Bioinformatic tools for metagenomic data analysis
|
|
|
- Jonathan Jenkins
- 6 years ago
- Views:
Transcription
1 Bioinformatic tools for metagenomic data analysis MEGAN - blast-based tool for exploring taxonomic content MG-RAST (SEED, FIG) - rapid annotation of metagenomic data, phylogenetic classification and metabolic reconstruction CAMERA (JCVI, Calit2, UCSD) - metagenomic data repository and blast server 1
2 Getting started Download MEGAN and metagenomic datasets BLAST files: 2
3 MEGAN (Metagenome analyzer) D.H. Huson et al., MEGAN analysis of metagenome data. Genome Research, 17: (2007). Software designed to analyze metagenomic datasets and assign reads to a known taxon. What organisms are present and what are their relative proportions? What types of types of genes are present? 3
4 Input files: - BLAST files; BLASTX, BLASTN or BLASTZ - NCBI-NR, NCBI-NT, NCBI-ENV-NR, NCBI-ENV-NT or genome specific databases Image from Huson et al., 2007 MEGAN uses NCBI taxonomy to summarize and order results. 4
5 MEGAN (Metagenomics analyzer) Lowest common ancestor (LCA) assignment algorithm: Filters set by user: - Min. Support for Taxa (default = 2) - Min. Score (default = 30.0) - Top Percentage (default = 10.0) - Win ( Winner ) Score (default = 0) 5
6 Comparison of metagenomic samples Single metagenomic sample 6
7 MEGAN (Metagenomics analyzer) Let s start playing with MEGAN! 7
8 MEGAN on startup screen: - will take a minute or so to load NCBI tree and taxonomy Default tree showing NCBI taxonomy 8
9 Step 1: Importing a BLAST file: - File Import. Choose import file type (e.g. BLASTX, BLASTN) 9
10 Step 2: Choosing MEGAN output format: - Full Dataset OR Summary (choose Full Dataset - greater control over filters) 10
11 Step 3: Importing and parsing the BLAST file: - Note: some taxa will remain unidentified! Summary of blast results 11
12 Step 4: Generating tree showing taxonomic distribution of reads: - Note: NCBI tree replaced by taxonomy of assigned reads Displays tree at the species level 12
13 Step 5: Setting filtering criteria - Options Min Support, Min Score, Top Percentage and Win Score 13
14 Step 6: Show the number of reads assigned to each taxa/node - Tree Show number of Reads Assigned 14
15 Step 7: Collapsing/expanding the tree to different taxonomic levels -Tree Collapse at Taxonomical Level - choose higher level to collapse, or lower level to expand 15
16 Step 8: Changing the vertical and horizontal view of the tree Expand Expand vertically horizontally Contract Contract vertically horizontally 16
17 Step 9: Taxon discovery rate - Window Chart Taxon Discovery Rate 17
18 Step 9: Exporting and saving MEGAN output - File Save As - File Export Image 18
19 Exercise: Comparing metagenomic samples using MEGAN 1) Import the BLASTX against NR file for the OMZ 20m dataset. **Note: this should be done already** (File name: OMZ_20m_allDNA.fna.5000VSnr.FF.b10.v10.c250.blastx) 2) Import the BLASTX against NR files for the OMZ 45m OR 60m dataset. (File name: OMZ_45m_allDNA.fna.5000VSnr.FF.b10.v10.c250.blastx OR OMZ_60m_allDNA.fna.5000VSnr.FF.b10.v10.c250.blastx) 3) Change the Min Score filter to a bitscore of 40 for each dataset. 4) Using the Compare tool (found under File--->Compare), select both datasets and choose the normalize over all reads option. 5) Collapse the resulting tree to the phylum level. Note any major differences in the representation of difference taxonomic groups. 19
20 MG-RAST MetaGenome Rapid Annotation using Subsystems Technology Login: CMORE Password: CMORE - Accurate and consistent annotations of metagenomic data (12-24 hours) - Currently accepts 454 and Sanger sequences - Automatic metabolic reconstruction - Phylogenetic classification of rrnas and proteins - Comparison tools 20
21 MG-RAST Subsystem a generalization of pathway as a collection of functional roles jointly involved in a biological process or complex * Submit fasta nucleotide sequences * Project name and description * Metadata (location, depth, altitude, time of collection, habitat) 21
22 CAMERA Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis The aim of this project is to serve the needs of the microbial ecology research community by creating a rich, distinctive data repository and a bioinformatics tools resource that will address many of the unique challenges of metagenomic analysis. 22
23 CAMERA Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis CAMERA requires registration: CMORE_2008 CMORE
24 USEFUL LINKS MARINE MICRO SPECIFIC GENERIC TOOLS AND MICROBIAL GENOME EXPLORATION RIBOSOMAL RNA DATABASE AND PROBE RESOURCES AND TOOLS TARGETED PROTEIN DATABASES AND SEARCH TOOLS ONLINE
Bioinformatics for Microbial Biology
 Bioinformatics for Microbial Biology Chaochun Wei ( 韦朝春 ) ccwei@sjtu.edu.cn http://cbb.sjtu.edu.cn/~ccwei Fall 2013 1 Outline Part I: Visualization tools for microbial genomes Tools: Gbrowser Part II:
Bioinformatics for Microbial Biology Chaochun Wei ( 韦朝春 ) ccwei@sjtu.edu.cn http://cbb.sjtu.edu.cn/~ccwei Fall 2013 1 Outline Part I: Visualization tools for microbial genomes Tools: Gbrowser Part II:
CBC Data Therapy. Metagenomics Discussion
 CBC Data Therapy Metagenomics Discussion General Workflow Microbial sample Generate Metaomic data Process data (QC, etc.) Analysis Marker Genes Extract DNA Amplify with targeted primers Filter errors,
CBC Data Therapy Metagenomics Discussion General Workflow Microbial sample Generate Metaomic data Process data (QC, etc.) Analysis Marker Genes Extract DNA Amplify with targeted primers Filter errors,
METAGENOME ANALYSIS USING MEGAN
 METAGENOME ANALYSIS USING MEGAN DANIEL H. HUSON and ALEXANDER F. AUCH Center for Bioinformatics, Tübingen University, Sand 14, 72076 Tübingen, Germany QI JI and STEPHAN C. SCHUSTER 310 Wartik Laboratories,
METAGENOME ANALYSIS USING MEGAN DANIEL H. HUSON and ALEXANDER F. AUCH Center for Bioinformatics, Tübingen University, Sand 14, 72076 Tübingen, Germany QI JI and STEPHAN C. SCHUSTER 310 Wartik Laboratories,
Functional annotation of metagenomes
 Functional annotation of metagenomes Jeroen F. J. Laros Leiden Genome Technology Center Department of Human Genetics Center for Human and Clinical Genetics Introduction Functional analysis Objectives:
Functional annotation of metagenomes Jeroen F. J. Laros Leiden Genome Technology Center Department of Human Genetics Center for Human and Clinical Genetics Introduction Functional analysis Objectives:
Assigning Sequences to Taxa CMSC828G
 Assigning Sequences to Taxa CMSC828G Outline Objective (1 slide) MEGAN (17 slides) SAP (33 slides) Conclusion (1 slide) Objective Given an unknown, environmental DNA sequence: Make a taxonomic assignment
Assigning Sequences to Taxa CMSC828G Outline Objective (1 slide) MEGAN (17 slides) SAP (33 slides) Conclusion (1 slide) Objective Given an unknown, environmental DNA sequence: Make a taxonomic assignment
CBC Data Therapy. Metatranscriptomics Discussion
 CBC Data Therapy Metatranscriptomics Discussion Metatranscriptomics Extract RNA, subtract rrna Sequence cdna QC Gene expression, function Institute for Systems Genomics: Computational Biology Core bioinformatics.uconn.edu
CBC Data Therapy Metatranscriptomics Discussion Metatranscriptomics Extract RNA, subtract rrna Sequence cdna QC Gene expression, function Institute for Systems Genomics: Computational Biology Core bioinformatics.uconn.edu
From DNA Sequences to Genome-Scale Metabolic Models (to Network Projects in CellNetAnalyzer)
 From DNA Sequences to Genome-Scale Metabolic Models (to Network Projects in CellNetAnalyzer) Sven Thiele Max Planck Institute for Dynamics of Complex Technical Systems, Germany Introduction A genome-scale
From DNA Sequences to Genome-Scale Metabolic Models (to Network Projects in CellNetAnalyzer) Sven Thiele Max Planck Institute for Dynamics of Complex Technical Systems, Germany Introduction A genome-scale
Computational Aspects of Metagenome Analysis
 Computational Aspects of Metagenome Analysis http://soils.usda.gov Daniel H. Huson Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or modify this document under the terms
Computational Aspects of Metagenome Analysis http://soils.usda.gov Daniel H. Huson Copyright (c) 2008 Daniel Huson. Permission is granted to copy, distribute and/or modify this document under the terms
TECHNIQUES FOR STUDYING METAGENOME DATASETS METAGENOMES TO SYSTEMS.
 TECHNIQUES FOR STUDYING METAGENOME DATASETS METAGENOMES TO SYSTEMS. Ian Jeffery I.Jeffery@ucc.ie What is metagenomics Metagenomics is the study of genetic material recovered directly from environmental
TECHNIQUES FOR STUDYING METAGENOME DATASETS METAGENOMES TO SYSTEMS. Ian Jeffery I.Jeffery@ucc.ie What is metagenomics Metagenomics is the study of genetic material recovered directly from environmental
Supplementary Figure 1. Design of the control microarray. a, Genomic DNA from the
 Supplementary Information Supplementary Figures Supplementary Figure 1. Design of the control microarray. a, Genomic DNA from the strain M8 of S. ruber and a fosmid containing the S. ruber M8 virus M8CR4
Supplementary Information Supplementary Figures Supplementary Figure 1. Design of the control microarray. a, Genomic DNA from the strain M8 of S. ruber and a fosmid containing the S. ruber M8 virus M8CR4
Download the Lectin sequence output from
 Computer Analysis of DNA and Protein Sequences Over the Internet Part I. IN CLASS Download the Lectin sequence output from http://stan.cropsci.uiuc.edu/courses/cpsc265/ Open these in BioEdit (free software).
Computer Analysis of DNA and Protein Sequences Over the Internet Part I. IN CLASS Download the Lectin sequence output from http://stan.cropsci.uiuc.edu/courses/cpsc265/ Open these in BioEdit (free software).
Prokaryotic Annotation Pipeline SOP HGSC, Baylor College of Medicine
 1 Abstract A prokaryotic annotation pipeline was developed to automatically annotate draft and complete bacterial genomes. The protein coding genes in the genomes are predicted by the combination of Glimmer
1 Abstract A prokaryotic annotation pipeline was developed to automatically annotate draft and complete bacterial genomes. The protein coding genes in the genomes are predicted by the combination of Glimmer
Practical Bioinformatics for Life Scientists. Week 14, Lecture 27. István Albert Bioinformatics Consulting Center Penn State
 Practical Bioinformatics for Life Scientists Week 14, Lecture 27 István Albert Bioinformatics Consulting Center Penn State No homework this week Project to be given out next Thursday (Dec 1 st ) Due following
Practical Bioinformatics for Life Scientists Week 14, Lecture 27 István Albert Bioinformatics Consulting Center Penn State No homework this week Project to be given out next Thursday (Dec 1 st ) Due following
Leonardo Mariño-Ramírez, PhD NCBI / NLM / NIH. BIOL 7210 A Computational Genomics 2/18/2015
 Leonardo Mariño-Ramírez, PhD NCBI / NLM / NIH BIOL 7210 A Computational Genomics 2/18/2015 The $1,000 genome is here! http://www.illumina.com/systems/hiseq-x-sequencing-system.ilmn Bioinformatics bottleneck
Leonardo Mariño-Ramírez, PhD NCBI / NLM / NIH BIOL 7210 A Computational Genomics 2/18/2015 The $1,000 genome is here! http://www.illumina.com/systems/hiseq-x-sequencing-system.ilmn Bioinformatics bottleneck
BIOINFORMATICS TO ANALYZE AND COMPARE GENOMES
 BIOINFORMATICS TO ANALYZE AND COMPARE GENOMES We sequenced and assembled a genome, but this is only a long stretch of ATCG What should we do now? 1. find genes What are the starting and end points for
BIOINFORMATICS TO ANALYZE AND COMPARE GENOMES We sequenced and assembled a genome, but this is only a long stretch of ATCG What should we do now? 1. find genes What are the starting and end points for
What will be covered?
 What will be covered? 1. Annotation overview 2. Using the RAST family for genome annotation: Optimizing RAST for phages Command line/ Batch options 3. Introducing PATRIC and resources in development Therapeutic
What will be covered? 1. Annotation overview 2. Using the RAST family for genome annotation: Optimizing RAST for phages Command line/ Batch options 3. Introducing PATRIC and resources in development Therapeutic
GPU-Meta-Storms: Computing the similarities among massive microbial communities using GPU
 GPU-Meta-Storms: Computing the similarities among massive microbial communities using GPU Xiaoquan Su $, Xuetao Wang $, JianXu, Kang Ning* Shandong Key Laboratory of Energy Genetics, CAS Key Laboratory
GPU-Meta-Storms: Computing the similarities among massive microbial communities using GPU Xiaoquan Su $, Xuetao Wang $, JianXu, Kang Ning* Shandong Key Laboratory of Energy Genetics, CAS Key Laboratory
ELE4120 Bioinformatics. Tutorial 5
 ELE4120 Bioinformatics Tutorial 5 1 1. Database Content GenBank RefSeq TPA UniProt 2. Database Searches 2 Databases A common situation for alignment is to search through a database to retrieve the similar
ELE4120 Bioinformatics Tutorial 5 1 1. Database Content GenBank RefSeq TPA UniProt 2. Database Searches 2 Databases A common situation for alignment is to search through a database to retrieve the similar
Carl Woese. Used 16S rrna to developed a method to Identify any bacterium, and discovered a novel domain of life
 METAGENOMICS Carl Woese Used 16S rrna to developed a method to Identify any bacterium, and discovered a novel domain of life His amazing discovery, coupled with his solitary behaviour, made many contemporary
METAGENOMICS Carl Woese Used 16S rrna to developed a method to Identify any bacterium, and discovered a novel domain of life His amazing discovery, coupled with his solitary behaviour, made many contemporary
Carl Woese. Used 16S rrna to develop a method to Identify any bacterium, and discovered a novel domain of life
 METAGENOMICS Carl Woese Used 16S rrna to develop a method to Identify any bacterium, and discovered a novel domain of life His amazing discovery, coupled with his solitary behaviour, made many contemporary
METAGENOMICS Carl Woese Used 16S rrna to develop a method to Identify any bacterium, and discovered a novel domain of life His amazing discovery, coupled with his solitary behaviour, made many contemporary
Exercise I, Sequence Analysis
 Exercise I, Sequence Analysis atgcacttgagcagggaagaaatccacaaggactcaccagtctcctggtctgcagagaagacagaatcaacatgagcacagcaggaaaa gtaatcaaatgcaaagcagctgtgctatgggagttaaagaaacccttttccattgaggaggtggaggttgcacctcctaaggcccatgaagt
Exercise I, Sequence Analysis atgcacttgagcagggaagaaatccacaaggactcaccagtctcctggtctgcagagaagacagaatcaacatgagcacagcaggaaaa gtaatcaaatgcaaagcagctgtgctatgggagttaaagaaacccttttccattgaggaggtggaggttgcacctcctaaggcccatgaagt
Introduction to iplant Collaborative Jinyu Yang Bioinformatics and Mathematical Biosciences Lab
 Introduction to iplant Collaborative Jinyu Yang Bioinformatics and Mathematical Biosciences Lab May/27 th /2016 About iplant What is iplant Collaborative? 1. A virtual organization 2. Created by a cooperative
Introduction to iplant Collaborative Jinyu Yang Bioinformatics and Mathematical Biosciences Lab May/27 th /2016 About iplant What is iplant Collaborative? 1. A virtual organization 2. Created by a cooperative
Fungal ITS Bioinformatics Efforts in Alaska
 Fungal ITS Bioinformatics Efforts in Alaska D. Lee Taylor ltaylor@iab.alaska.edu Institute of Arctic Biology University of Alaska Fairbanks Shawn Houston Minnesota Supercomputing Institute University of
Fungal ITS Bioinformatics Efforts in Alaska D. Lee Taylor ltaylor@iab.alaska.edu Institute of Arctic Biology University of Alaska Fairbanks Shawn Houston Minnesota Supercomputing Institute University of
Grundlagen der Bioinformatik Summer Lecturer: Prof. Daniel Huson
 Grundlagen der Bioinformatik, SoSe 11, D. Huson, April 11, 2011 1 1 Introduction Grundlagen der Bioinformatik Summer 2011 Lecturer: Prof. Daniel Huson Office hours: Thursdays 17-18h (Sand 14, C310a) 1.1
Grundlagen der Bioinformatik, SoSe 11, D. Huson, April 11, 2011 1 1 Introduction Grundlagen der Bioinformatik Summer 2011 Lecturer: Prof. Daniel Huson Office hours: Thursdays 17-18h (Sand 14, C310a) 1.1
Metagenome Analysis With MG- RAST
 Metagenome Analysis With MG- RAST Folker Meyer, PhD Argonne National Laboratory and University of Chicago http://metagenomics.anl.gov Palm Springs, March 2013 Acknowledgements Team: Dion Antonopoulos Daniela
Metagenome Analysis With MG- RAST Folker Meyer, PhD Argonne National Laboratory and University of Chicago http://metagenomics.anl.gov Palm Springs, March 2013 Acknowledgements Team: Dion Antonopoulos Daniela
DiScRIBinATE: a rapid method for accurate taxonomic classification of metagenomic sequences
 PROCEEDINGS Open Access DiScRIBinATE: a rapid method for accurate taxonomic classification of metagenomic sequences Tarini Shankar Ghosh, Monzoorul Haque M, Sharmila S Mande * From Asia Pacific Bioinformatics
PROCEEDINGS Open Access DiScRIBinATE: a rapid method for accurate taxonomic classification of metagenomic sequences Tarini Shankar Ghosh, Monzoorul Haque M, Sharmila S Mande * From Asia Pacific Bioinformatics
Methods for the phylogenetic inference from whole genome sequences and their use in Prokaryote taxonomy. M. Göker, A.F. Auch, H. P.
 Methods for the phylogenetic inference from whole genome sequences and their use in Prokaryote taxonomy M. Göker, A.F. Auch, H. P. Klenk Contents 1. Short introduction into the role of DNA DNA hybridization
Methods for the phylogenetic inference from whole genome sequences and their use in Prokaryote taxonomy M. Göker, A.F. Auch, H. P. Klenk Contents 1. Short introduction into the role of DNA DNA hybridization
Experimental Design Microbial Sequencing
 Experimental Design Microbial Sequencing Matthew L. Settles Genome Center Bioinformatics Core University of California, Davis settles@ucdavis.edu; bioinformatics.core@ucdavis.edu General rules for preparing
Experimental Design Microbial Sequencing Matthew L. Settles Genome Center Bioinformatics Core University of California, Davis settles@ucdavis.edu; bioinformatics.core@ucdavis.edu General rules for preparing
INTRODUCTION A clear cultivation bias exists in microbial phylogenetics. As of 2010, half of all
 Rachel L. Harris 1 Insights into the phylogeny and coding potential of microbial dark matter: a replication of phylogenetic anchoring methods described by Rinke et al., 2013 Rachel Harris, David Zhao,
Rachel L. Harris 1 Insights into the phylogeny and coding potential of microbial dark matter: a replication of phylogenetic anchoring methods described by Rinke et al., 2013 Rachel Harris, David Zhao,
Chapter 7. Motif finding (week 11) Chapter 8. Sequence binning (week 11)
 Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Bioinformatic methods for the analysis and comparison of metagenomes and metatranscriptomes
 Bioinformatic methods for the analysis and comparison of metagenomes and metatranscriptomes Ph.D. Thesis submitted to the Faculty of Technology, Bielefeld University, Germany for the degree of Dr. rer.
Bioinformatic methods for the analysis and comparison of metagenomes and metatranscriptomes Ph.D. Thesis submitted to the Faculty of Technology, Bielefeld University, Germany for the degree of Dr. rer.
Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G
![Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G](/thumbs/95/123686269.jpg) Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G Introduction: A genome is the total genetic content of
Annotation Practice Activity [Based on materials from the GEP Summer 2010 Workshop] Special thanks to Chris Shaffer for document review Parts A-G Introduction: A genome is the total genetic content of
MARINE BIOINFORMATICS & NANOBIOTECHNOLOGY - PBBT305
 MARINE BIOINFORMATICS & NANOBIOTECHNOLOGY - PBBT305 UNIT-1 MARINE GENOMICS AND PROTEOMICS 1. Define genomics? 2. Scope and functional genomics? 3. What is Genetics? 4. Define functional genomics? 5. What
MARINE BIOINFORMATICS & NANOBIOTECHNOLOGY - PBBT305 UNIT-1 MARINE GENOMICS AND PROTEOMICS 1. Define genomics? 2. Scope and functional genomics? 3. What is Genetics? 4. Define functional genomics? 5. What
Outline. Evolution. Adaptive convergence. Common similarity problems. Chapter 7: Similarity searches on sequence databases
 Chapter 7: Similarity searches on sequence databases All science is either physics or stamp collection. Ernest Rutherford Outline Why is similarity important BLAST Protein and DNA Interpreting BLAST Individualizing
Chapter 7: Similarity searches on sequence databases All science is either physics or stamp collection. Ernest Rutherford Outline Why is similarity important BLAST Protein and DNA Interpreting BLAST Individualizing
A Prac'cal Guide to NCBI BLAST
 A Prac'cal Guide to NCBI BLAST Leonardo Mariño-Ramírez NCBI, NIH Bethesda, USA June 2018 1 NCBI Search Services and Tools Entrez integrated literature and molecular databases Viewers BLink protein similarities
A Prac'cal Guide to NCBI BLAST Leonardo Mariño-Ramírez NCBI, NIH Bethesda, USA June 2018 1 NCBI Search Services and Tools Entrez integrated literature and molecular databases Viewers BLink protein similarities
Lecture 8: Predicting and analyzing metagenomic composition from 16S survey data
 Lecture 8: Predicting and analyzing metagenomic composition from 16S survey data What can we tell about the taxonomic and functional stability of microbiota? Why? Nature. 2012; 486(7402): 207 214. doi:10.1038/nature11234
Lecture 8: Predicting and analyzing metagenomic composition from 16S survey data What can we tell about the taxonomic and functional stability of microbiota? Why? Nature. 2012; 486(7402): 207 214. doi:10.1038/nature11234
Korilog. high-performance sequence similarity search tool & integration with KNIME platform. Patrick Durand, PhD, CEO. BIOINFORMATICS Solutions
 KLAST high-performance sequence similarity search tool & integration with KNIME platform Patrick Durand, PhD, CEO Sequence analysis big challenge DNA sequence... Context 1. Modern sequencers produce huge
KLAST high-performance sequence similarity search tool & integration with KNIME platform Patrick Durand, PhD, CEO Sequence analysis big challenge DNA sequence... Context 1. Modern sequencers produce huge
CloudLCA: finding the lowest common ancestor in metagenome analysis using cloud computing
 Protein Cell 2012, 3(2): 148 152 DOI 10.1007/s13238-012-2015-8 RESEARCH ARTICLE CloudLCA: finding the lowest common ancestor in metagenome analysis using cloud computing Guoguang Zhao 1,4*, Dechao Bu 1,4*,
Protein Cell 2012, 3(2): 148 152 DOI 10.1007/s13238-012-2015-8 RESEARCH ARTICLE CloudLCA: finding the lowest common ancestor in metagenome analysis using cloud computing Guoguang Zhao 1,4*, Dechao Bu 1,4*,
New Sequencing Technologies and Metagenomics
 New Sequencing Technologies and Metagenomics http://soils.usda.gov Daniel H. Huson Algorithms in Bioinformatics, 21.6.2007 Genomics Contents Metagenomics Metagenomics and ancient DNA 2 Genomics Contents
New Sequencing Technologies and Metagenomics http://soils.usda.gov Daniel H. Huson Algorithms in Bioinformatics, 21.6.2007 Genomics Contents Metagenomics Metagenomics and ancient DNA 2 Genomics Contents
BIO4342 Lab Exercise: Detecting and Interpreting Genetic Homology
 BIO4342 Lab Exercise: Detecting and Interpreting Genetic Homology Jeremy Buhler March 15, 2004 In this lab, we ll annotate an interesting piece of the D. melanogaster genome. Along the way, you ll get
BIO4342 Lab Exercise: Detecting and Interpreting Genetic Homology Jeremy Buhler March 15, 2004 In this lab, we ll annotate an interesting piece of the D. melanogaster genome. Along the way, you ll get
GenePainter 2 - Documentation
 GenePainter 2 - Documentation Authors Stefanie Mühlhausen, Björn Hammesfahr, Florian Odronitz, Marcel Hellkamp, Stephan Waack, Martin Kollmar How to cite Stefanie Mühlhausen, Marcel Hellkamp & Martin Kollmar
GenePainter 2 - Documentation Authors Stefanie Mühlhausen, Björn Hammesfahr, Florian Odronitz, Marcel Hellkamp, Stephan Waack, Martin Kollmar How to cite Stefanie Mühlhausen, Marcel Hellkamp & Martin Kollmar
NCBI web resources I: databases and Entrez
 NCBI web resources I: databases and Entrez Yanbin Yin Most materials are downloaded from ftp://ftp.ncbi.nih.gov/pub/education/ 1 Homework assignment 1 Two parts: Extract the gene IDs reported in table
NCBI web resources I: databases and Entrez Yanbin Yin Most materials are downloaded from ftp://ftp.ncbi.nih.gov/pub/education/ 1 Homework assignment 1 Two parts: Extract the gene IDs reported in table
Microbiome: Metagenomics 4/4/2018
 Microbiome: Metagenomics 4/4/2018 metagenomics is an extension of many things you have already learned! Genomics used to be computationally difficult, and now that s metagenomics! Still developing tools/algorithms
Microbiome: Metagenomics 4/4/2018 metagenomics is an extension of many things you have already learned! Genomics used to be computationally difficult, and now that s metagenomics! Still developing tools/algorithms
I AM NOT A METAGENOMIC EXPERT. I am merely the MESSENGER. Blaise T.F. Alako, PhD EBI Ambassador
 I AM NOT A METAGENOMIC EXPERT I am merely the MESSENGER Blaise T.F. Alako, PhD EBI Ambassador blaise@ebi.ac.uk Hubert Denise Alex Mitchell Peter Sterk Sarah Hunter http://www.ebi.ac.uk/metagenomics Blaise
I AM NOT A METAGENOMIC EXPERT I am merely the MESSENGER Blaise T.F. Alako, PhD EBI Ambassador blaise@ebi.ac.uk Hubert Denise Alex Mitchell Peter Sterk Sarah Hunter http://www.ebi.ac.uk/metagenomics Blaise
Basic Bioinformatics: Homology, Sequence Alignment,
 Basic Bioinformatics: Homology, Sequence Alignment, and BLAST William S. Sanders Institute for Genomics, Biocomputing, and Biotechnology (IGBB) High Performance Computing Collaboratory (HPC 2 ) Mississippi
Basic Bioinformatics: Homology, Sequence Alignment, and BLAST William S. Sanders Institute for Genomics, Biocomputing, and Biotechnology (IGBB) High Performance Computing Collaboratory (HPC 2 ) Mississippi
LAST + MEGAN-LR Approach to the Oxford Nanopore Wiggle Space Challenge
 LAST + MEGAN-LR Approach to the Oxford Nanopore Wiggle Space Challenge Caner Bagci 1,4 Benjamin Albrecht 1 Dominic Boceck 1 Ania Gorska 1,5 Dino Jolic 4,5 Irina Bessarab 3 Rohan Williams 2,3 Daniel H.
LAST + MEGAN-LR Approach to the Oxford Nanopore Wiggle Space Challenge Caner Bagci 1,4 Benjamin Albrecht 1 Dominic Boceck 1 Ania Gorska 1,5 Dino Jolic 4,5 Irina Bessarab 3 Rohan Williams 2,3 Daniel H.
Genomic Annotation Lab Exercise By Jacob Jipp and Marian Kaehler Luther College, Department of Biology Genomics Education Partnership 2010
 Genomic Annotation Lab Exercise By Jacob Jipp and Marian Kaehler Luther College, Department of Biology Genomics Education Partnership 2010 Genomics is a new and expanding field with an increasing impact
Genomic Annotation Lab Exercise By Jacob Jipp and Marian Kaehler Luther College, Department of Biology Genomics Education Partnership 2010 Genomics is a new and expanding field with an increasing impact
UNIVERSITY OF KWAZULU-NATAL EXAMINATIONS: MAIN, SUBJECT, COURSE AND CODE: GENE 320: Bioinformatics
 UNIVERSITY OF KWAZULU-NATAL EXAMINATIONS: MAIN, 2010 SUBJECT, COURSE AND CODE: GENE 320: Bioinformatics DURATION: 3 HOURS TOTAL MARKS: 125 Internal Examiner: Dr. Ché Pillay External Examiner: Prof. Nicola
UNIVERSITY OF KWAZULU-NATAL EXAMINATIONS: MAIN, 2010 SUBJECT, COURSE AND CODE: GENE 320: Bioinformatics DURATION: 3 HOURS TOTAL MARKS: 125 Internal Examiner: Dr. Ché Pillay External Examiner: Prof. Nicola
Sequence Based Function Annotation. Qi Sun Bioinformatics Facility Biotechnology Resource Center Cornell University
 Sequence Based Function Annotation Qi Sun Bioinformatics Facility Biotechnology Resource Center Cornell University Usage scenarios for sequence based function annotation Function prediction of newly cloned
Sequence Based Function Annotation Qi Sun Bioinformatics Facility Biotechnology Resource Center Cornell University Usage scenarios for sequence based function annotation Function prediction of newly cloned
The use of bioinformatic analysis in support of HGT from plants to microorganisms. Meeting with applicants Parma, 26 November 2015
 The use of bioinformatic analysis in support of HGT from plants to microorganisms Meeting with applicants Parma, 26 November 2015 WHY WE NEED TO CONSIDER HGT IN GM PLANT RA Directive 2001/18/EC As general
The use of bioinformatic analysis in support of HGT from plants to microorganisms Meeting with applicants Parma, 26 November 2015 WHY WE NEED TO CONSIDER HGT IN GM PLANT RA Directive 2001/18/EC As general
Determining presence/absence threshold for your dataset
 Determining presence/absence threshold for your dataset In PanCGHweb there are two ways to determine the presence/absence calling threshold. One is based on Receiver Operating Curves (ROC) generated for
Determining presence/absence threshold for your dataset In PanCGHweb there are two ways to determine the presence/absence calling threshold. One is based on Receiver Operating Curves (ROC) generated for
Why learn sequence database searching? Searching Molecular Databases with BLAST
 Why learn sequence database searching? Searching Molecular Databases with BLAST What have I cloned? Is this really!my gene"? Basic Local Alignment Search Tool How BLAST works Interpreting search results
Why learn sequence database searching? Searching Molecular Databases with BLAST What have I cloned? Is this really!my gene"? Basic Local Alignment Search Tool How BLAST works Interpreting search results
BIOINFORMATICS ORIGINAL PAPER
 BIOINFORMATICS ORIGINAL PAPER Vol. 27 no. 2 2011, pages 196 203 doi:10.1093/bioinformatics/btq649 Sequence analysis Advance Access publication December 1, 2010 MTR: taxonomic annotation of short metagenomic
BIOINFORMATICS ORIGINAL PAPER Vol. 27 no. 2 2011, pages 196 203 doi:10.1093/bioinformatics/btq649 Sequence analysis Advance Access publication December 1, 2010 MTR: taxonomic annotation of short metagenomic
arxiv: v1 [q-bio.gn] 25 Nov 2015
![arxiv: v1 [q-bio.gn] 25 Nov 2015 arxiv: v1 [q-bio.gn] 25 Nov 2015](/thumbs/76/73490664.jpg) MetaScope - Fast and accurate identification of microbes in metagenomic sequencing data Benjamin Buchfink 1, Daniel H. Huson 1,2 & Chao Xie 2,3 arxiv:1511.08753v1 [q-bio.gn] 25 Nov 2015 1 Department of
MetaScope - Fast and accurate identification of microbes in metagenomic sequencing data Benjamin Buchfink 1, Daniel H. Huson 1,2 & Chao Xie 2,3 arxiv:1511.08753v1 [q-bio.gn] 25 Nov 2015 1 Department of
Textbook Reading Guidelines
 Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: May 1, 2009 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: May 1, 2009 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Tutorial for Stop codon reassignment in the wild
 Tutorial for Stop codon reassignment in the wild Learning Objectives This tutorial has two learning objectives: 1. Finding evidence of stop codon reassignment on DNA fragments. 2. Detecting and confirming
Tutorial for Stop codon reassignment in the wild Learning Objectives This tutorial has two learning objectives: 1. Finding evidence of stop codon reassignment on DNA fragments. 2. Detecting and confirming
Protein Sequence Analysis. BME 110: CompBio Tools Todd Lowe April 19, 2007 (Slide Presentation: Carol Rohl)
 Protein Sequence Analysis BME 110: CompBio Tools Todd Lowe April 19, 2007 (Slide Presentation: Carol Rohl) Linear Sequence Analysis What can you learn from a (single) protein sequence? Calculate it s physical
Protein Sequence Analysis BME 110: CompBio Tools Todd Lowe April 19, 2007 (Slide Presentation: Carol Rohl) Linear Sequence Analysis What can you learn from a (single) protein sequence? Calculate it s physical
Functional profiling of metagenomic short reads: How complex are complex microbial communities?
 Functional profiling of metagenomic short reads: How complex are complex microbial communities? Rohita Sinha Senior Scientist (Bioinformatics), Viracor-Eurofins, Lee s summit, MO Understanding reality,
Functional profiling of metagenomic short reads: How complex are complex microbial communities? Rohita Sinha Senior Scientist (Bioinformatics), Viracor-Eurofins, Lee s summit, MO Understanding reality,
HC70AL SUMMER 2014 PROFESSOR BOB GOLDBERG Gene Annotation Worksheet
 HC70AL SUMMER 2014 PROFESSOR BOB GOLDBERG Gene Annotation Worksheet NAME: DATE: QUESTION ONE Using primers given to you by your TA, you carried out sequencing reactions to determine the identity of the
HC70AL SUMMER 2014 PROFESSOR BOB GOLDBERG Gene Annotation Worksheet NAME: DATE: QUESTION ONE Using primers given to you by your TA, you carried out sequencing reactions to determine the identity of the
BGGN 213: Foundations of Bioinformatics (Fall 2017)
 BGGN 213: Foundations of Bioinformatics (Fall 2017) Course Instructor: Dr. Barry J. Grant ( bjgrant@ucsd.edu ) Course Website: https://bioboot.github.io/bggn213_f17/ DRAFT: 2017-08-10 (15:02:30 PDT on
BGGN 213: Foundations of Bioinformatics (Fall 2017) Course Instructor: Dr. Barry J. Grant ( bjgrant@ucsd.edu ) Course Website: https://bioboot.github.io/bggn213_f17/ DRAFT: 2017-08-10 (15:02:30 PDT on
HMP Data Set Documentation
 HMP Data Set Documentation Introduction This document provides detail about files available via the DACC website. The goal of the HMP consortium is to make the metagenomics sequence data generated by the
HMP Data Set Documentation Introduction This document provides detail about files available via the DACC website. The goal of the HMP consortium is to make the metagenomics sequence data generated by the
Infectious Disease Omics
 Infectious Disease Omics Metagenomics Ernest Diez Benavente LSHTM ernest.diezbenavente@lshtm.ac.uk Course outline What is metagenomics? In situ, culture-free genomic characterization of the taxonomic and
Infectious Disease Omics Metagenomics Ernest Diez Benavente LSHTM ernest.diezbenavente@lshtm.ac.uk Course outline What is metagenomics? In situ, culture-free genomic characterization of the taxonomic and
COMPUTER RESOURCES II:
 COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
COMPUTER RESOURCES II: Using the computer to analyze data, using the internet, and accessing online databases Bio 210, Fall 2006 Linda S. Huang, Ph.D. University of Massachusetts Boston In the first computer
Data Retrieval from GenBank
 Data Retrieval from GenBank Peter J. Myler Bioinformatics of Intracellular Pathogens JNU, Feb 7-0, 2009 http://www.ncbi.nlm.nih.gov (January, 2007) http://ncbi.nlm.nih.gov/sitemap/resourceguide.html Accessing
Data Retrieval from GenBank Peter J. Myler Bioinformatics of Intracellular Pathogens JNU, Feb 7-0, 2009 http://www.ncbi.nlm.nih.gov (January, 2007) http://ncbi.nlm.nih.gov/sitemap/resourceguide.html Accessing
Introduction to taxonomic analysis of metagenomic amplicon and shotgun data with QIIME. Peter Sterk EBI Metagenomics Course 2014
 Introduction to taxonomic analysis of metagenomic amplicon and shotgun data with QIIME Peter Sterk EBI Metagenomics Course 2014 1 Taxonomic analysis using next-generation sequencing Objective we want to
Introduction to taxonomic analysis of metagenomic amplicon and shotgun data with QIIME Peter Sterk EBI Metagenomics Course 2014 1 Taxonomic analysis using next-generation sequencing Objective we want to
Two Mark question and Answers
 1. Define Bioinformatics Two Mark question and Answers Bioinformatics is the field of science in which biology, computer science, and information technology merge into a single discipline. There are three
1. Define Bioinformatics Two Mark question and Answers Bioinformatics is the field of science in which biology, computer science, and information technology merge into a single discipline. There are three
GREG GIBSON SPENCER V. MUSE
 A Primer of Genome Science ience THIRD EDITION TAGCACCTAGAATCATGGAGAGATAATTCGGTGAGAATTAAATGGAGAGTTGCATAGAGAACTGCGAACTG GREG GIBSON SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc.
A Primer of Genome Science ience THIRD EDITION TAGCACCTAGAATCATGGAGAGATAATTCGGTGAGAATTAAATGGAGAGTTGCATAGAGAACTGCGAACTG GREG GIBSON SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc.
Discover the Microbes Within: The Wolbachia Project. Bioinformatics Lab
 Bioinformatics Lab ACTIVITY AT A GLANCE "Understanding nature's mute but elegant language of living cells is the quest of modern molecular biology. From an alphabet of only four letters representing the
Bioinformatics Lab ACTIVITY AT A GLANCE "Understanding nature's mute but elegant language of living cells is the quest of modern molecular biology. From an alphabet of only four letters representing the
Genome Annotation - 2. Qi Sun Bioinformatics Facility Cornell University
 Genome Annotation - 2 Qi Sun Bioinformatics Facility Cornell University Output from Maker GFF file: Annotated gene, transcripts, and CDS FASTA file: Predicted transcript sequences Predicted protein sequences
Genome Annotation - 2 Qi Sun Bioinformatics Facility Cornell University Output from Maker GFF file: Annotated gene, transcripts, and CDS FASTA file: Predicted transcript sequences Predicted protein sequences
FUNCTIONAL BIOINFORMATICS
 Molecular Biology-2018 1 FUNCTIONAL BIOINFORMATICS PREDICTING THE FUNCTION OF AN UNKNOWN PROTEIN Suppose you have found the amino acid sequence of an unknown protein and wish to find its potential function.
Molecular Biology-2018 1 FUNCTIONAL BIOINFORMATICS PREDICTING THE FUNCTION OF AN UNKNOWN PROTEIN Suppose you have found the amino acid sequence of an unknown protein and wish to find its potential function.
Identify pathogen organisms from a stream of RNA sequences
 UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGINEERING AND COMPUTING MASTERS S THESIS no. 740 Identify pathogen organisms from a stream of RNA sequences Matija Šošić Zagreb, June 2014. Many thanks to my
UNIVERSITY OF ZAGREB FACULTY OF ELECTRICAL ENGINEERING AND COMPUTING MASTERS S THESIS no. 740 Identify pathogen organisms from a stream of RNA sequences Matija Šošić Zagreb, June 2014. Many thanks to my
Diversity and Abundance of Arsenic Resistant Genes in Soil Microbiomes. Susanna Yeh icer ACRES Presentation, 7/27/17
 Diversity and Abundance of Arsenic Resistant Genes in Soil Microbiomes Susanna Yeh icer ACRES Presentation, 7/27/17 Arsenic is the most Ubiquitous Toxic Element Arsenic (As) terrorizes the metabolism process
Diversity and Abundance of Arsenic Resistant Genes in Soil Microbiomes Susanna Yeh icer ACRES Presentation, 7/27/17 Arsenic is the most Ubiquitous Toxic Element Arsenic (As) terrorizes the metabolism process
Microbiomes and metabolomes
 Microbiomes and metabolomes Michael Inouye Baker Heart and Diabetes Institute Univ of Melbourne / Monash Univ Summer Institute in Statistical Genetics 2017 Integrative Genomics Module Seattle @minouye271
Microbiomes and metabolomes Michael Inouye Baker Heart and Diabetes Institute Univ of Melbourne / Monash Univ Summer Institute in Statistical Genetics 2017 Integrative Genomics Module Seattle @minouye271
BME 110 Midterm Examination
 BME 110 Midterm Examination May 10, 2011 Name: (please print) Directions: Please circle one answer for each question, unless the question specifies "circle all correct answers". You can use any resource
BME 110 Midterm Examination May 10, 2011 Name: (please print) Directions: Please circle one answer for each question, unless the question specifies "circle all correct answers". You can use any resource
Day 3. Examine gels from PCR. Learn about more molecular methods in microbial ecology
 Day 3 Examine gels from PCR Learn about more molecular methods in microbial ecology Genes We Targeted 1: dsrab 1800bp 2: mcra 750bp 3: Bacteria 1450bp 4: Archaea 950bp 5: Archaea + 950bp 6: Negative control
Day 3 Examine gels from PCR Learn about more molecular methods in microbial ecology Genes We Targeted 1: dsrab 1800bp 2: mcra 750bp 3: Bacteria 1450bp 4: Archaea 950bp 5: Archaea + 950bp 6: Negative control
Lecture 8: Predicting metagenomic composition from 16S survey data
 Lecture 8: Predicting metagenomic composition from 16S survey data Taxonomic and functional stability of microbiota Nature. 2012; 486(7402): 207 214. doi:10.1038/nature11234 2 1 7/6/16 A model of functional
Lecture 8: Predicting metagenomic composition from 16S survey data Taxonomic and functional stability of microbiota Nature. 2012; 486(7402): 207 214. doi:10.1038/nature11234 2 1 7/6/16 A model of functional
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16088 DOI: 10.1038/NMICROBIOL.2016.88 Species-function relationships shape ecological properties of the human gut microbiome Sara Vieira-Silva 1,2*, Gwen Falony
SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16088 DOI: 10.1038/NMICROBIOL.2016.88 Species-function relationships shape ecological properties of the human gut microbiome Sara Vieira-Silva 1,2*, Gwen Falony
Day 3. Examine gels from PCR. Learn about more molecular methods in microbial ecology
 Day 3 Examine gels from PCR Learn about more molecular methods in microbial ecology 1: dsrab 1800bp 2: mcra 750bp 3: Bacteria 1450bp 4: Archaea 950bp 5: Archaea + 950bp 6: Negative control Genes We Targeted
Day 3 Examine gels from PCR Learn about more molecular methods in microbial ecology 1: dsrab 1800bp 2: mcra 750bp 3: Bacteria 1450bp 4: Archaea 950bp 5: Archaea + 950bp 6: Negative control Genes We Targeted
Functional analysis using EBI Metagenomics
 Functional analysis using EBI Metagenomics Contents Tutorial information... 2 Tutorial learning objectives... 2 An introduction to functional analysis using EMG... 3 What are protein signatures?... 3 Assigning
Functional analysis using EBI Metagenomics Contents Tutorial information... 2 Tutorial learning objectives... 2 An introduction to functional analysis using EMG... 3 What are protein signatures?... 3 Assigning
Sequence Based Function Annotation
 Sequence Based Function Annotation Qi Sun Bioinformatics Facility Biotechnology Resource Center Cornell University Sequence Based Function Annotation 1. Given a sequence, how to predict its biological
Sequence Based Function Annotation Qi Sun Bioinformatics Facility Biotechnology Resource Center Cornell University Sequence Based Function Annotation 1. Given a sequence, how to predict its biological
Grand Challenges in Computational Biology
 Grand Challenges in Computational Biology Kimmen Sjölander UC Berkeley Reconstructing the Tree of Life CITRIS-INRIA workshop 24 May, 2011 Prediction of biological pathways and networks Human microbiome
Grand Challenges in Computational Biology Kimmen Sjölander UC Berkeley Reconstructing the Tree of Life CITRIS-INRIA workshop 24 May, 2011 Prediction of biological pathways and networks Human microbiome
Applied Bioinformatics
 Applied Bioinformatics Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Course overview What is bioinformatics Data driven science: the creation and advancement
Applied Bioinformatics Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Course overview What is bioinformatics Data driven science: the creation and advancement
Report on database pre-processing
 Multiscale Immune System SImulator for the Onset of Type 2 Diabetes integrating genetic, metabolic and nutritional data Work Package 2 Deliverable 2.3 Report on database pre-processing FP7-600803 [D2.3
Multiscale Immune System SImulator for the Onset of Type 2 Diabetes integrating genetic, metabolic and nutritional data Work Package 2 Deliverable 2.3 Report on database pre-processing FP7-600803 [D2.3
Genome 373: Genomic Informatics. Elhanan Borenstein
 Genome 373: Genomic Informatics Elhanan Borenstein Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis of genomes and biological systems,
Genome 373: Genomic Informatics Elhanan Borenstein Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis of genomes and biological systems,
Introduction to OTU Clustering. Susan Huse August 4, 2016
 Introduction to OTU Clustering Susan Huse August 4, 2016 What is an OTU? Operational Taxonomic Units a.k.a. phylotypes a.k.a. clusters aggregations of reads based only on sequence similarity, independent
Introduction to OTU Clustering Susan Huse August 4, 2016 What is an OTU? Operational Taxonomic Units a.k.a. phylotypes a.k.a. clusters aggregations of reads based only on sequence similarity, independent
Development of NGS metabarcoding. characterization of aerobiological samples. Lucia Muggia
 Development of NGS metabarcoding for the characterization of aerobiological samples Lucia Muggia Alberto Pallavicini, Elisa Banchi, Claudio G. Ametrano, David Stankovic, Silvia Ongaro, Enrico Tordoni,
Development of NGS metabarcoding for the characterization of aerobiological samples Lucia Muggia Alberto Pallavicini, Elisa Banchi, Claudio G. Ametrano, David Stankovic, Silvia Ongaro, Enrico Tordoni,
From assembled genome to annotated genome
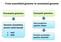 From assembled genome to annotated genome Procaryotic genomes Eucaryotic genomes Genome annotation servers (web based) 1. RAST 2. NCBI Gene prediction pipeline: Maker Function annotation pipeline: Blast2GO
From assembled genome to annotated genome Procaryotic genomes Eucaryotic genomes Genome annotation servers (web based) 1. RAST 2. NCBI Gene prediction pipeline: Maker Function annotation pipeline: Blast2GO
Advisors: Prof. Louis T. Oliphant Computer Science Department, Hiram College.
 Author: Sulochana Bramhacharya Affiliation: Hiram College, Hiram OH. Address: P.O.B 1257 Hiram, OH 44234 Email: bramhacharyas1@my.hiram.edu ACM number: 8983027 Category: Undergraduate research Advisors:
Author: Sulochana Bramhacharya Affiliation: Hiram College, Hiram OH. Address: P.O.B 1257 Hiram, OH 44234 Email: bramhacharyas1@my.hiram.edu ACM number: 8983027 Category: Undergraduate research Advisors:
 choose MBL-REGISTER user: dm00834 password: dm00834 http://register.mbl.edu/ stamps.mbl.edu this uses the username and password on your STAMPS name badge Strategies for Analysis of Microbial Population
choose MBL-REGISTER user: dm00834 password: dm00834 http://register.mbl.edu/ stamps.mbl.edu this uses the username and password on your STAMPS name badge Strategies for Analysis of Microbial Population
Last Update: 12/31/2017. Recommended Background Tutorial: An Introduction to NCBI BLAST
 BLAST Exercise: Detecting and Interpreting Genetic Homology Adapted by T. Cordonnier, C. Shaffer, W. Leung and SCR Elgin from Detecting and Interpreting Genetic Homology by Dr. J. Buhler Recommended Background
BLAST Exercise: Detecting and Interpreting Genetic Homology Adapted by T. Cordonnier, C. Shaffer, W. Leung and SCR Elgin from Detecting and Interpreting Genetic Homology by Dr. J. Buhler Recommended Background
Genetics and Bioinformatics
 Genetics and Bioinformatics Kristel Van Steen, PhD 2 Montefiore Institute - Systems and Modeling GIGA - Bioinformatics ULg kristel.vansteen@ulg.ac.be Lecture 1: Setting the pace 1 Bioinformatics what s
Genetics and Bioinformatics Kristel Van Steen, PhD 2 Montefiore Institute - Systems and Modeling GIGA - Bioinformatics ULg kristel.vansteen@ulg.ac.be Lecture 1: Setting the pace 1 Bioinformatics what s
AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE
 ACCELERATING PROGRESS IS IN OUR GENES AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE GENESPRING GENE EXPRESSION (GX) MASS PROFILER PROFESSIONAL (MPP) PATHWAY ARCHITECT (PA) See Deeper. Reach Further. BIOINFORMATICS
ACCELERATING PROGRESS IS IN OUR GENES AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE GENESPRING GENE EXPRESSION (GX) MASS PROFILER PROFESSIONAL (MPP) PATHWAY ARCHITECT (PA) See Deeper. Reach Further. BIOINFORMATICS
Applications of Next Generation Sequencing in Metagenomics Studies
 Applications of Next Generation Sequencing in Metagenomics Studies Francesca Rizzo, PhD Genomix4life Laboratory of Molecular Medicine and Genomics Department of Medicine and Surgery University of Salerno
Applications of Next Generation Sequencing in Metagenomics Studies Francesca Rizzo, PhD Genomix4life Laboratory of Molecular Medicine and Genomics Department of Medicine and Surgery University of Salerno
Transcription Start Sites Project Report
 Transcription Start Sites Project Report Student name: Student email: Faculty advisor: College/university: Project details Project name: Project species: Date of submission: Number of genes in project:
Transcription Start Sites Project Report Student name: Student email: Faculty advisor: College/university: Project details Project name: Project species: Date of submission: Number of genes in project:
Introduction to Bioinformatics CPSC 265. What is bioinformatics? Textbooks
 Introduction to Bioinformatics CPSC 265 Thanks to Jonathan Pevsner, Ph.D. Textbooks Johnathan Pevsner, who I stole most of these slides from (thanks!) has written a textbook, Bioinformatics and Functional
Introduction to Bioinformatics CPSC 265 Thanks to Jonathan Pevsner, Ph.D. Textbooks Johnathan Pevsner, who I stole most of these slides from (thanks!) has written a textbook, Bioinformatics and Functional
Transcriptomics analysis with RNA seq: an overview Frederik Coppens
 Transcriptomics analysis with RNA seq: an overview Frederik Coppens Platforms Applications Analysis Quantification RNA content Platforms Platforms Short (few hundred bases) Long reads (multiple kilobases)
Transcriptomics analysis with RNA seq: an overview Frederik Coppens Platforms Applications Analysis Quantification RNA content Platforms Platforms Short (few hundred bases) Long reads (multiple kilobases)
Evidence-Based Clustering of Reads and Taxonomic Analysis of Metagenomic Data
 Evidence-Based Clustering of Reads and Taxonomic Analysis of Metagenomic Data Gianluigi Folino 1,FabioGori 2, Mike S.M. Jetten 2,andElenaMarchiori 2, 1 ICAR-CNR,Rende,Italy 2 Radboud University, Nijmegen,
Evidence-Based Clustering of Reads and Taxonomic Analysis of Metagenomic Data Gianluigi Folino 1,FabioGori 2, Mike S.M. Jetten 2,andElenaMarchiori 2, 1 ICAR-CNR,Rende,Italy 2 Radboud University, Nijmegen,
Agenda. Web Databases for Drosophila. Gene annotation workflow. GEP Drosophila annotation projects 01/01/2018. Annotation adding labels to a sequence
 Agenda GEP annotation project overview Web Databases for Drosophila An introduction to web tools, databases and NCBI BLAST Web databases for Drosophila annotation UCSC Genome Browser NCBI / BLAST FlyBase
Agenda GEP annotation project overview Web Databases for Drosophila An introduction to web tools, databases and NCBI BLAST Web databases for Drosophila annotation UCSC Genome Browser NCBI / BLAST FlyBase
Agilent Genomic Workbench 7.0
 Agilent Genomic Workbench 7.0 Product Overview Guide Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual may be reproduced in any form or by any means (including electronic
Agilent Genomic Workbench 7.0 Product Overview Guide Agilent Technologies Notices Agilent Technologies, Inc. 2012, 2015 No part of this manual may be reproduced in any form or by any means (including electronic
Gene-centered resources at NCBI
 COURSE OF BIOINFORMATICS a.a. 2014-2015 Gene-centered resources at NCBI We searched Accession Number: M60495 AT NCBI Nucleotide Gene has been implemented at NCBI to organize information about genes, serving
COURSE OF BIOINFORMATICS a.a. 2014-2015 Gene-centered resources at NCBI We searched Accession Number: M60495 AT NCBI Nucleotide Gene has been implemented at NCBI to organize information about genes, serving
