Jana Voigt b, Jun-An Chen c, Mike Gilchrist a, Enrique Amaya c, Nancy Papalopulu b, *
|
|
|
- Thomas Harrell
- 6 years ago
- Views:
Transcription
1 Mechanisms of Development 122 (2005) Expression cloning screening of a unique and full-length set of cdna clones is an efficient method for identifying genes involved in Xenopus neurogenesis Jana Voigt b, Jun-An Chen c, Mike Gilchrist a, Enrique Amaya c, Nancy Papalopulu b, * a Wellcome Trust/Cancer Research UK Gurdon Institute, Tennis Court Road, Cambridge CB2 1QR, UK b Department of Anatomy, University of Cambridge, Downing Site, Cambridge CB2 3DY, UK c Department of Zoology, University of Cambridge, Downing Street, Cambridge CB2 3EJ, UK Received 28 October 2004; accepted 4 November 2004 Abstract Functional screens, where a large numbers of cdna clones are assayed for certain biological activity, are a useful tool in elucidating gene function. In Xenopus, gain of function screens are performed by pool screening, whereby RNA transcribed in vitro from groups of cdna clones, ranging from thousands to a hundred, are injected into early embryos. Once an activity is detected in a pool, the active clone is identified by sib-selection. Such screens are intrinsically biased towards potent genes, whose RNA is active at low quantities. To improve the sensitivity and efficiency of a gain of function screen we have bioinformatically processed an arrayed and EST sequenced set of 100,000 gastrula and neurula cdna clones, to create a unique and full-length set of approximately 2500 clones. Reducing the redundancy and excluding truncated clones from the starting clone set reduced the total number of clones to be screened, in turn allowing us to reduce the pool size to just eight clones per pool. We report that the efficiency of screening this clone set is five-fold higher compared to a redundant set derived from the same libraries. We have screened 960 cdna clones from this set, for genes that are involved in neurogenesis. We describe the overexpression phenotypes of 18 single clones, the majority of which show a previously uncharacterised phenotype and some of which are completely novel. In situ hybridisation analysis shows that a large number of these genes are specifically expressed in neural tissue. These results demonstrate the effectiveness of a unique full-length set of cdna clones for uncovering players in a developmental pathway. q 2004 Published by Elsevier Ireland Ltd. Keywords: Gain of function; Screen; Xenopus; Neurogenesis; Rab32; XER81; Frizzled 10B; Cullin1 1. Introduction Functional screens are valuable tools for identifying genes involved in specific developmental processes. In screens performed on a large scale, genes with similar functions can be grouped together, regulatory pathways suggested and interesting candidates selected for further study. Indeed, large-scale functional screens, involving either gain or loss of function, have been successfully carried out in a number of experimental model organisms. * Corresponding author. Address: Wellcome Trust/Cancer Research UK Gurdon Institute, Tennis Court Road, Cambridge CB2 1QR, UK. Tel.: C ; fax: C address: np209@mole.bio.cam.ac.uk (N. Papalopulu). Loss of function screens have been carried out in Drosophila, zebrafish and, more recently, in mouse and provided us with a wealth of information on developmental processes (Haffter et al., 1996; Nusslein-Volhard and Wieschaus, 1980; Driever et al., 1996; Kile et al., 2003; Mitchell et al., 2001; Skarnes et al., 2004). In recent times, the availability of genome sequence data and ORF prediction has allowed more direct, gene based, loss of function screens. For example, RNAi has been used in C. elegans and in Drosophila cell lines to knock down predicted genes on a genome wide basis (Boutros et al., 2004; Fraser et al., 2000; Gonczy et al., 2000; Kamath et al., 2003; Lum et al., 2003). On a smaller scale, antisense oligo (morpholino-mo) based knock-down screens have been carried out in Ciona and Xenopus and should also be /$ - see front matter q 2004 Published by Elsevier Ireland Ltd. doi: /j.mod
2 290 J. Voigt et al. / Mechanisms of Development 122 (2005) feasible in zebrafish (Chen et al., 2004a; Kenwrick et al., 2004; Stemple, 2004; Yamada et al., 2003). However, there are limitations to their effectiveness in uncovering gene function. Many genes do not show a loss of function phenotype, either because their phenotypes are too subtle to pick up in a large scale screen or because redundancy compensates for their loss of function (Gu et al., 2003). For example, in a MO screen in Xenopus 15 23% of genes had a detectable phenotype (Kenwrick et al., 2004), which is similar to the percentage found in a large-scale RNAi screen in C. elegans (10 14%; Fraser et al., 2000; Kamath et al., 2003) and in a MO screen in Ciona (20%; Yamada et al., 2003). Gain of function screens provide a complementary approach to loss of function screens in identifying gene function. In Drosophila, gain of function screens have been carried out by crossing lines that contain randomly inserted P elements with lines that carry the overexpression driver Gal4, under a tissue specific promoter (Rørth et al., 1998). However, P-element insertion events are not actually random, thus many areas of the genome might never be targeted by this approach. Moreover, genes are overexpressed, whether they are expressed during development or not, so the outcome may not be physiologically relevant. In vertebrates gain of function screens have been mainly carried out in Xenopus, an organism with a long history as a model system for the study of gene function in development. In Xenopus, gain of function experiments are based on injecting RNA made from cdna libraries of a developmental stage or tissue of interest. Therefore, genes that are being tested are transcribed during development and all parts of the genome that are transcribed in the stage or tissue of interest are represented, albeit with different frequency. As in loss of function screens, in gain of function screens many genes do not show a phenotype in the assay employed. Therefore, in order to be able to test a large number of cdna clones in a short period of time the cdnas are screened in pools. In vitro transcribed RNA from these pools are injected into embryos and once pools that cause phenotypes are found, are sib-selected by sub-dividing into smaller pools, until a single clone that causes the phenotype is identified (Lustig et al., 1997). In this way, several developmentally important molecules have been identified, such as noggin (Smith and Harland, 1992), Wnt-8 (Smith and Harland, 1991), b-catenin (Domingos et al., 2001), gremlin (Hsu et al., 1998), Xnr3 (Smith et al., 1995); (Glinka et al., 1996), Siamois (Lemaire et al., 1995), Xombi (Lustig et al., 1996), Twin (Laurent et al., 1997), Dickkopf-1 (Glinka et al., 1998), geminin (Kroll et al., 1998), Sizzled (Salic et al., 1997), Wise (Itasaki et al., 2003), Xath2 (Taelman et al., 2001), Sox17a/b (Zorn et al., 1999) and Mix.1 (Mead et al., 1996). Although this method has clearly led to the identification of important players in development, there are some significant limitations. First, there is the issue of the sensitivity of phenotype detection. When these gain of function screens were first developed, pool sizes of thousands of clones per pool were used (e.g. Smith and Harland, 1991; Lemaire et al., 1995; Mead et al., 1996). Since only a maximum of 5 10 ng of RNA can be injected into a X. laevis embryo without causing unspecific toxic effects, screening very large pools usually leads to the identification of only those genes that cause a phenotype when expressed in very small quantities. In order to make the screens more sensitive, pool sizes have been gradually reduced over time (e.g. Smith et al., 1995; Lustig et al., 1996; Salic et al., 1997; Hsu et al., 1998; Kroll et al., 1998, see Lustig et al., 1997). The smallest pool size so far published is 96 clones per pool (Grammer et al., 2000). However, the smaller the pool size, the more pools have to be screened to achieve the same gene coverage, which makes it a lot more time-consuming. A second drawback of this method is the redundancy of the utilised cdna libraries, thus, abundant clones are screened over and over again. Many of these abundant clones represent housekeeping genes, such as ef1a, which may have no function in specific developmental assays or contaminants such as ribosomal RNA. Moreover, there is a risk that potent genes can swamp any other activity in numerous pools. Finally, a large number of clones contained in an unprocessed cdna library are truncated clones. In oligo dt primed libraries these usually lack the initiation of translation at the 5 0 end. Although some truncated clones are active, as is the case for a truncated version of b-catenin (Funayama et al., 1995; Domingos et al., 2001) many are likely to be inactive and are screened unnecessarily. Here we present the results of a modified, small-pool, gain of function screen in Xenopus that was designed to overcome these limitations. To increase the efficiency and sensitivity of the small-pool screen we made use for the first time of a unique full-length (FL) cdna clone set, derived from gastrula and neurula stage Xenopus tropicalis cdna libraries. Unlike normal cdna libraries this unique fulllength cdna library contains only clones that are likely to be full length and each is represented by only one copy. Since less clones had to be screened we were able to reduce the pool size to eight clones per pool. We compare a gain of function screen carried out with this modified library (8 clones pool size) with a redundant small pool screen (96 clone pool size) of a redundant library, similar to those published before (Grammer et al., 2000), and we find that our modification made the screen a lot more sensitive and efficient. The modified screen we describe here was designed to discover genes involved in neurogenesis, a multistep process during which neuroepithelial cells make decisions between differentiation and continuation of a progenitor state. It is known that a genetic cascade is involved in controlling neurogenesis, which consists of neural inducers, pre-patterning, proneural and neurogenic genes (reviewed in Bertrand et al., 2002; Munoz-Sanjuan and Brivanlou, 2002). However, many gaps remain in our understanding of how cell fate decisions are regulated during neurogenesis
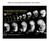
3 J. Voigt et al. / Mechanisms of Development 122 (2005) Fig. 1. Comparison of the phenotype penetrance between the redundant set and the unique full-length set screen. One hundred and six pools of a redundant neurula stage cdna library (pool size 96 clonesz cdnas) and 120 pools of the unique full-length (FL) gastrula/neurula cdna library (pool size eight clonesz960 unique and full-length cdnas) were screened by overexpressing z5 ng of in vitro transcribed RNA in one cell of two-cell stage embryos. Embryos were analysed at neurula and tadpole stages for changes in morphology and in the neural differentiation marker N-tubulin. Only phenotypes with a penetrance (percentage of affected embryos in a batch) of 40% or more were scored as real but analysis for lower penetrance is also shown. In the redundant set screen z10% (nz11) of the screened pools showed a phenotype (R40% penetrance) whereas this was raised to 50% (nz60) in the FL set screen (green star). At a higher penetrance (R80%) there is also a big difference in phenotype recovery between the redundant and FL set screen (3 vs. 28%). Thus the FL set screen was five-fold more sensitive in detecting gain-of-function phenotypes compared to an unmodified cdna library. and this functional screen was employed to increase our knowledge of genes that are involved in this process. We present the overexpression phenotypes of the single clones that were identified in this screen together with their expression patterns at some key developmental stages. Many of the single clones identified are novel or show a novel function when overexpressed. Moreover a large number of them are specifically expressed in neural tissue. These results demonstrate the effectiveness of the unique full length set as a resource for uncovering players in a developmental pathway. 2. Results 2.1. Identification of genes by overexpressing RNA from a redundant neurula stage cdna library We screened 10,176 clones of a neurula stage X. tropicalis cdna library by injecting 4 5 ng of pools of 96 RNAs in one half of two-cell stage X. laevis embryos. Ninety six clones per pool is a size substantially lower than used traditionally (Smith and Harland, 1991) and is the same pool size as in a recently published screen (Grammer et al., 2000). Screen analysis for phenotypes was carried out at neurula (open neural plate) and tadpole stages of development by the expression of the neuronal marker N-tubulin and morphology. N-tubulin marks the final stage of primary neurogenesis, thus any effect on neurogenesis should result in an effect on N-tubulin expression. In order to be able to distinguish phenotypes from background levels of malformations, phenotypes were only scored if they had a penetrance of a least 40% (i.e. percentage of affected embryos). We found that only 10% (nz11) of the pools screened (nz106) gave rise to a phenotype that was above the 40% penetrance threshold, based on N-tubulin and morphology (Fig. 1). This is similar to what has previously been reported (Grammer et al., 2000, 12%). We reasoned that this is likely to be due to the low amount of each RNA present in the pool (a maximum of approx. 50 pg). To test this hypothesis we injected RNA for a known gene involved in neurogenesis, Xiro3, at different concentrations. While at 1 ng the penetrance of the phenotype (suppression of neurogenesis) was 90%, as previously described (Bellefroid et al., 1998), at 50 pg (the equivalent amount in a pool size of 96) the penetrance was only 18% and 12% when Xiro3 was mixed in a 96-clone pool (see Table 1). Therefore, Xiro3 would have been missed in this screen. By contrast, Gremlin, for example, which was isolated in a 1000 clone pool screen, gives penetrance of 93% in concentrations as low as 50 pg (Hsu et al., 1998). Other genes cloned in previous screens are active at even lower amounts, in the range of 1 10 pg (Smith and Harland, 1991; Laurent et al., 1997). It is not clear what causes such variability in the dose required for each gene, although the combinatorial action of genes is certainly a factor, as has been shown for X-Wnt11 and Xnr-3, where a combination of the two genes dramatically reduces the amount required for each single gene (Glinka et al., 1996). These results indicate that carrying out a gain of function screen in this format is predominantly leading to the isolation of potent genes, which are more likely to have been already described.
4 292 J. Voigt et al. / Mechanisms of Development 122 (2005) Table 1 The effect of quantity of injected RNA on phenotype penetrance approximately five-fold higher than previously achieved with the 96-clone per pool redundant set screen, described above (Fig. 1)orin(Grammer et al., 2000) Unique full-length set screen summary In order to distinguish phenotypes from background malformations only phenotypes with a penetrance (number of affected embryos in a batch) of at least 40% are scored. Xiro3 RNA, whose overexpression leads to a reduction of N-tubulin at neurula stage (Bellefroid et al., 1998), was chosen as a representative gene to test if its activity can be detected in a 96 clones/ pool screen. Thus a range of doses of Xiro3 has been injected into one cell of two-cell stage Xenopus laevis embryos and the phenotype penetrance was scored. Five nanograms of an RNA pool containing Xiro3 RNA were also injected as a pool control. The approximate concentration of any individual RNA in a 96-clone pool is approximately 50 pg (due to a maximum of 5 ng that can be injected into embryos). At 50 pg (highlighted in red) or in the control pool, the phenotype penetrance of Xiro3 was below the scoring threshold A unique full-length cdna library allows the reduction of the pool size One way to increase the amount of injected RNA per clone is to inject the same total amount of RNA but decrease the pool size. To achieve the same overall clone coverage the number of pools to be screened would increase dramatically. Recently, 55,000 clones from each of the gastrula and neurula libraries have been sequenced in a large-scale X. tropicalis EST project ( Projects/X_tropicalis). It was possible to use this sequence information in order to create a unique and full-length cdna clone set by removing multiple as well as truncated presumably non-functional copies of cdnas from the collection of sequenced clones (see materials and methods). In this way, approximately 100,000 cdna clones from the gastrula and neurula libraries were condensed into a set of 2304 unique and full-length clones. This effectively reduced the number of cdnas to be screened, which in turn made it feasible to reduce the pool size and concomitantly increase the amount of RNA of each clone in a pool The unique full-length set makes the screen more sensitive We screened 960 clones of the unique full-length clone set in pools of eight clones (120 pools). Approximately 500 pg of RNA were injected per individual clone. As for the previous redundant clone set screen we analysed for phenotypes at neurula and tadpole stages using morphological changes and changes in the in situ pattern of N-tubulin. Interestingly, we now found that 50% (nz120) of the pools gave phenotypes of 40% penetrance or higher. This is The phenotypes caused by the overexpression of the pools fell into nine major categories: gastrulation defects, reduction of N-tubulin, ectopic tissue outgrowth, tissue loss, embryonic lethality, secondary axes/displacement of neural markers, ectopic N-tubulin, ectodermal blisters and anteriorisation (see Fig. 2). Most pools gave rise to complex phenotypes that fall into more than one of these categories, therefore the percentages sum up to more than 100%. This complexity might be due to the combined effects of different RNAs within a single pool. Since we wanted to identify novel players in neurogenesis, we were particularly interested in pools that alter the pattern of N-tubulin expression. Since neural tissue is derived from the ectoderm we also selected some pools with interesting ectodermal defects for further analysis. We selected 19 out of the 60 pools of R40% penetrance for phenotypes, which indicate a potential involvement in neural development. Seventeen out of the 19 pools (89.5%) showed a reproducible phenotype upon repeat injections. In order to identify single clones we took advantage of the sequence information that is available for each clone in a combination of candidate gene approach and injection of single clones. In the cases where the activity within a pool matched previously published observations for a gene present in this pool, we only injected the candidate gene. If the pool phenotype was reproduced, the activity was assigned to this clone. We picked candidate genes for five of the 17 reproducible pools and 4/5 matched the phenotype of the pool. For the remaining 13 pools (12 plus the nonmatching pool from the candidate approach), single clones were injected individually. With such a small pool size, the previously used step of sib-selection in gradually smaller pools was not necessary Description of the activities that have been identified Using both approaches we were able to identify the active clones for 15 of the 17 pools and from these, we identified 18 different genes with activity. Seven (39%) of the identified genes have been previously described, which indicates that our screen is able to identify known players during neurogenesis. Two (11%) have been cloned in Xenopus before but showed a novel function, 6 (33%) were novel in Xenopus (i.e. no NCBI database entry other than predicted genes exists for Xenopus), and three (17%) were completely novel (no entry in NCBI database, only predicted genes). The identified genes fell into the following categories: reduction or ectopic N-tubulin at neurula stage, secondary axis, anteriorisation, ectopic tissue outgrowth,
5 J. Voigt et al. / Mechanisms of Development 122 (2005) Fig. 2. Phenotypic categories found when screening the unique FL set pools. Sixty pools of the unique full-length gastrula/neurula stage cdna library gave rise to an overexpression phenotype (at least 40% penetrance). Nine major categories (x-axis) of phenotypes, for which representative images are shown below each bar, were observed and scored. Phenotypes were classified as secondary axes (rather than ectopic tissue outgrowth) if the ectopic tissue shows axial organisation, like secondary heads (at tadpole stage) or complete secondary neural tubes (at neurula stage). Ectopic tissue outgrowth comprises tumour-like growths, protrusions (which lack clear axial structures) and other abnormal thickenings observed. Blisters are oedema-like appearances, which do not contain tissue. Phenotypes such as loss/shedding of cells and or limited cell death were grouped into tissue loss. In contrast, embryos that showed global death are classified in death. Since most of the phenotypes observed were complex, many of the pools were included in more than one category (thus the sum of the percentages is greater than 100%). Of particular interest were phenotypes that affected the neural differentiation marker N-tubulin (stars), since they might contain an activity involved in the neurogenesis pathway. (Purple staining in images: N-tubulin, light blue staining: lineage tracer LacZ). epidermal defects, gastrulation defects and cell lethal (Table 2 and Fig. 3) Reduction/inhibition of N-tubulin Neurula stage The most common phenotype observed, was a reduction or inhibition of N-tubulin (Table 2, Fig. 3a,b). This was shown by 12 genes, three of which are novel, in that a BLAST search results only in predicted ORFs. The remaining nine are previously published genes although, with the exception of Zic3 (Nakata et al., 1997), their effect on neurogenesis has not been previously described. The reduction of N-tubulin seen here could be due to interference with an early step in neural development, namely neural induction, or a later failure of the neuroepithelial cells to differentiate. To distinguish these possibilities we examined the expression of the early neural marker of proliferating cells, Sox3 (Penzel et al., 1997; Hardcastle and Papalopulu, 2000) (data summarised in Table 2, column 3). Only two of the genes, Sox2 and Myocyte enhancer factor related 2D (SL1) reduced Sox3. For SL1 this is likely to indicate a general interference with neural development. SL1 is expressed in the somitic mesoderm (Fig. 4b; Chambers et al., 1992) and is thought to lie downstream of myod and Myf5 (Chambers et al., 1994). The suppression of Sox3 by Sox2 was somewhat surprising since Sox2 is a later neural marker than Sox3 (Rex et al., 1997). We suggest that suppression of Sox3 by Sox2 may reflect a negative transcriptional control of later acting genes (Sox2) on the earlier ones (Sox3) in the neurogenic cascade. Sox2 may also suppress later acting genes, reflected in the suppression of N-tubulin. It may be that such gene downregulation in both directions of the neurogenic pathway is an important step in the progression of cells through different stages of differentiation. A previous study has shown that a dominant negative form of Sox2 (dnsox2) inhibits early neural development in general (Kishi et al., 2000). Reduction of N-tubulin could also indicate an increase in proliferation of the ectoderm at the expense of differentiation. Indeed, five genes that caused a reduction of N-tubulin, (strabismus, Zic3, Frizzled 10B, cullin 1, XER81), caused a concomitant increase of Sox3, which marks proliferating neuroepithelial cells (Table 2, Fig. 3a,b). Four of these genes (designated with asterisks in Table 2) also showed a lateral displacement of N-tubulin. The most dramatic increase in Sox3 expression was shown by XER81, an ets-type transcription factor in the FGF pathway (Chen et al., 1999) (shown in Fig. 5A). Our observation
6 294 J. Voigt et al. / Mechanisms of Development 122 (2005) Table 2 Phenotype categories of the single clones identified in the screen Gene Sox 3 at neurula stage N-tubulin at tadpole stage I. Reduction of N-tubulin at neurula stage 1.2B (TGas002c02) X-dll3 No change No change 3.1G (TGas026h11) Transcription factor IIF, RAP74 No change No change subunit 3.2B (TGas021d05) Novel No change No change 3.4D (TGas023a22) Novel No change K 3.5H* (TGas027l08) Strabismus C C 3.9B* (TGas021i09) Zic3 C C 3.12E* (TGas025k08) Frizzled 10B C C 4.6D (TGas032i14) Novel No change No change 4.6H* (TGas038c21) Cullin 1 C C 19.11A (TNeu063l17) Sox2 K C 22.6D (TNeu107l17) Myocyte enhancer factor related K(Subtle) No change 2D (SL1) 22.9F (TNeu110d19) XER 81 C C II. Ectopic N-tubulin at neurula stage 3.12E (TGas025k08) Frizzled 10B C C III. Secondary axis 3.12E (TGas025k08) Frizzled 10B C C 4.6H (TGas038c21) Cullin 1 C C IV. Anteriorisation 2.8B (TGas012p20) Casein kinase 1 delta C No change V. Ectopic tissue outgrowth 1.4A (TGas001g08) Serine threonine kinase 17A No change No change 3.9B (TGas021i09) Zic3 C C 4.6D (TGas032i14) Novel No change No change 4.6H (TGas038c21) Cullin 1 C C 19.11A (TNeu063l17) Sox2 K C 22.6D (TNeu107l17) Myocyte enhancer factor related K(Subtle) No change 2D (SL1) 22.9F (TNeu110d19) XER 81 C C VI. Epidermal defects 1.2H (TGas009a06) Rab 32 No change No change 1.4A (TGas001g08) Serine/threonine kinase 17A No change No change 4.6D (TGas032i14) Novel No change No change VII. Gastrulation defect 1.2B (TGas002c02) X-dll3 No change No change 1.4A (TGas001g08) Serine/threonine kinase 17A No change No change 3.2B (TGas021d05) Novel No change No change 3.4D (TGas023a22) Novel No change K 3.5H (TGas027l08) Strabismus C C 3.12E (TGas025k08) Frizzled 10B C C 19.11A (TNeu063l17) Sox2 K C 22.6D (TNeu107l17) Myocyte enhancer factor related 2D (SL1) K(Subtle) No change VIII. Embryonic lethal/tissue loss 2.11E (TGas016h07) PKA, subunit beta K K 3.1C (TGas021n15) SFRS 5 K K 19.3F (TNeu070m22) SFRS 7 K K The phenotypes of the single clones identified in the unique full-length set screen were grouped into eight categories (I VIII.) Where embryo phenotypes fell into more than one category the clone was listed repeatedly. Neurula stage embryos were additionally analysed for changes of the proliferating neural tissue marker Sox3 and tadpoles were analysed for N-tubulin expression (alongside morphology analysis), all of which is summarised in column 3 and 4, respectively (C: marker expanded, K: marker reduced, no change : marker not affected). Asterisks in column 1 indicate genes that in addition to a reduction of N-tubulin also showed a displacement of the stripes (see Fig. 4b). The clones were coded as follows; bold: novel gene with no BLAST hit other than predicted genes, bold italic: novel gene in Xenopus; italic: cloned in Xenopus but novel function; normal: known gene in Xenopus with previous functional analysis carried out. The clone names (TNeu/Gas.) were given by the Sanger Xenopus tropicalis EST project and refer to the position of the clones in the original arrayed library. The gene name given to the clones is based on the highest BLAST hit that roughly aligns to the start methionine of the clone. Sequences, clusters and BLAST alignments are displayed on the website: The other clone name (1.2B, etc.) refers to their position on the full-length set plate.
7 J. Voigt et al. / Mechanisms of Development 122 (2005) Fig. 3. Overexpression phenotypes of the single clones identified in the screen. (acb) Approximately 400 pg in vitro transcribed RNA together with 200 pg beta-galactosidase RNA as a lineage tracer (light blue staining) were injected animally into one cell of two cell stage Xenopus laevis embryo. Embryos were analysed at neurula and tadpole stages for changes in the neural differentiation marker N-tubulin (purple) and morphology. Unless indicated otherwise, neurula stage embryos are viewed dorsally with anterior to the top and tadpoles are viewed laterally with anterior to the left. A transverse section is shown for clone 1.2B at tadpole stage.
8 296 J. Voigt et al. / Mechanisms of Development 122 (2005) Fig. 3 (continued)
9 J. Voigt et al. / Mechanisms of Development 122 (2005) complements previous findings where dorsal XER81 overexpression disrupts eye development (Chen et al., 1999). Moreover, FGF has been implicated in neural induction in Xenopus and chicken (e.g. Launay et al., 1996; Pera et al., 2003; Streit et al., 2000; Wilson et al., 2000) as well as the maintenance of neural progenitors cells (in chicken) (Diez del Corral et al., 2002; Mathis et al., 2001). In Xenopus, a role in enhancing ectopic neurogenesis has also been Fig. 4. (a,b) Expression patterns of the single clones identified in the unique full-length set screen. Xenopus tropicalis gastrula, neurula and tailbud stage embryos were in situ hybridised with antisense probes to the single clones identified in the unique full-length set screen. Gastrula stages embryos are shown vegetally with dorsal to the top (unless stated otherwise). At neurula stages an anterior and dorsal view is presented. Tailbud embryos are oriented with anterior to the left and dorsal to the top unless indicated otherwise. In come cases the wholemount images are supplemented by neurula and tailbud stage dissected embryos. If no specific staining was detected no image is shown.
10 298 J. Voigt et al. / Mechanisms of Development 122 (2005) Fig. 4 (continued) suggested (Hardcastle et al., 2000). The phenotype of XER81 described here, is consistent with XER81 mediatingat least in part-the proposed role of FGF in enhancing the formation of undifferentiating, proliferating neuroepithelium, but further experiments are needed to verify this hypothesis Tadpole stage We also analysed the expression of N-tubulin at tadpole stages (right column Table 2, Fig. 3a,b). We found that many of the genes that suppressed N-tubulin early on, caused ectopic N-tubulin at the tadpole stage. Interestingly, in almost all of these cases (5/6) (Strabismus, Zic3, Frizzled
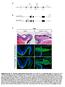
11 J. Voigt et al. / Mechanisms of Development 122 (2005) Fig. 5. Additional detail on the overexpression of 4 selected genes and Rab32 expression. (a) XER 81 RNA injections expand X-Sox3 (purple) on the injected side. (bcc) Frizzled10B RNA injections enhance the medial (motorneuron) stripe of N-tubulin expression (white arrowhead, in wholemount (B) and transverse (C) view), in addition to the ectopic expression of N-tubulin observed in the floorplate (shown in Fig. 4b). (dashed line-midline, l-lateral, i-intermediate, m-medial). (dce) 1 mm sections of Rab32 (1 ng) injected (E) and control Xenopus laevis (D) embryos. (D) In wild type embryos the melanosomes (darkly pigmented vesicles, arrow) are localised predominantly under the apical surface, but some can be found deeper within the cells (arrowhead). In contrast, when Rab32 was overexpressed the melanosomes were clustered tightly together and localised under the apical surface (arrows). (f) Transverse section (30 mm) of a bleached Xenopus tropicalis tailbud embryo in situ hybridised with an antisense probe to Rab32. Rab 32 is expressed in the retinal pigment epithelium (purple). 10B, Cullin 1, XER81), the expression of Sox3 was expanded at neurula stage. Thus, there appears to be a correlation between suppression of early neurogenesis and ectopic proliferation of neural tissue at neurula stage on the one hand, and ectopic neuronal differentiation at tadpole stage on the other. The genes that show this correlation may function in expanding the population of neural progenitor cells but their effect may be transient. We suggest that once the RNA concentration diminishes at the tadpole stage, the ectopic neural tissue is no longer repressed from differentiating, hence ectopic N-tubulin appears at the tadpole stage. Surprisingly, we observed reduced N-tubulin at tadpole stage only in one case, that of the novel gene TGas023a22 (3.2B). Since this gene had no effect on Sox3, we expect it to function quite late in the neurogenesis cascade. Finally, some of the clones that showed a reduction of N-tubulin at neurula stage did not show any effect on N-tubulin later (Xdll3, Transcription factor IIF, TGas023a22 3.2B and SL1) (Table 2). This could be due to a transient effect of the RNA or less interestingly, a developmental delay on the injected side. It could also be that a subtle reduction might be difficult to detect at the tadpole stage Ectopic N-tubulin at neurula stage Only one of the genes Frizzled 10B showed ectopic N-tubulin at neurula stage. This gene is of particular interest since it preferentially enhanced neurogenesis along the medial, motorneuron, stripe (Figs. 3b, 5B,C). The lateral (sensory neuron) stripe was not enhanced and in some cases it was suppressed, hence it is also classified under reduction of N-tubulin. Occasionally, Frizzled 10B overexpression resulted in N-tubulin expression along the floor plate (arrowhead in Fig. 3b). In these cases, the floor plate was expanded, causing a lateral shift of the N-tubulin stripes (Table 2, asterisk). The expression of Frizzled 10B is exclusively neural (Fig. 4b), consistent with previous reports (Moriwaki et al., 2000; Wheeler and Hoppler, 1999). The function of Xenopus Frizzled 10B has not been previously studied but it is likely to act as a Wnt receptor, similar to other members of the frizzled family. This clone would be interesting to investigate, as this is the first case of a gene that enhances specifically the primary motorneuron population when overexpressed in Xenopus embryos. The frequency of ectopic N-tubulin in this screen was lower than we might have expected (see Fig. 2). However,
12 300 J. Voigt et al. / Mechanisms of Development 122 (2005) we note that many published genes that result in ectopic N-tubulin when overexpressed (such as X-ngnr-1 and neurod (Ma et al., 1996; Lee et al., 1995) were not found in the EST database of the cdna libraries that we have used. These genes are expressed at the neural plate stage but we speculate that their abundance is still relatively low. As many such genes continue to be expressed later in development, we suspect that later stage libraries might have been more fruitful in uncovering neurogenesisinducing genes. Indeed, later stage libraries have since been added to the EST project ( Projects/X_tropicalis/) Secondary axis and anteriorisation Two genes, Frizzled 10B and cullin 1 gave rise to secondary axes while one gene, casein kinase (CK) 1 delta gave rise to anteriorised embryos at neural plate and tadpole stages, with an associated increase in Sox3 expression (Fig. 3a,b, Table 2). The phenotype of CK 1 delta, a kinase in the Wnt pathway, is slightly different than previously described (anteriorisation versus secondary axis; (Sakanaka et al., 1999; Peters et al., 1999; McKay et al., 2001) however, this is likely to be due to differences in the site of injection (animally, in our case). Cullin 1 is a ubiquitin ligase (reviewed in Tyers and Jorgensen, 2000) and is currently under investigation in our lab (JV and NP, in preparation) Ectopic tissue outgrowth Ectopic tissue outgrowth was observed at the tadpole stage and is quite common as it was observed for seven clones (Table 2, Fig. 3a,b). In all cases, clones that showed ectopic tissue outgrowth also fell into some other category. Six out of the seven genes that showed ectopic tissue outgrowth at the tadpole stage, 4.6D (TGas032i14), Sox2, SL1, XER81, Zic3 and cullin 1 also showed a reduction in N-tubulin at neurula stage, suggesting a role in increasing proliferation at the expense of differentiation. In the case of XER81, Zic3 and cullin 1, such role was further supported by an increase in Sox3 at neurula stages. In the case of cullin 1, the ectopic tissue outgrowths appeared distinct from the secondary axis and are likely to represent separate effects. It is likely that not all tissue outgrowths represent effects on proliferation. For example, in some embryos the effect of one clone, TGas001g08, which shows similarity to the human serine/threonine kinase 17A (Sanjo et al., 1998), appeared more like an ectodermal defect than a tissue outgrowth. Consistent with a change in ectodermal architecture rather than proliferation, this clone did not cause changes in Sox3 and N-tubulin expression Epidermal defects The distinction between epidermal outgrowths and other epidermal defects was not always clear and two clones were classified in both categories (see also above). Of particular interest in the epidermal defect category is the third clone, Rab32, encoding a small molecular weight Rab GTPase of the ras superfamily (Table 2, Fig. 3a,b). Overexpression of this clone caused a concentration of pigmentation into a spot like structure in each ectodermal cell (Fig. 3a). In sections, it was clear that Rab32 overexpression caused a tight clustering of the pigment organelles (melanosomes), which are normally evenly spread under the apical surface (Fig. 5D,E). In pigmented Xenopus cells, melanosomes are transported within the cell along microtubules and actin filaments (Gross et al., 2002). In apico-basally polarised early blastomeres (Chalmers et al., 2003), they have a clear enrichment on the apical side (Fig. 5D). Rab32 would be interesting to investigate further as it may be involved in directional melanosome transport, localisation or dispersion. The expression of this gene was not detected in early stages of development, perhaps indicating a ubiquitous low level of expression. However, Rab32 was strikingly upregulated in the pigmented epithelium of the retina at the tadpole stage, consistent with a role in melanocyte function (Fig. 5F). In cell lines, murine Rab32 co-localises with melanosomal proteins (Cohen-Solal et al., 2003) while the human protein is thought to act as a tether for campdependent protein kinase (PKA; Alto et al., 2002) Gastrulation defect Approximately 60% of all the pools showed some form of gastrulation defect (Fig. 2). When deciding which pools to follow up we excluded all the samples that showed only a gastrulation defect. Therefore, clones that cause gastrulation defect in the sib-selected list, also had some neural defect (Table 2). It is interesting to note here, that the morphogenetic defects of Strabismus and Frizzled 10B at the neural plate stage are remarkably similar (Fig. 3b). These two genes also have common elements in their effects on Sox3 and N-tubulin. However, Frizzled 10B has additional effects in inducing secondary axis formation and increasing specifically the number of primary motorneurons (Fig. 5B,C), as discussed above, which were not observed with Strabismus. This is what one would expect if Frizzled 10B, acting as a Wnt receptor, activated both the canonical and the non-canonical, PCP, Wnt pathway. Strabismus on the other hand, is known to act only on the PCP pathway (Park and Moon, 2002), which we believe explains why its overexpression phenotype is a subset of the Frizzled10B phenotype. The high incidence of gastrulation defects could mean that gastrulation is easily disrupted and/or that the concentration of some RNAs was too high. Alternatively, it is possible that our clone set was enriched for genes involved in gastrulation, as it derived from both gastrula and neurula stage libraries. As we were primarily interested in neurogenesis, we did not further study gastrulation defects and we made no attempt to distinguish between effects on
13 J. Voigt et al. / Mechanisms of Development 122 (2005) the morphogenetic movements of the mesoderm or of the ectoderm Embryonic lethal/tissue loss In this category, there are three clones, which were originally selected because they caused suppression of N-tubulin (Table 2, Fig. 3a,b). Upon further investigation, we concluded that this phenotype is due to cell loss, possibly due to excessive cell death. At the moment it is not clear whether this is a specific apoptotic effect and further investigation is needed to clarify this point. However, we are presenting these clones because they have specific expression patterns (Fig. 4a,b, Table 3), they are novel in Xenopus and their function in development is not known. The beta-subunit of PKA is specifically expressed in the notochord while two serine/arginine regulators of alternative RNA splicing, SFRS 5 and 7, are specifically expressed in neural tissue (Fig. 4a,b, Table 3) Expression pattern of single clones In addition to the overexpression phenotype we present the expression pattern for all of the identified clones (Fig. 4a,b, Table 3) in Xenopus tropicalis embryos. It was striking that 16/18 clones were expressed in neural tissue, which suggests that our screening strategy was successful in specifically identifying potential players in neural development. 3. Discussion 3.1. Functional screens in Xenopus We have created and used a unique full-length clone set from gastrula and neurula stage cdna libraries in a smallpool gain of function screen for genes involved in neurogenesis. Eliminating redundancy and truncated clones from the original cdna clone set allowed us to reduce Table 3 Expression analysis of the single clones identified in the screen Clone ID Gene Gastrula Neurula Tailbud Reference TGas002c02 1.2B X-dll3/Dlx5 Ubiquitous Anterior neural, cement gland, neural folds Anterior neural, cement gland, otic vesicle Papalopulu and Kintner (1993) TGas009a06 1.2H Rab32 None detected None detected Retina pigment epithelium TGas001g08 1.4A Serine/Threonine Ubiquitous None detected Eyes, pronephros Kinase 17A TGas012p20 2.8B Casein Kinase 1 delta Ubiquitous Anterior neural fold, Spinal cord low level spinal cord TGas016h E PKA, subunit Ubiquitous Neural enriched Eye, notochord TGas021n15 3.1C SFRS 5 Ubiquitous Anterior neural stripe, spinal cord Eye, branchial arches, brain TGas026h11 3.1G Transcription factor IIF, RAP 74 subunit None detected Anterior neural enriched, spinal cord Branchial arches and eye TGas021d05 3.2B Novel Dorsal Pan-neural, low level Eye, hindbrain, branchial arches TGas023a22 3.4D Novel None detected None detectable Cement gland TGas027l08 3.5H Strabismus Dorsal Anterior neural stripes, spinal cord Neural tube Darken et al. (2002) and Park and Moon (2002) TGas021i09 3.9B Zic3 Dorsal Lateral neural folds Neural tube Nakata et al. (1997) TGas025k E Frizzled 10B None detected Anterior neural stripes and lateral neural folds Neural tube Moriwaki et al. (2000) and Wheeler and Hoppler (1999) Eye TGas032i14 4.6D Novel Dorsal Neural enriched, low level TGas038c21 4.6H Cullin I Ubiquitous Anterior neural Eye, neural tube, enriched? branchial arches TNeu070m F SFRS 7 Ubiquitous Anterior neural, spinal Eye, neural tube, cord branchial arches TNeu063l A Sox2 Dorsal Pan neural,placodes Neural tube, eye Mizuseki et al. (1998) TNeu107l D SL-1 None detected Somites Tail somites, heart Chambers et al. (1992) TNeu110d F XER81 Blastopore Complex anterior Mid-hindbrain stripe, Chen et al. (1999) and neural, spinal cord, branchial arches, Munchberg and blastopore remnant tailbud Steinbeisser (1999) Table describing the expression pattern of the identified clones as determined by whole-mount in situ hybridisation. Only two genes (SL-1 and 3.4D) are not expressed in neural ectoderm. References are added where the expression pattern has been analysed previously in Xenopus.
14 302 J. Voigt et al. / Mechanisms of Development 122 (2005) the pool size to eight clones per pool without unreasonably increasing the amount of time needed to perform the screen. In total, 2304 FL clones were derived starting from 100,000 cdna clones and of those 960 were screened in this work. Overall, 18 genes have been identified out of this 960 clone set versus 37 out of a recent 55,000 redundant clone set (Grammer et al., 2000) a 30 fold improvement in the return of the screen. This is in fact an underestimate, if one takes into account that we only followed up pools that showed an effect on neural and/or ectodermal development. Of the 18 genes, three were entirely novel, in that they had no similarity to sequences in the sequence (NCBI) database The sensitivity and efficiency of this screen was also higher, with 50% of the screened pools showing a phenotype of 40% penetrance or higher, a five-fold improvement from the best return previously described from an unprocessed set of clones (Grammer et al., 2000). The cdna clones used here were derived from a X. tropicalis EST project, but the overexpression was carried out in X. laevis because the eggs are larger and more robust. Once candidate genes were identified, the expression analysis was carried out in X. tropicalis. Functional screens have been performed across more distant species, for example between Ciona or mouse and Xenopus (Davis and Smith, 2002; Baker and Harland, 1996). We expect that X. laevis and X. tropicalis are evolutionarily close enough for the results to be fully transferable between the two organisms. Indeed, a recent study examined the expression pattern of several genes in X. laevis and X. tropicalis and no significant differences were found (Khokha et al., 2002). We suggest that these two amphibian systems can be used in parallel, allowing one to exploit the advantages of both Identification of genes The scale of our screen was modest and therefore did not allow us to reach global conclusions about gene networks controlling neurogenesis. At this scale, such a screen is most useful in identifying single genes that can be followed up by more detailed functional analysis. Indeed, we have identified several genes, such as Frizzled 10B, cullin 1, XER81 and Rab32, which warrant further investigation, given our research interests. In addition, even this small scale analysis allowed some interesting correlations to emerge, such as the increase of Sox3/suppression of differentiation at neurula stage and the development of ectodermal growths at the tadpole stage (see Section 2) Future directions One of the great strengths of a functional screen is that it is highly adaptable to address specific questions (see Chen et al., 2005). Thus, one area of future improvements would be the development of diverse and imaginative assays. The Rab32 phenotype of melanosome aggregation within each cell, shows that such assays need not be limited to alterations in molecular markers or gross morphology but can be extended to a cellular level. As the sequence of the clones is known a priori, one could pre-select the gene set in such a way that only a certain class of molecules, for example, bhlh-motif containing genes, are screened. Finally, since beginning this work, many more ESTs have become available and the bioinformatic tools for FL prediction have been refined. As a result a larger unique FL set of 7000 clones, from three developmental stages, egg, neurula, gastrula and tadpole has been created (Gilchrist et al., 2004) and is available as a resource for the community ( These screens should provide yet another step on the way to assigning function to the large number of genes uncovered by the efforts in genome and cdna sequencing. 4. Experimental procedures 4.1. Creation and arraying of the cdna libraries The neurula (TNeu) and gastrula (TGas) stage Xenopus tropicalis cdna libraries were constructed as described (D Souza et al., 2003; Gilchrist et al., 2004). Approximately 55,000 clones of each cdna library were randomly picked, arrayed and sequenced from the 5 0 end, as part of a large scale X. tropicalis EST project ( Projects/X_tropicalis/). The individual clones are referred to by their position in the original arrayed library (e.g. TGas002c02). The sequence of each clone can be obtained by using this name, either at the above website or at the Xenopus tropicalis full-length EST database ( Furthermore, in the latter website, the sequences for only the clone set used in this work can be accessed by choosing Xt2:initial full length project. The cdna clones can be obtained from the MRC Geneservice: ( mrc.ac.uk/geneservice/reagents/products/descriptions/xtropest.shtml) Bioinformatics and re-arraying of the unique full-length set 95, EST sequences from the TNeu and the TGas library underwent the clustering process, which has been described in detail elsewhere (Gilchrist et al., 2004). Briefly, ESTs were clustered together that were 99% similar over a length of 100 nucleotides and aligned accordingly. We found that 74,683 ESTs fell into 11,078 clusters of two or more clones, whereas 20,536 clones remained as singletons (ESTs that fail to align with any other ESTs in the collection). In total, this led to 31,614 unique transcripts the unique clone set. For the purposes of the gain of function screen we wished to extract the clones that are likely to contain the translation
15 J. Voigt et al. / Mechanisms of Development 122 (2005) initiation site, referred to here as the full-length (FL) clones. An assumption was made that clones which contain the 5 0 start of translation also contain the 3 0 end, as the libraries were constructed with oligo dt priming. In order to identify full-length clones the following criteria were applied to all clusters of the unique clone set. First, a start codon preceded by an in frame stop codon must be present in at least 2 and at least 50% of the EST clones at the same position within a cluster to avoid misidentification of the start codon due to sequencing error. The 5 0 stop codon indicates that translation cannot start before the designated start codon Second, the ORF should be at least 210 nucleotides long (See Fig. 6). Unlike a more recently generated and more extensive fulllength clone set (Gilchrist et al., 2004) the criteria that we used to define the clone set used here did not include similarities in the database revealed by BLAST searches (except for the singletons, as described below). This was introduced to avoid biasing the selection towards genes that have been already described in the literature. This approach identified 1988 clusters, which are very likely to contain a full-length clone. To avoid potential negative translational control from sequences in the 5 0 UTR, the clone with the shortest 5 0 UTR was automatically chosen as the representative for its gene. These representatives were then re-arrayed onto 384 well plates. As we were using 384-well plates for the re-arraying, six plates were required, leaving 316 empty wells. These were allocated to singletons, which were selected using slightly different criteria than the clusters. Singletons were prioritised according to the length of their putative ORFs and those with long ORFs and a start codon preceded by an in frame stop were selected. In single read ESTs, the position of the putative initiator ATG cannot be verified by comparison to other ESTs. Therefore, BLAST information was used to confirm that the proposed initiator ATG corresponded to the start methionine of a matching protein in the NCBI protein database. In this way, 316 singletons were included in the full-length unique set of clones, eventually consisting of six 384-well plates or 2304 clones. The full-length cluster representative and the selected singletons were rearrayed onto 384-well plates at the MRC-Geneservice, UK. From these, a 96-well plate working copy was created for the gain-of-function screen. Since commencing this work, many more ESTs, including 3 0 sequences, have become available and the informatics programs have incorporated more sophisticated features and methods (Gilchrist et al., 2004). When comparing this initial full-length set against later data (Gilchrist et al., 2004) we found that approximately 75% of the clones from FL clusters described here were confirmed as almost certainly full length (not shown) Creation of mrna pools for the functional screen Clones were grown separately in 96-well plates and the cultures from each plate were pooled together, plasmid DNA was extracted and RNA was in vitro transcribed. These pools were used for the 96 clones/pool screen. To create the pools used for the unique full-length set the cultures were grown as above, but pools were created from the 12 columns (containing eight clones each) of each 96-well plate. The individual clones are referred to by their position in the original arrayed library ( Projects/X_tropicalis/) Embryo culture and RNA injections Eggs were collected from hormone-induced adult Xenopus laevis and in vitro fertilised with standard methods. Fig. 6. Creation of a unique full-length set. (a) The determination of the full-length representative in an EST cluster. 5 0 sequenced ESTs from the X. tropicalis neurula and gastrula stage cdna libraries that are 99% similar over a length of 100 nucleotides were aligned and clustered. Open reading frames of at least 210 base pair were identified. In order to ascertain that clusters contain full length EST clones in at least 2 and at least 50% of the EST clones at the same position a start codon (green) must be preceded by an in frame stop codon (red). To avoid 5 0 UTR dependent negative translational control an EST with the shortest 5 0 UTR (arrow) was chosen as a full-length representative of the cluster/gene and re-arrayed into the unique and full-length set. (b) Criteria for selecting singletons as full-length. EST clones that did not align with any other clones were classified as singletons. Only singletons that contained a long ORF with a start codon preceded by a stop codon were considered for the full-length set. However, since the 50% rule (see above) cannot be applied in this case, Blast information was also used to determine if a singleton is full length. Only if the translational start of a matching protein in the database matched the start codon of the singleton, the latter was included in the full-length set.
+ + Development and Evolution Dorsoventral axis. Developmental Readout. Foundations. Stem cells. Organ formation.
 Development and Evolution 7.72 9.11.06 Dorsoventral axis Human issues Organ formation Stem cells Developmental Readout Axes Growth control Axon guidance 3D structure Analysis Model + + organisms Foundations
Development and Evolution 7.72 9.11.06 Dorsoventral axis Human issues Organ formation Stem cells Developmental Readout Axes Growth control Axon guidance 3D structure Analysis Model + + organisms Foundations
Xenopus gastrulation. Dorsal-Ventral Patterning - The Spemann Organizer. Two mesoderm inducing signals
 Dorsal- atterning - The Spemann Organizer Two mesoderm inducing signals late-blastula early-gastrula nimal Blood Mesothelium Not Vegetal NC Endoderm Hans Spemann (1869-1941) Hilde Mangold (1898-1924) Dr
Dorsal- atterning - The Spemann Organizer Two mesoderm inducing signals late-blastula early-gastrula nimal Blood Mesothelium Not Vegetal NC Endoderm Hans Spemann (1869-1941) Hilde Mangold (1898-1924) Dr
Neural Induction. Chapter One
 Neural Induction Chapter One Fertilization Development of the Nervous System Cleavage (Blastula, Gastrula) Neuronal Induction- Neuroblast Formation Cell Migration Mesodermal Induction Lateral Inhibition
Neural Induction Chapter One Fertilization Development of the Nervous System Cleavage (Blastula, Gastrula) Neuronal Induction- Neuroblast Formation Cell Migration Mesodermal Induction Lateral Inhibition
Supplement Figure S1:
 Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
Supplement Figure S1: A, Sequence of Xcadherin-11 Morpholino 1 (Xcad-11MO) and 2 (Xcad-11 MO2) and control morpholino in comparison to the Xcadherin-11 sequence. The Xcad-11MO binding sequence spans the
7.22 Final Exam points
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 11 7.22 2004 FINAL FOR STUDY
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 11 7.22 2004 FINAL FOR STUDY
Mesoderm Formation. Fate map of early gastrula. Only two types of mesoderm are induced. Mesoderm induction by the vegetal hemisphere
 Fate map of early gastrula Mesoderm Formation Animal hemisphere forms ectoderm (lacks ) Sperm Entry Point dbl dbl brachyury Vegetal hemisphere forms endoderm (requires ) dbl goosecoid Marginal zone forms
Fate map of early gastrula Mesoderm Formation Animal hemisphere forms ectoderm (lacks ) Sperm Entry Point dbl dbl brachyury Vegetal hemisphere forms endoderm (requires ) dbl goosecoid Marginal zone forms
Neural Development. How does a single cell make a brain??? How are different brain regions specified??? Neural Development
 Neural Development How does a single cell make a brain??? How are different brain regions specified??? 1 Neural Development How do cells become neurons? Environmental factors Positional cues Genetic factors
Neural Development How does a single cell make a brain??? How are different brain regions specified??? 1 Neural Development How do cells become neurons? Environmental factors Positional cues Genetic factors
Fertilization. Animal hemisphere. Sperm entry point
 Fertilization Animal hemisphere Sperm entry point Establishes the dorsal/ventral axis Ventral side sperm entry Dorsal side gray crescent Organized by sperm centriole Cleavage Unequal radial holoblastic
Fertilization Animal hemisphere Sperm entry point Establishes the dorsal/ventral axis Ventral side sperm entry Dorsal side gray crescent Organized by sperm centriole Cleavage Unequal radial holoblastic
Patterning of the Brain
 Patterning of the Brain Clemens.Kiecker@kcl.ac.uk IoPPN 27 Oct 2015 Hundreds of billions of cells, hundreds of cell types Egg = single cell, some 20,000 genes Egg = single cell, some 20,000 genes How is
Patterning of the Brain Clemens.Kiecker@kcl.ac.uk IoPPN 27 Oct 2015 Hundreds of billions of cells, hundreds of cell types Egg = single cell, some 20,000 genes Egg = single cell, some 20,000 genes How is
Lecture 3 MOLECULAR REGULATION OF DEVELOPMENT
 Lecture 3 E. M. De Robertis, M.D., Ph.D. August 16, 2016 MOLECULAR REGULATION OF DEVELOPMENT GROWTH FACTOR SIGNALING, HOX GENES, AND THE BODY PLAN Two questions: 1) How is dorsal-ventral (D-V) cell differentiation
Lecture 3 E. M. De Robertis, M.D., Ph.D. August 16, 2016 MOLECULAR REGULATION OF DEVELOPMENT GROWTH FACTOR SIGNALING, HOX GENES, AND THE BODY PLAN Two questions: 1) How is dorsal-ventral (D-V) cell differentiation
LIFE Formation III. Morphogenesis: building 3D structures START FUTURE. How-to 2 PROBLEMS FORMATION SYSTEMS SYSTEMS.
 7.013 4.6.07 Formation III Morphogenesis: building 3D structures VIRUSES CANCER HUMAN DISEASE START SYSTEMS BIOLOGY PROBLEMS FUTURE LIFE IMMUNE NERVOUS SYSTEMS How-to 1 FOUNDATIONS How-to 2 REC. DNA BIOCHEM
7.013 4.6.07 Formation III Morphogenesis: building 3D structures VIRUSES CANCER HUMAN DISEASE START SYSTEMS BIOLOGY PROBLEMS FUTURE LIFE IMMUNE NERVOUS SYSTEMS How-to 1 FOUNDATIONS How-to 2 REC. DNA BIOCHEM
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes
 OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
Later Development. Caenorhabditis elegans. Later Processes 10/06/12. Cytoplasmic Determinants Fate Mapping & Cell Fate Limb Development
 Later Development Cytoplasmic Determinants Fate Mapping & Cell Fate Limb Development Caenorhabditis elegans Nematoda 10,000 worms/petri dish in cultivation short life cycle (~ 3 days egg to egg) wild-type
Later Development Cytoplasmic Determinants Fate Mapping & Cell Fate Limb Development Caenorhabditis elegans Nematoda 10,000 worms/petri dish in cultivation short life cycle (~ 3 days egg to egg) wild-type
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and
 Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
The competence of Xenopus blastomeres to produce neural and retinal progeny is repressed by two endo-mesoderm promoting pathways
 Developmental Biology 305 (2007) 103 119 www.elsevier.com/locate/ydbio The competence of Xenopus blastomeres to produce neural and retinal progeny is repressed by two endo-mesoderm promoting pathways Bo
Developmental Biology 305 (2007) 103 119 www.elsevier.com/locate/ydbio The competence of Xenopus blastomeres to produce neural and retinal progeny is repressed by two endo-mesoderm promoting pathways Bo
CAP BIOINFORMATICS Su-Shing Chen CISE. 10/5/2005 Su-Shing Chen, CISE 1
 CAP 5510-9 BIOINFORMATICS Su-Shing Chen CISE 10/5/2005 Su-Shing Chen, CISE 1 Basic BioTech Processes Hybridization PCR Southern blotting (spot or stain) 10/5/2005 Su-Shing Chen, CISE 2 10/5/2005 Su-Shing
CAP 5510-9 BIOINFORMATICS Su-Shing Chen CISE 10/5/2005 Su-Shing Chen, CISE 1 Basic BioTech Processes Hybridization PCR Southern blotting (spot or stain) 10/5/2005 Su-Shing Chen, CISE 2 10/5/2005 Su-Shing
7.22 Example Problems for Exam 1 The exam will be of this format. It will consist of 2-3 sets scenarios.
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Introduction to Microarray Analysis
 Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Early Development and Axis Formation in Amphibians
 Biology 4361 Early Development and Axis Formation in Amphibians October 25, 2006 Overview Cortical rotation Cleavage Gastrulation Determination the Organizer mesoderm induction Setting up the axes: dorsal/ventral
Biology 4361 Early Development and Axis Formation in Amphibians October 25, 2006 Overview Cortical rotation Cleavage Gastrulation Determination the Organizer mesoderm induction Setting up the axes: dorsal/ventral
Role of Bmi-1 in Epigenetic Regulation. During Early Neural Crest Development
 Role of Bmi-1 in Epigenetic Regulation During Early Neural Crest Development Thesis by Jane I. Khudyakov In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy California Institute
Role of Bmi-1 in Epigenetic Regulation During Early Neural Crest Development Thesis by Jane I. Khudyakov In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy California Institute
TRANSGENIC ANIMALS. transient. stable. - Two methods to produce transgenic animals:
 Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Only for teaching purposes - not for reproduction or sale CELL TRANSFECTION transient stable TRANSGENIC ANIMALS - Two methods to produce transgenic animals: 1- DNA microinjection 2- embryonic stem cell-mediated
Supporting Information
 Supporting Information Lu et al. 10.1073/pnas.1106801108 Fig. S1. Analysis of spatial localization of mrnas coding for 23 zebrafish Wnt genes by whole mount in situ hybridization to identify those present
Supporting Information Lu et al. 10.1073/pnas.1106801108 Fig. S1. Analysis of spatial localization of mrnas coding for 23 zebrafish Wnt genes by whole mount in situ hybridization to identify those present
1. Supplementary Tables
 SUPPLEMENTARY INFORMATION doi:10.1038/nature14215 1. Supplementary Tables Supplementary Table 1 Phenotype of medaka YAP MO-injected embryos Morpholinos Host Amount of MOs Human YAP mrna Total survived
SUPPLEMENTARY INFORMATION doi:10.1038/nature14215 1. Supplementary Tables Supplementary Table 1 Phenotype of medaka YAP MO-injected embryos Morpholinos Host Amount of MOs Human YAP mrna Total survived
Neural Induction. Steven McLoon Department of Neuroscience University of Minnesota
 Neural Induction Steven McLoon Department of Neuroscience University of Minnesota 1 Course News Coffee Hour (with Dr. Nakagawa) Friday, Sept 22 8:30-9:30am Surdyks Café in Northrop Auditorium Stop by for
Neural Induction Steven McLoon Department of Neuroscience University of Minnesota 1 Course News Coffee Hour (with Dr. Nakagawa) Friday, Sept 22 8:30-9:30am Surdyks Café in Northrop Auditorium Stop by for
Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27 XIC1 and imparting a neural fate
 Development 127, 1303-1314 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV1503 1303 Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27 XIC1 and imparting a
Development 127, 1303-1314 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV1503 1303 Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27 XIC1 and imparting a
We are walking and standing with parts of our bodies which could have been used for thinking had they developed in another part of the embryo.
 We are walking and standing with parts of our bodies which could have been used for thinking had they developed in another part of the embryo. Hans Spemann, 1943 Reading from Chapter 3 - types of cell
We are walking and standing with parts of our bodies which could have been used for thinking had they developed in another part of the embryo. Hans Spemann, 1943 Reading from Chapter 3 - types of cell
We are walking and standing with parts of our bodies which could have been used for thinking had they developed in another part of the embryo.
 We are walking and standing with parts of our bodies which could have been used for thinking had they developed in another part of the embryo. Hans Spemann, 1943 Reading from Chapter 3 - types of cell
We are walking and standing with parts of our bodies which could have been used for thinking had they developed in another part of the embryo. Hans Spemann, 1943 Reading from Chapter 3 - types of cell
ANAT2341 Embryology Introduction: Steve Palmer
 ANAT2341 Embryology Introduction: Steve Palmer Course overview Course lecturers specializations and roles Steve Palmer 9385 2957 Course overview Summary, aims and expected outcomes Course overview Graduate
ANAT2341 Embryology Introduction: Steve Palmer Course overview Course lecturers specializations and roles Steve Palmer 9385 2957 Course overview Summary, aims and expected outcomes Course overview Graduate
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Chapter 3. Two Hoxc6 transcripts are differentially expressed and regulate. primary neurogenesis in Xenopus laevis
 Chapter 3 Two Hoxc6 transcripts are differentially expressed and regulate primary neurogenesis in Xenopus laevis Nabila Bardine 1, Cornelia Donow 1, Brigitte Korte 1, Antony J. Durston 2, Walter Knöchel
Chapter 3 Two Hoxc6 transcripts are differentially expressed and regulate primary neurogenesis in Xenopus laevis Nabila Bardine 1, Cornelia Donow 1, Brigitte Korte 1, Antony J. Durston 2, Walter Knöchel
Name AP Biology Mrs. Laux Take home test #11 on Chapters 14, 15, and 17 DUE: MONDAY, DECEMBER 21, 2009
 MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
Lecture 22 Eukaryotic Genes and Genomes III
 Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
Lecture 22 Eukaryotic Genes and Genomes III In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand full of genes.
a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 252 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/91/106088469.jpg) Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2018 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo.
 Gus Frangou, Stephanie Palmer & Mark Groudine Fred Hutchinson Cancer Research Center, Seattle- USA A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo. During mammalian development and differentiation
Gus Frangou, Stephanie Palmer & Mark Groudine Fred Hutchinson Cancer Research Center, Seattle- USA A Genetic Screen to Identify Mammalian Chromatin Modifiers In Vivo. During mammalian development and differentiation
Enhancer genetics Problem set E
 Enhancer genetics Problem set E How are specific cell types generated during development? There are thought to be 10,000 different types of neurons in the human nervous system. How are all of these neural
Enhancer genetics Problem set E How are specific cell types generated during development? There are thought to be 10,000 different types of neurons in the human nervous system. How are all of these neural
Genome annotation & EST
 Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Genome annotation & EST What is genome annotation? The process of taking the raw DNA sequence produced by the genome sequence projects and adding the layers of analysis and interpretation necessary
Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3
 The EMBO Journal Vol.17 No.12 pp.3413 3427, 1998 Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3 Harry V.Isaacs 1, Mary Elizabeth Pownall and Jonathan
The EMBO Journal Vol.17 No.12 pp.3413 3427, 1998 Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3 Harry V.Isaacs 1, Mary Elizabeth Pownall and Jonathan
Readings. Lecture IV. Mechanisms of Neural. Neural Development. September 10, Bio 3411 Lecture IV. Mechanisms of Neural Development
 Readings Lecture IV. Mechanisms of Neural NEUROSCIENCE: References : 5 th ed, pp 477-506 (sorta) 4 th ed, pp 545-575 (sorta) Fainsod, A., Steinbeisser, H., & De Robertis, E. M. (1994). EMBO J, 13(21),
Readings Lecture IV. Mechanisms of Neural NEUROSCIENCE: References : 5 th ed, pp 477-506 (sorta) 4 th ed, pp 545-575 (sorta) Fainsod, A., Steinbeisser, H., & De Robertis, E. M. (1994). EMBO J, 13(21),
regression t value two sample t values regression line
 Suppl. Table 1: Testing SCRE sequence information without the structural constraint SCRE t test on difference of coefficients SCRE t value sequence only t value Dm1 6.62 6.84 1.29 Dm2 7.06 7.46 1.84 Dm3
Suppl. Table 1: Testing SCRE sequence information without the structural constraint SCRE t test on difference of coefficients SCRE t value sequence only t value Dm1 6.62 6.84 1.29 Dm2 7.06 7.46 1.84 Dm3
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Characterization of a novel gene involved in border cell migration
 Characterization of a novel gene involved in border cell migration Wei-Hao Li, He-Yen Chou and Li-Mei Pai Graduate Institute of Basic Medical Science Chang-Gung University, Tao-Yuan, Taiwan, R.O.C. Abstract
Characterization of a novel gene involved in border cell migration Wei-Hao Li, He-Yen Chou and Li-Mei Pai Graduate Institute of Basic Medical Science Chang-Gung University, Tao-Yuan, Taiwan, R.O.C. Abstract
Thyroid hormone transcriptome and cerebellum development.
 Thyroid hormone transcriptome and cerebellum development. 1) Developmental biology: General concepts 2) Genomic data: mouse and human genomes global view 3) A biological problem: mouse cerebellum and thyroid
Thyroid hormone transcriptome and cerebellum development. 1) Developmental biology: General concepts 2) Genomic data: mouse and human genomes global view 3) A biological problem: mouse cerebellum and thyroid
Trasposable elements: Uses of P elements Problem set B at the end
 Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Trasposable elements: Uses of P elements Problem set B at the end P-elements have revolutionized the way Drosophila geneticists conduct their research. Here, we will discuss just a few of the approaches
Gene Identification in silico
 Gene Identification in silico Nita Parekh, IIIT Hyderabad Presented at National Seminar on Bioinformatics and Functional Genomics, at Bioinformatics centre, Pondicherry University, Feb 15 17, 2006. Introduction
Gene Identification in silico Nita Parekh, IIIT Hyderabad Presented at National Seminar on Bioinformatics and Functional Genomics, at Bioinformatics centre, Pondicherry University, Feb 15 17, 2006. Introduction
a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.
![a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males. a) Stock 4: 126 flies total: 120 are P[w+]: 64 are CyO; 56 are TM3; all are females. 132 are w-: 68 are CyO; 64 are TM3; all are males.](/thumbs/88/116538069.jpg) Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Drosophila Problem Set 2012 1) You have created P element transformants of a construct that contains the mini-white gene, which confers an orange eye color in a homozygous white mutant background. For
Il trascrittoma dei mammiferi
 29 Novembre 2005 Il trascrittoma dei mammiferi dott. Manuela Gariboldi Gruppo di ricerca IFOM: Genetica molecolare dei tumori (responsabile dott. Paolo Radice) Copyright 2005 IFOM Fondazione Istituto FIRC
29 Novembre 2005 Il trascrittoma dei mammiferi dott. Manuela Gariboldi Gruppo di ricerca IFOM: Genetica molecolare dei tumori (responsabile dott. Paolo Radice) Copyright 2005 IFOM Fondazione Istituto FIRC
The Ensembl Database. Dott.ssa Inga Prokopenko. Corso di Genomica
 The Ensembl Database Dott.ssa Inga Prokopenko Corso di Genomica 1 www.ensembl.org Lecture 7.1 2 What is Ensembl? Public annotation of mammalian and other genomes Open source software Relational database
The Ensembl Database Dott.ssa Inga Prokopenko Corso di Genomica 1 www.ensembl.org Lecture 7.1 2 What is Ensembl? Public annotation of mammalian and other genomes Open source software Relational database
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl
 Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
Supplementary Fig. S1. Schematic representation of mouse lines Pax6 fl/fl and mrx-cre used in this study. (A) To generate Pax6 fl/ fl, loxp sites flanking exons 3-6 (red arrowheads) were introduced into
RNAs were transcribed from described expression conhibit a similar capacity to synergize with neural-inducing
 DEVELOPMENTAL BIOLOGY 172, 337 342 (1995) RAPID COMMUNICATION Specification of the Anteroposterior Neural Axis through Synergistic Interaction of the Wnt Signaling Cascade with noggin and follistatin L.
DEVELOPMENTAL BIOLOGY 172, 337 342 (1995) RAPID COMMUNICATION Specification of the Anteroposterior Neural Axis through Synergistic Interaction of the Wnt Signaling Cascade with noggin and follistatin L.
7 Gene Isolation and Analysis of Multiple
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 7 Gene Isolation and Analysis of Multiple
4 Mutant Hunts - To Select or to Screen (Perhaps Even by Brute Force)
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Understanding embryonic head development. ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute
 Understanding embryonic head development ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute Head malformations among the most common category of congenital malformations in
Understanding embryonic head development ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute Head malformations among the most common category of congenital malformations in
Pregnenolone Stabilizes Microtubules and Promotes Zebrafish Embryonic Cell Movement
 Pregnenolone Stabilizes Microtubules and Promotes Zebrafish Embryonic Cell Movement Hwei-Jan Hsu 1,2, Ming-Ren Liang 3, Chao-Tsen Chen 3, and Bon-Chu Chung 1 * Abstract Embryonic cell movement is essential
Pregnenolone Stabilizes Microtubules and Promotes Zebrafish Embryonic Cell Movement Hwei-Jan Hsu 1,2, Ming-Ren Liang 3, Chao-Tsen Chen 3, and Bon-Chu Chung 1 * Abstract Embryonic cell movement is essential
3. human genomics clone genes associated with genetic disorders. 4. many projects generate ordered clones that cover genome
 Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
Lectures 30 and 31 Genome analysis I. Genome analysis A. two general areas 1. structural 2. functional B. genome projects a status report 1. 1 st sequenced: several viral genomes 2. mitochondria and chloroplasts
A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos
 Development 126, 4715-4728 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV4207 4715 A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus
Development 126, 4715-4728 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV4207 4715 A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
(a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for
 Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
Supplementary Figure 1 Generation of Diaph3 ko mice. (a) Scheme depicting the strategy used to generate the ko and conditional alleles. (b) RT-PCR for different regions of Diaph3 mrna from WT, heterozygote
MODULE 5: TRANSLATION
 MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
MODULE 5: TRANSLATION Lesson Plan: CARINA ENDRES HOWELL, LEOCADIA PALIULIS Title Translation Objectives Determine the codons for specific amino acids and identify reading frames by looking at the Base
beta-galactosidase (A420) ctrl
 0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
0.6 beta-galactosidase (A420) 0.4 0.2 ctrl 2A 22A 35B 36B 58A 64A 68E 75C 86Fa 86Fb 102F Fig. S1. Levels of induced expression at various attp sites. The induced β-galactosidase activity was measured from
Branches of Genetics
 Branches of Genetics 1. Transmission genetics Classical genetics or Mendelian genetics 2. Molecular genetics chromosomes, DNA, regulation of gene expression recombinant DNA, biotechnology, bioinformatics,
Branches of Genetics 1. Transmission genetics Classical genetics or Mendelian genetics 2. Molecular genetics chromosomes, DNA, regulation of gene expression recombinant DNA, biotechnology, bioinformatics,
CRISPR GENOMIC SERVICES PRODUCT CATALOG
 CRISPR GENOMIC SERVICES PRODUCT CATALOG DESIGN BUILD ANALYZE The experts at Desktop Genetics can help you design, prepare and manufacture all of the components needed for your CRISPR screen. We provide
CRISPR GENOMIC SERVICES PRODUCT CATALOG DESIGN BUILD ANALYZE The experts at Desktop Genetics can help you design, prepare and manufacture all of the components needed for your CRISPR screen. We provide
AP Biology Gene Expression/Biotechnology REVIEW
 AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
AP Biology Gene Expression/Biotechnology REVIEW Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Gene expression can be a. regulated before transcription.
Lecture 8: Transgenic Model Systems and RNAi
 Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
Lecture 8: Transgenic Model Systems and RNAi I. Model systems 1. Caenorhabditis elegans Caenorhabditis elegans is a microscopic (~1 mm) nematode (roundworm) that normally lives in soil. It has become one
COMPUTER RESOURCES II:
 COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
Transcriptomics. Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona
 Transcriptomics Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Central dogma of molecular biology Central dogma of molecular biology Genome Complete DNA content of
Transcriptomics Marta Puig Institut de Biotecnologia i Biomedicina Universitat Autònoma de Barcelona Central dogma of molecular biology Central dogma of molecular biology Genome Complete DNA content of
efgf, Xcad3 and Hox genes form a molecular pathway that establishes the
 Development 122, 3881-3892 (1996) Printed in Great Britain The Company of Biologists Limited 1996 DEV3573 3881 efgf, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis
Development 122, 3881-3892 (1996) Printed in Great Britain The Company of Biologists Limited 1996 DEV3573 3881 efgf, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis
The F-box protein Cdc4/Fbxw7 is a novel regulator of neural crest development in Xenopus laevis
 RESEARCH ARTICLE Open Access The F-box protein Cdc4/Fbxw7 is a novel regulator of neural crest development in Xenopus laevis Alexandra D Almeida 1, Helen M Wise 2, Christopher J Hindley 1, Michael K Slevin
RESEARCH ARTICLE Open Access The F-box protein Cdc4/Fbxw7 is a novel regulator of neural crest development in Xenopus laevis Alexandra D Almeida 1, Helen M Wise 2, Christopher J Hindley 1, Michael K Slevin
7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV
 1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
1 Fall 2006 7.03 7.03, 2006, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV
 7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
7.05, 2005, Lecture 23 Eukaryotic Genes and Genomes IV In the last three lectures we have thought a lot about analyzing a regulatory system in S. cerevisiae, namely Gal regulation that involved a hand
GENES AND CHROMOSOMES V. Lecture 7. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 1 GENES AND CHROMOSOMES V Lecture 7 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University 2 CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION An Overview of Gene Regulation in Eukaryotes
1 GENES AND CHROMOSOMES V Lecture 7 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University 2 CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION An Overview of Gene Regulation in Eukaryotes
Genomic Annotation Lab Exercise By Jacob Jipp and Marian Kaehler Luther College, Department of Biology Genomics Education Partnership 2010
 Genomic Annotation Lab Exercise By Jacob Jipp and Marian Kaehler Luther College, Department of Biology Genomics Education Partnership 2010 Genomics is a new and expanding field with an increasing impact
Genomic Annotation Lab Exercise By Jacob Jipp and Marian Kaehler Luther College, Department of Biology Genomics Education Partnership 2010 Genomics is a new and expanding field with an increasing impact
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Gene Expression: Transcription
 Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Reviewers' Comments: Reviewer #1 (Remarks to the Author)
 Reviewers' Comments: Reviewer #1 (Remarks to the Author) In this study, Rosenbluh et al reported direct comparison of two screening approaches: one is genome editing-based method using CRISPR-Cas9 (cutting,
Reviewers' Comments: Reviewer #1 (Remarks to the Author) In this study, Rosenbluh et al reported direct comparison of two screening approaches: one is genome editing-based method using CRISPR-Cas9 (cutting,
Two classes of genetic pathways I) Developmental/synthetic pathways II) Regulatory/binary switch pathways
 Two classes of genetic pathways I) Developmental/synthetic pathways II) Regulatory/binary switch pathways I) Developmental pathways Synthesis and/or assembly of molecule(s), subcellular, cellular or multicellular
Two classes of genetic pathways I) Developmental/synthetic pathways II) Regulatory/binary switch pathways I) Developmental pathways Synthesis and/or assembly of molecule(s), subcellular, cellular or multicellular
 Background Wikipedia Lee and Mahadavan, JCB, 2009 History (Platform Comparison) P Park, Nature Review Genetics, 2009 P Park, Nature Reviews Genetics, 2009 Rozowsky et al., Nature Biotechnology, 2009
Background Wikipedia Lee and Mahadavan, JCB, 2009 History (Platform Comparison) P Park, Nature Review Genetics, 2009 P Park, Nature Reviews Genetics, 2009 Rozowsky et al., Nature Biotechnology, 2009
Neural crest induction in Xenopus: evidence for a two-signal model
 Development 125, 2403-2414 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV4980 2403 Neural crest induction in Xenopus: evidence for a two-signal model Carole LaBonne* and Marianne
Development 125, 2403-2414 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV4980 2403 Neural crest induction in Xenopus: evidence for a two-signal model Carole LaBonne* and Marianne
Technical Advancement
 INVITED ARTICLE Acta Histochem. Cytochem. 35 (5): 361 365, 2002 Technical Advancement Using Antisense Morpholino Oligos to Knockdown Gene Expression in the Chicken Embryo Richard P. Tucker 1 1 Department
INVITED ARTICLE Acta Histochem. Cytochem. 35 (5): 361 365, 2002 Technical Advancement Using Antisense Morpholino Oligos to Knockdown Gene Expression in the Chicken Embryo Richard P. Tucker 1 1 Department
Expression Pattern of Xtshz1 and Xtshz3 During Xenopus Early Eye Formation
 SUNY College of Environmental Science and Forestry Digital Commons @ ESF Honors Theses 5-2012 Expression Pattern of Xtshz1 and Xtshz3 During Xenopus Early Eye Formation Katherine L. McKissick Follow this
SUNY College of Environmental Science and Forestry Digital Commons @ ESF Honors Theses 5-2012 Expression Pattern of Xtshz1 and Xtshz3 During Xenopus Early Eye Formation Katherine L. McKissick Follow this
Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. Nature Methods: doi: /nmeth.3360
 Supplementary Figure 1 Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. (a) Wild-type, 2-dpf slc24a5 b1 embryos and a series of slc24a5 CRISPR-injected embryos; anterior is
Supplementary Figure 1 Optimizing Cas9 and sgrna concentrations for increasing mutation efficiency. (a) Wild-type, 2-dpf slc24a5 b1 embryos and a series of slc24a5 CRISPR-injected embryos; anterior is
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway
 Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Estradiol-Estrogen Receptor α Mediates the Expression of the CXXC5 Gene through the Estrogen Response Element-Dependent Signaling Pathway Pelin Yaşar, Gamze Ayaz and Mesut Muyan SUPPLEMENTARY INFORMATION
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling
 Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling Go-Woon Kim, Jong-Hoon Won, Ok-Kyung Lee, Sang-Soo Lee, Jeong-Hoon Han, Orkhon Tsogtbaatar, Sujin Nam, Yeon Kim, and Kyung-Ok Cho* Supplementary
Basics of RNA-Seq. (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly, PhD Team Lead, NCI Single Cell Analysis Facility
 2018 ABRF Meeting Satellite Workshop 4 Bridging the Gap: Isolation to Translation (Single Cell RNA-Seq) Sunday, April 22 Basics of RNA-Seq (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly,
2018 ABRF Meeting Satellite Workshop 4 Bridging the Gap: Isolation to Translation (Single Cell RNA-Seq) Sunday, April 22 Basics of RNA-Seq (With a Focus on Application to Single Cell RNA-Seq) Michael Kelly,
Non-coding Function & Variation, MPRAs II. Mike White Bio /5/18
 Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Non-coding Function & Variation, MPRAs II Mike White Bio 5488 3/5/18 MPRA Review Problem 1: Where does your CRE DNA come from? DNA synthesis Genomic fragments Targeted regulome capture Problem 2: How do
Homework 4. Due in class, Wednesday, November 10, 2004
 1 GCB 535 / CIS 535 Fall 2004 Homework 4 Due in class, Wednesday, November 10, 2004 Comparative genomics 1. (6 pts) In Loots s paper (http://www.seas.upenn.edu/~cis535/lab/sciences-loots.pdf), the authors
1 GCB 535 / CIS 535 Fall 2004 Homework 4 Due in class, Wednesday, November 10, 2004 Comparative genomics 1. (6 pts) In Loots s paper (http://www.seas.upenn.edu/~cis535/lab/sciences-loots.pdf), the authors
PLNT2530 (2018) Unit 6b Sequence Libraries
 PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
Bi8 Lecture 19. Review and Practice Questions March 8th 2016
 Bi8 Lecture 19 Review and Practice Questions March 8th 2016 Common Misconceptions Where Words Matter Common Misconceptions DNA vs RNA vs Protein Su(H) vs Su(H) Replication Transcription Transcribed vs
Bi8 Lecture 19 Review and Practice Questions March 8th 2016 Common Misconceptions Where Words Matter Common Misconceptions DNA vs RNA vs Protein Su(H) vs Su(H) Replication Transcription Transcribed vs
Supplementary Figure 1. Retinogenesis during human embryonic development and in 3-D retinal cups (RCs) derived from hipsc. a, Main steps leading to
 Supplementary Figure 1. Retinogenesis during human embryonic development and in 3-D retinal cups (RCs) derived from hipsc. a, Main steps leading to the formation of the human retina in vivo. b, Diagram
Supplementary Figure 1. Retinogenesis during human embryonic development and in 3-D retinal cups (RCs) derived from hipsc. a, Main steps leading to the formation of the human retina in vivo. b, Diagram
Hemidesmosome. Focal adhesion
 BIOLOGY 52 - - CELL AND DEVELOPMENTAL BIOLOGY - - FALL 2002 Third Examination - - November, 2002 -------------------------------------------------- Put your name at the top of each page. Please read each
BIOLOGY 52 - - CELL AND DEVELOPMENTAL BIOLOGY - - FALL 2002 Third Examination - - November, 2002 -------------------------------------------------- Put your name at the top of each page. Please read each
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Expressed genes profiling (Microarrays) Overview Of Gene Expression Control Profiling Of Expressed Genes
 Expressed genes profiling (Microarrays) Overview Of Gene Expression Control Profiling Of Expressed Genes Genes can be regulated at many levels Usually, gene regulation, are referring to transcriptional
Expressed genes profiling (Microarrays) Overview Of Gene Expression Control Profiling Of Expressed Genes Genes can be regulated at many levels Usually, gene regulation, are referring to transcriptional
Genome Biology and Biotechnology
 Genome Biology and Biotechnology Functional Genomics Prof. M. Zabeau Department of Plant Systems Biology Flanders Interuniversity Institute for Biotechnology (VIB) University of Gent International course
Genome Biology and Biotechnology Functional Genomics Prof. M. Zabeau Department of Plant Systems Biology Flanders Interuniversity Institute for Biotechnology (VIB) University of Gent International course
Pharmacogenetics: A SNPshot of the Future. Ani Khondkaryan Genomics, Bioinformatics, and Medicine Spring 2001
 Pharmacogenetics: A SNPshot of the Future Ani Khondkaryan Genomics, Bioinformatics, and Medicine Spring 2001 1 I. What is pharmacogenetics? It is the study of how genetic variation affects drug response
Pharmacogenetics: A SNPshot of the Future Ani Khondkaryan Genomics, Bioinformatics, and Medicine Spring 2001 1 I. What is pharmacogenetics? It is the study of how genetic variation affects drug response
Culture and differentiation of embryonic stem cells. Hong-Lin Su Department of Life Sciences, National Chung-Hsing University
 Culture and differentiation of embryonic stem cells Hong-Lin Su Department of Life Sciences, National Chung-Hsing University Topics-ES cell maintenance Establishment Culture condition, including the serum,
Culture and differentiation of embryonic stem cells Hong-Lin Su Department of Life Sciences, National Chung-Hsing University Topics-ES cell maintenance Establishment Culture condition, including the serum,
Sydney Brenner: Present
 Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
Sydney Brenner: 1927 - Present Born in the small town of Germiston, South Africa Won a scholarship to the University of Witwatersrand at the age of 15 Received his PhD from Exeter College, Oxford Made
There are four major types of introns. Group I introns, found in some rrna genes, are self-splicing: they can catalyze their own removal.
 1 2 Continuous genes - Intron: Many eukaryotic genes contain coding regions called exons and noncoding regions called intervening sequences or introns. The average human gene contains from eight to nine
1 2 Continuous genes - Intron: Many eukaryotic genes contain coding regions called exons and noncoding regions called intervening sequences or introns. The average human gene contains from eight to nine
Reading. Lecture III. Nervous System Embryology. Biology. Brain Diseases. September 5, Bio 3411 Lecture III. Nervous System Embryology
 Reading NEUROSCIENCE: 5 th ed, pp. 477-506 NEUROSCIENCE: 4 th ed, pp. 545-575 Bio 3411 Wednesday 2 Summary from Lecture II Biology Understanding the brain is THE major question in biology and science.
Reading NEUROSCIENCE: 5 th ed, pp. 477-506 NEUROSCIENCE: 4 th ed, pp. 545-575 Bio 3411 Wednesday 2 Summary from Lecture II Biology Understanding the brain is THE major question in biology and science.
Review Article On becoming neural: what the embryo can tell us about differentiating neural stem cells
 Am J Stem Cells 2013;2(2):74-94 www.ajsc.us /ISSN:2160-4150/AJSC1303003 Review Article : what the embryo can tell us about differentiating neural stem cells Sally A Moody 1,3, Steven L Klein 1, Beverley
Am J Stem Cells 2013;2(2):74-94 www.ajsc.us /ISSN:2160-4150/AJSC1303003 Review Article : what the embryo can tell us about differentiating neural stem cells Sally A Moody 1,3, Steven L Klein 1, Beverley
Concepts and Methods in Developmental Biology
 Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
Biology 4361 Developmental Biology Concepts and Methods in Developmental Biology June 16, 2009 Conceptual and Methodological Tools Concepts Genomic equivalence Differential gene expression Differentiation/de-differentiation
