Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman 1 *, Jerard Hurwitz 1 and Mike O Donnell 2
|
|
|
- Lambert Howard
- 6 years ago
- Views:
Transcription
1 Minireview 121 Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman 1 *, Jerard Hurwitz 1 and Mike O Donnell 2 Replicative DNA polymerases are highly processive enzymes that polymerize thousands of nucleotides without dissociating from the DNA template. The recently determined structure of the Escherichia coli bacteriophage T7 DNA polymerase suggests a unique mechanism that underlies processivity, and this mechanism may generalize to other replicative polymerases. Addresses: 1 Department of Molecular Biology, Memorial Sloan- Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA and 2 Laboratory of DNA Replication, Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA. *Corresponding author. z-kelman@ski.mskcc.org Structure 15 February 1998, 6: Current Biology Ltd ISSN DNA polymerases are a group of enzymes that use singlestranded DNA as a template for the synthesis of the complementary DNA strand. This family of enzymes plays an essential role in nucleic acid metabolism, including the processes of DNA replication, repair and recombination. DNA polymerases are ubiquitous in nature, having been identified in all cellular organisms from bacteria to humans as well as in many eukaryotic viruses and bacteriophages [1]. Furthermore, the proteins from different organisms share amino acid sequence similarities as well as a similar three-dimensional appearance (for examples see [2]). Replicative DNA polymerases, or replicases, form a subset of the DNA polymerase family. These polymerases are generally multisubunit complexes that replicate the chromosomes of cellular organisms and viruses. Another general feature of replicases is their high processivity of DNA synthesis; they are capable of polymerizing thousands of nucleotides without dissociating from the DNA template [1]. During replication of cellular chromosomes, DNA synthesis is continuous on the leading strand and discontinuous on the lagging strand. The discontinuous strand is synthesized as a series of short fragments (Okazaki fragments) that are joined later. Due to the intracellular scarcity of the replicase, the enzyme must be recycled and some specific mechanism must exist to accomplish this phenomenon. Thus, the polymerase has two apparently contradictory activities: it tightly associates with the DNA during elongation, but rapidly dissociates from DNA upon the completion of each Okazaki fragment. Processive DNA synthesis by cellular replicases and the bacteriophage T4 replicase Until recently, the only mechanism for high processivity that was understood in detail was that utilized by cellular replicases and the replicase of bacteriophage T4. This mechanism involves a ring-shaped protein called a DNA sliding clamp that encircles the DNA and tethers the polymerase catalytic unit to the DNA [3,4]. The threedimensional structures of several sliding clamps have been determined: the eukaryotic proliferating cell nuclear antigen (PCNA) [5,6]; the β subunit of the prokaryotic DNA polymerase III [7]; and the bacteriophage T4 gene 45 protein (gp45) (J Kuriyan, personal communication) (Figure 1). The overall structure of these clamps is very similar; the PCNA, β subunit and gp45 rings are superimposable [8]. Each ring has similar dimensions and a central cavity large enough to accommodate duplex DNA (Figure 1). The sliding clamp cannot assemble around DNA by itself, but must be loaded onto the DNA by a protein complex referred to as a clamp loader (Figure 2a) [9]. In prokaryotes and eukaryotes the clamp loaders are five-subunit complexes called the γ complex and replication factor C (RFC), respectively. The clamp loader recognizes the 3 end of the single-strand duplex (primer template) junction and utilizes ATP hydrolysis to assemble the clamp around the DNA primed site (Figure 2a; step I). The sliding clamp, encircling the DNA, then interacts with the polymerase for rapid and processive DNA synthesis (Figure 2a; steps II IV). Upon completion of an Okazaki fragment, the polymerase is rapidly ejected from the clamp (Figure 2a; step V), thus freeing the polymerase to associate with another clamp for the synthesis of the next Okazaki fragment. The sliding clamp is left behind assembled around the duplex DNA. During lagging strand synthesis, a new sliding clamp is needed for the synthesis of each Okazaki fragment. Ten times more Okazaki fragments are formed during replication than the number of sliding clamps present within the cell, therefore, clamps must also be recycled. In prokaryotes and eukaryotes the clamp loader has a dual function: besides its role as a clamp loader it also functions as a clamp unloader to remove used clamps from the DNA [10] (Figure 2a; step VII). Processive DNA synthesis by the bacteriophage T7 replicase The replicases of several viruses and bacteriophages utilize a different mechanism to achieve high processivity. The T7 DNA polymerase (also called gene 5 protein [gp5]) is an 80 kda protein which has low processivity and
2 122 Structure 1998, Vol 6 No 2 Figure 1 Computer generated images of two DNA sliding clamps. The figure shows ribbon representations of the polypeptide backbone of (a) a dimer of the E. coli DNA polymerase III holoenzyme β subunit and (b) a trimer of human PCNA. The strands of β sheets are shown as flat ribbons and α helices are shown as spirals. The subunits within each ring are distinguished by different colors. dissociates from the DNA after incorporation of only a few nucleotides. To become processive, T7 DNA polymerase recruits a host-encoded protein, thioredoxin (12 kda) [11,12] (Figure 2b). The T7 DNA replicase is a 1:1 heterodimer of these two subunits. Thioredoxin is not a ringshaped protein but nevertheless its association with T7 polymerase results in an ~80-fold increase in the affinity of the polymerase for the primer terminus. The resulting polymerase thioredoxin complex acts processively over several thousands of nucleotides [12]. The T7 replicase, therefore, utilizes neither a symmetric ring-shaped sliding clamp (although the T7 polymerase thioredoxin complex may function like a sliding clamp) nor a clamp loader, yet is highly processive. The structure of the large fragment of Escherichia coli DNA polymerase I (Klenow fragment), determined several years ago [13,14], contains grooves to accommodate the DNA and its structure resembles that of a right hand with thumb, palm and fingers subdomains. T7 DNA polymerase is homologous to the Klenow fragment [15] and has been hypothesized to have a similar structure. Amino acid sequence alignment of T7 DNA polymerase with the Klenow fragment suggests that the T7 enzyme contains an extra 71 amino acids in the thumb region located between α helices H and H1 of the Klenow fragment structure. It was shown biochemically that this extension interacts with thioredoxin and it was suggested that thioredoxin forms a cap over the DNA groove thus Figure 2 (a) E. coli γ complex β ADP ATP Polymerase Clamp loading I Preinitiation complex II Initiation complex III Elongation IV Termination V Orphaned β VI Clamp unloading VII VIII (b) Bacteriophage T7 Thioredoxin gp5 Initiation I Elongation II Termination III IV Structure Two mechanisms to assemble a processive polymerase. (a) The diagram illustrates the stages of DNA replication on the lagging strand by the E. coli DNA polymerase III holoenzyme. The γ complex recognizes a primer template (step I) and couples ATP hydrolysis to the assembly of the β clamp around the DNA (step II). The polymerase assembles with the β clamp (step III) to form a processive polymerase (step IV). Upon completion of an Okazaki fragment (step V) the polymerase dissociates from the clamp (step VI) leaving the β clamp around the DNA. The γ complex assembles with the clamp (step VII) and removes it from the DNA (step VIII). (b) A putative mode of action for bacteriophage T7 DNA polymerase. T7 polymerase (gp5) in complex with thioredoxin recognizes and assembles around the primer template (step I) to form a processive polymerase (step II). Upon completion of an Okazaki fragment (step III) the polymerase dissociates from the DNA (step IV).
3 Minireview T7 DNA polymerase Kelman, Hurwitz and O Donnell 123 locking the DNA inside the polymerase for processive DNA synthesis [16] (Figure 2b). Indeed mutations in the gene encoding T7 DNA polymerase (gene 5), that suppress mutations in thioredoxin, lie on the other side of the groove and could be indicative of thioredoxin binding across the groove [17]. Figure 3 These hypotheses, based on biochemical results, were recently examined with the determination of the threedimensional structure of T7 DNA polymerase in complex with thioredoxin, a primer template and a deoxynucleoside triphosphate [18]. The overall architecture of the complex is the same as that of the Klenow fragment, but the structure also shows that the palm ligates two metal ions in the polymerase active site and the incoming nucleotide is sandwiched between the fingers and the 3 end of the primer. The thumb presses on the DNA product as it exits the polymerase, creating an S-shaped bend in the DNA and helping to secure it in the active site. The structure also shows that thioredoxin, as anticipated, is associated with the extended loop of the thumb between α helices H and H1, located near the duplex DNA within the polymerase (Figure 3). Although the three-dimensional structure of the T7 polymerase (Figure 3) does not show it encircling the DNA, the structure suggests that the enzyme may be capable of opening and closing around the DNA as the biochemical results indicated. The thioredoxin is rotated away from the primer template in the crystal and thus may be in the open conformation, a prerequisite to assembling onto DNA. The thioredoxin and the loop of T7 polymerase to which it binds appear to be flexibly attached to the thumb and could swing across the DNA-binding groove to encircle the DNA. Further studies will be required to test these ideas. Advantages and disadvantages of the mechanism adopted by T7 replicase to achieve processivity The mechanism adopted by bacteriophage T7 to replicate its DNA is more economic than that used by cellular organisms. In T7 only three proteins are needed for processive DNA elongation: the polymerase (gp5), E. coli thioredoxin, and a single-stranded DNA-binding protein (SSB; gene 2.5 protein). In T7 there is no need for a sliding clamp protein or a clamp loader. However, there may be several advantages for an organism to utilize a sliding clamp which is not part of the catalytic unit. While replication is in progress on the lagging strand the clamp loader may assemble a new clamp on the primer of the next Okazaki fragment. When replication of an Okazaki fragment is completed the polymerase can rapidly recycle upon dissociating from the DNA by reassociating with the new clamp that was preassembled on the next primer. A precise regulation of DNA replication with other cellular events is essential. In eukaryotes, several cell-cycle Structure of the bacteriophage T7 DNA replicase. (a) Schematic representation in which α helices are shown as cylinders and β strands as arrows; the polymerase is shown in purple and thioredoxin is in orange. (b) Molecular surface representation of T7 polymerase in complex with thioredoxin and a primer template DNA. Proteins are colored according to electrostatic potential with regions of intense positive charge appearing blue and electronegative regions in red. The primer strand is shown in purple and the template strand is shown in yellow. In orange are 11 base pairs that are present in the crystal but are disordered. The fingers, thumb and exonuclease domains are labeled. (Figure adapted from [18] with permission.)
4 124 Structure 1998, Vol 6 No 2 regulators (e.g. p21/cip1 and Gadd45) have been shown to interact with the PCNA clamp and they affect its function during the cell cycle and DNA repair [8]. PCNA also interacts with the enzymes needed for Okazaki fragment maturation (e.g. Fen-1 and DNA ligase I) [19] and postreplication events (e.g. DNA methyltransferase) [20]. In bacteriophage T4, the gp45 sliding clamp was shown to be important for late gene transcription. In association with two other virus-encoded proteins, the gene 33 product (gp33) and gene 55 product (gp55), gp45 binds to the E. coli RNA polymerase and directs it to the promoters of the bacteriophage genes needed to be expressed in the late stages of infection [21]. The T7 replicase is a stable heterodimer. Therefore, if the replicase had a closed structure in solution, it would be expected to have a slow rate of association with the DNA template (slow k on ), which would have the net effect of slowing DNA replication. The three-dimensional structure of T7 replicase (Figure 3) suggests that the enzyme may be stable in an open conformation. The replicase complex may, therefore, assume an open structure in solution which allows it to rapidly associate with the template DNA. On the lagging strand the polymerase has to recycle between Okazaki fragments. Thioredoxin increases the affinity of the polymerase for DNA and thus may be predicted to slow its dissociation from DNA during lagging strand synthesis. It has been demonstrated, however, that whereas thioredoxin increases the intrinsic affinity of T7 polymerase for the primer template junction, it does not increase the intrinsic affinity of the polymerase for double-stranded DNA [22]. This may resolve the recycling dilemma by allowing the polymerase to dissociate from the DNA upon completing synthesis of an Okazaki fragment (C Richardson, personal communication). The nick in the DNA may also cause a structural change within the polymerase, resulting in the opening of the structure (e.g. lifting of the thioredoxin cap as depicted in Figure 2b; step III) and allowing it to transfer to a new primer. Alternatively, if the bacteriophage in infected E. coli cells synthesize sufficient numbers of polymerase molecules to satisfy the number of Okazaki fragments produced, then the polymerase need not recycle and could dissociate even very slowly from the DNA without a detrimental effect on replication. In addition, it has been proposed that the interaction between the T7 primase/ helicase protein and the polymerase at the replication fork may facilitate the dissociation and recycling of the polymerase [23]. Do other polymerases achieve processivity by a mechanism similar to that of bacteriophage T7? Can the mechanism used by bacteriophage T7 to achieve processive DNA synthesis be generalized to other polymerases? T7 DNA polymerase belongs to the E. coli DNA polymerase I family [15]. Two other members of this family contain a putative domain in a location similar to the one in T7 polymerase between helices H and H1. The polymerase of the E. coli bacteriophage T3 contains a thioredoxin-binding domain and thus may use thioredoxin as a processivity factor in a similar manner to T7. Similarly, the DNA polymerase of the Bacillus subtilis bacteriophage Spo1 also contains an insertion of 45 amino acids between α helices H and H1 [15]. This region is shorter than the one found in T3 and T7 and does not have significant similarities to the thioredoxin-binding domain. The region may interact, however, with a yet unidentified protein with the same function as thioredoxin in the T7 replicase. Bacteriophage T5 is also a member of the DNA polymerase I family. In contrast to the polymerases mentioned above, the T5 enzyme is processive by itself. Interestingly, the T5 polymerase has an extension of 75 amino acids at its C terminus [15]. In the three-dimensional structures of other members of the DNA polymerase I family, the polymerase C terminus is located near the thumb domain. It is possible that the C-terminal amino acid extension in T5 polymerase forms a domain that functions to lock the polymerase around the DNA in a manner similar to that of thioredoxin. Several eukaryotic viral DNA polymerases have also been shown to associate with a smaller protein to become processive enzymes. The 116 kda vaccinia virus DNA polymerase was shown to be a non-processive enzyme, but it becomes processive when associated with a 48 kda virally encoded protein [24]. The 136 kda polymerase of the herpes simplex virus associates with the 51 kda doublestranded DNA-binding protein UL42 for high processivity [25]. Similarly, the DNA-binding protein (DBP) of adenovirus (59 kda protein) stimulates the processivity of the adenovirus DNA polymerase (140 kda) [26]. The 125 kda mitochondrial DNA polymerase γ is a processive enzyme and has been shown to be associated with a 35 kda protein [27]. This small protein may serve as a processivity factor for the polymerase. In all these examples, the polymerase and its associated protein may achieve processivity by a mechanism similar to that of the T7 polymerase thioredoxin complex. DNA polymerases and sliding clamps are not the only proteins that encircle DNA. Like DNA polymerases, RNA polymerases are highly processive enzymes. Based on the structural analysis of RNA polymerases from E. coli and yeast, it was proposed that these enzymes encircle the DNA template to anchor the polymerase to the DNA [28]. Furthermore, several other enzymes involved in DNA metabolic processes (e.g. DNA helicases) have been shown to form ring-shaped structures that encircle DNA [29]. Concluding remarks The two mechanisms known to date by which replicative DNA polymerases achieve their high processivity involve
5 Minireview T7 DNA polymerase Kelman, Hurwitz and O Donnell 125 encircling the DNA. The tools used in these mechanisms are different, however. Cellular replicases and bacteriophage T4 utilize a ring-shaped processivity factor that encircles the DNA but is not a part of the catalytic unit. Bacteriophage T7 polymerase, on the other hand, forms a structure that might surround the DNA like a sliding clamp. Although the general theme of surrounding the DNA may be common to all replicative DNA polymerases, it may be achieved by different means in different organisms. It will be of great interest to determine whether different structures are employed by other eukaryotic viruses and bacteriophages to anchor their polymerase to the DNA. Future structural studies of replicases and other proteins involved in chromosome replication are sure to bring to light new and exiting mechanisms for handling the DNA helix. Acknowledgements We are grateful to David Shechter for his contribution to Figure 1 and Sylvie Doublié and Tom Ellenberger for assistance in providing Figure 3. We wish to thank Tom Ellenberger, Daochun Kong, and Charles Richardson for their comments on the manuscript. We would like to apologize to colleagues whose primary work has not been cited due to space limitations. ZK is a postdoctoral fellow of the Helen Hay Whitney Foundation. JH is a Professor of the American Cancer Society. This work was supported by grants GM38839 (MO D) and GM34559 (JH). References 1. Kornberg, A. & Baker, T.A. (1991). DNA Replication. WH Freeman, New York, NY. 2. Wang, J., Sattar, A.K.M.A., Wang, C.C., Karam, J.D., Konigsberg, W.H. & Steitz, T.A. (1997). Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell 89, Stukenberg, P.T., Studwell-Vaughan, P.S. & O Donnell, M. (1991). Mechanism of the β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 266, Kuriyan, J. & O Donnell, M. (1993). Sliding clamps of DNA polymerases. J. Mol. Biol. 234, Krishna, T.S.R., Kong, X.-P., Gary, S., Burgers, P.M. & Kuriyan, J. (1994). Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, Gulbis, J.M., Kelman, Z., Hurwitz, J., O Donnell, M. & Kuriyan, J. (1996). Structure of the C-terminal region of p21 WAF1/CIP1 complexed with human PCNA. Cell 87, Kong, X.-P., Onrust, R., O Donnell, M. & Kuriyan, J. (1992). Three dimensional structure of the β subunit of Escherichia coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69, Kelman, Z. & O Donnell, M. (1995). Structural and functional similarities of prokaryotic and eukaryotic sliding clamps. Nucleic Acids Res. 23, Kelman, Z. & O Donnell, M. (1994). DNA replication enzymology and mechanisms. Curr. Opin. Genet. Dev. 4, Yao, N., et al., & O Donnell, M. (1996). Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes to Cell 1, Richardson, C.C. (1983). Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell 33, Nakai, H., Beauchamp, B.B., Bernstein, J., Huber, H.E., Tabor, S. & Richardson, C.C. (1988). Formation and propagation of the bacteriophage T7 replication fork. In DNA Replication and Mutagenesis. (Moses, R.E. & Summers, W.C., eds), pp , American Society for Microbiology, Washington, DC. 13. Ollis, D.L., Hamlin, B.R., Xuong, N.G. & Steitz, T.A. (1985). Structure of large fragment of Escherichia coli DNA polymerase I complexed with dtmp. Nature 313, Beese, L.S., Derbyshire, V. & Steitz, T.A. (1993). Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science 260, Braithwaite, D.K. & Ito, J. (1993). Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21, Bedford, E., Tabor, S. & Richardson, C.C. (1997). The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. USA 94, Himawan, J.S. & Richardson, R.R. (1996). Amino acid residues critical for the interaction between bacteriophage T7 DNA polymerase and Escherichia coli thioredoxin. J. Biol. Chem. 271, Doublié, S., Tabor, S., Long, A.M., Richardson, C.C. & Ellenberger, T. (1998). Crystal structure of bacteriophage T7 DNA polymerase complexed to a primer-template, a nucleoside triphosphate, and its processivity factor thioredoxin. Nature 391, Levin, D.S., Bai, W., Yao, N., O Donnell, M. & Tomkinson, A.E. (1997). An interaction between DNA ligase I and proliferating cell nuclear antigen: implication for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. USA 94, Chuang, L.S.-H., Ian, H.-I., Koh, T.-W., Ng, H.-H., Xu, G. & Li, B.F.L. (1997). Human DNA-(cytosine-5) methyltransferase-pcna complex as a target for p21 waf1. Science 277, Geiduschek, E.P. (1995). Connecting a viral DNA replication apparatus with gene expression. Semin. Virol. 6, Huber, H.E., Tabor, S. & Richardson, C.C. (1987). Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 262, Debyser, Z., Tabor, S. & Richardson, C.C. (1994). Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell 77, McDonald, W.F., Klemperer, N. & Traktman, P. (1997). Characterization of a processive form of the vaccinia virus DNA polymerase. Virology 234, Boehmer, P.E. & Lehman, I.R. (1997). Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66, Hay, R.T. (1996). Adenovirus DNA replication. In DNA Replication in Eukaryotic Cells. (DePamphilis, M.L., ed), pp , CSH Laboratory Press, Cold Spring Harbor. 27. Olson, M.W., Wang, Y., Elder, R.H. & Kaguni, L.S. (1995). Subunit structure of mitochondrial DNA polymerase from Drosophila embryos. Physical and immunological studies. J. Biol. Chem. 270, Polyakov, A., Severinova, E. & Darst, S.A. (1995). Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell 83, Hingorani, M.M. & O Donnell, M. (1998). Toroidal proteins: running rings around DNA. Curr. Biol. 8, R83 R86.
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
DNA Replication II Biochemistry 302. Bob Kelm January 28, 2004
 DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
Fidelity of DNA polymerase
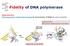 Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Questions from chapters in the textbook that are relevant for the final exam
 Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Polymerase III Holoenzyme
 Molecular Biology of the Cell Vol. 3, 953-957, September 1992 The Sliding Clamp of DNA Polymerase III Holoenzyme Encircles DNA Mike O'Donnell,*t John Kuriyan,* Xiang-Peng Kong,* P. Todd Stukenberg,* and
Molecular Biology of the Cell Vol. 3, 953-957, September 1992 The Sliding Clamp of DNA Polymerase III Holoenzyme Encircles DNA Mike O'Donnell,*t John Kuriyan,* Xiang-Peng Kong,* P. Todd Stukenberg,* and
Biochemistry. DNA Polymerase. Structure and Function of Biomolecules II. Principal Investigator
 Paper : 03 Module: 14 Principal Investigator Paper Coordinator and Content Writer Dr. Sunil Kumar Khare, Professor, Department of Chemistry, IIT-Delhi Dr. Sunil Kumar Khare, Professor, Department of Chemistry,
Paper : 03 Module: 14 Principal Investigator Paper Coordinator and Content Writer Dr. Sunil Kumar Khare, Professor, Department of Chemistry, IIT-Delhi Dr. Sunil Kumar Khare, Professor, Department of Chemistry,
DNA Replication II Biochemistry 302. January 25, 2006
 DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
DNA Replication II Biochemistry 302. Bob Kelm January 26, 2005
 DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication I Biochemistry 302. Bob Kelm January 24, 2005
 DNA Replication I Biochemistry 302 Bob Kelm January 24, 2005 Watson Crick prediction: Each stand of parent DNA serves as a template for synthesis of a new complementary daughter strand Fig. 4.12 Proof
DNA Replication I Biochemistry 302 Bob Kelm January 24, 2005 Watson Crick prediction: Each stand of parent DNA serves as a template for synthesis of a new complementary daughter strand Fig. 4.12 Proof
Molecular Biology (2)
 Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
The flow of Genetic information
 The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
Enzymes used in DNA Replication
 Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function. Gary Peter
 Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
Name 10 Molecular Biology of the Gene Test Date Study Guide You must know: The structure of DNA. The major steps to replication.
 Name 10 Molecular Biology of the Gene Test Date Study Guide You must know: The structure of DNA. The major steps to replication. The difference between replication, transcription, and translation. How
Name 10 Molecular Biology of the Gene Test Date Study Guide You must know: The structure of DNA. The major steps to replication. The difference between replication, transcription, and translation. How
IDTutorial: DNA Replication
 IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
Storage and Expression of Genetic Information
 Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA
 RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
Replication of DNA Virus Genomes. Lecture 7 Virology W3310/4310 Spring 2013
 Replication of DNA Virus Genomes Lecture 7 Virology W3310/4310 Spring 2013 It s all about Initiation Problems faced by DNA replication machinery Viruses must replicate their genomes to make new progeny
Replication of DNA Virus Genomes Lecture 7 Virology W3310/4310 Spring 2013 It s all about Initiation Problems faced by DNA replication machinery Viruses must replicate their genomes to make new progeny
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 11 DNA Replication
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
The replication forks Summarising what we know:
 When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
The Size and Packaging of Genomes
 DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Metabolism. I. DNA Replication. A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding
 DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
All This For Four Letters!?! DNA and Its Role in Heredity
 All This For Four Letters!?! DNA and Its Role in Heredity What Is the Evidence that the Gene Is DNA? By the 1920s, it was known that chromosomes consisted of DNA and proteins. A new dye stained DNA and
All This For Four Letters!?! DNA and Its Role in Heredity What Is the Evidence that the Gene Is DNA? By the 1920s, it was known that chromosomes consisted of DNA and proteins. A new dye stained DNA and
DNA replication: Enzymes link the aligned nucleotides by phosphodiester bonds to form a continuous strand.
 DNA replication: Copying genetic information for transmission to the next generation Occurs in S phase of cell cycle Process of DNA duplicating itself Begins with the unwinding of the double helix to expose
DNA replication: Copying genetic information for transmission to the next generation Occurs in S phase of cell cycle Process of DNA duplicating itself Begins with the unwinding of the double helix to expose
Genetic Information: DNA replication
 Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Gene Expression- Protein Synthesis
 Gene Expression- Protein Synthesis Protein synthesis is the key to expression of biological information - Structural Protein: bones, cartilage - Contractile Proteins: myosin, actin - Enzymes - Transport
Gene Expression- Protein Synthesis Protein synthesis is the key to expression of biological information - Structural Protein: bones, cartilage - Contractile Proteins: myosin, actin - Enzymes - Transport
DNA: Structure & Replication
 DNA Form & Function DNA: Structure & Replication Understanding DNA replication and the resulting transmission of genetic information from cell to cell, and generation to generation lays the groundwork
DNA Form & Function DNA: Structure & Replication Understanding DNA replication and the resulting transmission of genetic information from cell to cell, and generation to generation lays the groundwork
DNA Replication in Eukaryotes
 OpenStax-CNX module: m44517 1 DNA Replication in Eukaryotes OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
OpenStax-CNX module: m44517 1 DNA Replication in Eukaryotes OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
Viral DNA replication
 Viral DNA replication Lecture 8 Biology 3310/4310 Virology Spring 2017 The more the merrier --ANONYMOUS Viral DNA genomes must be replicated to make new progeny Parvovirus Retrovirus Poliovirus VII Hepatitis
Viral DNA replication Lecture 8 Biology 3310/4310 Virology Spring 2017 The more the merrier --ANONYMOUS Viral DNA genomes must be replicated to make new progeny Parvovirus Retrovirus Poliovirus VII Hepatitis
Student name ID # Second Mid Term Exam, Biology 2020, Spring 2002 Scores Total
 Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Replication. Obaidur Rahman
 Replication Obaidur Rahman DIRCTION OF DNA SYNTHESIS How many reactions can a DNA polymerase catalyze? So how many reactions can it catalyze? So 4 is one answer, right, 1 for each nucleotide. But what
Replication Obaidur Rahman DIRCTION OF DNA SYNTHESIS How many reactions can a DNA polymerase catalyze? So how many reactions can it catalyze? So 4 is one answer, right, 1 for each nucleotide. But what
DNA replication. - proteins for initiation of replication; - proteins for polymerization of nucleotides.
 DNA replication Replication represents the duplication of the genetic information encoded in DNA that is the crucial step in the reproduction of living organisms and the growth of multicellular organisms.
DNA replication Replication represents the duplication of the genetic information encoded in DNA that is the crucial step in the reproduction of living organisms and the growth of multicellular organisms.
Proposed Models of DNA Replication. Conservative Model. Semi-Conservative Model. Dispersive model
 5.2 DNA Replication Cell Cycle Life cycle of a cell Cells can reproduce Daughter cells receive an exact copy of DNA from parent cell DNA replication happens during the S phase Proposed Models of DNA Replication
5.2 DNA Replication Cell Cycle Life cycle of a cell Cells can reproduce Daughter cells receive an exact copy of DNA from parent cell DNA replication happens during the S phase Proposed Models of DNA Replication
Delve AP Biology Lecture 7: 10/30/11 Melissa Ko and Anne Huang
 Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
Today s Agenda: I. DNA Structure II. DNA Replication III. DNA Proofreading and Repair IV. The Central Dogma V. Transcription VI. Post-transcriptional Modifications Delve AP Biology Lecture 7: 10/30/11
Requirements for the Genetic Material
 Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
3.A.1 DNA and RNA: Structure and Replication
 3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
M1 - Biochemistry. Nucleic Acid Structure II/Transcription I
 M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
DNA metabolism. DNA Replication DNA Repair DNA Recombination
 DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
Figure A summary of spontaneous alterations likely to require DNA repair.
 DNA Damage Figure 5-46. A summary of spontaneous alterations likely to require DNA repair. The sites on each nucleotide that are known to be modified by spontaneous oxidative damage (red arrows), hydrolytic
DNA Damage Figure 5-46. A summary of spontaneous alterations likely to require DNA repair. The sites on each nucleotide that are known to be modified by spontaneous oxidative damage (red arrows), hydrolytic
NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses)
 NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses) Consist of chemically linked sequences of nucleotides Nitrogenous base Pentose-
NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses) Consist of chemically linked sequences of nucleotides Nitrogenous base Pentose-
The highly processive DNA polymerases that replicate cellular
 Colloquium Interaction of the sliding clamp with MutS, ligase, and DNA polymerase I Francisco J. López de Saro* and Mike O Donnell Howard Hughes Medical Institute and The Rockefeller University, 1230 York
Colloquium Interaction of the sliding clamp with MutS, ligase, and DNA polymerase I Francisco J. López de Saro* and Mike O Donnell Howard Hughes Medical Institute and The Rockefeller University, 1230 York
Chapter Fundamental Molecular Genetic Mechanisms
 Chapter 5-2 - Fundamental Molecular Genetic Mechanisms 5.1 Structure of Nucleic Acids 5.2 Transcription of Protein-Coding Genes and Formation of Fun ctional mrna 5.3 The Decoding of mrna by trnas 5.4 Stepwise
Chapter 5-2 - Fundamental Molecular Genetic Mechanisms 5.1 Structure of Nucleic Acids 5.2 Transcription of Protein-Coding Genes and Formation of Fun ctional mrna 5.3 The Decoding of mrna by trnas 5.4 Stepwise
The 5 end problem. 3 5 Now what? RNA primers. DNA template. elongate. excise primers, elongate, ligate
 1 2 3 DNA Replication Viruses must replicate their genomes to make new progeny This always requires expression of at least one virus protein, sometimes many (hence always delayed after infection) DNA is
1 2 3 DNA Replication Viruses must replicate their genomes to make new progeny This always requires expression of at least one virus protein, sometimes many (hence always delayed after infection) DNA is
DNA, RNA, Replication and Transcription
 Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College DNA, RNA, Replication and Transcription The metabolic processes described earlier (glycolysis, cellular respiration, photophosphorylation,
Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College DNA, RNA, Replication and Transcription The metabolic processes described earlier (glycolysis, cellular respiration, photophosphorylation,
Chapter Twelve: DNA Replication and Recombination
 This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
Replisome mechanics: insights into a twin DNA polymerase machine
 Review TRENDS in Microbiology Vol.15 No.4 Molecular Machines of the Cell Replisome mechanics: insights into a twin DNA polymerase machine Richard T. Pomerantz and Mike O Donnell Rockefeller University,
Review TRENDS in Microbiology Vol.15 No.4 Molecular Machines of the Cell Replisome mechanics: insights into a twin DNA polymerase machine Richard T. Pomerantz and Mike O Donnell Rockefeller University,
BIO 311C Spring Lecture 34 Friday 23 Apr.
 BIO 311C Spring 2010 1 Lecture 34 Friday 23 Apr. Summary of DNA Replication in Prokaryotes origin of replication initial double helix origin of replication new growing polynucleotide chains Circular molecule
BIO 311C Spring 2010 1 Lecture 34 Friday 23 Apr. Summary of DNA Replication in Prokaryotes origin of replication initial double helix origin of replication new growing polynucleotide chains Circular molecule
Chapter 16 The Molecular Basis of Inheritance
 Chapter 16 The Molecular Basis of Inheritance Chromosomes and DNA Morgan s experiments with Drosophila were able to link hereditary factors to specific locations on chromosomes. The double-helical model
Chapter 16 The Molecular Basis of Inheritance Chromosomes and DNA Morgan s experiments with Drosophila were able to link hereditary factors to specific locations on chromosomes. The double-helical model
DNA Model Stations. For the following activity, you will use the following DNA sequence.
 Name: DNA Model Stations DNA Replication In this lesson, you will learn how a copy of DNA is replicated for each cell. You will model a 2D representation of DNA replication using the foam nucleotide pieces.
Name: DNA Model Stations DNA Replication In this lesson, you will learn how a copy of DNA is replicated for each cell. You will model a 2D representation of DNA replication using the foam nucleotide pieces.
III. Detailed Examination of the Mechanism of Replication A. Initiation B. Priming C. Elongation D. Proofreading and Termination
 Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
RNA synthesis/transcription I Biochemistry 302. February 6, 2004 Bob Kelm
 RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
MBioS 503: Section 1 Chromosome, Gene, Translation, & Transcription. Gene Organization. Genome. Objectives: Gene Organization
 Overview & Recap of Molecular Biology before the last two sections MBioS 503: Section 1 Chromosome, Gene, Translation, & Transcription Gene Organization Joy Winuthayanon, PhD School of Molecular Biosciences
Overview & Recap of Molecular Biology before the last two sections MBioS 503: Section 1 Chromosome, Gene, Translation, & Transcription Gene Organization Joy Winuthayanon, PhD School of Molecular Biosciences
Essential Question. What is the structure of DNA, and how does it function in genetic inheritance?
 DNA Dr. Bertolotti Essential Question What is the structure of DNA, and how does it function in genetic inheritance? What is the role of DNA in hereditary? Transformation Transformation is the process
DNA Dr. Bertolotti Essential Question What is the structure of DNA, and how does it function in genetic inheritance? What is the role of DNA in hereditary? Transformation Transformation is the process
DNA REPLICATION. DNA structure. Semiconservative replication. DNA structure. Origin of replication. Replication bubbles and forks.
 DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
Zoo-342 Molecular biology Lecture 2. DNA replication
 Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
CH 4 - DNA. DNA = deoxyribonucleic acid. DNA is the hereditary substance that is found in the nucleus of cells
 CH 4 - DNA DNA is the hereditary substance that is found in the nucleus of cells DNA = deoxyribonucleic acid» its structure was determined in the 1950 s (not too long ago).» scientists were already investigating
CH 4 - DNA DNA is the hereditary substance that is found in the nucleus of cells DNA = deoxyribonucleic acid» its structure was determined in the 1950 s (not too long ago).» scientists were already investigating
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI REPLICATION - ENZYMES.
 ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
Chapter 16 DNA: The Genetic Material. The Nature of Genetic Material. Chemical Nature of Nucleic Acids. Chromosomes - DNA and protein
 Chapter 16 DNA: The Genetic Material The Nature of Genetic Material Chromosomes - DNA and protein Genes are subunits DNA = 4 similar nucleotides C(ytosine) A(denine) T(hymine) G(uanine) Proteins = 20 different
Chapter 16 DNA: The Genetic Material The Nature of Genetic Material Chromosomes - DNA and protein Genes are subunits DNA = 4 similar nucleotides C(ytosine) A(denine) T(hymine) G(uanine) Proteins = 20 different
DNA Replication AP Biology
 DNA Replication 2007-2008 Watson and Crick 1953 1953 article in Nature Directionality of DNA You need to number the carbons! u it matters! u 3 refers to the 3 carbon on the sugar u 5 refers to the 5 carbon
DNA Replication 2007-2008 Watson and Crick 1953 1953 article in Nature Directionality of DNA You need to number the carbons! u it matters! u 3 refers to the 3 carbon on the sugar u 5 refers to the 5 carbon
Nucleic Acid Structure:
 Nucleic Acid Structure: Purine and Pyrimidine nucleotides can be combined to form nucleic acids: 1. Deoxyribonucliec acid (DNA) is composed of deoxyribonucleosides of! Adenine! Guanine! Cytosine! Thymine
Nucleic Acid Structure: Purine and Pyrimidine nucleotides can be combined to form nucleic acids: 1. Deoxyribonucliec acid (DNA) is composed of deoxyribonucleosides of! Adenine! Guanine! Cytosine! Thymine
DNA REPLICATION. Third Stage. Lec. 12 DNA Replication. Lecture No.: 12. A. Watson & Crick (1952) C. Cairns (1963) autoradiographic experiment
 Lec. 12 DNA Replication A. Watson & Crick (1952) Proposed a model where hydrogen bonds break, the two strands separate, and DNA synthesis occurs semi-conservatively in the same net direction. While a straightforward
Lec. 12 DNA Replication A. Watson & Crick (1952) Proposed a model where hydrogen bonds break, the two strands separate, and DNA synthesis occurs semi-conservatively in the same net direction. While a straightforward
Expression of the genome. Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell
 Expression of the genome Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell 1 Transcription 1. Francis Crick (1956) named the flow of information from DNA RNA protein the
Expression of the genome Books: 1. Molecular biology of the gene: Watson et al 2. Genetics: Peter J. Russell 1 Transcription 1. Francis Crick (1956) named the flow of information from DNA RNA protein the
Answers to Module 1. An obligate aerobe is an organism that has an absolute requirement of oxygen for growth.
 Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
Answers to Module 1 Short Answers 1) What is an obligate aerobe? An obligate aerobe is an organism that has an absolute requirement of oxygen for growth. What about facultative anaerobe? 2) Distinguish
CELL BIOLOGY: DNA. Generalized nucleotide structure: NUCLEOTIDES: Each nucleotide monomer is made up of three linked molecules:
 BIOLOGY 12 CELL BIOLOGY: DNA NAME: IMPORTANT FACTS: Nucleic acids are organic compounds found in all living cells and viruses. Two classes of nucleic acids: 1. DNA = ; found in the nucleus only. 2. RNA
BIOLOGY 12 CELL BIOLOGY: DNA NAME: IMPORTANT FACTS: Nucleic acids are organic compounds found in all living cells and viruses. Two classes of nucleic acids: 1. DNA = ; found in the nucleus only. 2. RNA
Welcome to Class 18! Lecture 18: Outline and Objectives. Replication is semiconservative! Replication: DNA DNA! Introductory Biochemistry!
 Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
DNA Replication. DNA Replication Prof. Smita Patel. DNA replication. Chemical composition of DNA. Smita Patel
 DA Replication Smita Patel UMDJ-Robert Wood Johnson Medical School 1 DA replication DA, the genetic material of the living cell needs to be duplicated every time the cell divides 2 Chemical composition
DA Replication Smita Patel UMDJ-Robert Wood Johnson Medical School 1 DA replication DA, the genetic material of the living cell needs to be duplicated every time the cell divides 2 Chemical composition
Recitation CHAPTER 9 DNA Technologies
 Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
Recitation CHAPTER 9 DNA Technologies DNA Cloning: General Scheme A cloning vector and eukaryotic chromosomes are separately cleaved with the same restriction endonuclease. (A single chromosome is shown
DNA stands for deoxyribose nucleic acid
 DNA DNA stands for deoxyribose nucleic acid This chemical substance is present in the nucleus of all cells in all living organisms DNA controls all the chemical changes which take place in cells DNA Structure
DNA DNA stands for deoxyribose nucleic acid This chemical substance is present in the nucleus of all cells in all living organisms DNA controls all the chemical changes which take place in cells DNA Structure
The Structure of DNA
 The Structure of DNA Questions to Ponder 1) How is the genetic info copied? 2) How does DNA store the genetic information? 3) How is the genetic info passed from generation to generation? The Structure
The Structure of DNA Questions to Ponder 1) How is the genetic info copied? 2) How does DNA store the genetic information? 3) How is the genetic info passed from generation to generation? The Structure
5 -GAC-3 5 -GTC-3 5 -CAG Which of these are NOT important for RNA Polymerase interacting with DNA?
 Name This exam is schedule for 75 minutes and I anticipate it to take the full time allotted. You are free to leave if you finish. The exam is split into two sections. Part 1 is multiple choice select
Name This exam is schedule for 75 minutes and I anticipate it to take the full time allotted. You are free to leave if you finish. The exam is split into two sections. Part 1 is multiple choice select
Fundamental Process. Unit Map. Unit
 Unit 3 Unit Map 3.A DNA replication, repair and recombination 120 3.B RNA synthesis and processing 137 3.C Protein synthesis and processing 151 3.D Control of gene expression at transcription and translation
Unit 3 Unit Map 3.A DNA replication, repair and recombination 120 3.B RNA synthesis and processing 137 3.C Protein synthesis and processing 151 3.D Control of gene expression at transcription and translation
DNA Replication AP Biology
 DNA Replication 2007-2008 Watson and Crick 1953 article in Nature Double helix structure of DNA It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible
DNA Replication 2007-2008 Watson and Crick 1953 article in Nature Double helix structure of DNA It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible
Biochemistry 302, February 11, 2004 Exam 1 (100 points) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed DNA
 1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
1 Biochemistry 302, February 11, 2004 Exam 1 (100 points) Name I. Structural recognition (very short answer, 2 points each) 1. What form of DNA is shown on this Nature Genetics cover? Z-DNA or left-handed
DNA Replication AP Biology
 DNA Replication 2007-2008 Double helix structure of DNA It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
DNA Replication 2007-2008 Double helix structure of DNA It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
4) separates the DNA strands during replication a. A b. B c. C d. D e. E. 5) covalently connects segments of DNA a. A b. B c. C d. D e.
 1) Chargaff's analysis of the relative base composition of DNA was significant because he was able to show that a. the relative proportion of each of the four bases differs from species to species. b.
1) Chargaff's analysis of the relative base composition of DNA was significant because he was able to show that a. the relative proportion of each of the four bases differs from species to species. b.
DNA is the genetic material. DNA structure. Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test
 DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
University of Groningen. Single-molecule studies of DNA replication Geertsema, Hylkje
 University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen. Single-molecule studies of DNA replication Geertsema, Hylkje
 University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
University of Groningen Single-molecule studies of DNA replication Geertsema, Hylkje IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please
Chapter 12. DNA Replication and Recombination
 Chapter 12 DNA Replication and Recombination I. DNA replication Three possible modes of replication A. Conservative entire original molecule maintained B. Semiconservative one strand is template for new
Chapter 12 DNA Replication and Recombination I. DNA replication Three possible modes of replication A. Conservative entire original molecule maintained B. Semiconservative one strand is template for new
AP Biology Chapter 16 Notes:
 AP Biology Chapter 16 Notes: I. Chapter 16: The Molecular Basis of Inheritance a. Overview: i. April 1953 James Watson and Francis Crick great the double helix model of DNA- deoxyribonucleic acid ii. DNA
AP Biology Chapter 16 Notes: I. Chapter 16: The Molecular Basis of Inheritance a. Overview: i. April 1953 James Watson and Francis Crick great the double helix model of DNA- deoxyribonucleic acid ii. DNA
Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology
 Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology - The central dogma (principle) of molecular biology: Information from DNA are transcribed to mrna which will be further translated to synthesize
Unit IX Problem 3 Genetics: Basic Concepts in Molecular Biology - The central dogma (principle) of molecular biology: Information from DNA are transcribed to mrna which will be further translated to synthesize
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
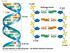 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
BCMB Chapters 34 & 35 DNA Replication and Repair
 BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
BCMB Chapters 34 & 35 DNA Replication and Repair
 BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
Chapter 30. Replication. Meselson Stahl Experiment. BCH 4054 Chapter 30 Lecture Notes. Slide 1. Slide 2 Conceptual Mechanism of.
 BCH 4054 Chapter 30 Lecture Notes 1 Chapter 30 DNA Replication and Repair 2 Conceptual Mechanism of Replication Strand separation, with copying of each strand by Watson-Crick base pairing Fig 30.2 Three
BCH 4054 Chapter 30 Lecture Notes 1 Chapter 30 DNA Replication and Repair 2 Conceptual Mechanism of Replication Strand separation, with copying of each strand by Watson-Crick base pairing Fig 30.2 Three
Feedback D. Incorrect! No, although this is a correct characteristic of RNA, this is not the best response to the questions.
 Biochemistry - Problem Drill 23: RNA No. 1 of 10 1. Which of the following statements best describes the structural highlights of RNA? (A) RNA can be single or double stranded. (B) G-C pairs have 3 hydrogen
Biochemistry - Problem Drill 23: RNA No. 1 of 10 1. Which of the following statements best describes the structural highlights of RNA? (A) RNA can be single or double stranded. (B) G-C pairs have 3 hydrogen
Chapter 12: Molecular Biology of the Gene
 Biology Textbook Notes Chapter 12: Molecular Biology of the Gene p. 214-219 The Genetic Material (12.1) - Genetic Material must: 1. Be able to store information that pertains to the development, structure,
Biology Textbook Notes Chapter 12: Molecular Biology of the Gene p. 214-219 The Genetic Material (12.1) - Genetic Material must: 1. Be able to store information that pertains to the development, structure,
Bio 366: Biological Chemistry II Test #3, 100 points
 Bio 366: Biological Chemistry II Test #3, 100 points READ THIS: Take a numbered test and sit in the seat with that number on it. Remove the numbered sticker from the desk, and stick it on the back of the
Bio 366: Biological Chemistry II Test #3, 100 points READ THIS: Take a numbered test and sit in the seat with that number on it. Remove the numbered sticker from the desk, and stick it on the back of the
Chapter 9. Topics - Genetics - Flow of Genetics - Regulation - Mutation - Recombination
 Chapter 9 Topics - Genetics - Flow of Genetics - Regulation - Mutation - Recombination 1 Genetics Genome Chromosome Gene Protein Genotype Phenotype 2 Terms and concepts gene Fundamental unit of heredity
Chapter 9 Topics - Genetics - Flow of Genetics - Regulation - Mutation - Recombination 1 Genetics Genome Chromosome Gene Protein Genotype Phenotype 2 Terms and concepts gene Fundamental unit of heredity
 www.iqeducation.in Name : Roll No. :.... Invigilator s Signature :.. CS/B.Sc(H)/MOL.BIO/SEM-3/GNO-304/2012-13 2012 GENOME ORGANIZATION Time Allotted : 3 Hours Full Marks : 70 The figures in the margin
www.iqeducation.in Name : Roll No. :.... Invigilator s Signature :.. CS/B.Sc(H)/MOL.BIO/SEM-3/GNO-304/2012-13 2012 GENOME ORGANIZATION Time Allotted : 3 Hours Full Marks : 70 The figures in the margin
Genetic material must be able to:
 Genetic material must be able to: Contain the information necessary to construct an entire organism Pass from parent to offspring and from cell to cell during cell division Be accurately copied Account
Genetic material must be able to: Contain the information necessary to construct an entire organism Pass from parent to offspring and from cell to cell during cell division Be accurately copied Account
Chapter 8. Microbial Genetics. Lectures prepared by Christine L. Case. Copyright 2010 Pearson Education, Inc.
 Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
Chapter 8 Microbial Genetics Lectures prepared by Christine L. Case Structure and Function of Genetic Material Learning Objectives 8-1 Define genetics, genome, chromosome, gene, genetic code, genotype,
DNA Replication semiconservative replication conservative replication dispersive replication DNA polymerase
 DNA Replication DNA Strands are templates for DNA synthesis: Watson and Crick suggested that the existing strands of DNA served as a template for the producing of new strands, with bases being added to
DNA Replication DNA Strands are templates for DNA synthesis: Watson and Crick suggested that the existing strands of DNA served as a template for the producing of new strands, with bases being added to
BIOLOGY 101. CHAPTER 16: The Molecular Basis of Inheritance: Life s Operating Instructions
 BIOLOGY 101 CHAPTER 16: The Molecular Basis of Inheritance: Life s Operating Instructions Life s Operating Instructions CONCEPTS: 16.1 DNA is the genetic material 16.2 Many proteins work together in DNA
BIOLOGY 101 CHAPTER 16: The Molecular Basis of Inheritance: Life s Operating Instructions Life s Operating Instructions CONCEPTS: 16.1 DNA is the genetic material 16.2 Many proteins work together in DNA
Molecular Biology, Lecture 3 DNA Replication
 Molecular Biology, Lecture 3 DNA Replication We will continue talking about DNA replication. We have previously t discussed the structure of DNA. DNA replication is the copying of the whole DNA content
Molecular Biology, Lecture 3 DNA Replication We will continue talking about DNA replication. We have previously t discussed the structure of DNA. DNA replication is the copying of the whole DNA content
Flow of Genetic Information
 Flow of Genetic Information DNA Replication Links to the Next Generation Standards Scientific and Engineering Practices: Asking Questions (for science) and Defining Problems (for engineering) Developing
Flow of Genetic Information DNA Replication Links to the Next Generation Standards Scientific and Engineering Practices: Asking Questions (for science) and Defining Problems (for engineering) Developing
