in the cells, but the three populations were not clearly distinguishable. Quantification of the fluorescence intensities of the dots indicated that the regenerated Golgi contained a relatively uniform level of both green and red NtP4H1.1 mkikgr proteins (Fig. 4B). In contrast, a control experiment without BFA did not cause the formation of uniformly yellow Golgi; in this case, red, yellow and green populations of the Golgi were distinctly observed (Fig. 4B). These observations confirmed that the discrete, differently colored populations of Golgi apparatus that we observed during the time-course analysis represented the newly synthesized green Golgi; yellow Golgi containing both pre-existing and newly synthesized protein; and pre-existing red Golgi. 544 Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author 2013.
5 Proliferation of Golgi apparatus in BY-2 cells Fig. 3 Time-course analysis of NtP4H1.1 mkikgr-expressing cells. (A) Three-day old BY-2 cells, expressing the NtP4H1.1 mkikgr fusion, were exposed to purple light for 3 h to convert the green fluorescence of mkikgr to red. Cells were analyzed for red and green fluorescence with a confocal microscope at the time points indicated. The control cells were analyzed before exposure to light. Scale bar = 5 mm. (B) Intensities of green and red fluorescent dots from time-course confocal images were analyzed separately for each time point and the ratio of green to red fluorescence was calculated using data of 40 puncta from five randomly selected cells. Percentages of green, yellow and red Golgi were calculated from the logarithm values of the green/red ratios as described in the Materials and Methods using the defined value range of green, red and yellow Golgi apparatus. (C) The relative Golgi number per culture (RelGolgiN/Culture) was determined by calculating the volume of individual cells (n = 50 cells). The actual volume of cells measured after centrifugation was divided by the calculated volume of cells and then multiplied by the average Golgi number per cell (determined by counting for each time point indicated). The values were expressed relative to the 0 h time value. Left axis: relative Golgi number per cell (RelGolgiN/Cell) was calculated by counting the puncta of Golgi as seen in the confocal images (n = 10 cells per time point). The values were presented as relative to the 0 h values for each time point. Right axis: cell growth determination. Photoconverted cells were cultured in fresh medium, and a known volume of culture was centrifuged and the sedimented volume of cell was measured. This was repeated every 12 h for 48 h. (D and E) The intensity of both the green (D) and red (E) fluorescence in the cell and culture. The intensity per culture was calculated by multiplying the average intensity per time point by the calculated volume of the cells and then the values were expressed relative to the 0 h time value. The intensity per cell was measured directly and expressed as relative values of (continued) Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author
6 M. O. Abiodun and K. Matsuoka Fig. 3 Continued the 0 h value. The two experiments are included to compare variations among experiments. (F I) The percentage distribution of green, red and yellow Golgi at 12 h intervals was determined from the values of intensities as defined in the legend to Fig. 2B. This was compared with three models, i.e. the Golgi fission (G), de novo formation from ER (H) and Mixed model of both de novo formation from ER and Golgi fission (I) that were calculated using the same intensity distribution values for 0 h in the actual data (F) as the starting point. Table 1 Lag and half-time of the degradation of red-converted fusion protein and calculated contribution of the de novo synthesis to Golgi apparatus Lag time (h) Half-time (h) Experiment Experiment Average Contribution from de novo synthesis (%) Effect of cycloheximide on the population distribution To investigate the biogenesis of Golgi apparatus in BY-2 cells, NtP4H1.1 mkikgr-expressing cells were treated with a protein synthesis inhibitor, cycloheximide (CHX), for 12 h immediately after color conversion. The control sample was also treated in the same manner, but without CHX. In the presence of CHX, the number of green and yellow dots did not increase during the 12 h incubation, whereas in the control, numerous green and yellow puncta were clearly observed (Fig. 5A, B). We also observed that the total number of Golgi stacks per cell was significantly higher after 12 h of CHX treatment (54/cell, n = 10) compared with the control (44/cell, n = 10), although both numbers were higher than at 0 h after photoconversion (28/cell, n = 10). This showed that, generally, the number of Golgi increased with time even in the presence of a protein synthesis inhibitor. The distribution of the log ratio of the intensity of fluorescence of the 0 h, non-treated photoconverted cells was nearly identical to that of the CHX-treated cells (Fig. 5B). This observation also confirmed that the green Golgi that we observed were the Golgi apparatus containing the newly synthesized protein. The higher Golgi number in CHX-treated cells can be attributed to the fact that protein synthesis is required for the turnover of NtP4H1.1 mkikgr, but generation of Golgi apparatus from ER-retained NtP4H1.1 mkikgr can take place without further supply of newly synthesized proteins. Cell cycle and Golgi proliferation As described above, we found that the two distinct mechanisms of Golgi proliferation are taking place during the growth of BY-2 cells. To address whether the operation of these two systems in tobacco cells takes place at different stages of the cell cycle, we analyzed the change in the color distribution of Golgi in cells at different stages of the cell cycle. We chose specific phases of cells for the comparison as cells at these phases can easily be identified by simple microscopic observation. The phases of the cell cycle that we chose are: G 1 /S phase, G 2 phase and late M phase (telophase to cytokinesis phase). Among these phases of the cell cycle, telophase and cytokinesis phase cells can easily be identified under 546 Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author 2013.
7 Proliferation of Golgi apparatus in BY-2 cells Fig. 4 Effect of brefeldin A on the color distribution of Golgi apparatus in tobacco BY-2 cells. (A) Log phase photoconverted cells were washed, cultured in fresh medium for 12 h and then treated with 14.8 mm BFA for 60 min. The BFA was washed off and the cells cultured in fresh medium for another 2 h. Green and red fluorescence signals were analyzed by confocal microscopy at the end of BFA treatment (+BFA), at the end of the 2 h BFA washout (BFA washout) and for untreated transformed cells ( BFA). Scale bar = 10 mm. (B) Intensities of green and red fluorescent dots were analyzed separately for each treatment including 0 h, immediately after photoconversion using the methods described in the legend to Fig. 2B. Fig. 5 Effect of the protein synthesis inhibitor cycloheximide on the distribution of Golgi apparatus in BY-2 cells. (A) Log phase BY-2 cells expressing the NtP4H1.1 mkikgr fusion protein were exposed to purple light for 3 h, washed three times with fresh medium, subcultured in fresh medium, immediately treated or not with 100 mg ml 1 CHX and cultured in the dark for 12 h. Samples of treated (+CHX) and untreated ( CHX) cells were then analyzed by confocal microscopy for green and red fluorescence. Scale bars = 10 mm. (B) Intensities of green and red fluorescent dots were analyzed separately for each treatment including 0 h, or immediately after photoconversion as described in the legend to Fig. 2B. the microscope. In terms of the difference between G 1 /S and G 2 cells, we had an impression though our routine observation of dividing BY-2 cells that the length of the nucleus is longer in G 2 than in G 1 and S phase cells. Before we analyzed the Golgi number, we confirmed our impression about the dimension of the nucleus by comparing the length, breadth and the intensity of DNA in the nuclei (or condensed chromosome region at telophase) after 4 0,6-diamidino-2-phenylindole (DAPI) staining. The intensity of the DAPI stain was highest at the G 2 phase and was approximately halved at late M phase (Fig. 6A, Table 2). The possible G 1 /S phase cells, having globular nuclei, showed an intensity between these phases. We also measured the length and breadth of the nucleus as seen with DAPI stain and confirmed that the longest nucleus was at the G 2 phase (Table 2). A scatter plot of DAPI signal intensity vs. the length of nuclei (or the condensed chromosome region at telophase) (Supplementary Fig. S5) clearly indicated a correlation between the length of the nucleus and the DNA content in the nuclear region. Based on these observations, we classified cells having a nucleus 11.4 mm or longer as G 2 phase cells, cells with nearly globular nuclei of around 8 mm in diameter as G 1 /S phase cells and cells having two condensed nuclei and a cell plate as late M phase cells. Transformed cells were photoconverted to red, washed with fresh medium, and confocal images were taken at 0, 6 and 12 h after the cells were transferred into fresh medium. The numbers of Golgi stacks (green, red and yellow) were counted from the Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author
8 M. O. Abiodun and K. Matsuoka Fig. 6 Analysis of the cell cycle for Golgi increase in tobacco BY-2 cells. (A) Three-day old cells were killed at 75 C, immediately treated with DAPI at a concentration 2.0 mg ml 1 for 5 min and epifluorescence images were collected (upper). Scale bar = 10 mm. The intensity of the nucleus was measured using Image J software. Bar charts indicate the average level of the DAPI intensities (n = 10). Error bar, SD. The length and breadth of the nucleus were also measured (indicated with white arrows) from the DAPI stains for each of the phases and the values are represented in Table 2. (B) Log phase transformed cells were photoconverted, washed with fresh medium and immediately cultivated in the dark using fresh medium. Images of cells at different phases of the cell cycle, namely G 1 /S, G 2 and late M phase (M: telophase and cytokinesis phase), were collected. White arrowheads point to the cells of interest. Scale bar = 10 mm. (C E) For each phase of the cell cycle, the green, red and yellow Golgi apparatus in the form of dots in the confocal images were counted (n = 10 cells) for each time points including 0 h (C), 6 h (D) and 12 h (E). The total number of Golgi apparatus per phase was included. 0 h means immediately after photoconversion; 6 h and 12 h mean 6 and 12 h after photoconversion. 548 Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author 2013.
9 Proliferation of Golgi apparatus in BY-2 cells Table 2 Length, breadth and intensity of DNA content in the nucleus as measured by DAPI treatment G 1 /S G 2 M Length (mm) Breadth (mm) Intensity 784, ,216, ,056.3 confocal images taken at the different time points and different phases of the cell cycle. In the case of late M phase cells, the number of Golgi apparatus between the cell plate and one of the ends of the cell were counted as this value represents the number of Golgi during or just after cytokinesis in daughter cell. At all the time points tested, the distribution of green, yellow and red Golgi in the different phases of cells were not clearly distinguishable (Fig. 6B). Generally, there was an increase in the total number of Golgi stacks from G 1 /S to G 2, becoming half of the G 2 value at late M phase as a result of cell division (Fig. 6B). The trend in the numbers of Golgi correlated well with the content of nuclear DNA measured by DAPI stain (Fig. 6) regardless of the time points analyzed. Analyses of traffic and other inhibitors that affect the proliferation of Golgi To investigate the mechanism of the proliferation of the Golgi apparatus, we analyzed the effect of different inhibitors on the growth rate and color change of the Golgi apparatus (Fig. 7). Immediately after conversion of the green fluorescence of Golgi into red fluorescence, different inhibitors were added to the cell suspension and the cells were cultured for 12 h. The images of the cells were collected and analyzed. The cell volume was also measured at the end of the treatment for each inhibitor tested. We used 2,3,5-triiodobenzoic acid (TIBA; 17.8 mm), which is a compound that stabilizes the actin bundle (Dhonukshe et al. 2008), ikarugamycin (20 mm), which is an inhibitor of clathrin-mediated endocytosis in tobacco (Onelli et al. 2008), MG-132 (200 mm), which is an inhibitor of proteasomes at low concentration and also inhibits other proteases, wortmannin (6.5 mm), which is an inhibitor of phosphatidylinositol kinases and also inhibits phospholipid biosynthesis in tobacco BY-2 cells (Matsuoka et al. 1995), 3-methyl adenine (3MA; 5 mm), which inhibits autophagy (Takatsuka et al. 2004), and E-64 (20 mm), which is an inhibitor of cysteine proteases and prevents autophagic degradation in tobacco BY-2 cells (Moriyasu and Ohsumi, 1996). After 12 h of treatment, all the inhibitors tested prevented the formation of green Golgi as shown by confocal microscopy images and their analyses (Fig. 7A, B). Ikarugamycin showed a different pattern of fluorescence compared with the other inhibitors and seemed to prevent formation of yellow Golgi. Other inhibitors did not prevent the formation of yellow Golgi (Fig. 7B). More red Golgi were found in ikarugamycintreated cells (similar to 0 h immediately after photoconversion) compared with the control (Fig. 7B, C). To reaffirm the effectiveness of ikarugamycin, the distribution of the intensity of fluorescence at 12 h after color conversion with this inhibitor was compared with the intensities immediately after photoconversion. The patterns of the intensities were nearly identical (Fig. 7C), suggesting an effective inhibitory activity of ikarugamycin on the transport of newly synthesized proteins into the Golgi apparatus. The effect of inhibitors on the growth of the cells revealed that growth was significantly inhibited by ikarugamycin treatment compared with the control and other inhibitors tested (Fig. 7D). As the fluorescence pattern of ikarugamycin-treated cells resembled that of CHX-treated cells and all the inhibitors tested prevented the formation of green Golgi, we tested whether the treatment of these inhibitors prevented the synthesis of NtP4H1.1 mkikgr. Proteins were extracted from cells that had been incubated with or without inhibitors for 12 h after color conversion, separated by SDS PAGE after partial denaturation and the bands of NtP4H1.1 mkikgr were visualized by fluorescence recording. The intensity of the green fluorescence of NtP4H1.1 mkikgr was then quantified. Control protein from cells just after color conversion was included. The result of this analysis showed a relatively high level of green NtP4H1.1 mkikgr protein in the ikarugamycin-treated cells followed by MG132 when compared with the control (Fig. 7E). This observation indicated that all the inhibitors, including ikarugamycin, did not prevent the synthesis of NtP4H1.1 mkikgr. We tried to quantify the red converted NtP4H1.1 mkikgr using the same method, but for unknown reasons we could not visualize red NtP4H1.1 mkikgr bands by this method. Discussion NtP4H1.1 is a type II membrane protein localizing in the cis- Golgi of BY-2 cells (Yuasa et al. 2005). When fused with mkikgr, this protein was targeted to the Golgi apparatus. Using this marker and exploiting the nature of the color conversion of mkikgr (Habuchi et al. 2008), three different types of Golgi population were identified in BY-2 cells (Fig. 2). These Golgi stacks correspond to the pre-existing Golgi (red), newly synthesized Golgi (green) and Golgi containing both the pre-existing and newly synthesized proteins (yellow). The newly synthesized population increased progressively over time (Fig. 3B) in order to ensure an equal distribution of Golgi apparatus between the two daughter cells during cell division (Garcia-Herdugo et al. 1988, Segui-Simarro and Staehelin, 2006, Toyooka et al. 2009) and to maintain the active secretion essential for cell growth. On the other hand, the pre-existing red Golgi decreased progressively over time (Fig. 3B, Supplementary S4a), possibly due to the division of Golgi after acquiring newly synthesized protein which depleted the intensity of the red signal. Also, the reduction might be partially due to the turnover of the protein over Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author
10 M. O. Abiodun and K. Matsuoka Fig. 7 Inhibitors that prevent the proliferation of Golgi apparatus. Photoconverted cells were incubated for 12 h with or without inhibitors at the following final concentrations: E-64, 20 mm; ikarugamycin, 20 mm; wortmannin, 16.5 mm; 3MA, 5 mm; MG-132, 200 mm; and TIBA, 50 mm. (A) Changes in the fluorescence pattern after 12 h of treatment with inhibitors. Images of non-transformed cells were included for each treatment to compare the background fluorescence of the cells. Fluorescence dots were analyzed using a confocal microscope. Scale bars = 5 mm. (B) Intensities of green and red fluorescent dots were analyzed separately for each inhibitor treatment as described in the legend to Fig. 2B. (C) The intensity distribution of an effective inhibitor, ikarugamycin, was compared with the distribution of intensity immediately after photoconversion to red (0 h). (D) The growth of inhibitor-treated cells was determined by measuring the volume of the cells after 12 h of treatment. 0 h is the volume of cells immediately after photoconversion without inhibitor, while control is the volume of cells after 12 h without inhibitor. The average of three independent experiments is shown. Error bar, SD. (E) Inhibitor-treated cells were centrifuged, sonicated in an equal volume of PBS, homogenates were centrifuged at 3,000 r.p.m. for 10 min at 4 C, the supernatant was added to SDS sample buffer, separated on a 12.5% polyacrylamide gel and the gel was scanned for green fluorescent signal. A 20 mg aliquot of protein was electrophoressed for each sample including freshly photoconverted cells without inhibitor (0 h) and cells treated without inhibitor (control). A bar chart indicating the level of the intensities was represented. The average of three independent experiments is shown. Error bar, SD. 550 Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author 2013.
11 Proliferation of Golgi apparatus in BY-2 cells time. The yellow Golgi also decreased after approximately 10 h of lag time and steadily decreased thereafter. The difference in the rate of decrease of red and yellow Golgi and the increase of green Golgi at earlier time points of analysis (Fig. 3) indicated that both the de novo synthesis of Golgi from the ER and the division of pre-existing Golgi after accepting newly synthesized protein from the ER are taking place in proliferating BY-2 cells at the same time. This observation is clearly different from the Golgi regeneration events in BY-2 cells observed at the stage of recovery from BFA treatment (Langhans et al. 2007, Ito et al. 2012). In this case, Golgi proteins that are localized in both the ER and small vesicles by the action of BFA first regenerated relatively wide Golgi apparatus and then divided to form individual Golgi with regular size (Langhans et al. 2007). In addition, the Golgi regenerated after BFA washout did not show any segregation of old (red) and newly synthesized (green) NtP4H1.1 mkikgr (Fig. 4). This situation is completely different from our observation without inhibitors as both the Golgi division and de novo formation take place at the same time (Figs. 2, 3). As a result, different Golgi apparatus might have a different proportion of proteins with different ages of proteins. In the future, it will be interesting to analyze whether Golgi apparatus at different ages (as observed here) can show different trends of movement in the cell as observed in Arabidopsis root cells (Akkerman et al. 2011). The kinetic difference in the decrease of red and yellow Golgi might indicate that red Golgi were converted to yellow Golgi after accepting green NtP4H1.1 mkikgr and then dividing into new Golgi. Actually, the Golgi division model partly explains the decrease in the red and yellow population of the Golgi over time (Fig. 3I) but cannot explain the rapid increase in green Golgi (Fig. 3F). An alternative model to explain the increase in green Golgi is that the Golgi apparatus was generated de novo from the newly synthesized protein (Fig. 3H). However, this mechanism alone cannot explain the kinetics of the decrease of red and yellow Golgi. The actual situation appeared to be a mixed case of these two extreme models as well as the turnover of NtP4H1.1 mkikgr (Fig. 3G, H) as the mixed model in Fig. 3I fits well to the actual data. This mixed model allowed for the explanation of the decrease in both red and yellow Golgi over time as well as the increase in green Golgi. In other words, this analysis indicated that tobacco BY-2 cells have two mechanisms/pathways to increase the number of Golgi during cell proliferation. One is the Golgi division after accepting Golgi proteins from the ER and the other is the de novo formation from the ER. During proliferation of BY-2 cells, about half of the Golgi stacks were found to be contributed by the de novo formation pathway while the rest were as a result of division of the pre-existing Golgi. The reduction in the red Golgi stacks might be as a result of both division of Golgi and the turnover of NtP4H1.1 mkikgr with relatively slow (24 h) half-life. The presence of both the de novo synthesis and Golgi division mechanisms in tobacco BY-2 cells is also supported by the analysis using inhibitors (Fig. 6). All the inhibitors tested prevented the formation of green Golgi, suggesting that the de novo formation of Golgi is sensitive to many inhibitors. In contrast, all except ikarugamycin did not prevent the increase of yellow Golgi formation (Fig. 6B). Thus ikarugamycin inhibits both the de novo synthesis and Golgi division. These difference in the sensitivity also supported that the mechanism to generate new Golgi and Golgi division are distinct. The effect of ikarugamycin, which is a known inhibitor of clathrin-mediated trafficking, is particularly interesting as this inhibitor prevented both the yellow and red Golgi but did not prevent the synthesis of NtP4H1.1 mkikgr (Fig. 7). It was reported recently that seeds of Golgi (that might be small vesicles) which are too small to visualize with a regular optical microscope, are present in tobacco BY-2 cells (Ito et al. 2012). Numbers of such vesicles increase the ability of cells to recover from BFA treatment than regular, non-bfa-treated cells (Ito et al. 2012). It was also reported recently that clathrin is required for the reassembly of fragmented Golgi-derived vesicles at the post-mitotic phase of the cell cycle in mammalian cells (Radulescu and Shields 2012). Thus it is possible that in ikarugamycin-treated cells, the newly synthesized NtP4H1.1 mkikgr with green fluorescence, which could only be detected by SDS PAGE and fluorescence recording (Fig. 7E) but not with confocal microscopic image collection (Fig. 7A), localized in vesicles such as the seed of the Golgi. Assembly of such vesicles into Golgi might be inhibited by ikarugamycin. Thus it will be interesting in the future to address this possibility with both super-resolution fluorescence microscopic and electron microscopic analyses. As described above, we found many inhibitors that prevented the formation of green Golgi. These included compounds known to inhibit several biochemical steps, including phospholipid metabolism and polypeptide hydrolysis. We have not analyzed whether the increase in yellow Golgi in inhibitortreated cells occurs solely as a result of the acquisition of newly synthesized protein or whether it is the result of both the acquisition and Golgi division. We also have not analyzed whether the target of the inhibitors that we used here is the known target of the inhibitors for other already reported cellular events. Further morphological characterization using electron microscope and biochemical analysis of the de novo formation of the Golgi will be needed to determine whether the inhibitors tested here prevent Golgi division, and this will lead to a better understanding of the de novo formation of the Golgi apparatus. The higher number of Golgi stacks at the G 2 phase of the cell cycle (Fig. 6) might have resulted from the need for the synthesis of new green Golgi and division of pre-existing red ones in the cellular response to preparation for cell division and to have an equal sustainable number of Golgi stacks in the daughter cells. This underscores the fact that new Golgi can be produced in the cell to assist the pre-existing Golgi when needed. The higher numbers of Golgi in G 1 /S phase cells than in late M phase cells suggested the possibility that Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author
12 M. O. Abiodun and K. Matsuoka the proliferation of Golgi stacks occurs in synchrony with nuclear DNA synthesis during the cell cycle. Although synchronization of the tobacco BY-2 cell cycle as established by Nagata and Kumagai (1999) would have helped more in comparing the exact timing of Golgi proliferation and DNA synthesis, we did not take such an approach because it is basically the treatment of stationary phase cells with aphidicolin. This decision was based on the fact that authophagy takes place in BY-2 cells at stationary phase (Toyooka et al. 2006), during which cells are under nutrient starvation conditions. Since a subtle change in nutritional conditions, i.e. changing cells into fresh medium and a subsequent higher level of aeration (opening the cover of the culture flask frequently for sampling), caused a transient increase in Golgi number and kept the relatively high number up to the end of the 48 h of the experiment (Fig. 3), analysis of the Golgi increase using the established protocol of cell cycle synchronization will not be able to dissect whether the increase in Golgi number after the release of cell cycle arrest is the result of cell cycle progression or release from starvation. Actually, our preliminary analyses suggested that the level of nutrients affects the nature and number of Golgi apparatus in BY-2 cells (S. Asatsuma, M.O. Abiodun and K. Matsuoka, unpublished result). Thus future development of a cell cycle synchronization protocol without nutrient starvation will be necessary to compare the timing of the increase in Golgi apparatus and other events in cell cycle progression. In this work, we used NtP4H1.1 as a Golgi marker and mkikgr as a reporter protein. Because we had to fuse the fluorescent protein to the luminal domain of NtP4H1.1, we could not apply the same method to other type II membrane proteins localized on the trans-side of the Golgi based on the difference in the ph in the different lumen of the Golgi where the trans-side is more acidic than the pka of the fluorescent protein. Thus, our result is an example of the analysis, and future confirmation using other proteins will be necessary to clarify the whole process of the maintenance of Golgi in proliferating plant cells. For this purpose, alternative and more ph-resistant color-convertible fluorescent proteins will be necessary when using a type II membrane protein as a reporter, since many of the Golgi proteins including glycosyltransferases are type II membrane proteins. In addition, future analysis using different proteins with a similar approach will be necessary to determine which of the mechanisms reported here is the predominant mechanism of Golgi biogenesis in proliferating plant cells. Materials and Methods Construction of plasmid and transformation into BY-2 cells NtP4H1.1 plasmid was constructed as described previously (Yuasa et al. 2005; formerly called tobacco PH). The cdna encoding NtP4H1.1 was amplified by PCR using primers that contain BamHI and NcoI sites. The resulting fragment was fused to BamHI- and NcoI-digested sites of photoconvertible fluorescent protein, mkikgr (MBL, Japan). The NtP4H1.1 mkikgr fragments were then cloned into BglII- and HindIII-digested binary vector, pmat 137 (Yuasa et al. 2005). These were then used to transform tobacco BY-2 cells, using Agrobacterium as described previously (Matsuoka and Nakamura, 1991). Cell culture Cultures of tobacco BY-2 cells were carried out as described previously (Matsuoka and Nakamura 1991). The cell lines used were wild-type BY-2 cells and BY-2 cells expressing NtP4H1.1 mkikgr. Three-day-old (log phase) cells were used in this experiment. Photoconversion of NtP4H1.1-mKikGR-expressing cells A 100 ml aliquot of the culture of log-phase-transformed cells, in an 300 ml Erlenmeyer flask, was exposed to purple light from a 100 W black light bulb (H100BL-L, Toshiba), which emits violet light capable of irreversibly converting the green fluorescence of the mkikgr protein to red fluorescence at a wavelength between 300 and 450 nm. This exposure, carried out with care to minimize various stresses on the cells, lasted for 3 h with gentle shaking. After 3 h, the cells were collected by sedimentation, washed with fresh medium three times and then cultured in fresh medium under dark conditions for the indicated periods of time for subsequent analyses. Treatment with chemicals For BFA treatment, 5 mg ml 1 stock solution of BFA in dimethylsulfoxide (DMSO) was prepared. This was used at a final concentration of 3.3 mgl 1 for 60 min at room temperature with gentle shaking. The BFA was added after 12 h of subculturing the photoconverted cells. Control experiments were performed similarly with an equal amount of DMSO in place of BFA. The BFA was washed off with fresh medium three times and dark-cultured in fresh medium for another 2 h. CHX dissolved in DMSO was added to the color-converted cells at a final concentration of 100 mg ml 1 just when transferring the cells into fresh medium. These were dark-cultured at room temperature for 12 h and images were collected. The control experiment was performed similarly but with DMSO. An equal volume of log phase photoconverted cells was washed with fresh medium three times, cultured in fresh medium and immediately treated with different inhibitors for 12 h. We used TIBA at a concentration of 17.8 mm (Aldrich Chem. Co.), ikarugamycin at a concentration of 20 mm (Biomol Research Labs Inc.), MG-132 at a concentration of 200 mm (Calbiochem Inc.), wortmannin at a concentration of 16.5 mm (Calbiochem Inc.), 3MA at a concentration of 5 mm (Sigma-Aldrich, Inc.) and E-64 at a concentration of 20 mm (Peptide Institute, Inc.). For growth rate measurement, equal 552 Plant Cell Physiol. 54(4): (2013) doi: /pcp/pct014! The Author 2013.
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
Practice Problems 5. Location of LSA-GFP fluorescence
 Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Yeast Nuclei Isolation Kit
 Yeast Nuclei Isolation Kit Catalog Number KA3951 50 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Yeast Nuclei Isolation Kit Catalog Number KA3951 50 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
BIO 315 Lab Exam I. Section #: Name:
 Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
Section #: Name: Also provide this information on the computer grid sheet given to you. (Section # in special code box) BIO 315 Lab Exam I 1. In labeling the parts of a standard compound light microscope
MEFs were treated with the indicated concentrations of LLOMe for three hours, washed
 Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
Supplementary Materials and Methods Cell Fractionation MEFs were treated with the indicated concentrations of LLOMe for three hours, washed with ice-cold PBS, collected by centrifugation, and then homogenized
ab CFSE Fluorescent Cell Labeling Kit
 ab113853 CFSE Fluorescent Cell Labeling Kit Instructions for Use For the durable fluorescent labeling of live cells for fluorescent microscopy and flow cytometry, population growth studies and within sample
ab113853 CFSE Fluorescent Cell Labeling Kit Instructions for Use For the durable fluorescent labeling of live cells for fluorescent microscopy and flow cytometry, population growth studies and within sample
 DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
DOI: 10.1038/ncb3259 A Ismail et al. Supplementary Figure 1 B 60000 45000 SSC 30000 15000 Live cells 0 0 15000 30000 45000 60000 FSC- PARR 60000 45000 PARR Width 30000 FSC- 15000 Single cells 0 0 15000
ab Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
CytoPainter Golgi Staining Kit Green Fluorescence
 ab139483 CytoPainter Golgi Staining Kit Green Fluorescence Instructions for Use Designed for the detection of Golgi bodies by microscopy This product is for research use only and is not intended for diagnostic
ab139483 CytoPainter Golgi Staining Kit Green Fluorescence Instructions for Use Designed for the detection of Golgi bodies by microscopy This product is for research use only and is not intended for diagnostic
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
SUPPLEMENTARY NOTE 3. Supplememtary Note 3, Wehr et al., Monitoring Regulated Protein-Protein Interactions Using Split-TEV 1
 SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
SUPPLEMENTARY NOTE 3 Fluorescent Proteolysis-only TEV-Reporters We generated TEV reporter that allow visualizing TEV activity at the membrane and in the cytosol of living cells not relying on the addition
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Gene specific primers. Left border primer (638) ala3-4 (Salk_082157) Gene specific primers. Left border primer (1343)
 Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Supplemental Data. Poulsen et al. (2008) The Arabidopsis P4-ATPase pump ALA3 localizes to the Golgi and requires a β subunit to function in lipid translocation and secretory vesicle formation. A ala3-1
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Rodríguez-Fraticelli et al., http://www.jcb.org/cgi/content/full/jcb.201203075/dc1 Figure S1. Cell spreading and lumen formation in confined
Synchronisation of BY-2 cell culture
 Synchronisation of BY-2 cell culture Introduction Nicotiana tobacum cell suspension culture Bright Yellow 2 (BY-2) Synchronisation with aphidicolin to achieve up to 70% synchrony Aphidicolin arrests cells
Synchronisation of BY-2 cell culture Introduction Nicotiana tobacum cell suspension culture Bright Yellow 2 (BY-2) Synchronisation with aphidicolin to achieve up to 70% synchrony Aphidicolin arrests cells
Supplementary Information
 Supplementary Information ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis Supplemental Figures a 8 6 4 2 Absolute Expression b Absolute Expression
Supplementary Information ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis Supplemental Figures a 8 6 4 2 Absolute Expression b Absolute Expression
ab CFSE Fluorescent Cell Labeling Kit
 ab113853 CFSE Fluorescent Cell Labeling Kit Instructions for Use For the durable fluorescent labeling of live cells for fluorescent microscopy and flow cytometry, population growth studies and within sample
ab113853 CFSE Fluorescent Cell Labeling Kit Instructions for Use For the durable fluorescent labeling of live cells for fluorescent microscopy and flow cytometry, population growth studies and within sample
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Supplementary Information. c d e
 Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
Supplementary Information a b c d e f Supplementary Figure 1. atabcg30, atabcg31, and atabcg40 mutant seeds germinate faster than the wild type on ½ MS medium supplemented with ABA (a and d-f) Germination
ab Autophagy/ Cytotoxicity Dual Staining Kit
 ab133075 Autophagy/ Cytotoxicity Dual Staining Kit Instructions for Use For the study of the regulation of autophagy and cytotoxicity at the cellular level. This product is for research use only and is
ab133075 Autophagy/ Cytotoxicity Dual Staining Kit Instructions for Use For the study of the regulation of autophagy and cytotoxicity at the cellular level. This product is for research use only and is
Dr: RAWIA BADR Associate Professor of Microbiology&Immunology
 Dr: RAWIA BADR Associate Professor of Microbiology&Immunology Cell culture Commonly refers to the culture of animal cells and tissues, while the more specific term plant tissue.culture is used only for
Dr: RAWIA BADR Associate Professor of Microbiology&Immunology Cell culture Commonly refers to the culture of animal cells and tissues, while the more specific term plant tissue.culture is used only for
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
< Supporting Information >
 SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
SUPPORTING INFORMATION 1 < Supporting Information > Discovery of autophagy modulators through the construction of high-content screening platform via monitoring of lipid droplets Sanghee Lee, Eunha Kim,
Supplementary Information
 Supplementary Information COP1 E3 ligase protects HYL1 to retain microrna biogenesis Seok Keun Cho 1, Samir Ben Chaabane 1, Pratik Shah 1, Christian Peter Poulsen 1, and Seong Wook Yang 1 * 1 Laboratory
Supplementary Information COP1 E3 ligase protects HYL1 to retain microrna biogenesis Seok Keun Cho 1, Samir Ben Chaabane 1, Pratik Shah 1, Christian Peter Poulsen 1, and Seong Wook Yang 1 * 1 Laboratory
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supporting Information
 Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Supporting Information Stavru et al. 0.073/pnas.357840 SI Materials and Methods Immunofluorescence. For immunofluorescence, cells were fixed for 0 min in 4% (wt/vol) paraformaldehyde (Electron Microscopy
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot-Marie-Tooth disease
 Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein mutant that causes type 1A Charcot-Marie-Tooth disease Taichi Hara, Yukiko Hashimoto, Tomoko Akuzawa, Rika Hirai,
Supplemental Information. Dynamic Organization of Chromatin Domains. Revealed by Super-Resolution Live-Cell Imaging
 Molecular Cell, Volume 67 Supplemental Information Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging Tadasu Nozaki, Ryosuke Imai, Mai Tanbo, Ryosuke Nagashima, Sachiko
Molecular Cell, Volume 67 Supplemental Information Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging Tadasu Nozaki, Ryosuke Imai, Mai Tanbo, Ryosuke Nagashima, Sachiko
Detection of bacterial aggregation by flow cytometry
 Detection of bacterial aggregation by flow cytometry Jack Coleman 1 and Melis McHenry 2 1 Enzo Life Sciences, Farmingdale NY, USA, 2 Beckman Coulter Inc., Miami FL, USA PROTEOSTAT Aggresome Detection Kit
Detection of bacterial aggregation by flow cytometry Jack Coleman 1 and Melis McHenry 2 1 Enzo Life Sciences, Farmingdale NY, USA, 2 Beckman Coulter Inc., Miami FL, USA PROTEOSTAT Aggresome Detection Kit
Chapter 20 Recombinant DNA Technology. Copyright 2009 Pearson Education, Inc.
 Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
Chapter 20 Recombinant DNA Technology Copyright 2009 Pearson Education, Inc. 20.1 Recombinant DNA Technology Began with Two Key Tools: Restriction Enzymes and DNA Cloning Vectors Recombinant DNA refers
2. Know the parts of a light microscope and general rules for using and focusing a microscope, such as:
 SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry
 Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
INDIAN INSTITUTE OF TECHNOLOGY ROORKEE NPTEL NPTEL ONLINE CERTIFICATION COURSE. Biomedical Nanotechnology
 INDIAN INSTITUTE OF TECHNOLOGY ROORKEE NPTEL NPTEL ONLINE CERTIFICATION COURSE Biomedical Nanotechnology Lec-19 In Vitro Methods of Study Antibacterial and Anticancer Properties of Nanomaterials Dr. P.
INDIAN INSTITUTE OF TECHNOLOGY ROORKEE NPTEL NPTEL ONLINE CERTIFICATION COURSE Biomedical Nanotechnology Lec-19 In Vitro Methods of Study Antibacterial and Anticancer Properties of Nanomaterials Dr. P.
ab CytoPainter ER Staining Kit Red Fluorescence
 ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
SUPPLEMENTARY INFORMATION
 Local auxin metabolism regulates environment-induced hypocotyl elongation Zuyu Zheng 1,2, Yongxia Guo 3, Ondřej Novák 4,5, William Chen 2, Karin Ljung 4, Joseph P. Noel 1,3, *, and Joanne Chory 1,2, *
Local auxin metabolism regulates environment-induced hypocotyl elongation Zuyu Zheng 1,2, Yongxia Guo 3, Ondřej Novák 4,5, William Chen 2, Karin Ljung 4, Joseph P. Noel 1,3, *, and Joanne Chory 1,2, *
PROTOCOL. Live Cell Imaging of 3D Cultures Grown on Alvetex Scaffold Using Confocal Microscopy. Introduction
 Page 1 Introduction Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell
Page 1 Introduction Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell
OPPF-UK Standard Protocols: Mammalian Expression
 OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
OPPF-UK Standard Protocols: Mammalian Expression Joanne Nettleship joanne@strubi.ox.ac.uk Table of Contents 1. Materials... 3 2. Cell Maintenance... 4 3. 24-Well Transient Expression Screen... 5 4. DNA
Autophagy/Cytotoxicity Dual Staining Kit
 Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
1. Carry the microscope in an upright position with both hands and place the base of the microscope 5cm from the edge of the bench
 The Microscope Operating the compound light microscope 1. Carry the microscope in an upright position with both hands and place the base of the microscope 5cm from the edge of the bench 2. Check that lenses
The Microscope Operating the compound light microscope 1. Carry the microscope in an upright position with both hands and place the base of the microscope 5cm from the edge of the bench 2. Check that lenses
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
SUPPLEMENTARY INFORMATION 1 Supplementary Figure S1. Proportion of HEK293T transfectants showing fluorescent foci after heat sho Blinded slides were counted for the appearance of fluorescent foci of tagrfp-t
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Chapter 10: Classification of Microorganisms
 Chapter 10: Classification of Microorganisms 1. The Taxonomic Hierarchy 2. Methods of Identification 1. The Taxonomic Hierarchy Phylogenetic Tree of the 3 Domains Taxonomic Hierarchy 8 successive taxa
Chapter 10: Classification of Microorganisms 1. The Taxonomic Hierarchy 2. Methods of Identification 1. The Taxonomic Hierarchy Phylogenetic Tree of the 3 Domains Taxonomic Hierarchy 8 successive taxa
Autophagy/Cytotoxicity Dual Staining Kit
 Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
CytoPainter Nucleolar Staining Kit - Green Fluorescence
 ab139475 CytoPainter Nucleolar Staining Kit - Green Fluorescence Instructions for Use Specifically designed for visualizing microscopically nucleoli in living cells. This product is for research use only
ab139475 CytoPainter Nucleolar Staining Kit - Green Fluorescence Instructions for Use Specifically designed for visualizing microscopically nucleoli in living cells. This product is for research use only
Shehab. Yousef... Omar. Yousef Omar. Anas
 3 Shehab Yousef Omar Yousef... Omar Anas Bacterial Growth and Survival After discussing the structure of a Bacteria, we must know how it survive and grow in a specific media. Firstly, the survival of any
3 Shehab Yousef Omar Yousef... Omar Anas Bacterial Growth and Survival After discussing the structure of a Bacteria, we must know how it survive and grow in a specific media. Firstly, the survival of any
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 1 SUPPLEMENTARY INFORMATION 2 WWW.NATURE.COM/NATURECELLBIOLOGY SUPPLEMENTARY INFORMATION WWW.NATURE.COM/NATURECELLBIOLOGY 3 SUPPLEMENTARY INFORMATION
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
RayBio Apoptotic DNA Ladder Extraction Kit
 RayBio Apoptotic DNA Ladder Extraction Kit User Manual Version 1.1 March 1, 2016 RayBio Apoptotic DNA Ladder Extraction (Cat#: 68SO-DNAL-S50) RayBiotech, Inc. We Provide You With Excellent Support And
RayBio Apoptotic DNA Ladder Extraction Kit User Manual Version 1.1 March 1, 2016 RayBio Apoptotic DNA Ladder Extraction (Cat#: 68SO-DNAL-S50) RayBiotech, Inc. We Provide You With Excellent Support And
Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg
![Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg](/thumbs/84/90687567.jpg) Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Supplementary information Supplementary methods PCNA antibody and immunodepletion Antibodies against PCNA were previously described [1]. To deplete PCNA from Xenopus egg extracts, one volume of protein
Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy
 Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
Supplementary Materials Self-labelling enzymes as universal tags for fluorescence microscopy, superresolution microscopy and electron microscopy Viktoria Liss, Britta Barlag, Monika Nietschke and Michael
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured
 Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
Supplementary Figure 1. Co-localization of GLUT1 and DNAL4 in BeWo cells cultured under static conditions. Cells were seeded in the chamber area of the device and cultured overnight without medium perfusion.
EXPERIMENT 2 Cell Fractionation and DNA Isolation
 EXPERIMENT 2 Cell Fractionation and DNA Isolation Background Information A. The Eukaryotic Cell Cells are frequently classified into two basic types: prokaryotic and eukar yotic. The prokaryotic cell,
EXPERIMENT 2 Cell Fractionation and DNA Isolation Background Information A. The Eukaryotic Cell Cells are frequently classified into two basic types: prokaryotic and eukar yotic. The prokaryotic cell,
Apoptosis And Anti-tumor Effect Induced By Mtor Inhibitor And Autophagy Inhibitor In Human Osteosarcoma Cells
 Apoptosis And Anti-tumor Effect Induced By Mtor Inhibitor And Autophagy Inhibitor In Human Osteosarcoma Cells Ryosuke Horie. Kagawa University of medecine, Kita-gun, Japan. Disclosures: R. Horie: None.
Apoptosis And Anti-tumor Effect Induced By Mtor Inhibitor And Autophagy Inhibitor In Human Osteosarcoma Cells Ryosuke Horie. Kagawa University of medecine, Kita-gun, Japan. Disclosures: R. Horie: None.
2. Know the parts of a light microscope and general rules for using and focusing a microscope, such as:
 SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
SNC 2DI Exam Review: Biology Unit 1. Understand the meaning of the following terms. Be able to recognize their definitions: Biology Mounting medium Telophase Organelle Cell Theory Cell cycle Cytokinesis
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supporting Information
 Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
Supporting Information Deng et al. 10.1073/pnas.1515692112 SI Materials and Methods FPLC. All fusion proteins were expressed and purified through a three-step FPLC purification protocol, as described (20),
FIVEphoton Biochemicals
 Human Insulin Degrading Enzyme (IDE) ELISA Kit Biotin Detection Antibody Format 96T FIVEphoton Biochemicals. For research use only. Not for diagnostics. Part No. hide-biotin(96t) FIVEphoton Biochemicals
Human Insulin Degrading Enzyme (IDE) ELISA Kit Biotin Detection Antibody Format 96T FIVEphoton Biochemicals. For research use only. Not for diagnostics. Part No. hide-biotin(96t) FIVEphoton Biochemicals
Proteasome Activity Fluorometric Assay Kit II (Cat. # J4120)
 Proteasome Activity Fluorometric Assay Kit II (Cat. # J4120) Each supplied substrate is sufficient for use in 250 X 100 µl reactions to monitor the chymotrypsin-like (Suc-LLVY- AMC), trypsin-like (Boc-LRR-AMC)
Proteasome Activity Fluorometric Assay Kit II (Cat. # J4120) Each supplied substrate is sufficient for use in 250 X 100 µl reactions to monitor the chymotrypsin-like (Suc-LLVY- AMC), trypsin-like (Boc-LRR-AMC)
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Introduction to Cells
 Introduction to Cells Key terms: Cell Microscopy Nucleus Organelle Micrograph Cytoplasm Define: Cell Microscopy Cell structure Today, all of biology is based on an understanding of what is going on within
Introduction to Cells Key terms: Cell Microscopy Nucleus Organelle Micrograph Cytoplasm Define: Cell Microscopy Cell structure Today, all of biology is based on an understanding of what is going on within
Cells and Tissues. Overview CELLS
 Cells and Tissues WIll The basic unit of structure and function in the human body is the cell. Each of a cell's parts, or organelles, as well as the entire cell, is organized to perform a specific function.
Cells and Tissues WIll The basic unit of structure and function in the human body is the cell. Each of a cell's parts, or organelles, as well as the entire cell, is organized to perform a specific function.
Supplementary Information
 Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Supplementary Information Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells Taeko Sasaki 1, Yoshikatsu Sato 2, Tetsuya Higashiyama 1,2,
Lab Exercise 13: Growth Curve
 Lab Exercise 13: Growth Curve OBJECTIVES 1. Know the different phases of a standard growth curve. 2. Understand and perform direct measurement of bacterial growth through serial dilutions and standard
Lab Exercise 13: Growth Curve OBJECTIVES 1. Know the different phases of a standard growth curve. 2. Understand and perform direct measurement of bacterial growth through serial dilutions and standard
BIO 121 LAB 10 - DNA I
 BIO 121 LAB 10 - DNA I All cellular organisms store their hereditary information as the precise sequence of nucleotides in DNA, just as written information is stored as the precise sequence of letters
BIO 121 LAB 10 - DNA I All cellular organisms store their hereditary information as the precise sequence of nucleotides in DNA, just as written information is stored as the precise sequence of letters
Heat-resistant microalgae are good materials for algae cultivation integrating into the sustainable built environment
 Heat-resistant microalgae are good materials for algae cultivation integrating into the sustainable built environment Tzan-Chain Lee 1, Ding-Chyu Liu 2 1 Department of Life Science, National Tsing Hua
Heat-resistant microalgae are good materials for algae cultivation integrating into the sustainable built environment Tzan-Chain Lee 1, Ding-Chyu Liu 2 1 Department of Life Science, National Tsing Hua
DNA REPLICATION & BIOTECHNOLOGY Biology Study Review
 DNA REPLICATION & BIOTECHNOLOGY Biology Study Review DNA DNA is found in, in the nucleus. It controls cellular activity by regulating the production of, which includes It is a very long molecule made up
DNA REPLICATION & BIOTECHNOLOGY Biology Study Review DNA DNA is found in, in the nucleus. It controls cellular activity by regulating the production of, which includes It is a very long molecule made up
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Sperm cells are passive cargo of the pollen tube in plant fertilization
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 17079 Sperm cells are passive cargo of the pollen tube in plant fertilization Jun Zhang 1, Qingpei
ab CytoPainter ER Staining Kit Red Fluorescence
 ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
ab139482 CytoPainter ER Staining Kit Red Fluorescence Instructions for Use Designed to detect Human endoplasmic reticulum by microscopy. This product is for research use only and is not intended for diagnostic
FIVEphoton Biochemicals
 Human Cyclophilin B (CYPB) ELISA Kit Protocol Protocol for other species is identical except for dilutions of species specific standard. Use the protocol shipped with the kit for your experiment. FIVEphoton
Human Cyclophilin B (CYPB) ELISA Kit Protocol Protocol for other species is identical except for dilutions of species specific standard. Use the protocol shipped with the kit for your experiment. FIVEphoton
Cell Division. embryo: an early stage of development in organisms
 Over the past several years, a debate has been brewing over the use of stem cells. Stem cells can be used to treat certain diseases and conditions such as spinal cord injuries, diabetes, arthritis, and
Over the past several years, a debate has been brewing over the use of stem cells. Stem cells can be used to treat certain diseases and conditions such as spinal cord injuries, diabetes, arthritis, and
ATCC ANIMAL CELL CULTURE GUIDE
 ATCC ANIMAL CELL CULTURE GUIDE tips and techniques for continuous cell lines The Essentials of Life Science Research Globally Delivered Table of Contents This guide contains general technical information
ATCC ANIMAL CELL CULTURE GUIDE tips and techniques for continuous cell lines The Essentials of Life Science Research Globally Delivered Table of Contents This guide contains general technical information
The highlighted CTE Model Standards are covered in Unit 2 on diabetes.
 The highlighted CTE Model Standards are covered in Unit 2 on diabetes. A1.0 Define and assess biotechnology and recognize the diverse applications and impact on society. A1.1 Use data to explain how biotechnology
The highlighted CTE Model Standards are covered in Unit 2 on diabetes. A1.0 Define and assess biotechnology and recognize the diverse applications and impact on society. A1.1 Use data to explain how biotechnology
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
λ N -GFP: an RNA reporter system for live-cell imaging
 λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
λ N -GFP: an RNA reporter system for live-cell imaging Nathalie Daigle & Jan Ellenberg Supplementary Figures and Text: Supplementary Figure 1 localization in the cytoplasm. 4 λ N22-3 megfp-m9 serves as
Maintenance of Tobacco TBY2-AtRER1B Callus Culture
 Introduction C-TBY2-AtRER1B-005 Maintenance of Tobacco TBY2-AtRER1B Callus Culture The Tobacco TBY2-AtRER1B cell line is a transgenic BY-2 cell line expressing Green Fluorescent Protein (GFP) fused with
Introduction C-TBY2-AtRER1B-005 Maintenance of Tobacco TBY2-AtRER1B Callus Culture The Tobacco TBY2-AtRER1B cell line is a transgenic BY-2 cell line expressing Green Fluorescent Protein (GFP) fused with
LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic
 information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
information MATERIAL AND METHODS Lysosome staining LysoTracker Red DND-99 (Invitrogen) was used as a marker of lysosome or acidic compartments, according to the manufacturer s protocol. Plasmid independent
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Jan 25, 05 His Bind Kit (Novagen)
 Jan 25, 05 His Bind Kit (Novagen) (1) Prepare 5ml of 1X Charge buffer (stock is 8X= 400mM NiSO4): 0.625ml of the stock + 4.375ml DH2O. (2) Prepare 13ml of 1X Binding buffer (stock is 8X = 40mM imidazole,
Jan 25, 05 His Bind Kit (Novagen) (1) Prepare 5ml of 1X Charge buffer (stock is 8X= 400mM NiSO4): 0.625ml of the stock + 4.375ml DH2O. (2) Prepare 13ml of 1X Binding buffer (stock is 8X = 40mM imidazole,
Physical characterization and in vitro cytotoxicity screening of single-walled carbon nanotubes (SWNTs)
 Physical characterization and in vitro cytotoxicity screening of single-walled carbon nanotubes (SWNTs) Ruhung Wang, Ph.D. (ruhung.wang@utdallas.edu) Bionano Science Group University of Texas at Dallas
Physical characterization and in vitro cytotoxicity screening of single-walled carbon nanotubes (SWNTs) Ruhung Wang, Ph.D. (ruhung.wang@utdallas.edu) Bionano Science Group University of Texas at Dallas
SensoLyte AMC Calpain Activity Assay Kit *Fluorimetric*
 SensoLyte AMC Calpain Activity Assay Kit *Fluorimetric* Catalog # 72150 Kit Size 100 Assays (96-well plate) Optimized Performance: This kit is optimized to detect calpain activity Enhanced Value: It provides
SensoLyte AMC Calpain Activity Assay Kit *Fluorimetric* Catalog # 72150 Kit Size 100 Assays (96-well plate) Optimized Performance: This kit is optimized to detect calpain activity Enhanced Value: It provides
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Cell Cycle Phase Determination Kit
 Cell Cycle Phase Determination Kit Catalog Number KA1301 100 assays Version: 04 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General
Cell Cycle Phase Determination Kit Catalog Number KA1301 100 assays Version: 04 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General
MaxPar Cytoplasmic/Secreted Antigen Staining Protocol
 MaxPar Cytoplasmic/Secreted Antigen Staining Protocol CHEMICAL HAZARD: Before handling any chemicals, refer to the Material Safety Data Sheet (MSDS) provided by the manufacturer, and observe all relevant
MaxPar Cytoplasmic/Secreted Antigen Staining Protocol CHEMICAL HAZARD: Before handling any chemicals, refer to the Material Safety Data Sheet (MSDS) provided by the manufacturer, and observe all relevant
Pre-Lab: Molecular Biology
 Pre-Lab: Molecular Biology Name 1. What are the three chemical parts of a nucleotide. Draw a simple sketch to show how the three parts are arranged. 2. What are the rules of base pairing? 3. In double
Pre-Lab: Molecular Biology Name 1. What are the three chemical parts of a nucleotide. Draw a simple sketch to show how the three parts are arranged. 2. What are the rules of base pairing? 3. In double
Data Sheet IDO2 - HEK293 Recombinant Cell Line Cat #: 60533
 Data Sheet IDO2 - HEK293 Recombinant Cell Line Cat #: 60533 Description Recombinant HEK293 cell line expressing tetracycline-inducible human indoleamine 2,3- dioxygenase (IDO2), Genbank accession number
Data Sheet IDO2 - HEK293 Recombinant Cell Line Cat #: 60533 Description Recombinant HEK293 cell line expressing tetracycline-inducible human indoleamine 2,3- dioxygenase (IDO2), Genbank accession number
These molecules make up the ladder of the DNA Bound by weak hydrogen bonds. 4 Different Types (2 specific matches) look at the
 B.A.T. Review DNA & Cell Cycle Test is 11/3/16 NAME: PERIOD DNA STRUCTURE VOCABULARY Mitosis Chromosomes Interphase G1,S,G2,M Prophase Metaphase Anaphase Telophase Cytokinesis Cleavage Furrow Spindle Fibers
B.A.T. Review DNA & Cell Cycle Test is 11/3/16 NAME: PERIOD DNA STRUCTURE VOCABULARY Mitosis Chromosomes Interphase G1,S,G2,M Prophase Metaphase Anaphase Telophase Cytokinesis Cleavage Furrow Spindle Fibers
