Reducing the number of animals used in behavioural genetic experiments using chromosome substitution strains
|
|
|
- Damon Rodgers
- 6 years ago
- Views:
Transcription
1 1 The Old School, Brewhouse Hill, Wheathampstead, Hertfordshire AL4 8AN, UK Animal Welfare 2006, 15: xxx-xxx ISSN Reducing the number of animals used in behavioural genetic experiments using chromosome substitution strains MC Laarakker*, F Ohl and HA van Lith Department of Animals, Science & Society, Division of Laboratory Animal Science, Faculty of Veterinary Medicine, Utrecht University, PO Box , 3508 TD Utrecht, The Netherlands * Contact for correspondence and requests for reprints: M.C.Laarakker@vet.uu.nl Abstract Chromosome substitution strains (also called consomic lines or strains) are strains in which a single, full-length chromosome from one inbred strain the donor strain has been transferred onto the genetic background of a second inbred strain the host strain. Based on the results obtained from behavioural tests with the two parental strains, the minimum number of animals from each of the host and consomic strains that are required for a successful behavioural genetic analysis can be estimated. Correct application of statistical knowledge can lead to a further reduction in the number of animals used in behavioural genetic experiments using chromosome substitution strains. Keywords: animal welfare, behavioural genetics, chromosome substitution strain, mice, QTL, reduction Introduction A quantitative trait locus (QTL) is a position on the genome that is associated with genetic differences for a quantitative trait. Over the past decade, methods for genome analysis of animal models have been developed to identify and locate QTLs. Chromosome substitution strains, also called consomic strains, can accelerate the identification and mapping of QTLs. Chromosome substitution strains are produced by transferring a single, full-length chromosome from one inbred strain the donor strain onto the genetic background of a second strain the host strain by repeated backcrossing (Singer et al 2004). Because the host and donor strain are genetically very diverse, the consomic panels (a set of chromosome substitution strains) can be used as a general genetic discovery tool. Therefore, panels of chromosome substitution strains are an advantage to researchers studying the genes affecting developmental, physiological and behavioural processes. The Division of Laboratory Animal Science, Utrecht University, is specifically interested in behavioural genetic research and plans to use a commercially available set of mouse chromosome substitution strains (van Lith 2005). Determination of the number of animals required per strain of both the host strain and the consomic strain for the genetic analysis is one of the most important and difficult decisions one has to make. Based on the results obtained from behavioural tests with the two parental strains, the minimum number of animals from the host and consomic strains that are required for a successful behavioural genetic analysis can be estimated. This paper demonstrates that, by using statistical Universities Federation for Animal Welfare knowledge in a correct way, it is possible to reduce the number of laboratory animals used in behavioural genetic experiments using chromosome substitution strains. Materials and methods The protocol of the experiment was approved by the Animal Experiments Committee of the Academic Biomedical Centre, Utrecht, and peer-reviewed by the scientific and ethical committee of the Department of Animals, Science & Society, Utrecht University. Animals This study was performed using naive male mice from two commercially available mouse inbred strains: A/J, the donor strain, and C57BL/6J, the host strain (n = 9 per strain; The Jackson Laboratory, Bar Harbor, USA). The mice were 4 6 weeks old at arrival, and were housed in a room of the laboratory animal facility at the Department of Animals, Science & Society (Utrecht University) for two weeks for habituation before the behavioural testing started; testing was carried out in the same room. The animals were socially housed in Macrolon 2 cages (three mice per cage) and maintained under a reversed light:dark cycle (white light: 1900h 0700h; red light: 0700h 1900h) with food and water available ad libitum. In each cage, animals were provided with a shelter, tissues (Kleenex : Kimberly Clark Professional BV, Ede, The Netherlands) and paper shreds (EnviroDri : Tecnilab BMI BV, Someren, The Netherlands) as environmental enrichment. Humidity was kept at a constant level of 50% and the ambient temperature was maintained at 21.0 ± 2.0ºC. During the habituation period all mice were handled at Science in the Service of Animal Welfare
2 2 Laarakker et al Figure 1 The modified hole board test. The experimental animal was placed in the testing compartment, with the hole board in the centre. For socially housed mice, group mates were placed in the group compartment behind the partition to prevent isolation stress. least four times per week for a few minutes by the person who performed the behavioural experiments (MC Laarakker), this included picking up the animal at the tail base and placing it on the hand or arm, and restraining it by hand for a few seconds at random times of the day. The modified hole board test The behavioural tests were performed using a modified hole board (mhb) test (Figure 1; Ohl 2003). The mhb test combines the features of an open field and a hole board test. In summary, the mhb consists of an opaque grey polyvinylchloride (PVC) box ( cm, length width height) that is divided into two compartments by a transparent partition perforated with 120 holes each 3 mm in diameter. The smaller compartment ( cm, length width height) is used as a group compartment and is enriched with a tissue (Kleenex : Kimberly Clark Professional BV, Ede, The Netherlands); all the cage mates of the tested animal are placed in this group compartment, allowing the tested animal both olfactory and visible contact in order to prevent isolation stress. The experimental compartment ( cm, length width height) consists of two areas, one protected area the box which is surrounded by the protective walls of the set-up, and an unprotected area the board. Black lines divide the box into 10 rectangles (20 15 cm, length width) and 2 squares (20 20 cm, length width). The board ( cm, length width height) is placed in the centre of the experimental compartment, and contains 23 PVC cylinders (3 3 cm, diameter height), positioned across the board in three rows. The board is lit with an additional red light lamp (80 W), such that the board is illuminated with approximately 35 lux, whereas the box is only illuminated with 1 3 lux. Behavioural testing was performed between 1000h and 1400h under red light conditions; all behavioural tests were videotaped from above the experimental compartment and from the side through the partition wall. At the start of the behavioural test, all three mice from one cage were placed in the group compartment of the mhb for 10 min in order to allow for habituation. The mice were subsequently placed in the experimental compartment one at a time and allowed free exploration for 5 min while a trained observer (MC Laarakker) scored the behaviour by hand, using the program Observer 4.1 (Noldus, Wageningen, The Netherlands). The test set-up was cleaned with water and a damp towel between each mouse. As in previous studies (Ohl et al 2001; references cited in Ohl 2003), several parameters for anxiety related behaviour (eg latency until the first board entry and number of board entries), locomotion, exploration, risk assessment, memory, arousal, immobility and social affinity were measured. However, only the results for the two parameters latency until the first board entry and number of board entries have been presented here. Statistical analyses All statistical analyses were carried out using the SPSS computer program. Two-tailed probabilities were estimated throughout. Continuous data (parameter latency until the first board entry ) were summarised as means ± standard error of the mean (SEM), whereas discrete data on the ordinal scale (parameter number of board entries ) were presented as medians with the interquartile range (IQR). The Kolmogorov-Smirnov onesample test was used to check normality of the continuous data; all continuous results within the A/J and C57BL/6J strains were normally distributed. Significant differences in the continuous data between A/J and C57BL/6J mice were calculated using the unpaired Student s t-test. The unpaired Student s t-tests were performed using pooled (for equal variances) or separate (for unequal variances) variance estimates. The equality of variances was tested using an F test. For the unpaired Student s t-test with separate variance estimates, SPSS uses the Welch correction. The significance of differences for the ordinal data between A/J and C57BL/6J mice was calculated using the Mann-Whitney U test. Results Parental strains Nine mice from the strains A/J and C57BL/6J were used as donor and host strains respectively. Based on former mouse strain comparisons (Ohl et al 2001) two discriminating parameters were selected: (1) latency until the first board entry and (2) number of board entries. Significant differences between the two parental strains were found for both parameters (see below). The obtained strain differences prompted the investigation into the chromosomal location of the QTL involved by testing a set of chromosome substitution strains between the A/J and C57BL/6J strains.
3 Reducing animal numbers in behavioural genetic experiments 3 Figure 2 Figure 3 Calculation of the mean value for a consomic strain containing one QTL using the behavioural data obtained for the parameter latency until the first board entry. Latency until first board entry The parameter latency until the first board entry was considered to be a continuous variable. The mean ± standard error of the mean [SEM] for this parameter were: C57BL/6J = 43.5 ± 13.7 s (n = 9, all male); A/J = ± 38.7 s (n = 9, all male). Using a two-tailed unpaired Student s t-test with Welch correction, there was a significant difference between the two strains and the parameter latency until the first board entry, t = 3.89, df = 9.97, P = Using two standard behavioural tests, the open field test and the dark light transmission test, Singer et al (2004) found that there are probably 3 4 non-linked (ie located on different chromosomes) QTLs present per parameter. If the effects of these QTLs are assumed to be additive and that every QTL has the same magnitude of effect, then this means that a chromosome substitution strain with n = 9 animals (all male) can have a mean of 83.5 ± 20.0 s, assuming that the four QTLs are non-linked. The mean value for a consomic strain containing one QTL was calculated using the following equation (and see Figure 2): The standard error of the mean (based on n = 9) for a consomic strain with one QTL (linear interpolation; Figure 3) was calculated as follows: In order to compare the chromosome substitution strain with its host strain, α has to be adjusted because of a greater probability of a Type 1 error attributable to multiple comparisons. Belknap (2003) proposes an α value of between and A chromosome substitution strain cannot be expected to differ significantly from the host strain (C57BL/6J) with respect to the parameter latency until the first board entry when n = 9 animals per strain are used (two-tailed Student s t-test, t = 1.65, df = 16, P = 0.119). Therefore, in order to obtain a significant result, the number of animals (n) was Calculation of the standard error of the mean for a consomic strain with one QTL using the behavioural data obtained for the parameter latency until the first board entry. increased. If n is doubled, the results for the host strain and the consomic strain (eg for chromosome 1) become: C57BL/6J = 43.5 ± 9.4 s (n = 18, all male); C57BL/6J Chr1 A /NaJ (the consomic strain = 83.5 ± 13.7 s (n = 18, all male). Using an unpaired two-tailed Student s t-test with Welch correction, P = (t = 2.40, df = 34), which, according to Belknap (2003), was not significant. The results were statistically significant when n = 27 animals per strain were used, when P = (t = 2.97, df = 52). The predicted results were: C57BL/6J = 43.5 ± 7.6 s (n = 27, all male); C57BL/6J Chr1 A /NaJ = 83.5 ± 11.1 s (n = 27, all male). This suggested that behavioural testing should start with 27 mice of the C57BL/6J host strain and 6 mice per chromosome substitution strain, that is, to start with a ratio of 4.5:1 (27:6), as suggested by Belknap (2003). This experiment predicted the following results: C57BL/6J = 43.5 ± 7.6 s (n = 27, all male); C57BL/6J Chr1 A /NaJ = 83.5 ± 25.3 s (n = 6, all male); P = (t = 2.02, df = 31). With a value of P < 0.053, then for the chromosome substitution strain for which this is the case (assuming 4 consomic strains are found, and using the parameter latency until the first board entry ), 21 additional male animals would be tested. After behavioural testing, the statistical analysis would be repeated, but with 27 animals for both the chromosome substitution strain and the host strain (C57BL/6J), it is most likely that a value close to P = will be obtained. Animal Welfare 2006, 15: xxx-xxx
4 4 Laarakker et al Table 1 The number of board entries made by A/J and C57BL/6J male mice. A/J C57BL/6J median 0 14 IQR Table 2 The number of board entries made by consomic mice (C57BL/6J Chr1 A /NaJ) and C57BL/6J male mice. C57BL/6J Chr1 A /NaJ C57BL/6J median 4 14 IQR Board entries The parameter board entries was considered to be a discrete variable on the ordinal scale. See Table 1 for the the number of board entries made by the A/J and C57BL/6J mice, and the median with the IQR. For this parameter the two inbred strains were significantly different (two-tailed Mann- Whitney U test, U = 7.5, P = ). Assuming there were also four QTLs with equal affects involved for this parameter then the results for the chromosome substitution strain and the host strain could be calculated (see Table 2). Estimating the median for a consomic strain with one QTL was calculated as follows: Because an odd number of animals were used, the median was either 3 or 4. Considering the most unfavourable situation (which means the difference between host strain and consomic strain is smaller) a median of 4 was chosen. The IQR was estimated as follows: Because the numbers of board entries were counts and nine animals were used, the IQR became 6.25 or 6.5; therefore, similar to Again, the most unfavourable situation was considered and the largest IQR was chosen, ie 6.5. Although it is not correct for discrete data, the mean and SEM for the parameter number of board entries were calculated to guarantee comparability of the shape of the distribution between the A/J, consomic and C57BL/6J strain: for the A/J strain, mean ± SEM = 3.1 ± 1.5 and for the C57BL/6J 12.3 ± 2.0; a chromosome substitution strain could be 5.4 ± 1.6. The mean for a consomic strain containing one QTL (Figure 4) was calculated as follows: The SEM for a consomic strain containing one QTL (linear interpolation; Figure 5) was calculated as follows: It appears that with the values presented in Table 2 for the C57BL/6J Chr1 A /NaJ consomic strain, the preferred results were obtained; Mann-Whitney U test, U = 15.5, P = Using 9 consomic animals and 27 host strain animals (median = 14; IQR = 6.0), P = If 27 consomic animals (median = 14, IQR = 3.0) and 27 host strain animals (median = 14, IQR = 6.0) were tested (see section Latency until the first board entry), P = (U = 139.5). For this parameter (data were analysed using the Mann-Whitney U test), α was also adjusted because of the increased probability of a Type 1 error. Here as well, the criteria of Belknap (2003) can be used (P < 0.004). Applying these rules, and using 27 consomic and 27 host strain animals for this parameter would satisfy the criteria. The criteria would not be satisfied using 9 consomic and 27 host strain animals (U = 46.5, P = 0.006,); therefore, our suggestion is to use 12 consomic mice for this parameter. Discussion When more than one statistical test is performed when analysing data from a study, statisticians demand that a more stringent criterion be used for statistical significance than the conventional P < As previously explained in the section Latency until the first board entry, multiple comparisons (comparing consomic strains with the host strain) were carried out, therefore there was an increased probability of a Type 1 error. A method to adjust for multiple tests is known as the Bonferroni adjustment: α is divided by the number of comparisons that are carried out, and, for example, Singer et al 2004 and Singer et al 2005 use this method. A complete set of chromosome substitution strains of the mouse contains of 21 different strains
5 Reducing animal numbers in behavioural genetic experiments 5 Table 3 Overview of the number of laboratory animals for the parameter latency until the first board entry using different statistical methods. Figure 4 α = α = α = multiple phase approach Singer et al 2005 Belknap 2003 This article (19 autosomes, the X and Y chromosome). Because we plan to compare all 21 available consomic strains with the host strain, the adapted α of Singer et al s (2004; 2005) method would give a value of Belknap s (2003) method is actually based on the method developed by Dunnett (1955) and applies to the situation where different test groups (in this case the consomic strains) are compared to only one control group (in this case the host strain). For example, the continuous parameter latency until the first board entry, an α of (suggested by Belknap 2003) when compared with α = (suggested by Singer et al 2004; Singer et al 2005) implies a reduction in the total number of animals that have to be tested (726 versus 594 animals). A further reduction in the number of laboratory animals used in such an experiment would be possible if the behavioural tests were started with 27 C57BL/6J host strain animals and 6 animals per consomic strain (according to Belknap [2003] a 4.5:1, or 27:6, ratio is the most efficient) and extra animals (21 in case of the parameter latency until the first board entry and 6 in case of the parameter board entry ) of the appropriate consomic strains were only tested if P < Table 3 shows an overview of the number of animals used for testing the parameter latency until the first board entry. The reduction in the number of laboratory animals can be derived from this. The two parameters latency until the first board entry and number of board entries are related to each other (Spearman s coefficient of rank correlation, R s = , n = 18, P = 0.000); therefore, a multivariate method, such as Hotelling s T 2 test, could have been used. Furthermore, Turri et al (2004) demonstrated that multivariate analysis, when compared with univariate analysis, has an increased power to detect QTLs when the genetic effects are correlated. However, several assumptions are necessary for proper application of the Hotelling s T 2 test. One of the assumptions is that dependent variables should have a multivariate normal distribution. Because latency until the first board entry is a continuous variable and number of board entries is a discrete variable, the joint distribution can never be bivariate normal; therefore we have not attempted to analyse these two parameters jointly. Based on the results obtained from the behavioural tests using the two parental strains, the minimum number of animals from the host and consomic strains that are needed for a successful behavioural genetic analysis can be estimated. By adjusting α to (Belknap 2003), and performing a multiple phase approach (our suggestion), a reduction in the number of animals used in these Calculation of the mean value for a consomic strain containing one QTL using the behavioural data from the parameter number of board entries. Figure 5 Calculation of the standard error of the mean for a consomic strain with one QTL using the behavioural data obtained for the parameter number of board entries. experiments can be obtained. The multiple phase approach that we suggest is in fact a form of sequential analysis, which Russell and Burch (1959, reprinted 1992) had already suggested in 1959 as one method of reducing the number of animals used in experiments. Acknowledgements The authors would like to thank two anonymous referees for their useful comments on the manuscript. References Belknap JK 2003 Chromosome substitution strains: some quantitative considerations for genome scans and fine mapping. Mammalian Genome 14: Dunnett CW 1955 A multiple comparison procedure for comparing several treatments with a control. Journal of the American Statistical Association 50: Ohl F 2003 Testing for anxiety. Clinical Neuroscience Research 3: Ohl F, Sillaber I, Binder E, Keck ME and Holsboer F 2001 Differential analysis of behavior and diazepam-induced alterations in C57BL/6N and BALB/c mice using the modified hole board test. Journal of Psychiatric Research 35: Animal Welfare 2006, 15: xxx-xxx
6 6 Laarakker et al Russell WMS and Burch RL 1959 (reprinted 1992) The Principles of Humane Experimental Technique. Universities Federation for Animal Welfare: Wheathampstead, UK Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O Brien WE, Conti DV, Witte JS, Lander ES and Nadeau JH 2004 Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304: Singer JB, Hill AE, Nadeau JH and Lander ES 2005 Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics 169: Turri MG, DeFries JC, Henderson ND and Flint J 2004 Multivariate analysis of quantitative trait loci influencing variation in anxiety-related behaviour in laboratory mice. Mammalian Genome 15: van Lith HA 2005 Mapping of quantitative trait loci underlying strain differences in treatment susceptibility in rodents. /main_frame_dtml?path=viavet_english/faculty/departments& skeleton_url= (accessed 2 November 2005)
Marijke C. Laarakker Æ Frauke Ohl Æ Hein A. van Lith
 Behav Genet (2008) 38:159 184 DOI 10.1007/s10519-007-9188-6 ORIGINAL RESEARCH Chromosomal Assignment of Quantitative Trait Loci Influencing Modified Hole Board Behavior in Laboratory Mice using Consomic
Behav Genet (2008) 38:159 184 DOI 10.1007/s10519-007-9188-6 ORIGINAL RESEARCH Chromosomal Assignment of Quantitative Trait Loci Influencing Modified Hole Board Behavior in Laboratory Mice using Consomic
The Mouse as Animal Model
 The Mouse as Animal Model 1. Genetic manipulation is possible Transgenic mice Gene targeted mice 2. Three month generation time 3. Behavioural test are available 4. Physiology is well established Reverse
The Mouse as Animal Model 1. Genetic manipulation is possible Transgenic mice Gene targeted mice 2. Three month generation time 3. Behavioural test are available 4. Physiology is well established Reverse
EFFECTS OF ENVIRONMENTAL ENRICHMENT ON THE BEHAVIOUR OF MICE IN OPEN FIELD TESTS
 Effects of enrichment on open field test behaviour EFFECTS OF ENVIRONMENTAL ENRICHMENT ON THE BEHAVIOUR OF MICE IN OPEN FIELD TESTS HA Van de Weerd 1, IAD Van Elderen 1,2, V Baumans 1, T Zethof 2, LFM
Effects of enrichment on open field test behaviour EFFECTS OF ENVIRONMENTAL ENRICHMENT ON THE BEHAVIOUR OF MICE IN OPEN FIELD TESTS HA Van de Weerd 1, IAD Van Elderen 1,2, V Baumans 1, T Zethof 2, LFM
Impact of transgenic procedures on behavioural and physiological responses in post-weaning mice
 Impact of transgenic procedures on behavioural and physiological responses in post-weaning mice Miriam van der Meer 1, Vera Baumans 1, Berend Olivier 2, 3 and Bert van Zutphen 1 1 Department of Laboratory
Impact of transgenic procedures on behavioural and physiological responses in post-weaning mice Miriam van der Meer 1, Vera Baumans 1, Berend Olivier 2, 3 and Bert van Zutphen 1 1 Department of Laboratory
QTL mapping in mice. Karl W Broman. Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA.
 QTL mapping in mice Karl W Broman Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA www.biostat.jhsph.edu/ kbroman Outline Experiments, data, and goals Models ANOVA at marker
QTL mapping in mice Karl W Broman Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA www.biostat.jhsph.edu/ kbroman Outline Experiments, data, and goals Models ANOVA at marker
QTL Analysis of Multiple Behavioral Measures of Anxiety in Mice
 Behavior Genetics, Vol. 34, No. 3, May 2004 ( 2004) QTL Analysis of Multiple Behavioral Measures of Anxiety in Mice Norman D. Henderson, 1,4 Maria Grazia Turri, 2 John C. DeFries, 3 and Jonathan Flint
Behavior Genetics, Vol. 34, No. 3, May 2004 ( 2004) QTL Analysis of Multiple Behavioral Measures of Anxiety in Mice Norman D. Henderson, 1,4 Maria Grazia Turri, 2 John C. DeFries, 3 and Jonathan Flint
QTL mapping in mice. Karl W Broman. Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA.
 QTL mapping in mice Karl W Broman Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA www.biostat.jhsph.edu/ kbroman Outline Experiments, data, and goals Models ANOVA at marker
QTL mapping in mice Karl W Broman Department of Biostatistics Johns Hopkins University Baltimore, Maryland, USA www.biostat.jhsph.edu/ kbroman Outline Experiments, data, and goals Models ANOVA at marker
Statistical Methods for Quantitative Trait Loci (QTL) Mapping
 Statistical Methods for Quantitative Trait Loci (QTL) Mapping Lectures 4 Oct 10, 011 CSE 57 Computational Biology, Fall 011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 1:00-1:0 Johnson
Statistical Methods for Quantitative Trait Loci (QTL) Mapping Lectures 4 Oct 10, 011 CSE 57 Computational Biology, Fall 011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday 1:00-1:0 Johnson
Lecture 2: Height in Plants, Animals, and Humans. Michael Gore lecture notes Tucson Winter Institute version 18 Jan 2013
 Lecture 2: Height in Plants, Animals, and Humans Michael Gore lecture notes Tucson Winter Institute version 18 Jan 2013 Is height a polygenic trait? http://en.wikipedia.org/wiki/gregor_mendel Case Study
Lecture 2: Height in Plants, Animals, and Humans Michael Gore lecture notes Tucson Winter Institute version 18 Jan 2013 Is height a polygenic trait? http://en.wikipedia.org/wiki/gregor_mendel Case Study
Animal Research Ethics Committee. Guidelines for Submitting Protocols for ethical approval of research or teaching involving live animals
 Animal Research Ethics Committee Guidelines for Submitting Protocols for ethical approval of research or teaching involving live animals AREC Document No: 3 Revised Guidelines for Submitting Protocols
Animal Research Ethics Committee Guidelines for Submitting Protocols for ethical approval of research or teaching involving live animals AREC Document No: 3 Revised Guidelines for Submitting Protocols
Experimental Design and Sample Size Requirement for QTL Mapping
 Experimental Design and Sample Size Requirement for QTL Mapping Zhao-Bang Zeng Bioinformatics Research Center Departments of Statistics and Genetics North Carolina State University zeng@stat.ncsu.edu 1
Experimental Design and Sample Size Requirement for QTL Mapping Zhao-Bang Zeng Bioinformatics Research Center Departments of Statistics and Genetics North Carolina State University zeng@stat.ncsu.edu 1
Why do we need statistics to study genetics and evolution?
 Why do we need statistics to study genetics and evolution? 1. Mapping traits to the genome [Linkage maps (incl. QTLs), LOD] 2. Quantifying genetic basis of complex traits [Concordance, heritability] 3.
Why do we need statistics to study genetics and evolution? 1. Mapping traits to the genome [Linkage maps (incl. QTLs), LOD] 2. Quantifying genetic basis of complex traits [Concordance, heritability] 3.
B6 Albino A ++ Mutant Mice as Embryo Donors for Efficient Germline Transmission of B6 ES Cells. Taconic Webinar Prof. Dr.
 B6 Albino A ++ Mutant Mice as Embryo Donors for Efficient Germline Transmission of B6 ES Cells Taconic Webinar 2014-05-14 Prof. Dr. Branko Zevnik A decade of gene targeting in B6 ES cells at TaconicArtemis
B6 Albino A ++ Mutant Mice as Embryo Donors for Efficient Germline Transmission of B6 ES Cells Taconic Webinar 2014-05-14 Prof. Dr. Branko Zevnik A decade of gene targeting in B6 ES cells at TaconicArtemis
Inflorescence QTL, Canalization, and Selectable Cryptic Variation. Patrick J. Brown Department of Crop Sciences UIUC
 Inflorescence QTL, Canalization, and Selectable Cryptic Variation Patrick J. Brown Department of Crop Sciences UIUC What is genetic architecture and why should we care? GENETIC ARCHITECTURE (ANY TRAIT
Inflorescence QTL, Canalization, and Selectable Cryptic Variation Patrick J. Brown Department of Crop Sciences UIUC What is genetic architecture and why should we care? GENETIC ARCHITECTURE (ANY TRAIT
GENETICS - CLUTCH CH.20 QUANTITATIVE GENETICS.
 !! www.clutchprep.com CONCEPT: MATHMATICAL MEASRUMENTS Common statistical measurements are used in genetics to phenotypes The mean is an average of values - A population is all individuals within the group
!! www.clutchprep.com CONCEPT: MATHMATICAL MEASRUMENTS Common statistical measurements are used in genetics to phenotypes The mean is an average of values - A population is all individuals within the group
Relaxed Significance Criteria for Linkage Analysis
 Genetics: Published Articles Ahead of Print, published on June 18, 2006 as 10.1534/genetics.105.052506 Relaxed Significance Criteria for Linkage Analysis Lin Chen and John D. Storey Department of Biostatistics
Genetics: Published Articles Ahead of Print, published on June 18, 2006 as 10.1534/genetics.105.052506 Relaxed Significance Criteria for Linkage Analysis Lin Chen and John D. Storey Department of Biostatistics
B6 Albino Mouse Models
 B6 Albino Mouse Models INTRODUCING TWO NEW B6 ALBINO MODELS ON THE C57BL/6NTac BACKGROUND FOR USE AS BLASTOCYST DONORS FOR B6 ES CELLS AND AS AN IMPROVED BACKGROUND FOR IMAGING OF B6 STRAINS: B6 ALBINO
B6 Albino Mouse Models INTRODUCING TWO NEW B6 ALBINO MODELS ON THE C57BL/6NTac BACKGROUND FOR USE AS BLASTOCYST DONORS FOR B6 ES CELLS AND AS AN IMPROVED BACKGROUND FOR IMAGING OF B6 STRAINS: B6 ALBINO
QTL Mapping Using Multiple Markers Simultaneously
 SCI-PUBLICATIONS Author Manuscript American Journal of Agricultural and Biological Science (3): 195-01, 007 ISSN 1557-4989 007 Science Publications QTL Mapping Using Multiple Markers Simultaneously D.
SCI-PUBLICATIONS Author Manuscript American Journal of Agricultural and Biological Science (3): 195-01, 007 ISSN 1557-4989 007 Science Publications QTL Mapping Using Multiple Markers Simultaneously D.
Lab 1: A review of linear models
 Lab 1: A review of linear models The purpose of this lab is to help you review basic statistical methods in linear models and understanding the implementation of these methods in R. In general, we need
Lab 1: A review of linear models The purpose of this lab is to help you review basic statistical methods in linear models and understanding the implementation of these methods in R. In general, we need
A Simple Method for Intracage Mouse Environmental Enrichment Device Assessment
 A Simple Method for Intracage Mouse Environmental Enrichment Device Assessment Wilma Lagerwerf; AHT, RVT, RMLAT Jane Cator, AHT, RLAT(R) Laura Balsdon Chloe Garner Animal Care and Veterinary Services Funded
A Simple Method for Intracage Mouse Environmental Enrichment Device Assessment Wilma Lagerwerf; AHT, RVT, RMLAT Jane Cator, AHT, RLAT(R) Laura Balsdon Chloe Garner Animal Care and Veterinary Services Funded
SUPPLEMENTARY INFORMATION
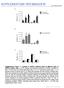 SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
SUPPLEMENTARY INFORMATION doi:10.1038/nature12474 Supplementary Figure 1 Analysis of mtdna mutation loads in different types of mtdna mutator mice. a, PCR, cloning, and sequencing analysis of mtdna mutations
Quantitative Genetics
 Quantitative Genetics Polygenic traits Quantitative Genetics 1. Controlled by several to many genes 2. Continuous variation more variation not as easily characterized into classes; individuals fall into
Quantitative Genetics Polygenic traits Quantitative Genetics 1. Controlled by several to many genes 2. Continuous variation more variation not as easily characterized into classes; individuals fall into
Feature Selection in Pharmacogenetics
 Feature Selection in Pharmacogenetics Application to Calcium Channel Blockers in Hypertension Treatment IEEE CIS June 2006 Dr. Troy Bremer Prediction Sciences Pharmacogenetics Great potential SNPs (Single
Feature Selection in Pharmacogenetics Application to Calcium Channel Blockers in Hypertension Treatment IEEE CIS June 2006 Dr. Troy Bremer Prediction Sciences Pharmacogenetics Great potential SNPs (Single
SNPs - GWAS - eqtls. Sebastian Schmeier
 SNPs - GWAS - eqtls s.schmeier@gmail.com http://sschmeier.github.io/bioinf-workshop/ 17.08.2015 Overview Single nucleotide polymorphism (refresh) SNPs effect on genes (refresh) Genome-wide association
SNPs - GWAS - eqtls s.schmeier@gmail.com http://sschmeier.github.io/bioinf-workshop/ 17.08.2015 Overview Single nucleotide polymorphism (refresh) SNPs effect on genes (refresh) Genome-wide association
Behavioral trait genetics in mice
 Behavioral trait genetics in mice Opportunities for translational research of psychiatric endophenotypes Annetrude de Mooij - van Malsen 1 De Mooij - van Malsen, J.G. Behavioral trait genetics in mice
Behavioral trait genetics in mice Opportunities for translational research of psychiatric endophenotypes Annetrude de Mooij - van Malsen 1 De Mooij - van Malsen, J.G. Behavioral trait genetics in mice
Mapping Toxicity Traits using Diversity Outbred Mice. Daniel M. Gatti, Ph.D. HESI Genomics Genetically Diverse Mouse Models Nov.
 Mapping Toxicity Traits using Diversity Outbred Mice Daniel M. Gatti, Ph.D. HESI Genomics Genetically Diverse Mouse Models Nov. 28, 2012 The Diversity Outbred (DO) mice are a mapping population Ideal for
Mapping Toxicity Traits using Diversity Outbred Mice Daniel M. Gatti, Ph.D. HESI Genomics Genetically Diverse Mouse Models Nov. 28, 2012 The Diversity Outbred (DO) mice are a mapping population Ideal for
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: RuRong Ji NNA59063A Article Reporting Checklist for Nature Neuroscience # Main ures: 8 # Supplementary ures: 15 # Supplementary Tables: 0 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: RuRong Ji NNA59063A Article Reporting Checklist for Nature Neuroscience # Main ures: 8 # Supplementary ures: 15 # Supplementary Tables: 0 # Supplementary
Mapping Toxicity Traits using Diversity Outbred Mice. Daniel M. Gatti, Ph.D. HESI Genomics Genetically Diverse Mouse Models Nov.
 Mapping Toxicity Traits using Diversity Outbred Mice Daniel M. Gatti, Ph.D. HESI Genomics Genetically Diverse Mouse Models Nov. 28, 2012 The Diversity Outbred (DO) mice are a mapping population Ideal for
Mapping Toxicity Traits using Diversity Outbred Mice Daniel M. Gatti, Ph.D. HESI Genomics Genetically Diverse Mouse Models Nov. 28, 2012 The Diversity Outbred (DO) mice are a mapping population Ideal for
STATISTICS PART Instructor: Dr. Samir Safi Name:
 STATISTICS PART Instructor: Dr. Samir Safi Name: ID Number: Question #1: (20 Points) For each of the situations described below, state the sample(s) type the statistical technique that you believe is the
STATISTICS PART Instructor: Dr. Samir Safi Name: ID Number: Question #1: (20 Points) For each of the situations described below, state the sample(s) type the statistical technique that you believe is the
Complex Genetic Architecture Revealed by Analysis of HDL in
 Genetics: Published Articles Ahead of Print, published on September 1, 26 as 1.1534/genetics.16.59717 1 Complex Genetic Architecture Revealed by Analysis of HDL in Chromosome Substitution Strains and F
Genetics: Published Articles Ahead of Print, published on September 1, 26 as 1.1534/genetics.16.59717 1 Complex Genetic Architecture Revealed by Analysis of HDL in Chromosome Substitution Strains and F
Outline of lectures 9-11
 GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 2011 Genetics of quantitative characters Outline of lectures 9-11 1. When we have a measurable (quantitative) character, we may not be able to discern
GENOME 453 J. Felsenstein Evolutionary Genetics Autumn, 2011 Genetics of quantitative characters Outline of lectures 9-11 1. When we have a measurable (quantitative) character, we may not be able to discern
This training module covers the following animals housed in vivaria: rodents rabbits birds amphibians reptiles non-human primates other mammals
 www.ccac.ca This training module is relevant to all animal users working with animals housed in vivaria (enclosed areas such as laboratories) in biomedical research This training module covers the following
www.ccac.ca This training module is relevant to all animal users working with animals housed in vivaria (enclosed areas such as laboratories) in biomedical research This training module covers the following
Association Mapping in Plants PLSC 731 Plant Molecular Genetics Phil McClean April, 2010
 Association Mapping in Plants PLSC 731 Plant Molecular Genetics Phil McClean April, 2010 Traditional QTL approach Uses standard bi-parental mapping populations o F2 or RI These have a limited number of
Association Mapping in Plants PLSC 731 Plant Molecular Genetics Phil McClean April, 2010 Traditional QTL approach Uses standard bi-parental mapping populations o F2 or RI These have a limited number of
Psych 3102 Introduction to Behavior genetics. Lecture 13 Identifying genes for behavioral traits
 Psych 3102 Introduction to Behavior genetics Lecture 13 Identifying genes for behavioral traits - towards behavioral genomics QUANTITATIVE GENETICS biometrical methods natural genetic variation complex
Psych 3102 Introduction to Behavior genetics Lecture 13 Identifying genes for behavioral traits - towards behavioral genomics QUANTITATIVE GENETICS biometrical methods natural genetic variation complex
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Flowchart illustrating the three complementary strategies for gene prioritization used in this study.. Supplementary Figure 2 Flow diagram illustrating calcaneal quantitative ultrasound
Supplementary Figure 1 Flowchart illustrating the three complementary strategies for gene prioritization used in this study.. Supplementary Figure 2 Flow diagram illustrating calcaneal quantitative ultrasound
David M. Rocke Division of Biostatistics and Department of Biomedical Engineering University of California, Davis
 David M. Rocke Division of Biostatistics and Department of Biomedical Engineering University of California, Davis Outline RNA-Seq for differential expression analysis Statistical methods for RNA-Seq: Structure
David M. Rocke Division of Biostatistics and Department of Biomedical Engineering University of California, Davis Outline RNA-Seq for differential expression analysis Statistical methods for RNA-Seq: Structure
Genetics of extreme body size evolution in mice from Gough Island
 Genetics of extreme body size evolution in mice from Gough Island Karl Broman Biostatistics & Medical Informatics, UW Madison kbroman.org github.com/kbroman @kwbroman bit.ly/bds2017-07 This is a collaboration
Genetics of extreme body size evolution in mice from Gough Island Karl Broman Biostatistics & Medical Informatics, UW Madison kbroman.org github.com/kbroman @kwbroman bit.ly/bds2017-07 This is a collaboration
Measurement and Scaling Concepts
 Business Research Methods 9e Zikmund Babin Carr Griffin Measurement and Scaling Concepts 13 Chapter 13 Measurement and Scaling Concepts 2013 Cengage Learning. All Rights Reserved. May not be scanned, copied
Business Research Methods 9e Zikmund Babin Carr Griffin Measurement and Scaling Concepts 13 Chapter 13 Measurement and Scaling Concepts 2013 Cengage Learning. All Rights Reserved. May not be scanned, copied
Wu et al., Determination of genetic identity in therapeutic chimeric states. We used two approaches for identifying potentially suitable deletion loci
 SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
selective breeding of a novel outbred mouse stock Matsumoto, Yuki Doctor of Philosophy Department of Genetics School of Life Science SOKENDAI
 Identification of genomic regions associated with tameness using selective breeding of a novel outbred mouse stock Matsumoto, Yuki Doctor of Philosophy Department of Genetics School of Life Science SOKENDAI
Identification of genomic regions associated with tameness using selective breeding of a novel outbred mouse stock Matsumoto, Yuki Doctor of Philosophy Department of Genetics School of Life Science SOKENDAI
B) You can conclude that A 1 is identical by descent. Notice that A2 had to come from the father (and therefore, A1 is maternal in both cases).
 Homework questions. Please provide your answers on a separate sheet. Examine the following pedigree. A 1,2 B 1,2 A 1,3 B 1,3 A 1,2 B 1,2 A 1,2 B 1,3 1. (1 point) The A 1 alleles in the two brothers are
Homework questions. Please provide your answers on a separate sheet. Examine the following pedigree. A 1,2 B 1,2 A 1,3 B 1,3 A 1,2 B 1,2 A 1,2 B 1,3 1. (1 point) The A 1 alleles in the two brothers are
AN EVALUATION OF POWER TO DETECT LOW-FREQUENCY VARIANT ASSOCIATIONS USING ALLELE-MATCHING TESTS THAT ACCOUNT FOR UNCERTAINTY
 AN EVALUATION OF POWER TO DETECT LOW-FREQUENCY VARIANT ASSOCIATIONS USING ALLELE-MATCHING TESTS THAT ACCOUNT FOR UNCERTAINTY E. ZEGGINI and J.L. ASIMIT Wellcome Trust Sanger Institute, Hinxton, CB10 1HH,
AN EVALUATION OF POWER TO DETECT LOW-FREQUENCY VARIANT ASSOCIATIONS USING ALLELE-MATCHING TESTS THAT ACCOUNT FOR UNCERTAINTY E. ZEGGINI and J.L. ASIMIT Wellcome Trust Sanger Institute, Hinxton, CB10 1HH,
SPSS 14: quick guide
 SPSS 14: quick guide Edition 2, November 2007 If you would like this document in an alternative format please ask staff for help. On request we can provide documents with a different size and style of
SPSS 14: quick guide Edition 2, November 2007 If you would like this document in an alternative format please ask staff for help. On request we can provide documents with a different size and style of
STAT 2300: Unit 1 Learning Objectives Spring 2019
 STAT 2300: Unit 1 Learning Objectives Spring 2019 Unit tests are written to evaluate student comprehension, acquisition, and synthesis of these skills. The problems listed as Assigned MyStatLab Problems
STAT 2300: Unit 1 Learning Objectives Spring 2019 Unit tests are written to evaluate student comprehension, acquisition, and synthesis of these skills. The problems listed as Assigned MyStatLab Problems
Summary for BIOSTAT/STAT551 Statistical Genetics II: Quantitative Traits
 Summary for BIOSTAT/STAT551 Statistical Genetics II: Quantitative Traits Gained an understanding of the relationship between a TRAIT, GENETICS (single locus and multilocus) and ENVIRONMENT Theoretical
Summary for BIOSTAT/STAT551 Statistical Genetics II: Quantitative Traits Gained an understanding of the relationship between a TRAIT, GENETICS (single locus and multilocus) and ENVIRONMENT Theoretical
Genetic drift is change in allele frequencies due to chance fluctuations; its strength depends on population size.
 Roadmap Genetic drift is change in allele frequencies due to chance fluctuations; its strength depends on population size. Rate of fixation (recap) Proportion of homozygotes in population (genetic diversity)
Roadmap Genetic drift is change in allele frequencies due to chance fluctuations; its strength depends on population size. Rate of fixation (recap) Proportion of homozygotes in population (genetic diversity)
1 Genetic Strategies to Detect Genes Involved in Alcoholism and Alcohol Related Traits
 1 Genetic Strategies to Detect Genes Involved in Alcoholism and Alcohol Related Traits Danielle M. Dick, Ph.D., and Tatiana Foroud, Ph.D. Danielle M. Dick, Ph.D., is a postdoctoral fellow, and Tatiana
1 Genetic Strategies to Detect Genes Involved in Alcoholism and Alcohol Related Traits Danielle M. Dick, Ph.D., and Tatiana Foroud, Ph.D. Danielle M. Dick, Ph.D., is a postdoctoral fellow, and Tatiana
Human SNP haplotypes. Statistics 246, Spring 2002 Week 15, Lecture 1
 Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Human SNP haplotypes Statistics 246, Spring 2002 Week 15, Lecture 1 Human single nucleotide polymorphisms The majority of human sequence variation is due to substitutions that have occurred once in the
Package qtldesign. R topics documented: February 20, Title Design of QTL experiments Version Date 03 September 2010
 Title Design of QTL experiments Version 0.941 Date 03 September 2010 Package qtldesign February 20, 2015 Author Saunak Sen, Jaya Satagopan, Karl Broman, and Gary Churchill Tools for the design of QTL experiments
Title Design of QTL experiments Version 0.941 Date 03 September 2010 Package qtldesign February 20, 2015 Author Saunak Sen, Jaya Satagopan, Karl Broman, and Gary Churchill Tools for the design of QTL experiments
1 why study multiple traits together?
 Multiple Traits & Microarrays why map multiple traits together? central dogma via microarrays diabetes case study why are traits correlated? close linkage or pleiotropy? how to handle high throughput?
Multiple Traits & Microarrays why map multiple traits together? central dogma via microarrays diabetes case study why are traits correlated? close linkage or pleiotropy? how to handle high throughput?
MAS refers to the use of DNA markers that are tightly-linked to target loci as a substitute for or to assist phenotypic screening.
 Marker assisted selection in rice Introduction The development of DNA (or molecular) markers has irreversibly changed the disciplines of plant genetics and plant breeding. While there are several applications
Marker assisted selection in rice Introduction The development of DNA (or molecular) markers has irreversibly changed the disciplines of plant genetics and plant breeding. While there are several applications
Linkage & Genetic Mapping in Eukaryotes. Ch. 6
 Linkage & Genetic Mapping in Eukaryotes Ch. 6 1 LINKAGE AND CROSSING OVER! In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even
Linkage & Genetic Mapping in Eukaryotes Ch. 6 1 LINKAGE AND CROSSING OVER! In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even
Xinmin Li 1,3, Richard J Quigg 1,4, Jian Zhou 1, Shizhong Xu 2, Godfred Masinde 3, Subburaman Mohan 3 and David J. Baylink 3
 Research Article Genetics and Molecular Biology, 29, 1, 166-173 (2006) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br A critical evaluation of the effect of population
Research Article Genetics and Molecular Biology, 29, 1, 166-173 (2006) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br A critical evaluation of the effect of population
Reciprocal backcross mice confirm major loci linked to hyperoxic acute lung injury survival time
 Physiol Genomics 38: 158 168, 2009. First published May 5, 2009; doi:10.1152/physiolgenomics.90392.2008. Reciprocal backcross mice confirm major loci linked to hyperoxic acute lung injury survival time
Physiol Genomics 38: 158 168, 2009. First published May 5, 2009; doi:10.1152/physiolgenomics.90392.2008. Reciprocal backcross mice confirm major loci linked to hyperoxic acute lung injury survival time
b. (3 points) The expected frequencies of each blood type in the deme if mating is random with respect to variation at this locus.
 NAME EXAM# 1 1. (15 points) Next to each unnumbered item in the left column place the number from the right column/bottom that best corresponds: 10 additive genetic variance 1) a hermaphroditic adult develops
NAME EXAM# 1 1. (15 points) Next to each unnumbered item in the left column place the number from the right column/bottom that best corresponds: 10 additive genetic variance 1) a hermaphroditic adult develops
1. You have isolated 20 new mutant yeast strains that are defective in synthesis of threonine, an amino acid. (a)
 ANSWERS TO Problem set questions from Exam 1 Unit Basic Genetic Tests, Setting up and Analyzing Crosses, and Genetic Mapping Basic genetic tests for complementation and/or dominance 1. You have isolated
ANSWERS TO Problem set questions from Exam 1 Unit Basic Genetic Tests, Setting up and Analyzing Crosses, and Genetic Mapping Basic genetic tests for complementation and/or dominance 1. You have isolated
Genetics or Genomics?
 Genetics or Genomics? genetics: study single genes or a few genes first identify mutant organism with change of interest characterize effects of mutation but only a fraction of 30k human genes directly
Genetics or Genomics? genetics: study single genes or a few genes first identify mutant organism with change of interest characterize effects of mutation but only a fraction of 30k human genes directly
Genetics - Fall 2004 Massachusetts Institute of Technology Professor Chris Kaiser Professor Gerry Fink Professor Leona Samson
 7.03 - Genetics - Fall 2004 Massachusetts Institute of Technology Professor Chris Kaiser Professor Gerry Fink Professor Leona Samson 1 1. Consider an autosomal recessive trait that occurs at a frequency
7.03 - Genetics - Fall 2004 Massachusetts Institute of Technology Professor Chris Kaiser Professor Gerry Fink Professor Leona Samson 1 1. Consider an autosomal recessive trait that occurs at a frequency
SECTION 11 ACUTE TOXICITY DATA ANALYSIS
 SECTION 11 ACUTE TOXICITY DATA ANALYSIS 11.1 INTRODUCTION 11.1.1 The objective of acute toxicity tests with effluents and receiving waters is to identify discharges of toxic effluents in acutely toxic
SECTION 11 ACUTE TOXICITY DATA ANALYSIS 11.1 INTRODUCTION 11.1.1 The objective of acute toxicity tests with effluents and receiving waters is to identify discharges of toxic effluents in acutely toxic
Identifying Genes Underlying QTLs
 Identifying Genes Underlying QTLs Reading: Frary, A. et al. 2000. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289:85-87. Paran, I. and D. Zamir. 2003. Quantitative
Identifying Genes Underlying QTLs Reading: Frary, A. et al. 2000. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289:85-87. Paran, I. and D. Zamir. 2003. Quantitative
energy usage summary (both house designs) Friday, June 15, :51:26 PM 1
 energy usage summary (both house designs) Friday, June 15, 18 02:51:26 PM 1 The UNIVARIATE Procedure type = Basic Statistical Measures Location Variability Mean 13.87143 Std Deviation 2.36364 Median 13.70000
energy usage summary (both house designs) Friday, June 15, 18 02:51:26 PM 1 The UNIVARIATE Procedure type = Basic Statistical Measures Location Variability Mean 13.87143 Std Deviation 2.36364 Median 13.70000
POPULATION GENETICS Winter 2005 Lecture 18 Quantitative genetics and QTL mapping
 POPULATION GENETICS Winter 2005 Lecture 18 Quantitative genetics and QTL mapping - from Darwin's time onward, it has been widely recognized that natural populations harbor a considerably degree of genetic
POPULATION GENETICS Winter 2005 Lecture 18 Quantitative genetics and QTL mapping - from Darwin's time onward, it has been widely recognized that natural populations harbor a considerably degree of genetic
High-density SNP Genotyping Analysis of Broiler Breeding Lines
 Animal Industry Report AS 653 ASL R2219 2007 High-density SNP Genotyping Analysis of Broiler Breeding Lines Abebe T. Hassen Jack C.M. Dekkers Susan J. Lamont Rohan L. Fernando Santiago Avendano Aviagen
Animal Industry Report AS 653 ASL R2219 2007 High-density SNP Genotyping Analysis of Broiler Breeding Lines Abebe T. Hassen Jack C.M. Dekkers Susan J. Lamont Rohan L. Fernando Santiago Avendano Aviagen
Welfare of Genetically Modified Mice
 Welfare of Genetically Modified Mice Garvan Institute of Medical Research Immunology diseases Neurological diseases Metabolic diseases Musculoskeletal diseases Cancer My perspective Veterinarian specialising
Welfare of Genetically Modified Mice Garvan Institute of Medical Research Immunology diseases Neurological diseases Metabolic diseases Musculoskeletal diseases Cancer My perspective Veterinarian specialising
Measurement and sampling
 Name: Instructions: (1) Answer questions in your blue book. Number each response. (2) Write your name on the cover of your blue book (and only on the cover). (3) You are allowed to use your calculator
Name: Instructions: (1) Answer questions in your blue book. Number each response. (2) Write your name on the cover of your blue book (and only on the cover). (3) You are allowed to use your calculator
Evolutionary Algorithms
 Evolutionary Algorithms Fall 2008 1 Introduction Evolutionary algorithms (or EAs) are tools for solving complex problems. They were originally developed for engineering and chemistry problems. Much of
Evolutionary Algorithms Fall 2008 1 Introduction Evolutionary algorithms (or EAs) are tools for solving complex problems. They were originally developed for engineering and chemistry problems. Much of
Design and Construction of Recombinant Inbred Lines
 Chapter 3 Design and Construction of Recombinant Inbred Lines Daniel A. Pollard Abstract Recombinant inbred lines (RILs) are a collection of strains that can be used to map quantitative trait loci. Parent
Chapter 3 Design and Construction of Recombinant Inbred Lines Daniel A. Pollard Abstract Recombinant inbred lines (RILs) are a collection of strains that can be used to map quantitative trait loci. Parent
Supplementary Figure 1 Genotyping by Sequencing (GBS) pipeline used in this study to genotype maize inbred lines. The 14,129 maize inbred lines were
 Supplementary Figure 1 Genotyping by Sequencing (GBS) pipeline used in this study to genotype maize inbred lines. The 14,129 maize inbred lines were processed following GBS experimental design 1 and bioinformatics
Supplementary Figure 1 Genotyping by Sequencing (GBS) pipeline used in this study to genotype maize inbred lines. The 14,129 maize inbred lines were processed following GBS experimental design 1 and bioinformatics
AcaStat How To Guide. AcaStat. Software. Copyright 2016, AcaStat Software. All rights Reserved.
 AcaStat How To Guide AcaStat Software Copyright 2016, AcaStat Software. All rights Reserved. http://www.acastat.com Table of Contents Frequencies... 3 List Variables... 4 Descriptives... 5 Explore Means...
AcaStat How To Guide AcaStat Software Copyright 2016, AcaStat Software. All rights Reserved. http://www.acastat.com Table of Contents Frequencies... 3 List Variables... 4 Descriptives... 5 Explore Means...
CHAPTER 23 THE EVOLUTIONS OF POPULATIONS. Section A: Population Genetics
 CHAPTER 23 THE EVOLUTIONS OF POPULATIONS Section A: Population Genetics 1. The modern evolutionary synthesis integrated Darwinian selection and Mendelian inheritance 2. A population s gene pool is defined
CHAPTER 23 THE EVOLUTIONS OF POPULATIONS Section A: Population Genetics 1. The modern evolutionary synthesis integrated Darwinian selection and Mendelian inheritance 2. A population s gene pool is defined
MSc specialization Animal Breeding and Genetics at Wageningen University
 MSc specialization Animal Breeding and Genetics at Wageningen University The MSc specialization Animal Breeding and Genetics focuses on the development and transfer of knowledge in the area of selection
MSc specialization Animal Breeding and Genetics at Wageningen University The MSc specialization Animal Breeding and Genetics focuses on the development and transfer of knowledge in the area of selection
Risperidone and NAP protect cognition and normalize gene expression in a schizophrenia. mouse model
 Risperidone and NAP protect cognition and normalize gene expression in a schizophrenia mouse model Short title: Risperidone and NAP protect Foxp2 in DISC1 mice Sinaya Vaisburd 1, Zeev Shemer 1, Adva Yeheskel
Risperidone and NAP protect cognition and normalize gene expression in a schizophrenia mouse model Short title: Risperidone and NAP protect Foxp2 in DISC1 mice Sinaya Vaisburd 1, Zeev Shemer 1, Adva Yeheskel
Mapping and Mapping Populations
 Mapping and Mapping Populations Types of mapping populations F 2 o Two F 1 individuals are intermated Backcross o Cross of a recurrent parent to a F 1 Recombinant Inbred Lines (RILs; F 2 -derived lines)
Mapping and Mapping Populations Types of mapping populations F 2 o Two F 1 individuals are intermated Backcross o Cross of a recurrent parent to a F 1 Recombinant Inbred Lines (RILs; F 2 -derived lines)
Monday, November 8 Shantz 242 E (the usual place) 5:00-7:00 PM
 Review Session Monday, November 8 Shantz 242 E (the usual place) 5:00-7:00 PM I ll answer questions on my material, then Chad will answer questions on his material. Test Information Today s notes, the
Review Session Monday, November 8 Shantz 242 E (the usual place) 5:00-7:00 PM I ll answer questions on my material, then Chad will answer questions on his material. Test Information Today s notes, the
Identification of an Acute Ethanol Response Quantitative Trait Locus on Mouse Chromosome 2
 The Journal of Neuroscience, January 15, 1999, 19(2):549 561 Identification of an Acute Ethanol Response Quantitative Trait Locus on Mouse Chromosome 2 Kristin Demarest, James McCaughran Jr, Elham Mahjubi,
The Journal of Neuroscience, January 15, 1999, 19(2):549 561 Identification of an Acute Ethanol Response Quantitative Trait Locus on Mouse Chromosome 2 Kristin Demarest, James McCaughran Jr, Elham Mahjubi,
Designing a Complex-Omics Experiments. Xiangqin Cui. Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham
 Designing a Complex-Omics Experiments Xiangqin Cui Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham 1/7/2015 Some slides are from previous lectures of Grier
Designing a Complex-Omics Experiments Xiangqin Cui Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham 1/7/2015 Some slides are from previous lectures of Grier
APPLICATION OF SEASONAL ADJUSTMENT FACTORS TO SUBSEQUENT YEAR DATA. Corresponding Author
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 APPLICATION OF SEASONAL ADJUSTMENT FACTORS TO SUBSEQUENT
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 APPLICATION OF SEASONAL ADJUSTMENT FACTORS TO SUBSEQUENT
THE EFFECTS OF FULL TRANSPARENCY IN SUPPLIER SELECTION ON SUBJECTIVITY AND BID QUALITY. Jan Telgen and Fredo Schotanus
 THE EFFECTS OF FULL TRANSPARENCY IN SUPPLIER SELECTION ON SUBJECTIVITY AND BID QUALITY Jan Telgen and Fredo Schotanus Jan Telgen, Ph.D. and Fredo Schotanus, Ph.D. are NEVI Professor of Public Procurement
THE EFFECTS OF FULL TRANSPARENCY IN SUPPLIER SELECTION ON SUBJECTIVITY AND BID QUALITY Jan Telgen and Fredo Schotanus Jan Telgen, Ph.D. and Fredo Schotanus, Ph.D. are NEVI Professor of Public Procurement
Genomic Selection in Breeding Programs BIOL 509 November 26, 2013
 Genomic Selection in Breeding Programs BIOL 509 November 26, 2013 Omnia Ibrahim omniyagamal@yahoo.com 1 Outline 1- Definitions 2- Traditional breeding 3- Genomic selection (as tool of molecular breeding)
Genomic Selection in Breeding Programs BIOL 509 November 26, 2013 Omnia Ibrahim omniyagamal@yahoo.com 1 Outline 1- Definitions 2- Traditional breeding 3- Genomic selection (as tool of molecular breeding)
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Liu NNA52890T Article Reporting Checklist for Nature Neuroscience # Main Figures: 4 # Supplementary Figures: 3 # Supplementary Tables: 1 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Liu NNA52890T Article Reporting Checklist for Nature Neuroscience # Main Figures: 4 # Supplementary Figures: 3 # Supplementary Tables: 1 # Supplementary
X-linked loci influence spatial navigation performance in Dahl rats
 Articles in PresS. Physiol Genomics (December 9, 2003). 10.1152/physiolgenomics.00162.2003 X-linked loci influence spatial navigation performance in Dahl rats Nelson Ruiz-Opazo 1,3 and John Tonkiss 2 1
Articles in PresS. Physiol Genomics (December 9, 2003). 10.1152/physiolgenomics.00162.2003 X-linked loci influence spatial navigation performance in Dahl rats Nelson Ruiz-Opazo 1,3 and John Tonkiss 2 1
Lesson Overview. What would happen when genetics answered questions about how heredity works?
 17.1 Darwin developed his theory of evolution without knowing how heritable traits passed from one generation to the next or where heritable variation came from. What would happen when genetics answered
17.1 Darwin developed his theory of evolution without knowing how heritable traits passed from one generation to the next or where heritable variation came from. What would happen when genetics answered
COMPUTER SIMULATIONS AND PROBLEMS
 Exercise 1: Exploring Evolutionary Mechanisms with Theoretical Computer Simulations, and Calculation of Allele and Genotype Frequencies & Hardy-Weinberg Equilibrium Theory INTRODUCTION Evolution is defined
Exercise 1: Exploring Evolutionary Mechanisms with Theoretical Computer Simulations, and Calculation of Allele and Genotype Frequencies & Hardy-Weinberg Equilibrium Theory INTRODUCTION Evolution is defined
Evolutionary Algorithms
 Evolutionary Algorithms Evolutionary Algorithms What is Evolutionary Algorithms (EAs)? Evolutionary algorithms are iterative and stochastic search methods that mimic the natural biological evolution and/or
Evolutionary Algorithms Evolutionary Algorithms What is Evolutionary Algorithms (EAs)? Evolutionary algorithms are iterative and stochastic search methods that mimic the natural biological evolution and/or
Behavioural and physiological effects of biotechnology procedures used for gene targeting in mice
 Behavioural and physiological effects of biotechnology procedures used for gene targeting in mice Miriam van der Meer 1, Vera Baumans 1, Berend Olivier 2, 3, Cas Kruitwagen 4, Jaap van Dijk 5 and Bert
Behavioural and physiological effects of biotechnology procedures used for gene targeting in mice Miriam van der Meer 1, Vera Baumans 1, Berend Olivier 2, 3, Cas Kruitwagen 4, Jaap van Dijk 5 and Bert
The genetic improvement of wheat and barley for reproductive frost tolerance
 The genetic improvement of wheat and barley for reproductive frost tolerance By Jason Reinheimer Bachelor of Agricultural Science, University of Adelaide A thesis submitted for the degree of Doctor of
The genetic improvement of wheat and barley for reproductive frost tolerance By Jason Reinheimer Bachelor of Agricultural Science, University of Adelaide A thesis submitted for the degree of Doctor of
Introduction to PLABQTL
 Introduction to PLABQTL H.F. Utz Institute of Plant Breeding, Seed Science and Population Genetics, University of Hohenheim Germany Utz Introduction to PLABQTL 1 PLABQTL is a computer program for 1. detection
Introduction to PLABQTL H.F. Utz Institute of Plant Breeding, Seed Science and Population Genetics, University of Hohenheim Germany Utz Introduction to PLABQTL 1 PLABQTL is a computer program for 1. detection
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Robert Froemke NNA7412T Article Reporting Checklist for Nature Neuroscience # Main s: 7 # Supplementary s: 14 # Supplementary Tables: 0 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Robert Froemke NNA7412T Article Reporting Checklist for Nature Neuroscience # Main s: 7 # Supplementary s: 14 # Supplementary Tables: 0 # Supplementary
Course Announcements
 Statistical Methods for Quantitative Trait Loci (QTL) Mapping II Lectures 5 Oct 2, 2 SE 527 omputational Biology, Fall 2 Instructor Su-In Lee T hristopher Miles Monday & Wednesday 2-2 Johnson Hall (JHN)
Statistical Methods for Quantitative Trait Loci (QTL) Mapping II Lectures 5 Oct 2, 2 SE 527 omputational Biology, Fall 2 Instructor Su-In Lee T hristopher Miles Monday & Wednesday 2-2 Johnson Hall (JHN)
Michelle Wang Department of Biology, Queen s University, Kingston, Ontario Biology 206 (2008)
 An investigation of the fitness and strength of selection on the white-eye mutation of Drosophila melanogaster in two population sizes under light and dark treatments over three generations Image Source:
An investigation of the fitness and strength of selection on the white-eye mutation of Drosophila melanogaster in two population sizes under light and dark treatments over three generations Image Source:
Linking Genetic Variation to Important Phenotypes
 Linking Genetic Variation to Important Phenotypes BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2018 Anthony Gitter gitter@biostat.wisc.edu These slides, excluding third-party material, are licensed under
Linking Genetic Variation to Important Phenotypes BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2018 Anthony Gitter gitter@biostat.wisc.edu These slides, excluding third-party material, are licensed under
A Statistical Analysis of the Mechanical Properties of the Beam 14 in Lines 630 and 650 of Iran National Steel Industrial Group
 Research Note International Journal of ISSI, Vol. 13 (2016), No. 2, pp. 40-45 A Statistical Analysis of the Mechanical Properties of the Beam 14 in Lines 630 and 650 of Iran National Steel Industrial Group
Research Note International Journal of ISSI, Vol. 13 (2016), No. 2, pp. 40-45 A Statistical Analysis of the Mechanical Properties of the Beam 14 in Lines 630 and 650 of Iran National Steel Industrial Group
Near-Balanced Incomplete Block Designs with An Application to Poster Competitions
 Near-Balanced Incomplete Block Designs with An Application to Poster Competitions arxiv:1806.00034v1 [stat.ap] 31 May 2018 Xiaoyue Niu and James L. Rosenberger Department of Statistics, The Pennsylvania
Near-Balanced Incomplete Block Designs with An Application to Poster Competitions arxiv:1806.00034v1 [stat.ap] 31 May 2018 Xiaoyue Niu and James L. Rosenberger Department of Statistics, The Pennsylvania
Chapter Six- Selecting the Best Innovation Model by Using Multiple Regression
 Chapter Six- Selecting the Best Innovation Model by Using Multiple Regression 6.1 Introduction In the previous chapter, the detailed results of FA were presented and discussed. As a result, fourteen factors
Chapter Six- Selecting the Best Innovation Model by Using Multiple Regression 6.1 Introduction In the previous chapter, the detailed results of FA were presented and discussed. As a result, fourteen factors
Association Mapping. Mendelian versus Complex Phenotypes. How to Perform an Association Study. Why Association Studies (Can) Work
 Genome 371, 1 March 2010, Lecture 13 Association Mapping Mendelian versus Complex Phenotypes How to Perform an Association Study Why Association Studies (Can) Work Introduction to LOD score analysis Common
Genome 371, 1 March 2010, Lecture 13 Association Mapping Mendelian versus Complex Phenotypes How to Perform an Association Study Why Association Studies (Can) Work Introduction to LOD score analysis Common
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Marcelo Coba Manuscript Number: NNRS3314D Manuscript Type: Resource Reporting Checklist for Nature Neuroscience # Main Figures: 7 # lementary Figures: 4 # lementary Tables: 11 # lementary
Corresponding Author: Marcelo Coba Manuscript Number: NNRS3314D Manuscript Type: Resource Reporting Checklist for Nature Neuroscience # Main Figures: 7 # lementary Figures: 4 # lementary Tables: 11 # lementary
Efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement
 Mol Breeding (21) 26:493 11 DOI 1.17/s1132-1-939-8 Efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement Yanping Sun Jiankang Wang Jonathan
Mol Breeding (21) 26:493 11 DOI 1.17/s1132-1-939-8 Efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement Yanping Sun Jiankang Wang Jonathan
Linkage Disequilibrium
 Linkage Disequilibrium Why do we care about linkage disequilibrium? Determines the extent to which association mapping can be used in a species o Long distance LD Mapping at the tens of kilobase level
Linkage Disequilibrium Why do we care about linkage disequilibrium? Determines the extent to which association mapping can be used in a species o Long distance LD Mapping at the tens of kilobase level
Trudy F C Mackay, Department of Genetics, North Carolina State University, Raleigh NC , USA.
 Question & Answer Q&A: Genetic analysis of quantitative traits Trudy FC Mackay What are quantitative traits? Quantitative, or complex, traits are traits for which phenotypic variation is continuously distributed
Question & Answer Q&A: Genetic analysis of quantitative traits Trudy FC Mackay What are quantitative traits? Quantitative, or complex, traits are traits for which phenotypic variation is continuously distributed
1. why study multiple traits together?
 Multiple Traits & Microarrays 1. why study multiple traits together? 2-10 diabetes case study 2. design issues 11-13 selective phenotyping 3. why are traits correlated? 14-17 close linkage or pleiotropy?
Multiple Traits & Microarrays 1. why study multiple traits together? 2-10 diabetes case study 2. design issues 11-13 selective phenotyping 3. why are traits correlated? 14-17 close linkage or pleiotropy?
