Navigating BioLuminate for PyMOL Users: A Practical Approach. 2nd European Life Science Bootcamp Ana Negri, PhD
|
|
|
- Samuel Copeland
- 6 years ago
- Views:
Transcription
1 Navigating BioLuminate for PyMOL Users: A Practical Approach 2nd European Life Science Bootcamp Ana Negri, PhD
2 Agenda Introduction to BioLuminate PyMOL-like features in the BioLuminate interface: Basic rendering and visualization using BioLuminate Beyond visualization: Antibody Modeling and Protein-Protein Docking Residue Mutation and Affinity Maturation Hands-on tutorial: Visualization, Sequence Analysis, and Modeling with BioLuminate: A case Study with Neuraminidase Questions
3 Introduction to BioLuminate BioLuminate is the Schrödinger biologics software platform A suitably low learning curve for new users, non expert users Focused on workflows and tasks, with multiple modeling features GUI modeled on PyMOL Antibody Antigen
4 Functionality groups in BioLuminate Antibody design Structure prediction from sequence, humanization Protein design Residue scanning (affinity / stability); Cysteine scanning; Protein crosslinking; In silico affinity maturation; Non-standard amino acids; Protein FEP Protein liability ID Aggregation propensity ID; Reactive residue ID (glycosylation, proteolysis, oxidation, deamination); Peptide QSAR; Titration curve / Isoelectric point; Solubility Protein modeling Homology modeling Protein-protein docking; Prime de novo loop modeling Protein interaction analysis; Chimeric model generation; Automated peptide docking Advanced sequence viewer (MSV); Automated protein quality report Molecular Dynamics, minimization, quantum mechanics, etc
5 Agenda Introduction to BioLuminate PyMOL-like features in the BioLuminate interface: Basic rendering and visualization using BioLuminate Beyond visualization: Antibody Modeling and Protein-Protein Docking Residue Mutation and Affinity Maturation Hands-on tutorial: Visualization, Sequence Analysis, and Modeling with BioLuminate: A case Study with Neuraminidase Questions
6 PyMOL-like features in the BioLuminate interface
7 PyMOL-like features in the BioLuminate interface
8 Agenda Introduction to BioLuminate PyMOL-like features in the BioLuminate interface: Basic rendering and visualization using BioLuminate Beyond visualization: Antibody Modeling and Protein-Protein Docking Residue Mutation and Affinity Maturation Hands-on tutorial: Visualization, Sequence Analysis, and Modeling with BioLuminate: A case Study with Neuraminidase Questions
9 Antibody Modeling and Protein-Protein Docking (1A43) HIV-1 Capsid Protein (P24) FAB13B5 Green=xray Blue/Maroon=predict Piper/Cluspro (in BioLuminate) CAPRI rankings (Nir London, Rosetta Design Group, 2010) Orange=xray (1E6J) Blue/Green/Red=predict Third ranked complex shown
10 Antibody Modeling CDR Prediction: Input & Framework Antibody-specific workflow Search public or in-house structures for templates Framework selection: Separate control over L/H chain framework templates Control over framework used to align chains
11 Antibody Modeling CDR Prediction: Loop Grafting Start with fast loop lookup: Find largest cluster with maximum sequence similarity Minimize grafted loop Optionally perform advanced loop prediction using Prime
12 Antibody Modeling Database Management Multiple databases can be created and later selected New structures can be added to existing or new DB
13 Protein-Protein Docking Licensed from Vajda group at Boston University Kozakov et al. (2006) Proteins: Struct, Funct, Bioinf #1 server in most recent CAPRI competition Competitive with human groups CAPRI rankings (Nir London, Rosetta Design Group, 2010) Piper/Cluspro: #1 group #1 server
14 Protein-Protein Docking is Easy to Run
15 Antibody/Antigen Complex: Predicted Versus Experiment Modeled FAB13B5 CDR docked with crystal structure of unbound antigen P24 (orange) versus x-ray complex 1E6J. 3 rd ranked complex shown.
16 Residue Scanning: ID mutations for improved affinity / stability Residue scanning is a technique used to determine the contribution of a specific residue to the stability, binding or function of a given protein Walk mutation R Ala along sequence Measure Δ Affinity Δ Stability Tells us what mutations may be beneficial, and what may be harmful Can be a very laborious and difficult task to do in the lab.
17 MM-GBSA residue mutation calculations Prime energy function Select residues to mutate Calculate relevant energies Apply equations from statistical mechanics Implicit solvent (Generalized Born) Rigorous all atom force field model Extensively validated for protein structure prediction Collaborative effort: Schrödinger, Rich Friesner (Columbia) and Matt Jacobson (UCSF) (Output) Free energies from mutation
18 From Residue Scanning to Affinity Maturation Residue Scanning (single mutations): Protein 1 Protein 1 Protein 2 Protein 2 Affinity Maturation/Protein Design (multiple simultaneous mutations): Protein 1 Protein 1 Protein 2 Protein 2
19 Affinity Maturation in BioLuminate
20 Affinity Maturation in BioLuminate Search multiple residue positions simultaneously for changes Implicit solvent MM-GBSA model Uses Monte Carlo sampling due to combinatorial explosion Use to suggest new sequences, or to influence random library design Reveals positions and types of beneficial mutations
21 Agenda Introduction to BioLuminate PyMOL-like features in the BioLuminate interface: Basic rendering and visualization using BioLuminate Beyond visualization: Antibody Modeling and Protein-Protein Docking Residue Mutation and Affinity Maturation Hands-on tutorial: Visualization, Sequence Analysis, and Modeling with BioLuminate: A case Study with Neuraminidase Questions
22 Useful Video Links Related to Today s Webinar Biologics modeling tutorial series: Intro to BioLuminate Homology Modeling (Antibody modeling) Residue Scanning Protein-Protein Docking, etc
23 Other Useful Resources Schrödinger support: Knowledge Base: Support Center and Training Videos: Schrödinger Seminar Series: Script Center:
24 Questions? Thanks for Joining Us!
Introduction to BioLuminate
 Introduction to BioLuminate Janet Paulsen Stanford University 10/16/17 BioLuminate has Broad Functionality Antibody design Structure prediction from sequence, humanization Protein design Residue scanning
Introduction to BioLuminate Janet Paulsen Stanford University 10/16/17 BioLuminate has Broad Functionality Antibody design Structure prediction from sequence, humanization Protein design Residue scanning
static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, i
 BIOLUMINATE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
BIOLUMINATE static MM_Index snap(mm_index corect, MM_Index ligct, int imatch0, int *moleatoms, int *refcoreatoms){int ncoreat = :vector mappings; PhpCoreMapping mapping; for COMMON(glidelig).
What s New in Discovery Studio 2.5.5
 What s New in Discovery Studio 2.5.5 Discovery Studio takes modeling and simulations to the next level. It brings together the power of validated science on a customizable platform for drug discovery research.
What s New in Discovery Studio 2.5.5 Discovery Studio takes modeling and simulations to the next level. It brings together the power of validated science on a customizable platform for drug discovery research.
4/10/2011. Rosetta software package. Rosetta.. Conformational sampling and scoring of models in Rosetta.
 Rosetta.. Ph.D. Thomas M. Frimurer Novo Nordisk Foundation Center for Potein Reseach Center for Basic Metabilic Research Breif introduction to Rosetta Rosetta docking example Rosetta software package Breif
Rosetta.. Ph.D. Thomas M. Frimurer Novo Nordisk Foundation Center for Potein Reseach Center for Basic Metabilic Research Breif introduction to Rosetta Rosetta docking example Rosetta software package Breif
Self-financed Coursework Advanced Workshop on Molecular Docking, Virtual Screening & Computational Biology 13 th - 15 th March 2019
 Self-financed Coursework Advanced Workshop on Molecular Docking, Virtual Screening & Computational Biology 13 th - 15 th March 2019 Indian Institute of Information Technology- Allahabad Jhalwa Campus,
Self-financed Coursework Advanced Workshop on Molecular Docking, Virtual Screening & Computational Biology 13 th - 15 th March 2019 Indian Institute of Information Technology- Allahabad Jhalwa Campus,
Prediction of Protein-Protein Binding Sites and Epitope Mapping. Copyright 2017 Chemical Computing Group ULC All Rights Reserved.
 Prediction of Protein-Protein Binding Sites and Epitope Mapping Epitope Mapping Antibodies interact with antigens at epitopes Epitope is collection residues on antigen Continuous (sequence) or non-continuous
Prediction of Protein-Protein Binding Sites and Epitope Mapping Epitope Mapping Antibodies interact with antigens at epitopes Epitope is collection residues on antigen Continuous (sequence) or non-continuous
Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions
 Introduction Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke, Rochester Institute of Technology Mentor: Carlos Camacho, University
Introduction Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke, Rochester Institute of Technology Mentor: Carlos Camacho, University
Worked Example of Humanized Fab D3h44 in Complex with Tissue Factor
 Worked Example of Humanized Fab D3h44 in Complex with Tissue Factor Here we provide an example worked in detail from antibody sequence and unbound antigen structure to a docked model of the antibody antigen
Worked Example of Humanized Fab D3h44 in Complex with Tissue Factor Here we provide an example worked in detail from antibody sequence and unbound antigen structure to a docked model of the antibody antigen
Antibodies to Challenging Receptor Targets through NGS and Cell-Based Antibody Phage Panning. Mark Tornetta
 Antibodies to Challenging Receptor Targets through NGS and Cell-Based Antibody Phage Panning Mark Tornetta Points to convey regarding receptor targets Antigen diversity- cells and engineered ECDs Be creative
Antibodies to Challenging Receptor Targets through NGS and Cell-Based Antibody Phage Panning Mark Tornetta Points to convey regarding receptor targets Antigen diversity- cells and engineered ECDs Be creative
Comparative Modeling Part 1. Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center
 Comparative Modeling Part 1 Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center Function is the most important feature of a protein Function is related to structure Structure is
Comparative Modeling Part 1 Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center Function is the most important feature of a protein Function is related to structure Structure is
Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction
 Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction Conformational Analysis 2 Conformational Analysis Properties of molecules depend on their three-dimensional
Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction Conformational Analysis 2 Conformational Analysis Properties of molecules depend on their three-dimensional
Enhancing the Quality of Biologics Patents:
 Enhancing the Quality of Biologics Patents: Computational Simulations as Evidence BCP Customer Partnership Meeting: April 26, 2016 Brian K. Lathrop, Ph.D., J.D., Sole Practitioner 4/22/2016 LAW OFFICE
Enhancing the Quality of Biologics Patents: Computational Simulations as Evidence BCP Customer Partnership Meeting: April 26, 2016 Brian K. Lathrop, Ph.D., J.D., Sole Practitioner 4/22/2016 LAW OFFICE
Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity
 Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity Stanley G. Kimani 1 *, Sushil Kumar 1 *, Nitu Bansal 2 *, Kamalendra Singh 3, Vladyslav Kholodovych
Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity Stanley G. Kimani 1 *, Sushil Kumar 1 *, Nitu Bansal 2 *, Kamalendra Singh 3, Vladyslav Kholodovych
Partnered Discovery of High-quality Antibody Drug Candidates COMPANY PRESENTATION DECEMBER 2016
 Partnered Discovery of High-quality Antibody Drug Candidates COMPANY PRESENTATION DECEMBER 2016 Corporate Overview A unique source of therapeutic human antibodies with one of the industry's most versatile
Partnered Discovery of High-quality Antibody Drug Candidates COMPANY PRESENTATION DECEMBER 2016 Corporate Overview A unique source of therapeutic human antibodies with one of the industry's most versatile
Docking. Why? Docking : finding the binding orientation of two molecules with known structures
 Docking : finding the binding orientation of two molecules with known structures Docking According to the molecules involved: Protein-Ligand docking Protein-Protein docking Specific docking algorithms
Docking : finding the binding orientation of two molecules with known structures Docking According to the molecules involved: Protein-Ligand docking Protein-Protein docking Specific docking algorithms
Computer Aided Drug Design. Protein structure prediction. Qin Xu
 Computer Aided Drug Design Protein structure prediction Qin Xu http://cbb.sjtu.edu.cn/~qinxu/cadd.htm Course Outline Introduction and Case Study Drug Targets Sequence analysis Protein structure prediction
Computer Aided Drug Design Protein structure prediction Qin Xu http://cbb.sjtu.edu.cn/~qinxu/cadd.htm Course Outline Introduction and Case Study Drug Targets Sequence analysis Protein structure prediction
Introduction to Bioinformatics
 Introduction to Bioinformatics http://1.51.212.243/bioinfo.html Dr. rer. nat. Jing Gong Cancer Research Center School of Medicine, Shandong University 2011.10.19 1 Chapter 4 Structure 2 Protein Structure
Introduction to Bioinformatics http://1.51.212.243/bioinfo.html Dr. rer. nat. Jing Gong Cancer Research Center School of Medicine, Shandong University 2011.10.19 1 Chapter 4 Structure 2 Protein Structure
Discovery Studio. Life Science Modeling and Simulations. Platform
 Discovery Studio is a singleunified, easy-to-use, graphical interface for powerful drug design and protein modeling research. Discovery Studio contains both established gold-standard applications (e.g.,
Discovery Studio is a singleunified, easy-to-use, graphical interface for powerful drug design and protein modeling research. Discovery Studio contains both established gold-standard applications (e.g.,
Applied Protein Services
 Applied Protein Services Applied Protein Services A Window into the Future Development risk and attrition rates remain two of the greatest challenges to a successful biopharmaceutical pipeline. To help
Applied Protein Services Applied Protein Services A Window into the Future Development risk and attrition rates remain two of the greatest challenges to a successful biopharmaceutical pipeline. To help
Computational methods in drug discovery
 Computational methods in drug discovery COURSE HIGHLIGHTS v Structure-based drug discovery Ø Binding site identification Ø Molecular docking v Biologics Ø Molecular modelling Ø Protein-Protein docking
Computational methods in drug discovery COURSE HIGHLIGHTS v Structure-based drug discovery Ø Binding site identification Ø Molecular docking v Biologics Ø Molecular modelling Ø Protein-Protein docking
Structure guided homology model based design and engineering of mouse antibodies for humanization
 www.bioinformation.net Hypothesis Volume 10(4) Structure guided homology model based design and engineering of mouse antibodies for humanization Vinodh B Kurella & Reddy Gali* Center for Biomedical Informatics
www.bioinformation.net Hypothesis Volume 10(4) Structure guided homology model based design and engineering of mouse antibodies for humanization Vinodh B Kurella & Reddy Gali* Center for Biomedical Informatics
Ligand docking. CS/CME/Biophys/BMI 279 Oct. 22 and 27, 2015 Ron Dror
 Ligand docking CS/CME/Biophys/BMI 279 Oct. 22 and 27, 2015 Ron Dror 1 Outline Goals of ligand docking Defining binding affinity (strength) Computing binding affinity: Simplifying the problem Ligand docking
Ligand docking CS/CME/Biophys/BMI 279 Oct. 22 and 27, 2015 Ron Dror 1 Outline Goals of ligand docking Defining binding affinity (strength) Computing binding affinity: Simplifying the problem Ligand docking
Products & Services. Customers. Example Customers & Collaborators
 Products & Services Eidogen-Sertanty is dedicated to delivering discovery informatics technologies that bridge the target-tolead knowledge gap. With a unique set of ligand- and structure-based drug discovery
Products & Services Eidogen-Sertanty is dedicated to delivering discovery informatics technologies that bridge the target-tolead knowledge gap. With a unique set of ligand- and structure-based drug discovery
This practical aims to walk you through the process of text searching DNA and protein databases for sequence entries.
 PRACTICAL 1: BLAST and Sequence Alignment The EBI and NCBI websites, two of the most widely used life science web portals are introduced along with some of the principal databases: the NCBI Protein database,
PRACTICAL 1: BLAST and Sequence Alignment The EBI and NCBI websites, two of the most widely used life science web portals are introduced along with some of the principal databases: the NCBI Protein database,
What s New in LigandScout 4.4
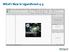 What s New in LigandScout 4.4 What's New in LigandScout 4.4? 2D Molecule editor for convenient structure editing Binding affinity prediction and atom contribution display Automated protein binding site
What s New in LigandScout 4.4 What's New in LigandScout 4.4? 2D Molecule editor for convenient structure editing Binding affinity prediction and atom contribution display Automated protein binding site
MCB 4211, Fall 2018, Practice Exam 1 Last, First name Student ID # Seat No. ***NOTE: Exam will have 40 multiple choice questions.
 MCB 4211, Fall 2018, Practice Exam 1 Last, First name Student ID # Seat No. ***NOTE: Exam 1 2018 will have 40 multiple choice questions. READ ALL THE CHOICES AND SELECT THE BEST 1. Which of the following
MCB 4211, Fall 2018, Practice Exam 1 Last, First name Student ID # Seat No. ***NOTE: Exam 1 2018 will have 40 multiple choice questions. READ ALL THE CHOICES AND SELECT THE BEST 1. Which of the following
a. Hypoxanthine was present in the media. MCB 4211, Fall 2018, Practice Exam 1 Last, First name Student ID # Seat No.
 MCB 4211, Fall 2018, Practice Exam 1 Last, First name Student ID # Seat No. ***NOTE: Exam 1 2018 will have 40 multiple choice questions. READ ALL THE CHOICES AND SELECT THE BEST 1. Which of the following
MCB 4211, Fall 2018, Practice Exam 1 Last, First name Student ID # Seat No. ***NOTE: Exam 1 2018 will have 40 multiple choice questions. READ ALL THE CHOICES AND SELECT THE BEST 1. Which of the following
The generation of lymphocyte antigen receptors (Chapter 5):
 The generation of lymphocyte antigen receptors (Chapter 5): 1. Ig Gene Rearrangement (somatic recombination). 2. Ig Somatic Hypermutation. 3. Ig Class Switching 4. T cell Receptor Gene Rearragement 1.
The generation of lymphocyte antigen receptors (Chapter 5): 1. Ig Gene Rearrangement (somatic recombination). 2. Ig Somatic Hypermutation. 3. Ig Class Switching 4. T cell Receptor Gene Rearragement 1.
Partnered Discovery of High-quality Antibody Drug Candidates
 Partnered Discovery of High-quality Antibody Drug Candidates COMPANY PRESENTATION SEPTEMBER 2017 Corporate Overview A unique source of therapeutic human antibodies with one of the industry's most versatile
Partnered Discovery of High-quality Antibody Drug Candidates COMPANY PRESENTATION SEPTEMBER 2017 Corporate Overview A unique source of therapeutic human antibodies with one of the industry's most versatile
Protein design. CS/CME/BioE/Biophys/BMI 279 Oct. 24, 2017 Ron Dror
 Protein design CS/CME/BioE/Biophys/BMI 279 Oct. 24, 2017 Ron Dror 1 Outline Why design proteins? Overall approach: Simplifying the protein design problem Protein design methodology Designing the backbone
Protein design CS/CME/BioE/Biophys/BMI 279 Oct. 24, 2017 Ron Dror 1 Outline Why design proteins? Overall approach: Simplifying the protein design problem Protein design methodology Designing the backbone
Homology Modelling. Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen
 Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
COMPAS for the Analysis of SELEX Experiments
 COMPAS for the Analysis of SELEX Experiments COMPAS (COMmon PAtternS) is a software tool that was especially developed to harness the technology of next generation sequencing (NGS) to bring light into
COMPAS for the Analysis of SELEX Experiments COMPAS (COMmon PAtternS) is a software tool that was especially developed to harness the technology of next generation sequencing (NGS) to bring light into
Partnered Discovery of High-Quality Antibody Drug Candidates. Company Presentation
 Partnered Discovery of High-Quality Antibody Drug Candidates Company Presentation AbCheck s Unique Offering A unique source of human antibodies with one of the industry's most versatile technology platforms
Partnered Discovery of High-Quality Antibody Drug Candidates Company Presentation AbCheck s Unique Offering A unique source of human antibodies with one of the industry's most versatile technology platforms
Protein design. CS/CME/BioE/Biophys/BMI 279 Oct. 24, 2017 Ron Dror
 Protein design CS/CME/BioE/Biophys/BMI 279 Oct. 24, 2017 Ron Dror 1 Outline Why design proteins? Overall approach: Simplifying the protein design problem < this step is really key! Protein design methodology
Protein design CS/CME/BioE/Biophys/BMI 279 Oct. 24, 2017 Ron Dror 1 Outline Why design proteins? Overall approach: Simplifying the protein design problem < this step is really key! Protein design methodology
Homology Modelling. Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen
 Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Homology Modelling Thomas Holberg Blicher NNF Center for Protein Research University of Copenhagen Why are Protein Structures so Interesting? They provide a detailed picture of interesting biological features,
Molecular Modeling 9. Protein structure prediction, part 2: Homology modeling, fold recognition & threading
 Molecular Modeling 9 Protein structure prediction, part 2: Homology modeling, fold recognition & threading The project... Remember: You are smarter than the program. Inspecting the model: Are amino acids
Molecular Modeling 9 Protein structure prediction, part 2: Homology modeling, fold recognition & threading The project... Remember: You are smarter than the program. Inspecting the model: Are amino acids
Peptide libraries: applications, design options and considerations. Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST
 Peptide libraries: applications, design options and considerations Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST Overview 1 2 3 4 5 Introduction Peptide library basics Peptide library design considerations
Peptide libraries: applications, design options and considerations Laura Geuss, PhD May 5, 2015, 2:00-3:00 pm EST Overview 1 2 3 4 5 Introduction Peptide library basics Peptide library design considerations
Immunogenicity Prediction Where are we?
 Pharma&Biotech Immunogenicity Prediction Where are we? European Immunogenicity Platform, 24th February 2016 Immunogenicity of Biopharmaceuticals Potential causes 2 Pre-clinical Immunogenicity Prediction
Pharma&Biotech Immunogenicity Prediction Where are we? European Immunogenicity Platform, 24th February 2016 Immunogenicity of Biopharmaceuticals Potential causes 2 Pre-clinical Immunogenicity Prediction
Antibody-Antigen recognition. Structural Biology Weekend Seminar Annegret Kramer
 Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
Antibody-Antigen recognition Structural Biology Weekend Seminar 10.07.2005 Annegret Kramer Contents Function and structure of antibodies Features of antibody-antigen interfaces Examples of antibody-antigen
What can SharePoint do?
 What can SharePoint do? Lots!! Collaboration Document management Internal Social Network Forms Process Automation (Workflows) Business Intelligence Reporting Search Metadata Management Super secret The
What can SharePoint do? Lots!! Collaboration Document management Internal Social Network Forms Process Automation (Workflows) Business Intelligence Reporting Search Metadata Management Super secret The
Immunoglobulins. Even variable chain differ in variability of amino acid sequences:
 Revision: Immunoglobulin structure 2 light chain 25KDa and 2 heavy chain 50KDa (total=150kda) Heavy chain one quarter variable three quarter constant Even variable chain differ in variability of amino
Revision: Immunoglobulin structure 2 light chain 25KDa and 2 heavy chain 50KDa (total=150kda) Heavy chain one quarter variable three quarter constant Even variable chain differ in variability of amino
Can self-inhibitory peptides be derived from protein interfaces?
 Can self-inhibitory peptides be derived from protein interfaces? Ora Schueler-Furman RosettaCon 2010 Outline of today s talk 1. Short introduction on peptide-protein interactions and their modeling 2.
Can self-inhibitory peptides be derived from protein interfaces? Ora Schueler-Furman RosettaCon 2010 Outline of today s talk 1. Short introduction on peptide-protein interactions and their modeling 2.
Structural bioinformatics
 Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Chapter 17. Prediction, Engineering, and Design of Protein Structures
 Chapter 17. Prediction, Engineering, and Design of Protein Structures Homologous proteins have similar structure and function Branden & Tooze (1998), Introduction to protein structure, 2 nd ed., p.349.
Chapter 17. Prediction, Engineering, and Design of Protein Structures Homologous proteins have similar structure and function Branden & Tooze (1998), Introduction to protein structure, 2 nd ed., p.349.
How to Screen a Billion Drug Candidates?
 How to Screen a Billion Drug Candidates? Single Cell Functional Assays: Ultra-sensitive, Ultra-fast, Ultrahigh-throughput Next-Generation Drug Discovery The advantages of de novo protein engineering for
How to Screen a Billion Drug Candidates? Single Cell Functional Assays: Ultra-sensitive, Ultra-fast, Ultrahigh-throughput Next-Generation Drug Discovery The advantages of de novo protein engineering for
Building an Antibody Homology Model
 Application Guide Antibody Modeling: A quick reference guide to building Antibody homology models, and graphical identification and refinement of CDR loops in Discovery Studio. Francisco G. Hernandez-Guzman,
Application Guide Antibody Modeling: A quick reference guide to building Antibody homology models, and graphical identification and refinement of CDR loops in Discovery Studio. Francisco G. Hernandez-Guzman,
Protein design. CS/CME/Biophys/BMI 279 Oct. 20 and 22, 2015 Ron Dror
 Protein design CS/CME/Biophys/BMI 279 Oct. 20 and 22, 2015 Ron Dror 1 Optional reading on course website From cs279.stanford.edu These reading materials are optional. They are intended to (1) help answer
Protein design CS/CME/Biophys/BMI 279 Oct. 20 and 22, 2015 Ron Dror 1 Optional reading on course website From cs279.stanford.edu These reading materials are optional. They are intended to (1) help answer
Streamline Your Antibody Enrichment Using Scalable Magnetic Bead-Based Chemistries
 Streamline Your Antibody Enrichment Using Scalable Magnetic Bead-Based Chemistries Samantha Lewis, Ph.D. 2013. Webinar Outline Introduction Magnetic Bead-Based Method Overview Scalability Chemistries o
Streamline Your Antibody Enrichment Using Scalable Magnetic Bead-Based Chemistries Samantha Lewis, Ph.D. 2013. Webinar Outline Introduction Magnetic Bead-Based Method Overview Scalability Chemistries o
Project ideas. CS/BioE/CME/Biophys/BMI 279 Nov. 9, 2017 Ron Dror
 Project ideas CS/BioE/CME/Biophys/BMI 279 Nov. 9, 2017 Ron Dror Project Ideas 2 Project Ideas You are welcome (and encouraged) to pick something that is not on this list. 2 Project Ideas You are welcome
Project ideas CS/BioE/CME/Biophys/BMI 279 Nov. 9, 2017 Ron Dror Project Ideas 2 Project Ideas You are welcome (and encouraged) to pick something that is not on this list. 2 Project Ideas You are welcome
Protein-Protein Interactions II
 Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Biochemistry 412 Protein-Protein Interactions II March 28, 2008 Delano (2002) Curr. Opin. Struct. Biol. 12, 14. Some ways that mutations can destabilize protein-protein interactions Delano (2002) Curr.
Molecular recognition
 Lecture 9 Molecular recognition Antoine van Oijen BCMP201 Spring 2008 Structural principles of binding 4 fundamental functions of proteins: 1) Binding 2) Catalysis 3) Switching 4) Structural All involve
Lecture 9 Molecular recognition Antoine van Oijen BCMP201 Spring 2008 Structural principles of binding 4 fundamental functions of proteins: 1) Binding 2) Catalysis 3) Switching 4) Structural All involve
Directed Evolution Methods for Protein Engineering
 Directed Evolution Methods for Protein Engineering XXX Biosciences, Life Sciences Solutions, Geneart AG, Regensburg, Germany The world leader in serving science 1 Directed Evolution 2 Random mutagenesis
Directed Evolution Methods for Protein Engineering XXX Biosciences, Life Sciences Solutions, Geneart AG, Regensburg, Germany The world leader in serving science 1 Directed Evolution 2 Random mutagenesis
CENTRE FOR BIOINFORMATICS M. D. UNIVERSITY, ROHTAK
 Program Specific Outcomes: The students upon completion of Ph.D. coursework in Bioinformatics will be able to: PSO1 PSO2 PSO3 PSO4 PSO5 Produce a well-developed research proposal. Select an appropriate
Program Specific Outcomes: The students upon completion of Ph.D. coursework in Bioinformatics will be able to: PSO1 PSO2 PSO3 PSO4 PSO5 Produce a well-developed research proposal. Select an appropriate
Problem Set #3
 20.320 Problem Set #3 Due on October 7th, 2011 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all of your assumptions, and provide step- by- step solutions
20.320 Problem Set #3 Due on October 7th, 2011 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all of your assumptions, and provide step- by- step solutions
Next-generation analysis of deep sequencing data: Bringing light into the black box of phage display experiments
 Next-generation analysis of deep sequencing data: Bringing light into the black box of phage display experiments Dr. Michael Blank (CSO, AptaIT) Biologics 2015; Berlin 02-03 February 1 Accelerating in
Next-generation analysis of deep sequencing data: Bringing light into the black box of phage display experiments Dr. Michael Blank (CSO, AptaIT) Biologics 2015; Berlin 02-03 February 1 Accelerating in
Characterization of Aptamer Binding using SensíQ SPR Platforms
 Characterization of Aptamer Binding using SensíQ SPR Platforms APPLICATION NOTE INTRODUCTION Aptamers have the potential to provide a better solution in diagnostics and other research areas than traditional
Characterization of Aptamer Binding using SensíQ SPR Platforms APPLICATION NOTE INTRODUCTION Aptamers have the potential to provide a better solution in diagnostics and other research areas than traditional
Bioinformatics & Protein Structural Analysis. Bioinformatics & Protein Structural Analysis. Learning Objective. Proteomics
 The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
Introduction to Bioinformatics Introduction to Bioinformatics
 Dr. rer. nat. Gong Jing Cancer Research center Medicine School of Shandong University 2012.11.09 1 Chapter 5 Structure 2 Protein Structure When you study a protein, you are usually interested in its function.
Dr. rer. nat. Gong Jing Cancer Research center Medicine School of Shandong University 2012.11.09 1 Chapter 5 Structure 2 Protein Structure When you study a protein, you are usually interested in its function.
CPC field-specific training
 CPC field-specific training G06F19/10 Bioinformatics, G06F19/70 Chemoinformatics, C40B30/02, C40B50/02 In silico Combinatorial Chemistry, G16H Healthcare Informatics Przemyslaw Godzina Examiner, European
CPC field-specific training G06F19/10 Bioinformatics, G06F19/70 Chemoinformatics, C40B30/02, C40B50/02 In silico Combinatorial Chemistry, G16H Healthcare Informatics Przemyslaw Godzina Examiner, European
Information Page. A List of Secondary Structure Prediction Servers
 Introduction to Bioinformatics Information Page Chapter 5, page 1/4, 2012/11/09 A List of Secondary Structure Prediction Servers Name APSSP PredictProtein GOR IV Jpred 3 CFSSP NNPREDICT JUFO BCM Search
Introduction to Bioinformatics Information Page Chapter 5, page 1/4, 2012/11/09 A List of Secondary Structure Prediction Servers Name APSSP PredictProtein GOR IV Jpred 3 CFSSP NNPREDICT JUFO BCM Search
Protein Structure Prediction
 Homology Modeling Protein Structure Prediction Ingo Ruczinski M T S K G G G Y F F Y D E L Y G V V V V L I V L S D E S Department of Biostatistics, Johns Hopkins University Fold Recognition b Initio Structure
Homology Modeling Protein Structure Prediction Ingo Ruczinski M T S K G G G Y F F Y D E L Y G V V V V L I V L S D E S Department of Biostatistics, Johns Hopkins University Fold Recognition b Initio Structure
Byos. A Clear Sky Solution for Analytical Scientists
 A Clear Sky Solution for Analytical Scientists One-touch Platform to Launch Seamless Workflows Raw File Report in Minutes Integrated application with templated workflows Workflows Step 1: Click to launch
A Clear Sky Solution for Analytical Scientists One-touch Platform to Launch Seamless Workflows Raw File Report in Minutes Integrated application with templated workflows Workflows Step 1: Click to launch
Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL*
 Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL* Site AA Sequence 3x10 6 3x10 6 20x10 6 20x10 6 100x10 6 100x10 6 Post-1 st Post-2 nd Post-1 st Post-2 nd Post-1 st Post-2
Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL* Site AA Sequence 3x10 6 3x10 6 20x10 6 20x10 6 100x10 6 100x10 6 Post-1 st Post-2 nd Post-1 st Post-2 nd Post-1 st Post-2
Immunoglobulins. Harper s biochemistry Chapter 49
 Immunoglobulins Harper s biochemistry Chapter 49 Immune system Detects and inactivates foreign molecules, viruses, bacteria and microorganisms Two components with 2 strategies B Lymphocytes (humoral immune
Immunoglobulins Harper s biochemistry Chapter 49 Immune system Detects and inactivates foreign molecules, viruses, bacteria and microorganisms Two components with 2 strategies B Lymphocytes (humoral immune
Globus Genomics at GSI Boston University. Dinanath Sulakhe, Alex Rodriguez
 Globus Genomics at GSI Boston University Dinanath Sulakhe, Alex Rodriguez July 2014 Agenda 1. Introduction to Globus Genomics - Key features of Globus Genomics - How to use Globus Transfer 2. Introduce
Globus Genomics at GSI Boston University Dinanath Sulakhe, Alex Rodriguez July 2014 Agenda 1. Introduction to Globus Genomics - Key features of Globus Genomics - How to use Globus Transfer 2. Introduce
Information Processing in Living Systems
 Information Processing in Living Systems http://upload.wikimedia.org/wikipedia/commons/f/f3/cavernous_hemangioma_t2.jpg Does the brain compute? http://www.cheniere.org/images/rife/rife20.jpg Does the
Information Processing in Living Systems http://upload.wikimedia.org/wikipedia/commons/f/f3/cavernous_hemangioma_t2.jpg Does the brain compute? http://www.cheniere.org/images/rife/rife20.jpg Does the
Attribution: University of Michigan Medical School, Department of Microbiology and Immunology
 Attribution: University of Michigan Medical School, Department of Microbiology and Immunology License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution
Attribution: University of Michigan Medical School, Department of Microbiology and Immunology License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution
Supplementary Material for
 Supplementary Material for Engineering IgGs to aggregation resistance by two independent mechanisms: lessons from comparison of Pichia pastoris and mammalian cell expression Jonas V. Schaefer and Andreas
Supplementary Material for Engineering IgGs to aggregation resistance by two independent mechanisms: lessons from comparison of Pichia pastoris and mammalian cell expression Jonas V. Schaefer and Andreas
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I
 BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
Identifying the antibody specificity repertoire
 Identifying the antibody specificity repertoire Rafael Smith, Joel Bozekowski, Michael Paul, Patrick Daugherty Department of Chemical Engineering Daugherty Group Gorman Scholar You have a unique antibody
Identifying the antibody specificity repertoire Rafael Smith, Joel Bozekowski, Michael Paul, Patrick Daugherty Department of Chemical Engineering Daugherty Group Gorman Scholar You have a unique antibody
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools News About NCBI Site Map
National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools News About NCBI Site Map
systemsdock Operation Manual
 systemsdock Operation Manual Version 2.0 2016 April systemsdock is being developed by Okinawa Institute of Science and Technology http://www.oist.jp/ Integrated Open Systems Unit http://openbiology.unit.oist.jp/_new/
systemsdock Operation Manual Version 2.0 2016 April systemsdock is being developed by Okinawa Institute of Science and Technology http://www.oist.jp/ Integrated Open Systems Unit http://openbiology.unit.oist.jp/_new/
Module 2 overview SPRING BREAK
 1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
CS612 - Algorithms in Bioinformatics
 Spring 2012 Class 6 February 9, 2012 Backbone and Sidechain Rotational Degrees of Freedom Protein 3-D Structure The 3-dimensional fold of a protein is called a tertiary structure. Many proteins consist
Spring 2012 Class 6 February 9, 2012 Backbone and Sidechain Rotational Degrees of Freedom Protein 3-D Structure The 3-dimensional fold of a protein is called a tertiary structure. Many proteins consist
Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD
 Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD Department of Biopharmacy, Faculty of Pharmacy, Silpakorn University Example of critical checkpoints
Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD Department of Biopharmacy, Faculty of Pharmacy, Silpakorn University Example of critical checkpoints
Extracting Pure Proteins from Cells
 Extracting Pure Proteins from Cells 0 Purification techniques focus mainly on size & charge 0 The first step is homogenization (grinding, Potter Elvejhem homogenizer, sonication, freezing and thawing,
Extracting Pure Proteins from Cells 0 Purification techniques focus mainly on size & charge 0 The first step is homogenization (grinding, Potter Elvejhem homogenizer, sonication, freezing and thawing,
Bioinformatic Tools. So you acquired data.. But you wanted knowledge. So Now What?
 Bioinformatic Tools So you acquired data.. But you wanted knowledge So Now What? We have a series of questions What the Heck is That Ion? How come my MW does not match? How do I make a DB to search against?
Bioinformatic Tools So you acquired data.. But you wanted knowledge So Now What? We have a series of questions What the Heck is That Ion? How come my MW does not match? How do I make a DB to search against?
Molecular Modeling Lecture 8. Local structure Database search Multiple alignment Automated homology modeling
 Molecular Modeling 2018 -- Lecture 8 Local structure Database search Multiple alignment Automated homology modeling An exception to the no-insertions-in-helix rule Actual structures (myosin)! prolines
Molecular Modeling 2018 -- Lecture 8 Local structure Database search Multiple alignment Automated homology modeling An exception to the no-insertions-in-helix rule Actual structures (myosin)! prolines
Digital Display Libraries using High Density Peptide Arrays Max Bergmann Konferenz XXXVI
 Digital Display Libraries using High Density Peptide Arrays Max Bergmann Konferenz XXXVI - 2015 Klaus-Peter Stengele, Jigar Patel Roche NimbleGen Inc, Madison, WI www.nimblegen.com Roche NimbleGen Peptide
Digital Display Libraries using High Density Peptide Arrays Max Bergmann Konferenz XXXVI - 2015 Klaus-Peter Stengele, Jigar Patel Roche NimbleGen Inc, Madison, WI www.nimblegen.com Roche NimbleGen Peptide
Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs
 Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Pharma&Biotech. Developability Assessment Platform Reduce Attrition Rates, Improve Clinical Safety and Avoid the Valley of Death
 Pharma&Biotech Developability Assessment Platform Reduce Attrition Rates, Improve Clinical Safety and Avoid the Valley of Death The Valley of Death Development risk, safety and high attrition rates remain
Pharma&Biotech Developability Assessment Platform Reduce Attrition Rates, Improve Clinical Safety and Avoid the Valley of Death The Valley of Death Development risk, safety and high attrition rates remain
Western-GUARANTEED Antibody Service FAQ
 Western-GUARANTEED Antibody Service FAQ Content Q 1: When do I need a Western GUARANTEED Peptide Antibody Package?...2 Q 2: Can GenScript provide a Western blot guaranteed antibody?...2 Q 3: Does GenScript
Western-GUARANTEED Antibody Service FAQ Content Q 1: When do I need a Western GUARANTEED Peptide Antibody Package?...2 Q 2: Can GenScript provide a Western blot guaranteed antibody?...2 Q 3: Does GenScript
Creation of a PAM matrix
 Rationale for substitution matrices Substitution matrices are a way of keeping track of the structural, physical and chemical properties of the amino acids in proteins, in such a fashion that less detrimental
Rationale for substitution matrices Substitution matrices are a way of keeping track of the structural, physical and chemical properties of the amino acids in proteins, in such a fashion that less detrimental
Discovery and Humanization of Novel High Affinity Neutralizing Monoclonal Antibodies to Human IL-17A
 Discovery and Humanization of Novel High Affinity Neutralizing Monoclonal Antibodies to Human IL-17A Contacts: Marty Simonetti martysimonetti@gmail.com Kirby Alton kirby.alton@abeomecorp.com Rick Shimkets
Discovery and Humanization of Novel High Affinity Neutralizing Monoclonal Antibodies to Human IL-17A Contacts: Marty Simonetti martysimonetti@gmail.com Kirby Alton kirby.alton@abeomecorp.com Rick Shimkets
Immune Epitope Database NEWSLETTER Volume 1, Issue 2 August 2012
 Immune Epitope Database NEWSLETTER Volume 1, Issue 2 http://www.iedb.org August 2012 The IEDB at AAI 2012 The IEDB exhibit booth was present at the Annual Meeting of the American Association of Immunologists
Immune Epitope Database NEWSLETTER Volume 1, Issue 2 http://www.iedb.org August 2012 The IEDB at AAI 2012 The IEDB exhibit booth was present at the Annual Meeting of the American Association of Immunologists
ProteoGenix. Life Sciences Services and Products. From gene to biotherapeutics Target Validation to Lead optimisation
 ProteoGenix Life Sciences Services and Products From gene to biotherapeutics Target Validation to Lead optimisation ProteoGenix Philippe FUNFROCK, founder and CEO French company located in Strasbourg,
ProteoGenix Life Sciences Services and Products From gene to biotherapeutics Target Validation to Lead optimisation ProteoGenix Philippe FUNFROCK, founder and CEO French company located in Strasbourg,
Box 7905, Engineering Building I, North Carolina State University, Raleigh, NC 27695, Phone: Fax:
 Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
" Rational drug design; a Structural Genomics approach"
 " Rational drug design; a Structural Genomics approach" Meeting the need of time; Dr Sanjan K. Das, PhD. Chief Scientific Officer (CSO), BBUK. Institute of Cancer Therapeutics University of Bradford, UK.
" Rational drug design; a Structural Genomics approach" Meeting the need of time; Dr Sanjan K. Das, PhD. Chief Scientific Officer (CSO), BBUK. Institute of Cancer Therapeutics University of Bradford, UK.
This place covers: Methods or systems for genetic or protein-related data processing in computational molecular biology.
 G16B BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY Methods or systems for genetic
G16B BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY Methods or systems for genetic
Agilent AssayMAP Bravo Platform AUTOMATED PROTEIN AND PEPTIDE SAMPLE PREPARATION FOR MASS SPEC ANALYSIS
 Agilent AssayMAP Bravo Platform AUTOMATED PROTEIN AND PEPTIDE SAMPLE PREPARATION FOR MASS SPEC ANALYSIS GET FAST, ACCURATE, REPRODUCIBLE RESULTS The Agilent AssayMAP platform is an easy to use yet flexible
Agilent AssayMAP Bravo Platform AUTOMATED PROTEIN AND PEPTIDE SAMPLE PREPARATION FOR MASS SPEC ANALYSIS GET FAST, ACCURATE, REPRODUCIBLE RESULTS The Agilent AssayMAP platform is an easy to use yet flexible
Antibody Generation: challenges and solutions. Glen Marszalowicz, PHD May 10, AM
 Antibody Generation: challenges and solutions Glen Marszalowicz, PHD May 10, 2015 9AM GenScript the most cited biology CRO CRISPR Services Gene Services Peptide Services Protein Services Antibody Services
Antibody Generation: challenges and solutions Glen Marszalowicz, PHD May 10, 2015 9AM GenScript the most cited biology CRO CRISPR Services Gene Services Peptide Services Protein Services Antibody Services
Custom Antibody Services. Antibodies Designed Just for You. HuCAL Recombinant Monoclonal Antibody Generation Service
 Custom Antibody Services Antibodies Designed Just for You HuCAL Recombinant Monoclonal Antibody Generation Service Antibodies Designed Just for You What if there was a technology that would allow you to
Custom Antibody Services Antibodies Designed Just for You HuCAL Recombinant Monoclonal Antibody Generation Service Antibodies Designed Just for You What if there was a technology that would allow you to
Streamlined and Complete LC-MS Workflow for MAM
 Streamlined and Complete LC-MS Workflow for MAM Multiple Attribute Methodology (MAM) for Biotherapeutic Attribute Monitoring and Purity Testing BV MAM WORKFLOW Define Track Quantify Purity New Peaks Report
Streamlined and Complete LC-MS Workflow for MAM Multiple Attribute Methodology (MAM) for Biotherapeutic Attribute Monitoring and Purity Testing BV MAM WORKFLOW Define Track Quantify Purity New Peaks Report
User interface technologies for Manufacturing Execution Systems
 User interface technologies for Manufacturing Execution Systems VALTER SANDSTRÖM SEMINAR REPORT 13.11.2015 Contents Manufacturing Execution Systems (MES) MES User Interfaces MES Reports Function levels
User interface technologies for Manufacturing Execution Systems VALTER SANDSTRÖM SEMINAR REPORT 13.11.2015 Contents Manufacturing Execution Systems (MES) MES User Interfaces MES Reports Function levels
Engineering the Medicines of Tomorrow The Path to Platinum: The Evolution of Human Combinatorial Antibody Libraries (HuCAL )
 Engineering the Medicines of Tomorrow The Path to Platinum: The Evolution of Human Combinatorial Antibody Libraries (HuCAL ) Antibody Engineering December 2008, San Diego Dr. Stefanie Urlinger, Associate
Engineering the Medicines of Tomorrow The Path to Platinum: The Evolution of Human Combinatorial Antibody Libraries (HuCAL ) Antibody Engineering December 2008, San Diego Dr. Stefanie Urlinger, Associate
Bioinformatics Practical Course. 80 Practical Hours
 Bioinformatics Practical Course 80 Practical Hours Course Description: This course presents major ideas and techniques for auxiliary bioinformatics and the advanced applications. Points included incorporate
Bioinformatics Practical Course 80 Practical Hours Course Description: This course presents major ideas and techniques for auxiliary bioinformatics and the advanced applications. Points included incorporate
Oracle Prime Projects Cloud Service
 Oracle Prime Projects Cloud Service Oracle Prime Projects Cloud Service is a complete, cloud-first success platform for all stages of the project lifecycle. It empowers executives, project managers, and
Oracle Prime Projects Cloud Service Oracle Prime Projects Cloud Service is a complete, cloud-first success platform for all stages of the project lifecycle. It empowers executives, project managers, and
Chapter 8. One-Dimensional Structural Properties of Proteins in the Coarse-Grained CABS Model. Sebastian Kmiecik and Andrzej Kolinski.
 Chapter 8 One-Dimensional Structural Properties of Proteins in the Coarse-Grained CABS Model Abstract Despite the significant increase in computational power, molecular modeling of protein structure using
Chapter 8 One-Dimensional Structural Properties of Proteins in the Coarse-Grained CABS Model Abstract Despite the significant increase in computational power, molecular modeling of protein structure using
Increase throughput: spend less time cutting and. any CDS into Agilent's ELN
 Increase throughput: spend less time cutting and pasting. Smart Import your results directly from any CDS into Agilent's ELN e-seminar, November 18 th, 2009 Agenda Introduction on Agilent Software and
Increase throughput: spend less time cutting and pasting. Smart Import your results directly from any CDS into Agilent's ELN e-seminar, November 18 th, 2009 Agenda Introduction on Agilent Software and
Spectrum Mill MS Proteomics Workbench. Comprehensive tools for MS proteomics
 Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
