Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
|
|
|
- Sabrina Evans
- 6 years ago
- Views:
Transcription
1 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by a one step PCR based gene replacement method (S1) with the pfa6a-pclb2-3ha- KanMX6 plasmid as template. The pfa6a-pclb2-3ha-kanmx6 plasmid was constructed by replacing the GAL1 promoter fragment in pfa6a-gal-3ha-kanmx6 (S1) with a 1kb PCR fragment of the CLB2 promoter. The pscc1-3ha-pds1 strains were constructed by PCR replacing the PDS1 promoter with the SCC1 promoter with pfa6a-pscc1-3ha-kanmx6 as template. The pfa6a-pscc1-3ha-kanmx6 plasmid was constructed as described above except that a 1kb PCR fragment containing the SCC1 promoter was used instead of the CLB2 promoter. REC8-3HA and URA-GFP dots were described in (S2). NDC1-13MYC, UBR1 and MAM1 deletions were constructed by a one-step PCR based gene replacement method (S1). MAM1-9MYC, NDC1-6HA, prec8-scc1-3ha, SPO11 and REC8 deletion as well as the CEN-GFP dots were described in (S3). PDS1-18MYC was described in (S4) 3HA-CDC14, the SPO12 deletion construct and the temperature sensitive cdc14-1 mutant were described in (S5). Sporulation condition: Cells were grown to saturation in YPD (YEP + 2% glucose) for 24 hours, diluted into YPA (YEP + 2% KAc) at OD 6 =.25 and grown over night. The cells were then washed with water and resuspended in SPO medium (.3% KAc, ph = 7.) at OD 6 = 1.85 to induce sporulation. Cells were grown at 3 C unless otherwise indicated. CIP phosphatase treatment: Extracts were prepared as described in (S6). Extracts were split in half and subjected to immunoprecipitation using appropriate antibodies. Imunoprecipitates were incubated with either buffer alone or buffer containing 8 units of Calf Intestinal Alkaline Phosphatase (CIP) at 37 C for 3min and were then analyzed by Western blot analysis.
2 Immunofluorescence techniques: Indirect in situ immunofluorescence was carried out as described in (S7). Rat -tubulin antibodies (Oxford Biotechnology) were use at a 1:1 dilution and -rat FITC or -rat Rhodamine antibodies (Jackson ImmunoResearch) at a 1:1 dilution. To detect Mam1-9MYC, mouse -MYC antibodies (Babco) were used at a 1:25 and -mouse FITC antibodies (Jackson ImmunoResearch) at a 1:1 dilution. 3HA-Cdc14 was detected with mouse -HA antibody (Covance) at a1:5 dilution and mouse CY3 (Jackson ImmunoResearch) at 1:5 dilution. Metaphase I cells were defined as cells with one DNA mass spanned by a short thick meiotic spindle measuring 2 3 µm in length. Anaphase I cells were defined as cells with spindles measuring at least 5 µm. Metaphase II and anaphase II cells were defined in a similar manner. Binucleate cells were defined as cells with two distinct but not necessarily separated DNA masses. Tetranucleate cells were defined in a similar manner. GFP, green fluorescence protein, chromosome dots: Heterozygous URA3 GFP dot: An array of TET operator sites was integrated at the URA3 locus, 35kb from the centromere of one chromosome V (S2). A TET repressor GFP fusion was also express in this strain to visualized the location of the of TET operator sites. Heterozygous CEN GFP dots were constructed in a similar manner except the array of TET operator sites were integrated 1.4kb from the centromere of one homolog of chromosome V (S3). Meiotic Spreads: Chromosomes were spread as described by (S8). Rec8-3HA was detected using an -HA antibody at a 1:5 dilution and an -mouse CY3 antibody at a 1:1 dilution. Ndc1-13MYC was detected using a rabbit -MYC antibody (Gramsch) at a 1:2 dilution and an -rabbit FITC antibody (Jackson ImmunoResearch) at 1:3 dilution. Mam1-9MYC was detected using a rabbit anti-myc antibody at a 1:6 and an -rabbit FITC antibody at a 1:1 dilution. Ndc1-6HA was detected using a mouse -HA antibody at a 1:32 and an -mouse CY3 antibody at a 1:3 dilution.
3 Quantification of Mam1 localization: Mam1 was determined to localize to kinetochores when more than 6 percent of Mam1 dots co-localized with Ndc1 dots on chromosome spreads. Note to the readers: Throughout this manuscript we define cell cycle stages by meiotic spindle morphology. Using this definition both, Cdc2- and Cdc5-depleted cells arrest in metaphase I. It is, however, important to note that the metaphase I arrest exhibited by Cdc2-depleted cells differs from that of Cdc5-depleted cells at the molecular level. In Cdc2-depleted cells Pds1 is not degraded whereas the protein is eventually degraded in Cdc5-depleted cells. Furthermore, sister kinetochores are co-oriented in Cdc2-depleted cells but bi-oriented in Cdc5-depleted cells.
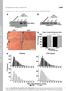
4 A Time (Hrs) Pds1-18Myc WT pclb2-cdc pclb2-cdc Rec8-3HA Pgk3 B % Cells spo11, pscc1-3ha-pds1, pclb2-3ha-cdc5 MI spo11, pscc1-3ha-pds1, pclb2-3ha-cdc5 MII spo11, pscc1-3ha-pds1 MI spo11, pscc1-3ha-pds1 MII spo11 MI spo11 MII % Metaphase I Spindles spo11, pscc1-3ha-pds1, pclb2-3ha-cdc5 spo11, pscc1-3ha-pds1 spo Time in Hrs Time in Hrs Time in Hrs % Anasphase I to Anaphase II Spindles spo11, pscc1-3ha-pds1, pclb2-3ha-cdc5 spo11, pscc1-3ha-pds1 spo11 Supplemental Figure 1
5 Figure S1: Pds1 is stabilized in Cdc5-depleted cells but Pds1 stabilization is not the sole reason for the metaphase I arrest exhibited by the mutant. (A) Wild type (A8188), pclb2-cdc5 (A8189) and pclb2-cdc2 (A819) carrying a PDS1-18MYC fusion and a REC8-3HA fusion were sporulated as described in Materials and Methods. Pds1-18MYC, Rec8-3HA, and Pgk3 (loading control) were monitored by Western blot analysis (S6). (B) spo11 (A2983), spo11 pscc1-3ha-pds1 (A7859) and spo11 pscc1-3ha- PDS1 pclb2-3ha-cdc5 (A785) were sporulated as described in Materials and Methods. The percentage of cells with two or more nuclei (closed symbols, MI) three or four nuclei (open symbols, MII, left graph) metaphase I spindles (middle graph) and the sum of anaphase I and meiosis II spindles (right graph) was determined. Pds1 produced from the Scc1 promoter is absent during meiosis as judged by Western blot analysis (S9). spo11 cells do not accumulate metaphase I spindles as the formation of stable metaphase I spindles requires tension provided by chiasmata which are absent in spo11 cells. Instead these cells elongate their spindle and enter anaphase I prematurely (S4). The fact that at least a fraction of Cdc5-depleted cells lacking PDS1 arrest with metaphase I spindles indicates that some Rec8 is retained on chromosomes. Thus, Cdc5 appears to be important not only for Pds1 degradation but also for Rec8 removal from chromosomes after Pds1 inactivation. During mitosis, Cdc5 s role in aiding in the cleavage of Scc1/Mcd1 is minor (S1). Our results suggest that the protein kinase is more important if not critical for this process during meiosis.
6 pclb2-cdc2 pclb2-cdc5 Wild type no cohesin Wild type Cen only Wild type Full Length Rec8-3HA Ndc1-13MYC DAPI Merged Supplemental Figure 2
7 Figure S2: Localization of Rec8 on chromosomes. Examples of Rec8 localization of strains described in Figure 2C. Rec8 localized along the entire length of the chromosome (full length localization) in wild-type cells (top panel), Rec8 localized to centromeric regions (Cen only) of anaphase I wild-type cells (second panel). No Rec8 staining is detected in wild-type anaphase II cells (no cohesion) in wild-type cells (third panel). Rec8 localization in pclb2-cdc5 and pclb2-cdc2 cells, respectively (forth and bottom panel). Rec8-3HA is shown in red, Ndc1-13MYC in green and DAPI in blue.
8 A % Metaphase I Spindles spo11 pclb2-3ha-cdc2 spo11 pclb2-3ha-cdc5 spo11 % Anaphase I Spindles % Metaphase II Spindles % Anaphase II Spindles Time in Hrs Time in Hrs Time in Hrs Time in Hrs B % Cells spo11, rec8, pclb2-3ha-cdc5 MI 7 spo11 rec8 pclb2-3ha-cdc5 spo11, rec8, pclb2-3ha-cdc5 MII spo11 pclb2-3ha-cdc5 spo11, pclb2-3ha-cdc5 MI 6 spo11 rec8 spo11, pclb2-3ha-cdc5 MII spo11, rec8 MI 5 spo11, rec8 MII % Metaphase I Spindles % Anaphase I Spindles % Meiosis II Spindles Time in Hrs Time in Hrs Time in Hrs Time in Hrs Supplemental Figure 3
9 Figure S3: Preventing recombination allows pclb2-cdc2 cells to enter anaphase I, whereas loss of REC8 allows anaphase I entry in CDC5-depleted cells. (A) spo11 (, A2983), spo11 pclb2-3ha-cdc5 (, A5764) and spo11 pclb2-3ha-cdc2 (, A5459) were sporulated as described in Materials and Methods. The percentage of cells with metaphase I spindles (first graph), anaphase I spindles (second graph), metaphase II spindles (third graph) and anaphase II spindles (fourth graph) was determined. spo11 cells do not accumulate metaphase I spindles as the formation of stable metaphase I spindles requires tension provided by chiasmata which is absent in spo11 cells. Instead these cells elongate their spindle and enter anaphase I prematurely (S4). (B) spo11 rec8 pclb2-3ha-cdc5 (A5457), spo11 pclb2-3ha-cdc5 (A5764) and spo11 rec8 (A389) were sporulated as described in Materials and Methods. The percentage of cells with two or more nuclei (closed symbols, MI, first graph) three or four nuclei (open symbols, MII, first graph) metaphase I spindles (C, second graph), anaphase I spindles (C, third graph) and meiosis II spindles (C, last graph) was determined.
10 Percentage of Anaphase I Cells Cdc14 Release from the Nucleolus full partial control pclb2-cdc5 Supplemental Figure 4
11 Figure S4: CDC5 is required for Cdc14 release from the nucleolus during anaphase I. spo11 rec8 (A8175) and pclb2-cdc5 spo11 rec8 ( 8157) containing a heterozygous 3HA-CDC14 fusion were sporulated as described in Materials and Methods. Cdc14 localization was determined in 15 anaphase I cells. Partial release was defined as 3HA-Cdc14 staining present in the nucleus but with nucleolar enrichment. Full released was defined as an even staining of 3HA-Cdc14 throughout the nucleus. CDC5 is required for exit from mitosis through its role in promoting the release of the protein phosphatase Cdc14 from the nucleolus during anaphase (reviewed in (S11). These results indicate that CDC5 is also essential for this process during anaphase I.
12 A pclb2-cdc2 pclb2-3ha-cdc5 pclb2-3ha-cdc5 CENV dot DAPI CENV dot DAPI CENV dot DAPI B % Metaphase I Spindles Time (Hrs) % Anaphase I to Anaphase II Spindles pclb2-cdc2 pclb2-3ha-cdc5 pclb2-cdc2 mam1 pclb2-cdc2 pclb2-3ha-cdc5 pclb2-cdc2 spo12 pclb2-cdc2 cdc14-1 pclb2-cdc2 spo12 mam Time (Hrs) % CENV GFP Separation Time (Hrs) Supplemental Figure 5
13 Figure S5: Cdc5-depleted cells but not FEAR network mutants are defective in sister kinetochore co-orientation during metaphase I. (A) Examples of centromere separation in Cdc5-depleted metaphase I cells. The left panel shows the behavior of co-oriented sister kinetochores during metaphase I in Cdc2- depleted cells carrying a CEN-GFP dot 1.4kb from the centromere of one homolog of chromosome V (heterozygous CEN-GFP dot). The middle and right panels show examples of sister kinetochore separation observed in Cdc5-depleted metaphase I cells carrying heterozygous CEN-GFP dots, which is indicative of sister kinetochores being bioriented. Genotypes of strains are described in Figure 3. CEN-GFP dots are shown in green, DNA in blue. Pictures of cells were taken 8 hours after the induction of sporulation. (B) pclb2-cdc2 (, A7118), pclb2-3ha-cdc5 (, A7151), pclb2-cdc2 mam1 (, A7316), pclb2-cdc2 pclb2-3ha-cdc5 (, A8328), pclb2-cdc2 spo12 (, A8326), pclb2-cdc2 cdc14-1 (, A8369) and pclb2-cdc2 spo12 mam1 (, A8327) containing heterozygous CEN-GFP dot were sporulated as described in Materials and Methods, except that cells were grown at room temperature in YPD and YPA and were then shifted to 3 C in SPO medium to inactivate cdc14-1. The percentage of cells with metaphase I spindles (left panel), sum of anaphase I and meiosis II spindles (middle panel) and separated CEN-GFP dots (right panel) was determined. Cdc5 is a component of the FEAR network, which is required to promote the release of the protein phosphatase Cdc14 from the nucleolus during anaphase (S12). Cdc14 released from the nucleolus then promotes exit from mitosis (reviewed in (S11)). During meiosis, inactivation of the FEAR network or CDC14 leads to an uncoupling of meiotic events. Cells carrying a FEAR network or CDC14 mutation initiate many aspects of meiosis II despite defects in anaphase I spindle disassembly (S5, 13). It was, therefore, possible that Cdc5-depleted cells exhibited this peculiar uncoupling phenotype and that sister kinetochore bi-orientation was not occurring during meiosis I but when meiosis II was scheduled to occur. To address this possibility we compared the behavior of sister kinetochores of Cdc5-depleted cells with that of another FEAR network mutant (spo12 )
14 and with that of a cdc14-1 mutant under two experimental conditions. In the first experiment we compared the segregation pattern of chromosome V of Cdc5-depleted cells with that of cells lacking SPO12 that had the REC8 open reading frame replaced by SCC1/MCD1. In Cdc5-depleted cells sister chromatids segregated from each other, whereas in spo12 cells homologs disjoined (Figure 3B). In the second experiment we compared the kinetics of centromere separation of Cdc5-depleted cells with that of cells lacking SPO12 or cells carrying a cdc14-1 mutation. As cdc14-1 mutants and spo12 mutants do not arrest in metaphase I, we conducted this experiment in cells depleted for Cdc2. Cdc5- Cdc2- double depleted cells separated sister kinetochores in metaphase I, with kinetics similar to that of pclb-cdc2 mam1 cells. In contrast, Cdc2-depleted cells carrying a cdc14-1 mutation or a SPO12 deletion did not separate sister kinetochores. This was not due to a failure to detect sister centromere separation in these cells as pclb2-cdc2 mam1 spo12 cells did separate sister centromeres. Together, these findings indicate that sister kinetochore bi-orientation in the metaphase I arrest of Cdc5-depleted cells is not a FEAR network mutant phenotype. The observation that sister kinetochores are bi-oriented even in Cdc5- Cdc2- doubly depleted cells further suggests that sister kinetochores are bi-oriented even under conditions when Pds1 degradation is prevented.
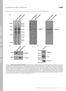
15 pclb2-cdc5 Wild type pclb2-cdc5 pclb2-cdc5 + CIP Wild type Wild type + CIP A Time (Hrs) Mam1-9MYC Pgk3 B Mam1-9MYC Wild type pclb2-cdc5 pclb2-cdc C Mam1-9MYC Supplemental Figure 6
16 Figure S6: CDC5 is required for Mam1 phosphorylation. (A) Wild type (A797), pclb2-cdc5 (A7449) and pclb2-cdc2 (A745) carrying a MAM1-9MYC and NDC1-6HA fusions were sporulated as described in Materials and Methods. Pgk3 and Mam1 protein level were monitored by Western blot analysis. (B) Analysis of Mam1 mobility in SDS PAGE in Cdc5-depleted (A7449) and wild-type (A797) cells 5hrs after the induction of sporulation. (C) Wild-type (A797) and Cdc5-depleted cells (A7449) cells were sporulated as described in Materials and Methods and samples were collected 5 hrs after the induction of sporulation. Mam1 was immunopreciptiated and either incubated with buffer or 8 units of Calf Intestinal Alkaline Phosphatase as described as in Materials and Methods.
17 Table S1: Strain List Strain number A512 A5128 A4964 A5844 A5823 A3685 A274 A6143 A6144 A4758 A6136 A6586 A6245 A6321 A6342 A8279 A7118 A7151 A7316 A7117 A5762 A746 A797 A7449 Relevant genotype MATa/ pclb2-3ha-cdc5/pclb2-3ha-cdc5 MATa/ pclb2-3ha-cdc2/pclb2-3ha-cdc2 MATa/ PDS1-18MYC/+ REC8-3HA/+ MATa/ PDS1-18MYC/+ REC8-3HA/+ pclb2-cdc5/pclb2-cdc5 MATa/ PDS1-18MYC/+ REC8-3HA/+ pclb2-cdc2/pclb2-cdc2 MATa/ PDS1-18MYC/+ MATa/ ubr1 /ubr1 REC8-3HA/REC8-3HA MATa/ ubr1 /ubr1 REC8-3HA/REC8-3HA pclb2-cdc5/pclb2- CDC5 MATa/ ubr1 /ubr1 REC8-3HA/REC8-3HA pclb2-cdc2/pclb2- CDC2 MATa/ NDC1-13MYC/+ REC8-3HA/+ MATa/ NDC1-13MYC/+ REC8-3HA/+ pclb2-cdc5/pclb2-cdc5 MATa/ NDC1-13MYC/+ REC8-3HA/+ pclb2-cdc2/pclb2-cdc2 MATa/ spo11 /spo11 rec8 /rec8 prec8::prec8-scc1-3ha// prec8::prec8-scc1-3ha LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ MATa/ spo11 /spo11 rec8 /rec8 prec8::prec8-scc1-3ha// prec8::prec8-scc1-3ha LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ pclb2-cdc2/pclb2-cdc2 MATa/ spo11 /spo11 rec8 /rec8 prec8::prec8-scc1-3ha// prec8::prec8-scc1-3ha LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ pclb2-cdc5/pclb2-cdc5 MATa/ spo11 /spo11 rec8 /rec8 prec8::prec8-scc1-3ha// prec8::prec8-scc1-3ha LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ spo12 /spo12 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2-cdc- 3HA-CDC5/pCLB2-3HA-CDC5 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 mam1 /mam1 MATa/ LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 MATa/ LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ pclb2-cdc- 3HA-CDC5/pCLB2-3HA-CDC5 MATa/ LEU2::pURA3-TetR-GFP /+ URA3::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 mam1 /mam1 MATa/ MAM1-9MYC/MAM1-9MYC NDC1-6HA/NDC1-6HA MATa/ MAM1-9MYC/MAM1-9MYC NDC1-6HA/NDC1-6HA pclb2-
18 A745 A2983 A7859 A785 A8188 A8189 A819 A5764 A5459 A5457 A389 A8175 A8157 A8328 A8326 A8369 A8327 CDC5/pCLB2-CDC5 MATa/ MAM1-9MYC/MAM1-9MYC NDC1-6HA/NDC1-6HA pclb2- CDC2/pCLB2-CDC2 MATa/ spo11 /spo11 MATa/ spo11 /spo11 pscc1-3ha-pds1/ pscc1-3ha-pds1 MATa/ spo11 /spo11 pscc1-3ha-pds1/ pscc1-3ha-pds1 pclb2- CDC-3HA-CDC5/pCLB2-3HA-CDC5 MATa/ PDS1-18MYC/ PDS1-18MYC REC8-3HA/+ MATa/ PDS1-18MYC/ PDS1-18MYC REC8-3HA/+ pclb2- CDC5/pCLB2-CDC5 MATa/ PDS1-18MYC/ PDS1-18MYC REC8-3HA/+ pclb2- CDC2/pCLB2-CDC2 MATa/ spo11 /spo11 pclb2-3ha-cdc5/pclb2-3ha-cdc5 MATa/ spo11 /spo11 pclb2-3ha-cdc2/pclb2-3ha-cdc2 MATa/ spo11 /spo11 rec8 /rec8 pclb2-3ha-cdc5/pclb2-3ha- CDC5 MATa/ spo11 /spo11 rec8 /rec8 MATa/ spo11 /spo11 rec8 /rec8 3HA-CDC14/3HA-CDC14 MATa/ spo11 /spo11 rec8 /rec8 3HA-CDC14/3HA-CDC14 pclb2- CDC5/pCLB2-CDC5 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 pclb2-cdc-3ha-cdc5/pclb2-3ha-cdc5 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 spo12 /spo12 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 cdc14 /cdc14 TRP1::cdc14-1/ TRP1::cdc14-1 MATa/ LEU2::pURA3-TetR-GFP /+ CENV::TetOx2243/+ pclb2- CDC2/pCLB2-CDC2 mam1 /mam1 spo12 /spo12 S1. M. S. Longtine et al., Yeast 14, 953 (1998). S2. F. Klein et al., Cell 98, 91 (1999). S3. A. Toth et al., Cell 13, 1155 (2). S4. M. A. Shonn, R. McCarroll, A. W. Murray, Science 289, 3 (2). S5. A. Marston, B. Lee, A. Amon, submitted. S6. T. Moll, G. Tebb, U. Surana, H. Robitsch, K. Nasmyth, Cell 66, 743 (1991). S7. R. Visintin et al., Mol Cell 2, 79 (1998). S8. K. Nairz, F. Klein, Genes Dev 11, 2272 (1997). S9. B. Lee, unpublished observations. S1. G. Alexandru, F. Uhlmann, K. Mechtler, M. A. Poupart, K. Nasmyth, Cell 15, 459 (21). S11. A. J. Bardin, A. Amon, Nat Rev Mol Cell Biol 2, 815 (21).
19 S12. F. Stegmeier, R. Visintin, A. Amon, Cell 18, 27 (22). S13. S. B. Buonomo et al., submitted.
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
The Multiple Roles of Cohesin in Meiotic Chromosome Morphogenesis and Pairing
 The Multiple Roles of Cohesin in Meiotic Chromosome Morphogenesis and Pairing The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
The Multiple Roles of Cohesin in Meiotic Chromosome Morphogenesis and Pairing The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7652 Supplementary Figure legends Figure 1. Diagrammatic representation of the rdna locus on yeast chromosome XII and the processing pathway from 35S full length nascent rrna to mature
doi: 1.138/nature7652 Supplementary Figure legends Figure 1. Diagrammatic representation of the rdna locus on yeast chromosome XII and the processing pathway from 35S full length nascent rrna to mature
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
7.06 Problem Set Four, 2006
 7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
7.06 Problem Set Four, 2006 1. Explain the molecular mechanism behind each of the following events that occur during the cell cycle, making sure to discuss specific proteins that are involved in making
MEIOSIS is a cellular division consisting of a single
 Copyright Ó 2010 by the Genetics Society of America DOI: 10.1534/genetics.110.115139 Condensins Promote Coorientation of Sister Chromatids During Meiosis I in Budding Yeast Ilana L. Brito,* Hong-Guo Yu
Copyright Ó 2010 by the Genetics Society of America DOI: 10.1534/genetics.110.115139 Condensins Promote Coorientation of Sister Chromatids During Meiosis I in Budding Yeast Ilana L. Brito,* Hong-Guo Yu
supplementary information
 DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
Cyclin-Specific Control of Ribosomal DNA Segregation
 MOLECULAR AND CELLULAR BIOLOGY, Sept. 2008, p. 5328 5336 Vol. 28, No. 17 0270-7306/08/$08.00 0 doi:10.1128/mcb.00235-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Cyclin-Specific
MOLECULAR AND CELLULAR BIOLOGY, Sept. 2008, p. 5328 5336 Vol. 28, No. 17 0270-7306/08/$08.00 0 doi:10.1128/mcb.00235-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Cyclin-Specific
Pds5p Is an Essential Chromosomal Protein Required for both Sister Chromatid Cohesion and Condensation in Saccharomyces cerevisiae
 Pds5p Is an Essential Chromosomal Protein Required for both Sister Chromatid Cohesion and Condensation in Saccharomyces cerevisiae Theresa Hartman,* Kristen Stead,* Douglas Koshland, and Vincent Guacci*
Pds5p Is an Essential Chromosomal Protein Required for both Sister Chromatid Cohesion and Condensation in Saccharomyces cerevisiae Theresa Hartman,* Kristen Stead,* Douglas Koshland, and Vincent Guacci*
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2% glucose,
 SUPPLEMENTARY MATERIAL MATERIALS AND METHODS Media and growth conditions. All strains were grown at 30 C (unless otherwise indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2%
SUPPLEMENTARY MATERIAL MATERIALS AND METHODS Media and growth conditions. All strains were grown at 30 C (unless otherwise indicated) in rich, 5-fluoroorotic acid (5FOA) or drop-out media containing 2%
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION Dynamic Phosphorylation of HP1 Regulates Mitotic Progression in Human Cells Supplementary Figures Supplementary Figure 1. NDR1 interacts with HP1. (a) Immunoprecipitation using
SUPPLEMENTARY INFORMATION
 Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
7.06 Cell Biology EXAM #2 March 20, 2003
 7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #2 March 20, 2003 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
SUPPLEMENTARY INFORMATION doi:10.1038/nature10331 Supplemental Figures and Legends Supplementary Figure 1. Pch2-dependent suppression of DSBs at the rdna borders. a, Example of a CHEF gel/southern blot
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Materials
 Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
Supplementary Materials Supplementary Figure 1. PKM2 interacts with MLC2 in cytokinesis. a, U87, U87/EGFRvIII, and HeLa cells in cytokinesis were immunostained with DAPI and an anti-pkm2 antibody. Thirty
SUPPLEMENTARY INFORMATION
 Supplementary table 1 S.pombe strains used in this study Fig. 1a to c PM158 h 90 leu1 ade6-m216 cen2-gfp Z::P adh15 -mcherry-atb2 +
Supplementary table 1 S.pombe strains used in this study Fig. 1a to c PM158 h 90 leu1 ade6-m216 cen2-gfp Z::P adh15 -mcherry-atb2 +
Gene disruption and construction of C-terminally tagged strains
 Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Supporting Online Material for
 Supporting Online Material for Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation Hengyi Shao, Yuejia Huang, Liangyu
Supporting Online Material for Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation Hengyi Shao, Yuejia Huang, Liangyu
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
doi:10.1038/nature11355 Supplemental Table 1. Gene targeting efficiency in mutants of nonessential genes. This table is presented as a separate Excel file. Supplemental Table 2. Genes with altered expression
supplementary information
 supplementary information Figure S1 (a) Alignment of the Sgf11 amino acid sequences from S. cerevisiae, Yarrowia lipolytica and S. pombe with Sgf73 sequences from S. cerevisiae, S. pombe and Candida albicans.
supplementary information Figure S1 (a) Alignment of the Sgf11 amino acid sequences from S. cerevisiae, Yarrowia lipolytica and S. pombe with Sgf73 sequences from S. cerevisiae, S. pombe and Candida albicans.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Nakajima and Tanoue, http://www.jcb.org/cgi/content/full/jcb.201104118/dc1 Figure S1. DLD-1 cells exhibit the characteristic morphology
Disjunction of Homologous Chromosomes in Meiosis I Depends on Proteolytic Cleavage of the Meiotic Cohesin Rec8 by Separin
 Cell, Vol. 103, 387 398, October 27, 2000, Copyright 2000 by Cell Press Disjunction of Homologous Chromosomes in Meiosis I Depends on Proteolytic Cleavage of the Meiotic Cohesin Rec8 by Separin Sara B.
Cell, Vol. 103, 387 398, October 27, 2000, Copyright 2000 by Cell Press Disjunction of Homologous Chromosomes in Meiosis I Depends on Proteolytic Cleavage of the Meiotic Cohesin Rec8 by Separin Sara B.
Supplemental Information. Plk1/Polo Phosphorylates Sas-4 at the Onset. of Mitosis for an Efficient Recruitment
 Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
Cell Reports, Volume 25 Supplemental Information Plk1/Polo Phosphorylates Sas-4 at the Onset of Mitosis for an Efficient Recruitment of Pericentriolar Material to Centrosomes Anand Ramani, Aruljothi Mariappan,
Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis
 Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis Philip Jordan, 1 Alice Copsey, 1 Louise Newnham, 1 Elizabeth
Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis Philip Jordan, 1 Alice Copsey, 1 Louise Newnham, 1 Elizabeth
Early Expressed Clb Proteins Allow Accumulation of Mitotic Cyclin by Inactivating Proteolytic Machinery during S Phase
 MOLECULAR AND CELLULAR BIOLOGY, Aug. 2001, p. 5071 5081 Vol. 21, No. 15 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.15.5071 5081.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, Aug. 2001, p. 5071 5081 Vol. 21, No. 15 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.15.5071 5081.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling
 Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Supplementary information to accompany: A novel role for the DNA repair gene Rad51 in Netrin-1 signalling Glendining KA 1, Markie D 2, Gardner RJM 4, Franz EA 3, Robertson SP 4, Jasoni CL 1 Supplementary
Materials and Methods
 Materials and Methods Strain construction, materials, and Net1 mutagenesis All strains used are in the W33 background (can1-1, leu2-3, his3-11, trp1-1, ura3-1, ade2-1) except where noted in the strain
Materials and Methods Strain construction, materials, and Net1 mutagenesis All strains used are in the W33 background (can1-1, leu2-3, his3-11, trp1-1, ura3-1, ade2-1) except where noted in the strain
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit
 Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
Cell Reports, Volume 5 Supplemental Information The Human Protein PRR14 Tethers Heterochromatin to the Nuclear Lamina During Interphase and Mitotic Exit Andrey Poleshko, Katelyn M. Mansfield, Caroline
indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) B. Male mice of
 SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
SUPPLEMENTRY FIGURE LEGENDS Figure S1. USP44 loss leads to chromosome missegregation.. Genotypes obtained from the indicated numbers of pups at day of life (DOL) 10, or embryonic day (ED) 13.5.. Male mice
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
Supplementary Figure 1 Collision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK2, PPK3 and PPK4 respectively.
 Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1 lision-induced dissociation (CID) mass spectra of peptides from PPK1, PPK, PPK3 and PPK respectively. % of nuclei with signal / field a 5 c ppif3:gus pppk1:gus 0 35 30 5 0 15 10
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric
 Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Supplementary Figure 1. Chromosome 3 is devoid of the telomere-proximal subtelomeric common sequences. (a) Schematic illustration of the telomere-proximal site of subtelomeres. The restriction map is based
Mps1 Kinase Promotes Sister-Kinetochore Bi-orientation by a Tension-Dependent Mechanism
 Current Biology 17, 2175 2182, December 18, 2007 ª2007 Elsevier Ltd. Open access under CC BY-NC-ND license. Mps1 Kinase Promotes Sister-Kinetochore Bi-orientation by a Tension-Dependent Mechanism DOI 10.1016/j.cub.2007.11.032
Current Biology 17, 2175 2182, December 18, 2007 ª2007 Elsevier Ltd. Open access under CC BY-NC-ND license. Mps1 Kinase Promotes Sister-Kinetochore Bi-orientation by a Tension-Dependent Mechanism DOI 10.1016/j.cub.2007.11.032
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
Supplemental Material THE JOURNAL OF CELL BIOLOGY Toso et al., http://www.jcb.org/cgi/content/full/jcb.200809055/dc1 Figure S1. Control experiments for sirna depletions used in Figs. 1 3. The phenotype
To investigate the heredity of the WFP gene, we selected plants that were homozygous
 Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Supplementary information Supplementary Note ST-12 WFP allele is semi-dominant To investigate the heredity of the WFP gene, we selected plants that were homozygous for chromosome 1 of Nipponbare and heterozygous
Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
 Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes Hannah G. Blitzblau 1,2, Clara S. Chan 1, Andreas Hochwagen 2,3, Stephen P. Bell 1 * 1 Department of Biology, Howard
Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes Hannah G. Blitzblau 1,2, Clara S. Chan 1, Andreas Hochwagen 2,3, Stephen P. Bell 1 * 1 Department of Biology, Howard
(phosphatase tensin) domain is shown in dark gray, the FH1 domain in black, and the
 Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Supplemental Figure 1. Predicted Domain Organization of the AFH14 Protein. (A) Schematic representation of the predicted domain organization of AFH14. The PTEN (phosphatase tensin) domain is shown in dark
Genetics Lecture Notes Lectures 6 9
 Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Genetics Lecture Notes 7.03 2005 Lectures 6 9 Lecture 6 Until now our analysis of genes has focused on gene function as determined by phenotype differences brought about by different alleles or by a direct
Autophagy induction [h]
![Autophagy induction [h] Autophagy induction [h]](/thumbs/80/80564099.jpg) A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of
 Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Figure S1. gfp tola and pal mcherry can complement deletion mutants of tola and pal respectively. (A)When strain LS4522 was grown in the presence of xylose, inducing expression of gfp-tola, localization
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA
 Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
Supplemental Material: Rev1 promotes replication through UV lesions in conjunction with DNA polymerases,, and, but not with DNA polymerase Jung-Hoon Yoon, Jeseong Park, Juan Conde, Maki Wakamiya, Louise
monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal antibody was examined in
 Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
Supplementary information Supplementary figures Supplementary Figure 1 Determination of the s pecificity of in-house anti-rhbdd1 mouse monoclonal antibody. (a) The specificity of the anti-rhbdd1 monoclonal
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
Supplemental material Ono et al., http://www.jcb.org/cgi/content/full/jcb.201208008/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Chromatin-binding properties of MCM2 and condensin II during
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
SUPPLEMENTARY INFORMATION Legends for Supplementary Tables. Supplementary Table 1. An excel file containing primary screen data. Worksheet 1, Normalized quantification data from a duplicated screen: valid
Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11( )::kanmx, his3::pgal1-
 Supplementary Table Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11(253-764)::kanmx, his3::pgal1-1 4HA-sld2 T84D::HIS3, bar1δ::trp1 YB291 MATa cdc7-4 leu2-3,
Supplementary Table Yeast strains used in this study Strain genotype source y2007 MATa sld3-600,609,622a-dpb11(253-764)::kanmx, his3::pgal1-1 4HA-sld2 T84D::HIS3, bar1δ::trp1 YB291 MATa cdc7-4 leu2-3,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Ricke et al., http://www.jcb.org/cgi/content/full/jcb.201205115/dc1 Figure S1. Kinetochore localization of mitotic regulators in wild-type
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Ricke et al., http://www.jcb.org/cgi/content/full/jcb.201205115/dc1 Figure S1. Kinetochore localization of mitotic regulators in wild-type
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
C. Incorrect! Second Law: Law of Independent Assortment - Genes for different traits sort independently of one another in the formation of gametes.
 OAT Biology - Problem Drill 20: Chromosomes and Genetic Technology Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as needed, (3) Pick
OAT Biology - Problem Drill 20: Chromosomes and Genetic Technology Question No. 1 of 10 Instructions: (1) Read the problem and answer choices carefully, (2) Work the problems on paper as needed, (3) Pick
At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in
 Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
Supplementary Materials and Methods Barrier function assays At E17.5, the embryos were rinsed in phosphate-buffered saline (PBS) and immersed in acidic X-gal mix (100 mm phosphate buffer at ph4.3, 3 mm
LETTERS. Fused sister kinetochores initiate the reductional division in meiosis I. Xuexian Li 1 and R. Kelly Dawe 1,2,3
 LETTERS Fused sister kinetochores initiate the reductional division in meiosis I uexian Li 1 and R. Kelly Dawe 1,2,3 During meiosis I the genome is reduced to the haploid content by a coordinated reductional
LETTERS Fused sister kinetochores initiate the reductional division in meiosis I uexian Li 1 and R. Kelly Dawe 1,2,3 During meiosis I the genome is reduced to the haploid content by a coordinated reductional
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
doi:1.138/nature11676 Replication template exchange Ori Rearrangement: acentric and/or dicentric Model for HR-dependent restart Restart on wrong template HJ resolution either restores original Restart
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
doi:10.1038/nature12119 SUPPLEMENTARY FIGURES AND LEGENDS pre-let-7a- 1 +14U pre-let-7a- 1 Ddx3x Dhx30 Dis3l2 Elavl1 Ggt5 Hnrnph 2 Osbpl5 Puf60 Rnpc3 Rpl7 Sf3b3 Sf3b4 Tia1 Triobp U2af1 U2af2 1 6 2 4 3
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Han et al., http://www.jcb.org/cgi/content/full/jcb.201311007/dc1 Figure S1. SIVA1 interacts with PCNA. (A) HEK293T cells were transiently
Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated
 Supplemntary figure legend Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated with methionine to repress transcription of genomic SEC63, and total protein extracts
Supplemntary figure legend Western blot showing expression of mutant Sec63p. The strains used in Figure 7 were treated with methionine to repress transcription of genomic SEC63, and total protein extracts
Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and
 Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
Supplementary Figure Legend: Supplementary Fig. 1 Proteomic analysis of ATR-interacting proteins. ATR, ARID1A and ATRIP protein peptides identified from our mass spectrum analysis were shown. Supplementary
plasmid shuffle strain
 Histone-Histone Interactions and Centromere Function LYNN GLOWCZEWSKI, PEIRONG YANG, TATYANA KALASHNIKOVA, MARIA SOLEDAD SANTISTEBAN, AND M. MITCHELL SMITH* Department of Microbiology and Cancer Center,
Histone-Histone Interactions and Centromere Function LYNN GLOWCZEWSKI, PEIRONG YANG, TATYANA KALASHNIKOVA, MARIA SOLEDAD SANTISTEBAN, AND M. MITCHELL SMITH* Department of Microbiology and Cancer Center,
Regulation and Function of the Ipll/Aurora Kinase. Seattle, Washington Approved for Public Release; Distribution Unlimited
 AD Award Number: DAMD17-02-1-0385 TITLE: Regulation and Function of the Ipll/Aurora Kinase PRINCIPAL INVESTIGATOR: Mr. Chitra Kotwaliwale Sue Biggins, Ph.D. CONTRACTING ORGANIZATION: Fred Hutchinson Cancer
AD Award Number: DAMD17-02-1-0385 TITLE: Regulation and Function of the Ipll/Aurora Kinase PRINCIPAL INVESTIGATOR: Mr. Chitra Kotwaliwale Sue Biggins, Ph.D. CONTRACTING ORGANIZATION: Fred Hutchinson Cancer
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supplemental material THE JOURNAL OF CELL BIOLOGY Gillespie et al., http://www.jcb.org/cgi/content/full/jcb.200907037/dc1 repressor complex induced by p38- Gillespie et al. Figure S1. Reduced fiber size
Supporting Information
 Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Bays et al., http://www.jcb.org/cgi/content/full/jcb.201309092/dc1 Figure S1. Specificity of the phospho-y822 antibody. (A) Total cell
Research Paper Address: Research Institute of Molecular Pathology, Dr Bohr-gasse 7, A-1030 Vienna, Austria.
 Research Paper 1557 Pds5 cooperates with cohesin in maintaining sister chromatid cohesion Silvia Panizza, Tomoyuki Tanaka, Andreas Hochwagen, Frank Eisenhaber and Kim Nasmyth Background: Sister chromatid
Research Paper 1557 Pds5 cooperates with cohesin in maintaining sister chromatid cohesion Silvia Panizza, Tomoyuki Tanaka, Andreas Hochwagen, Frank Eisenhaber and Kim Nasmyth Background: Sister chromatid
Cell Cycle. Trends in Cell Biology
 Cell Cycle Trends in Cell Biology Cyclic proteolysis Two distinct enzyme complexes proteolysis of cyclin-cdk complexes SCF (Skp1-Cul1-F box) Ubiquitylation and destruction of G1/S-cyclins and CKI proteins
Cell Cycle Trends in Cell Biology Cyclic proteolysis Two distinct enzyme complexes proteolysis of cyclin-cdk complexes SCF (Skp1-Cul1-F box) Ubiquitylation and destruction of G1/S-cyclins and CKI proteins
Supplementary information
 Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Supplementary information The E3 ligase RNF8 regulates KU80 removal and NHEJ repair Lin Feng 1, Junjie Chen 1 1 Department of Experimental Radiation Oncology, The University of Texas M. D. Anderson Cancer
Mitotic expression of Spo13 alters M-phase progression and nucleolar localization of Cdc14 in budding yeast
 Genetics: Published Articles Ahead of Print, published on April 20, 2010 as 10.1534/genetics.109.113746 1 2 3 4 5 6 7 Mitotic expression of Spo13 alters M-phase progression and nucleolar localization of
Genetics: Published Articles Ahead of Print, published on April 20, 2010 as 10.1534/genetics.109.113746 1 2 3 4 5 6 7 Mitotic expression of Spo13 alters M-phase progression and nucleolar localization of
SUPPLEMENTAL MATERIALS
 SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURES Supplemental Figure S1. Analyses of the CRY1-SPA1 interaction (A) An auxotrophy growth assay (left) and a filter-based -galactosidase colorimetric assay (right)
SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURES Supplemental Figure S1. Analyses of the CRY1-SPA1 interaction (A) An auxotrophy growth assay (left) and a filter-based -galactosidase colorimetric assay (right)
Supplemental Data. The Origin Recognition Complex. Functions in Sister-Chromatid Cohesion. in Saccharomyces cerevisiae
 Cell, Volume 128 Supplemental Data The Origin Recognition Complex Functions in Sister-Chromatid Cohesion in Saccharomyces cerevisiae Kenji Shimada and Susan M. Gasser Supplemental Experimental Procedures
Cell, Volume 128 Supplemental Data The Origin Recognition Complex Functions in Sister-Chromatid Cohesion in Saccharomyces cerevisiae Kenji Shimada and Susan M. Gasser Supplemental Experimental Procedures
Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
 Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation Karolina Peplowska., Andreas U. Wallek., Zuzana Storchova* Group Maintenance of Genome Stability, Max Planck Institute
Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation Karolina Peplowska., Andreas U. Wallek., Zuzana Storchova* Group Maintenance of Genome Stability, Max Planck Institute
Biological Sciences 50 Practice Final Exam. Allocate your time wisely.
 NAME: Fall 2005 TF: Biological Sciences 50 Practice Final Exam A. Be sure to write your name on the top of each of page of the examination. B. Write each answer only on the same page as the pertinent question.
NAME: Fall 2005 TF: Biological Sciences 50 Practice Final Exam A. Be sure to write your name on the top of each of page of the examination. B. Write each answer only on the same page as the pertinent question.
An in silico mathematical model of the initiation of DNA replication
 An in silico mathematical model of the initiation of DNA replication Rohan D. Gidvani a1, Brendan J. McConkey a, Bernard P. Duncker a and Brian P. Ingalls a,b a Department of Biology, University of Waterloo,
An in silico mathematical model of the initiation of DNA replication Rohan D. Gidvani a1, Brendan J. McConkey a, Bernard P. Duncker a and Brian P. Ingalls a,b a Department of Biology, University of Waterloo,
Figure S1. sporulation frequency (%) 80. sme2-5. sme2-3. sme2
 Figure S1 N WT -5-3 - DSRless mei4 100 ssm4 MS2loop () (kb) 1.0 0.5 sporulation frequency (%) 80 60 40 20 rrna 1 2 3 4 5 6 7 8 0 WT -5-3 -DSRless Figure S1. The DSR motifs in are crucial for its function.
Figure S1 N WT -5-3 - DSRless mei4 100 ssm4 MS2loop () (kb) 1.0 0.5 sporulation frequency (%) 80 60 40 20 rrna 1 2 3 4 5 6 7 8 0 WT -5-3 -DSRless Figure S1. The DSR motifs in are crucial for its function.
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
The mitotic spindle is required for loading of the DASH complex onto the kinetochore
 The mitotic spindle is required for loading of the DASH complex onto the kinetochore Yumei Li, 1 Jeff Bachant, 1 Annette A.Alcasabas, 1 Yanchang Wang, 1 Jun Qin, 1,2 and Stephen J.Elledge 1,3,4,5 1 Verna
The mitotic spindle is required for loading of the DASH complex onto the kinetochore Yumei Li, 1 Jeff Bachant, 1 Annette A.Alcasabas, 1 Yanchang Wang, 1 Jun Qin, 1,2 and Stephen J.Elledge 1,3,4,5 1 Verna
four chromosomes ` four chromosomes correct markers (sister chromatids identical!)
 Name KEY total=107 pts 1. Genes G and H are on one chromosome; gene F is on another chromosome. Assume the organism is diploid and that there is no crossing over in this species. You are examining the
Name KEY total=107 pts 1. Genes G and H are on one chromosome; gene F is on another chromosome. Assume the organism is diploid and that there is no crossing over in this species. You are examining the
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
SUPPLEMENTARY INFORMATION Supplementary figures Supplementary Figure 1: Suv39h1, but not Suv39h2, promotes HP1α sumoylation in vivo. In vivo HP1α sumoylation assay. Top: experimental scheme. Middle: we
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # C Component Size Shipping Temperature
 Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Histone H3K27 Methylation Antibody Panel Pack Base Catalog # PACK CONTENTS Component Size Shipping Temperature Upon Receipt Checklist 3K27M Histone H3K27me1 (H3K27 Monomethyl) Polyclonal Antibody 25 µg
Mannen et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Mannen et al., http ://www.jcb.org /cgi /content /full /jcb.201601024 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Characterization of SNB components. (A) SNB localization of Venus-tagged
Supplemental material JCB Mannen et al., http ://www.jcb.org /cgi /content /full /jcb.201601024 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Characterization of SNB components. (A) SNB localization of Venus-tagged
Cell division requires the precise coordination of an
 Control of Mitotic Events by Nap1 and the Gin4 Kinase Roger Altman and Douglas Kellogg Sinsheimer Laboratories, Department of Biology, University of California, Santa Cruz, California 95064 Abstract. Little
Control of Mitotic Events by Nap1 and the Gin4 Kinase Roger Altman and Douglas Kellogg Sinsheimer Laboratories, Department of Biology, University of California, Santa Cruz, California 95064 Abstract. Little
Figure S1. Figure S2. Figure S3 HB Anti-FSP27 (COOH-terminal peptide) Ab. Anti-GST-FSP27(45-127) Ab.
 / 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
/ 36B4 mrna ratio Figure S1 * 2. 1.6 1.2.8 *.4 control TNFα BRL49653 Figure S2 Su bw AT p iw Anti- (COOH-terminal peptide) Ab Blot : Anti-GST-(45-127) Ab β-actin Figure S3 HB2 HW AT BA T Figure S4 A TAG
R-loops cause replication impairment and genome instability during meiosis
 Manuscript EMBOR-2012-35976 R-loops cause replication impairment and genome instability during meiosis Maikel Castellano-Pozo, Tatiana Garcia-Muse and Andres Aguilera Corresponding author: Andres Aguilera,
Manuscript EMBOR-2012-35976 R-loops cause replication impairment and genome instability during meiosis Maikel Castellano-Pozo, Tatiana Garcia-Muse and Andres Aguilera Corresponding author: Andres Aguilera,
Cell division requires the precise coordination of an
 Published Online: 14 July, 1997 Supp Info: http://doi.org/10.1083/jcb.138.1.119 Downloaded from jcb.rupress.org on October 17, 2018 Control of Mitotic Events by Nap1 and the Gin4 Kinase Roger Altman and
Published Online: 14 July, 1997 Supp Info: http://doi.org/10.1083/jcb.138.1.119 Downloaded from jcb.rupress.org on October 17, 2018 Control of Mitotic Events by Nap1 and the Gin4 Kinase Roger Altman and
Supplementary Figure legends
 Supplementary Figure legends Figure S. Histone shuffle strain construction and characterization. A. Schematic representing the histone shuffle strain. Both genomic copies of the and genes are deleted (genes
Supplementary Figure legends Figure S. Histone shuffle strain construction and characterization. A. Schematic representing the histone shuffle strain. Both genomic copies of the and genes are deleted (genes
SUPPLEMENTARY INFORMATION
 Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
3.1 The Role of the Enzymatic Activity of Sir2 for Efficient. The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and
 35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
Supplementary Methods
 Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplementary Methods MARCM-based forward genetic screen We used ethylmethane sulfonate (EMS) to mutagenize flies carrying FRT 2A and FRT 82B transgenes, which are sites for the FLP-mediated recombination
Supplemental Material for Xue et al. List. Supplemental Figure legends. Figure S1. Related to Figure 1. Figure S2. Related to Figure 3
 Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
Supplemental Material for Xue et al List Supplemental Figure legends Figure S1. Related to Figure 1 Figure S. Related to Figure 3 Figure S3. Related to Figure 4 Figure S4. Related to Figure 4 Figure S5.
Novel Response to Microtubule Perturbation in Meiosis
 SUPPLEMENTAL MATERIAL REFERENCES CONTENT ALERTS Novel Response to Microtubule Perturbation in Meiosis Andreas Hochwagen, Gunnar Wrobel, Marie Cartron, Philippe Demougin, Christa Niederhauser-Wiederkehr,
SUPPLEMENTAL MATERIAL REFERENCES CONTENT ALERTS Novel Response to Microtubule Perturbation in Meiosis Andreas Hochwagen, Gunnar Wrobel, Marie Cartron, Philippe Demougin, Christa Niederhauser-Wiederkehr,
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
The Origin Recognition Complex Functions in Sister-Chromatid Cohesion in Saccharomyces cerevisiae
 The Origin Recognition Complex Functions in Sister-Chromatid Cohesion in Saccharomyces cerevisiae Kenji Shimada 1 and Susan M. Gasser 1, * 1 Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse
The Origin Recognition Complex Functions in Sister-Chromatid Cohesion in Saccharomyces cerevisiae Kenji Shimada 1 and Susan M. Gasser 1, * 1 Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse
