Title: A topological and conformational stability alphabet for multi-pass membrane proteins
|
|
|
- Karin Norma Hines
- 5 years ago
- Views:
Transcription
1 Supplementary Information Title: A topological and conformational stability alphabet for multi-pass membrane proteins Authors: Feng, X. 1 & Barth, P. 1,2,3 Correspondences should be addressed to: P.B. (patrickb@bcm.edu) 1 Department of Pharmacology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA. 2 Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA. 3 Structural and Computational Biology and Molecular Biophysics Graduate Program, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
2 Supplementary Results Supplementary Figure 1. A large majority of TMHs in multi-pass membrane proteins form binding interfaces with two other helices. Venn Diagram describing the fractions of TMHs in multi-pass membrane proteins: 1) interacting with two other helices, 2) interacting with two other helices with more than one residue forming contacts with the 2 helices simultaneously, 3) interacting with two other helices with more than five residues forming contacts with the 2 helices simultaneously. Contacts are defined by residues pairs with Cα distance less than 9Å.
3 Supplementary Figure 2. Uncovering universal sequence/structure principles governing multi-pass membrane protein topology and structure flexibility. a. The panel describes the process of identifying consensus sequence/structure motifs from the protein structure database. Elementary interacting TMH trimer units are extracted from unrelated protein structures, clustered into structurally similar families. Combinations of residues enriched within each trimer family creating consensus networks of stabilizing interhelical contacts are identified. b. The panel describes the stringent validation of the sequence/structure determinants of TMH trimer packing. If the sequence motifs are strong determinants of trimer conformations, prediction of trimer topology from sequence should be feasible. If motifs and associated interhelical contacts are strong determinants of trimer stability, they should guide the prediction of local conformational flexibility in TM proteins.
4 Supplementary Figure 3. Pi chart describing the clustering of TMH trimers into structurally similar families based on Cα RMSD. 56% of the trimer unit library can be classified into only 6 well-defined structural classes. The topology and percentage of each trimer type is labeled in the chart.
5 Supplementary Table 1. The geometry of the 6 most populated trimer structure clusters. The table describes the geometrical features of the 3 helical pairs constituting each trimer type. Helices are numbered (second column) according to a reference trimer structure in each class to assign helix pairs with specific topology. The same helix numbers are used to assign enriched motifs to specific helices in main text Figure 1 and Supplementary Figure 2.
6 Supplementary Table 2: Table of enriched sequence motifs at TMH trimer interfaces. For each trimer structure class (i.e. cluster), the p-value of enrichment of a motif on a specific helix is calculated using TMSTAT (see methods) and reported for two homology reduction thresholds: 60% sequence identity (60SI) removing close homologs for the structural analysis and one protein per superfamily (1/superfamily), a stringent homology reduction for training and testing the topology predictor (see methods). Motifs are classified as major if they form consensus contacts at trimer interfaces and are found in at least 3 protein families and superfamilies (see methods). P-values highlighted in red correspond to motifs that loose significant enrichment (pvalue 0.05) in the dataset composed of only one protein per superfamily.
7 Supplementary Figure 4. Proteins from the same superfamily display different sequence/structure determinants of the same trimer topology. Four examples of trimers from the all-left handed trimer structure class. a,b. Trimers are from two proteins of the Cytochrome c oxidase subunit I-like superfamily. These proteins share more than 30% sequence identity but bear distinct enriched sequence motif at the same trimer interface. c,d. Trimers are from two proteins of the MFS general substrate transporter superfamily but bear distinct enriched sequence motif at the same trimer interface. Trimers across protein superfamilies (a,c and b,d) share the same motifs.
8 Supplementary Table 3: Summary of consensus contact map and number of interacting helices for each trimer motif. The table describes the number of consensus contacts and the number of helices interacting with each position of the motif. A contact between one position of a motif and a residue on adjacent helices is defined as consensus if it is found in more than 50% of the trimers from the same structure class sharing the same sequence motif. For comparison, the average number of contacts established by the same position of the motif in all trimers sharing the same sequence motif is provided in parentheses. * Threonine is also observed at both positions. + Methionine is also observed in the second position, # Histidine is also observed in the first position.
9 Supplementary Table 4. Support Vector Classification (SVC) of trimer sequences into structures. The table reports the accuracy in correctly assigning trimer sequences into one of two possible trimer structure classes. The accuracy of classification is calculated using 5-fold cross validation (see method). Results are given for all possible 15 pairs of trimer structure classes. Random assignment would correspond to 50% accuracy. Below the table, the average accuracy for two-classes assignment is reported. * Average accuracy for two classes assignment is significantly higher than random assignment with P-value < using Welch s t-test. The accuracy in correctly assigning trimer sequences into one of six possible trimer structure classes (6-classes assignment) is reported below the table. Random assignment would correspond to 16.7% accuracy.
10 Supplementary Table 5. Trimers bearing sequence/contact motifs are less flexible. The left part of the Supplementary Table displays the name of the proteins and the PDB codes of different protein conformations (states) that differ by at least 0.5Å in Cα RMSD. The right part of the table indicates the extent of conformational changes of each trimer unit in the protein measured by Cα RMSD (Å). The trimer unit is defined by the trimer type (i.e. topology) and motif type. The topology is encoded as follows: AL: All left-handed Trimer; AR, All Right-handed Trimer; PL: Parallel & Left handed Trimer; PR: Parallel & Right handed Trimer; LR1: Left & Right Type 1 Trimer; LR2: Left & Right Type 2 Trimer. Motif types are encoded as follows: NA: no sequence/structure motif found in the trimer; (AL, M1): I/L/V/M-X 3 -G/A/S; (AL, M2): I/L/V/M- X 6 -I/L/V/M; (AL, M3): G/A/S-X 6 -G/A/S; (AL, M4): I/L/V/M-X 6 -F/W/Y; (AR, M1): G/A/S-X 3 -G/A/S; (AR, M2): G/A/S-X 7 -G/A/S; (PR, M1): I/L/V/M-X 3 -F/W/Y; (PR, M2): F/W/Y-X 2 -F/W/Y; (LR1, M1): F/W/Y-X 3 -G/A/S; (LR1, M2): F/W/Y-X 6 -G/A/S; (LR2, M1): F/W/Y-X 6 -G/A/S.
11
12 Supplementary Table 6. Trimers extracted only from distinct protein superfamilies bearing sequence/contact motifs are less flexible. Compared to Main Text Figure 6 and Supplementary Table 5, only one protein per superfamily was selected leading to 10 protein structures and 40 trimer units. The left part of the Supplementary Table displays the name of the proteins and the PDB codes of different protein conformations (states) that differ by at least 0.5Å in Cα RMSD. The right part of the table indicates the extent of conformational changes of each trimer unit in the protein measured by Cα RMSD (Å). The trimer unit is defined by the trimer type (i.e. topology) and motif type. The topology is encoded as follows: AL: All left-handed Trimer; AR, All Right-handed Trimer; PL: Parallel & Left handed Trimer; PR: Parallel & Right handed Trimer; LR1: Left & Right Type 1 Trimer; LR2: Left & Right Type 2 Trimer. Motif types are encoded as follows: NA: no sequence/structure motif found in the trimer; (AL, M1): I/L/V/M- X 3 -G/A/S; (AL, M2): I/L/V/M-X 6 -I/L/V/M; (AL, M3): G/A/S-X 6 -G/A/S; (AR, M1): G/A/S-X 3 -G/A/S; (AR, M2): G/A/S-X 7 -G/A/S; (PR, M1): I/L/V/M-X 3 -F/W/Y; (PR, M2): F/W/Y-X 2 -F/W/Y; (LR1, M1): F/W/Y-X 3 -G/A/S; (LR1, M2): F/W/Y-X 6 -G/A/S; (LR2, M1): F/W/Y-X 6 -G/A/S.
13
14 Supplementary Figure 5. Sequence/3D contact motifs are strong predictors of local conformational stability. Distribution of trimer unit structural changes (measured by Cα rmsd in Å) in multi-pass membrane proteins crystallized in distinct conformations. Only one protein per superfamily was selected leading to 10 protein structures and 40 trimer units. Data for trimers containing enriched sequence/contact motifs are in black, others are in grey. Trimers with sequence motifs and corresponding interaction patterns had substantially smaller Cα rmsd (P < , Welch s t-test) between distinct protein conformations and were therefore significantly more rigid than the trimers without such sequence/3d contact features.
Assessing a novel approach for predicting local 3D protein structures from sequence
 Assessing a novel approach for predicting local 3D protein structures from sequence Cristina Benros*, Alexandre G. de Brevern, Catherine Etchebest and Serge Hazout Equipe de Bioinformatique Génomique et
Assessing a novel approach for predicting local 3D protein structures from sequence Cristina Benros*, Alexandre G. de Brevern, Catherine Etchebest and Serge Hazout Equipe de Bioinformatique Génomique et
CSE : Computational Issues in Molecular Biology. Lecture 19. Spring 2004
 CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
Technical Seminar 22th Jan 2013 DNA Origami
 Technical Seminar 22th Jan 2013 DNA Origami Hitoshi Takizawa, PhD Agenda 1) Basis of structural DNA nanotechnology 2) DNA origami technique (2D, 3D, complex shape) 3) Programmable nanofactory 4) Application
Technical Seminar 22th Jan 2013 DNA Origami Hitoshi Takizawa, PhD Agenda 1) Basis of structural DNA nanotechnology 2) DNA origami technique (2D, 3D, complex shape) 3) Programmable nanofactory 4) Application
Bioinformatics & Protein Structural Analysis. Bioinformatics & Protein Structural Analysis. Learning Objective. Proteomics
 The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature16191 SUPPLEMENTARY DISCUSSION We analyzed the current database of solved protein structures to assess the uniqueness of our designed toroids in terms of global similarity and bundle
doi:10.1038/nature16191 SUPPLEMENTARY DISCUSSION We analyzed the current database of solved protein structures to assess the uniqueness of our designed toroids in terms of global similarity and bundle
A Protein Secondary Structure Prediction Method Based on BP Neural Network Ru-xi YIN, Li-zhen LIU*, Wei SONG, Xin-lei ZHAO and Chao DU
 2017 2nd International Conference on Artificial Intelligence: Techniques and Applications (AITA 2017 ISBN: 978-1-60595-491-2 A Protein Secondary Structure Prediction Method Based on BP Neural Network Ru-xi
2017 2nd International Conference on Artificial Intelligence: Techniques and Applications (AITA 2017 ISBN: 978-1-60595-491-2 A Protein Secondary Structure Prediction Method Based on BP Neural Network Ru-xi
Structural bioinformatics
 Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Structural bioinformatics Why structures? The representation of the molecules in 3D is more informative New properties of the molecules are revealed, which can not be detected by sequences Eran Eyal Plant
Textbook Reading Guidelines
 Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: May 1, 2009 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: May 1, 2009 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
CFSSP: Chou and Fasman Secondary Structure Prediction server
 Wide Spectrum, Vol. 1, No. 9, (2013) pp 15-19 CFSSP: Chou and Fasman Secondary Structure Prediction server T. Ashok Kumar Department of Bioinformatics, Noorul Islam College of Arts and Science, Kumaracoil
Wide Spectrum, Vol. 1, No. 9, (2013) pp 15-19 CFSSP: Chou and Fasman Secondary Structure Prediction server T. Ashok Kumar Department of Bioinformatics, Noorul Islam College of Arts and Science, Kumaracoil
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
Supplementary Figure 1 Domain architecture and conformational states of the decapping complex, as revealed by structural studies. (a) Domain organization of Schizosaccharomyces pombe (Sp) and Saccharomyces
BCH222 - Greek Key β Barrels
 BCH222 - Greek Key β Barrels Reading C.I. Branden and J. Tooze (1999) Introduction to Protein Structure, Second Edition, pp. 77-78 & 335-336 (look at the color figures) J.S. Richardson (1981) "The Anatomy
BCH222 - Greek Key β Barrels Reading C.I. Branden and J. Tooze (1999) Introduction to Protein Structure, Second Edition, pp. 77-78 & 335-336 (look at the color figures) J.S. Richardson (1981) "The Anatomy
Ab Initio SERVER PROTOTYPE FOR PREDICTION OF PHOSPHORYLATION SITES IN PROTEINS*
 COMPUTATIONAL METHODS IN SCIENCE AND TECHNOLOGY 9(1-2) 93-100 (2003/2004) Ab Initio SERVER PROTOTYPE FOR PREDICTION OF PHOSPHORYLATION SITES IN PROTEINS* DARIUSZ PLEWCZYNSKI AND LESZEK RYCHLEWSKI BiolnfoBank
COMPUTATIONAL METHODS IN SCIENCE AND TECHNOLOGY 9(1-2) 93-100 (2003/2004) Ab Initio SERVER PROTOTYPE FOR PREDICTION OF PHOSPHORYLATION SITES IN PROTEINS* DARIUSZ PLEWCZYNSKI AND LESZEK RYCHLEWSKI BiolnfoBank
Protein Structure Prediction
 Homology Modeling Protein Structure Prediction Ingo Ruczinski M T S K G G G Y F F Y D E L Y G V V V V L I V L S D E S Department of Biostatistics, Johns Hopkins University Fold Recognition b Initio Structure
Homology Modeling Protein Structure Prediction Ingo Ruczinski M T S K G G G Y F F Y D E L Y G V V V V L I V L S D E S Department of Biostatistics, Johns Hopkins University Fold Recognition b Initio Structure
Visualization of codon-dependent conformational rearrangements during translation termination
 Supplementary information for: Visualization of codon-dependent conformational rearrangements during translation termination Shan L. He 1 and Rachel Green 1 1 Howard Hughes Medical Institute, Department
Supplementary information for: Visualization of codon-dependent conformational rearrangements during translation termination Shan L. He 1 and Rachel Green 1 1 Howard Hughes Medical Institute, Department
Statistical Machine Learning Methods for Bioinformatics VI. Support Vector Machine Applications in Bioinformatics
 Statistical Machine Learning Methods for Bioinformatics VI. Support Vector Machine Applications in Bioinformatics Jianlin Cheng, PhD Computer Science Department and Informatics Institute University of
Statistical Machine Learning Methods for Bioinformatics VI. Support Vector Machine Applications in Bioinformatics Jianlin Cheng, PhD Computer Science Department and Informatics Institute University of
CS273: Algorithms for Structure Handout # 5 and Motion in Biology Stanford University Tuesday, 13 April 2004
 CS273: Algorithms for Structure Handout # 5 and Motion in Biology Stanford University Tuesday, 13 April 2004 Lecture #5: 13 April 2004 Topics: Sequence motif identification Scribe: Samantha Chui 1 Introduction
CS273: Algorithms for Structure Handout # 5 and Motion in Biology Stanford University Tuesday, 13 April 2004 Lecture #5: 13 April 2004 Topics: Sequence motif identification Scribe: Samantha Chui 1 Introduction
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 The mass accuracy of fragment ions is important for peptide recovery in wide-tolerance searches. The same data as in Figure 1B was searched with varying fragment ion tolerances (FIT).
Supplementary Figure 1 The mass accuracy of fragment ions is important for peptide recovery in wide-tolerance searches. The same data as in Figure 1B was searched with varying fragment ion tolerances (FIT).
Docking. Why? Docking : finding the binding orientation of two molecules with known structures
 Docking : finding the binding orientation of two molecules with known structures Docking According to the molecules involved: Protein-Ligand docking Protein-Protein docking Specific docking algorithms
Docking : finding the binding orientation of two molecules with known structures Docking According to the molecules involved: Protein-Ligand docking Protein-Protein docking Specific docking algorithms
Representation in Supervised Machine Learning Application to Biological Problems
 Representation in Supervised Machine Learning Application to Biological Problems Frank Lab Howard Hughes Medical Institute & Columbia University 2010 Robert Howard Langlois Hughes Medical Institute What
Representation in Supervised Machine Learning Application to Biological Problems Frank Lab Howard Hughes Medical Institute & Columbia University 2010 Robert Howard Langlois Hughes Medical Institute What
Protein Structure. Protein Structure Tertiary & Quaternary
 Lecture 4 Protein Structure Protein Structure Tertiary & Quaternary Dr. Sameh Sarray Hlaoui Primary structure: The linear sequence of amino acids held together by peptide bonds. Secondary structure: The
Lecture 4 Protein Structure Protein Structure Tertiary & Quaternary Dr. Sameh Sarray Hlaoui Primary structure: The linear sequence of amino acids held together by peptide bonds. Secondary structure: The
BIOINFORMATICS Introduction
 BIOINFORMATICS Introduction Mark Gerstein, Yale University bioinfo.mbb.yale.edu/mbb452a 1 (c) Mark Gerstein, 1999, Yale, bioinfo.mbb.yale.edu What is Bioinformatics? (Molecular) Bio -informatics One idea
BIOINFORMATICS Introduction Mark Gerstein, Yale University bioinfo.mbb.yale.edu/mbb452a 1 (c) Mark Gerstein, 1999, Yale, bioinfo.mbb.yale.edu What is Bioinformatics? (Molecular) Bio -informatics One idea
Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction
 Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction Conformational Analysis 2 Conformational Analysis Properties of molecules depend on their three-dimensional
Structural Bioinformatics (C3210) Conformational Analysis Protein Folding Protein Structure Prediction Conformational Analysis 2 Conformational Analysis Properties of molecules depend on their three-dimensional
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Supplementary Figure 1 Multiple sequence alignments of four Swi2/Snf2 subfamily proteins, ScChd1, SsoRad54 and the RNA helicase Vasa. The sequence alignments of the Swi2/Snf2 subfamily proteins, ScChd1
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
BIRKBECK COLLEGE (University of London)
 BIRKBECK COLLEGE (University of London) SCHOOL OF BIOLOGICAL SCIENCES M.Sc. EXAMINATION FOR INTERNAL STUDENTS ON: Postgraduate Certificate in Principles of Protein Structure MSc Structural Molecular Biology
BIRKBECK COLLEGE (University of London) SCHOOL OF BIOLOGICAL SCIENCES M.Sc. EXAMINATION FOR INTERNAL STUDENTS ON: Postgraduate Certificate in Principles of Protein Structure MSc Structural Molecular Biology
Sequence Analysis '17 -- lecture Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction
 Sequence Analysis '17 -- lecture 16 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction Alpha helix Right-handed helix. H-bond is from the oxygen at i to the nitrogen
Sequence Analysis '17 -- lecture 16 1. Secondary structure 3. Sequence similarity and homology 2. Secondary structure prediction Alpha helix Right-handed helix. H-bond is from the oxygen at i to the nitrogen
Figure S1
 Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Supplementary Figure 1 The distribution of chlorophyll containing complexes eluted from DEAE-cellulose column in a sucrose gradient tube Six pigment-containing bands were resolved and identified as: B1,
Predicting Protein Structure and Examining Similarities of Protein Structure by Spectral Analysis Techniques
 Predicting Protein Structure and Examining Similarities of Protein Structure by Spectral Analysis Techniques, Melanie Abeysundera, Krista Collins, Chris Field Department of Mathematics and Statistics Dalhousie
Predicting Protein Structure and Examining Similarities of Protein Structure by Spectral Analysis Techniques, Melanie Abeysundera, Krista Collins, Chris Field Department of Mathematics and Statistics Dalhousie
Supplementary Information. Structural basis for duplex RNA recognition and cleavage by A.
 Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
Supplementary Information Structural asis for duplex RNA recognition and cleavage y A. fulgidus C3PO Eneida arizotto 1, Edward D Lowe 1 & James S Parker 1 1 Department of Biochemistry University of Oxford
RNA does not adopt the classic B-DNA helix conformation when it forms a self-complementary double helix
 Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
What s New in Discovery Studio 2.5.5
 What s New in Discovery Studio 2.5.5 Discovery Studio takes modeling and simulations to the next level. It brings together the power of validated science on a customizable platform for drug discovery research.
What s New in Discovery Studio 2.5.5 Discovery Studio takes modeling and simulations to the next level. It brings together the power of validated science on a customizable platform for drug discovery research.
Supplementary Fig.1. Accuracy of LIPS predicted TMH binding surfaces.
 Wang, Y. & Barth, P. Supplementary Materials Supplementary Fig.1. Accuracy of LIPS predicted TMH binding surfaces. The percentage of residues predicted by LIPS 1 to be the least exposed to lipids that
Wang, Y. & Barth, P. Supplementary Materials Supplementary Fig.1. Accuracy of LIPS predicted TMH binding surfaces. The percentage of residues predicted by LIPS 1 to be the least exposed to lipids that
COMPAS for the Analysis of SELEX Experiments
 COMPAS for the Analysis of SELEX Experiments COMPAS (COMmon PAtternS) is a software tool that was especially developed to harness the technology of next generation sequencing (NGS) to bring light into
COMPAS for the Analysis of SELEX Experiments COMPAS (COMmon PAtternS) is a software tool that was especially developed to harness the technology of next generation sequencing (NGS) to bring light into
Name with Last Name, First: BIOE111: Functional Biomaterial Development and Characterization MIDTERM EXAM (October 7, 2010) 93 TOTAL POINTS
 BIOE111: Functional Biomaterial Development and Characterization MIDTERM EXAM (October 7, 2010) 93 TOTAL POINTS Question 0: Fill in your name and student ID on each page. (1) Question 1: What is the role
BIOE111: Functional Biomaterial Development and Characterization MIDTERM EXAM (October 7, 2010) 93 TOTAL POINTS Question 0: Fill in your name and student ID on each page. (1) Question 1: What is the role
SUPPLEMENTARY INFORMATION
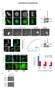 SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
SUPPLEMENTARY INFORMATION Supplementary Figure 1: Function of MICAL1 and dmical in cytokinesis (a) HeLa transfected with GFP- MICAL1 (green) were stained with Aurora B (red). Scale bars, 10 µm. (b) Western
Structure formation and association of biomolecules. Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München
 Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Structure formation and association of biomolecules Prof. Dr. Martin Zacharias Lehrstuhl für Molekulardynamik (T38) Technische Universität München Motivation Many biomolecules are chemically synthesized
Protein Structural Motifs Search in Protein Data Base
 Protein Structural Motifs Search in Protein Data Base Virginio Cantoni 1, Alessio Ferone 2, Ozlem Ozbudak 3, Alfredo Petrosino 2 1 Dept. of Electrical Engineering and Computer Science, Pavia Univ., Italy
Protein Structural Motifs Search in Protein Data Base Virginio Cantoni 1, Alessio Ferone 2, Ozlem Ozbudak 3, Alfredo Petrosino 2 1 Dept. of Electrical Engineering and Computer Science, Pavia Univ., Italy
All Rights Reserved. U.S. Patents 6,471,520B1; 5,498,190; 5,916, North Market Street, Suite CC130A, Milwaukee, WI 53202
 Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron
 Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Supplementary Fig. 1. Initial electron density maps for the NOX-D20:mC5a complex obtained after SAD-phasing. (a) Initial experimental electron density map obtained after SAD-phasing and density modification
Supplementary Material. for. A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis
 Supplementary Material for A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis Stephen Boulton 1, Madoka Akimoto 2, Rajeevan Selvaratnam 2, Amir Bashiri 2 and Giuseppe
Supplementary Material for A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis Stephen Boulton 1, Madoka Akimoto 2, Rajeevan Selvaratnam 2, Amir Bashiri 2 and Giuseppe
Sequence Determinants of a Conformational Switch in a Protein
 Sequence Determinants of a Conformational Switch in a Protein Thomas A. Anderson, Matthew H. J. Cordes, and Robert T. Sauer Kyle Skalenko 4/17/09 Introduction Protein folding is guided by the following
Sequence Determinants of a Conformational Switch in a Protein Thomas A. Anderson, Matthew H. J. Cordes, and Robert T. Sauer Kyle Skalenko 4/17/09 Introduction Protein folding is guided by the following
Protein structure. Wednesday, October 4, 2006
 Protein structure Wednesday, October 4, 2006 Introduction to Bioinformatics Johns Hopkins School of Public Health 260.602.01 J. Pevsner pevsner@jhmi.edu Copyright notice Many of the images in this powerpoint
Protein structure Wednesday, October 4, 2006 Introduction to Bioinformatics Johns Hopkins School of Public Health 260.602.01 J. Pevsner pevsner@jhmi.edu Copyright notice Many of the images in this powerpoint
Molecular Modeling Lecture 8. Local structure Database search Multiple alignment Automated homology modeling
 Molecular Modeling 2018 -- Lecture 8 Local structure Database search Multiple alignment Automated homology modeling An exception to the no-insertions-in-helix rule Actual structures (myosin)! prolines
Molecular Modeling 2018 -- Lecture 8 Local structure Database search Multiple alignment Automated homology modeling An exception to the no-insertions-in-helix rule Actual structures (myosin)! prolines
Exploring Similarities of Conserved Domains/Motifs
 Exploring Similarities of Conserved Domains/Motifs Sotiria Palioura Abstract Traditionally, proteins are represented as amino acid sequences. There are, though, other (potentially more exciting) representations;
Exploring Similarities of Conserved Domains/Motifs Sotiria Palioura Abstract Traditionally, proteins are represented as amino acid sequences. There are, though, other (potentially more exciting) representations;
M1 - Biochemistry. Nucleic Acid Structure II/Transcription I
 M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
Bacteriophages get a foothold on their prey
 Bacteriophages get a foothold on their prey Long and thin, the receptor-binding needle of bacteriophage T4 Bacterial viruses, bacteriophages or phages, have served as a tool to decipher principles of molecular
Bacteriophages get a foothold on their prey Long and thin, the receptor-binding needle of bacteriophage T4 Bacterial viruses, bacteriophages or phages, have served as a tool to decipher principles of molecular
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
J. DISTRIBUTION STATEMENT A Approved for Public Release. a. vm-mmm OHGANüATIOH. Reset
 REPORT DOCUMENTATION PAGE Foim Approved OMBNo. 0704-0183 Th» pub«.tboorhie burdon for «hre coitaraon of inform anon B «itimatad to anraga 1 hour par nssen». inoludina th» rim» for r«vi«imb initnicrinra.
REPORT DOCUMENTATION PAGE Foim Approved OMBNo. 0704-0183 Th» pub«.tboorhie burdon for «hre coitaraon of inform anon B «itimatad to anraga 1 hour par nssen». inoludina th» rim» for r«vi«imb initnicrinra.
Protein Structure Analysis
 BINF 731 Protein Structure Analysis http://binf.gmu.edu/vaisman/binf731/ Secondary Structure: Computational Problems Secondary structure characterization Secondary structure assignment Secondary structure
BINF 731 Protein Structure Analysis http://binf.gmu.edu/vaisman/binf731/ Secondary Structure: Computational Problems Secondary structure characterization Secondary structure assignment Secondary structure
CASP 13 Assembly assessment
 CASP 13 Assembly assessment Riviera Maya, Dec 2018 Jose Duarte, Dmytro Guzenko RCSB Protein Data Bank, UC San Diego Biological assembly of targets The Ground Truth is not always 100% clear when talking
CASP 13 Assembly assessment Riviera Maya, Dec 2018 Jose Duarte, Dmytro Guzenko RCSB Protein Data Bank, UC San Diego Biological assembly of targets The Ground Truth is not always 100% clear when talking
Molecular Biology Services. Make Research Easy
 Molecular Biology Services Make Research Easy Gene-on-Demand Technology Platform Any gene conceivable in any vector you desire GenScript s Gene-on-Demand technology platform combines patented OptimumGene
Molecular Biology Services Make Research Easy Gene-on-Demand Technology Platform Any gene conceivable in any vector you desire GenScript s Gene-on-Demand technology platform combines patented OptimumGene
Fundamentals of Biochemistry
 Donald Voet Judith G. Voet Charlotte W. Pratt Fundamentals of Biochemistry Second Edition Chapter 6: Proteins: Three-Dimensional Structure Copyright 2006 by John Wiley & Sons, Inc. 1958, John Kendrew Any
Donald Voet Judith G. Voet Charlotte W. Pratt Fundamentals of Biochemistry Second Edition Chapter 6: Proteins: Three-Dimensional Structure Copyright 2006 by John Wiley & Sons, Inc. 1958, John Kendrew Any
Molecules VII. Structures and Functions of Proteins
 Molecules VII Structures and Functions of Proteins The 20 Amino Acids Sequences from Protein Data Bank (PDB) The primary sequence can be indicated very compactly using the one letter abbreviations. This
Molecules VII Structures and Functions of Proteins The 20 Amino Acids Sequences from Protein Data Bank (PDB) The primary sequence can be indicated very compactly using the one letter abbreviations. This
Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions
 www.iba-biotagnology.com Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions Strep-tag and One-STrEP-tag PPI Analysis with the co-precipitation/ mass spectrometry approach 3 Background
www.iba-biotagnology.com Newsletter Issue 7 One-STrEP Analysis of Protein:Protein-Interactions Strep-tag and One-STrEP-tag PPI Analysis with the co-precipitation/ mass spectrometry approach 3 Background
Prot-SSP: A Tool for Amino Acid Pairing Pattern Analysis in Secondary Structures
 Mol2Net, 2015, 1(Section F), pages 1-6, Proceedings 1 SciForum Mol2Net Prot-SSP: A Tool for Amino Acid Pairing Pattern Analysis in Secondary Structures Miguel de Sousa 1, *, Cristian R. Munteanu 2 and
Mol2Net, 2015, 1(Section F), pages 1-6, Proceedings 1 SciForum Mol2Net Prot-SSP: A Tool for Amino Acid Pairing Pattern Analysis in Secondary Structures Miguel de Sousa 1, *, Cristian R. Munteanu 2 and
RNP purification, components and activity.
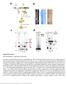 Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded
This place covers: Methods or systems for genetic or protein-related data processing in computational molecular biology.
 G16B BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY Methods or systems for genetic
G16B BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY Methods or systems for genetic
Gene Identification in silico
 Gene Identification in silico Nita Parekh, IIIT Hyderabad Presented at National Seminar on Bioinformatics and Functional Genomics, at Bioinformatics centre, Pondicherry University, Feb 15 17, 2006. Introduction
Gene Identification in silico Nita Parekh, IIIT Hyderabad Presented at National Seminar on Bioinformatics and Functional Genomics, at Bioinformatics centre, Pondicherry University, Feb 15 17, 2006. Introduction
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Molecular Modeling 9. Protein structure prediction, part 2: Homology modeling, fold recognition & threading
 Molecular Modeling 9 Protein structure prediction, part 2: Homology modeling, fold recognition & threading The project... Remember: You are smarter than the program. Inspecting the model: Are amino acids
Molecular Modeling 9 Protein structure prediction, part 2: Homology modeling, fold recognition & threading The project... Remember: You are smarter than the program. Inspecting the model: Are amino acids
A New Database of Genetic and. Molecular Pathways. Minoru Kanehisa. sequencing projects have been. Mbp) and for several bacteria including
 Toward Pathway Engineering: A New Database of Genetic and Molecular Pathways Minoru Kanehisa Institute for Chemical Research, Kyoto University From Genome Sequences to Functions The Human Genome Project
Toward Pathway Engineering: A New Database of Genetic and Molecular Pathways Minoru Kanehisa Institute for Chemical Research, Kyoto University From Genome Sequences to Functions The Human Genome Project
Protein Domain Boundary Prediction from Residue Sequence Alone using Bayesian Neural Networks
 Protein Domain Boundary Prediction from Residue Sequence Alone using Bayesian s DAVID SACHEZ SPIROS H. COURELLIS Department of Computer Science Department of Computer Science California State University
Protein Domain Boundary Prediction from Residue Sequence Alone using Bayesian s DAVID SACHEZ SPIROS H. COURELLIS Department of Computer Science Department of Computer Science California State University
Nature Methods: doi: /nmeth Supplementary Figure 1. Construction of a sensitive TetR mediated auxotrophic off-switch.
 Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
Supplementary Figure 1 Construction of a sensitive TetR mediated auxotrophic off-switch. A Production of the Tet repressor in yeast when conjugated to either the LexA4 or LexA8 promoter DNA binding sequences.
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
6-Foot Mini Toober Activity
 Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
Big Idea The interaction between the substrate and enzyme is highly specific. Even a slight change in shape of either the substrate or the enzyme may alter the efficient and selective ability of the enzyme
SUPPLEMENTARY INFORMATION Figures. Supplementary Figure 1 a. Page 1 of 30. Nature Chemical Biology: doi: /nchembio.2528
 SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
SUPPLEMENTARY INFORMATION Figures Supplementary Figure 1 a b c Page 1 of 0 11 Supplementary Figure 1: Biochemical characterisation and binding validation of the reversible USP inhibitor 1. a, Biochemical
Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions
 Introduction Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke, Rochester Institute of Technology Mentor: Carlos Camacho, University
Introduction Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke, Rochester Institute of Technology Mentor: Carlos Camacho, University
STRUCTURE, DYNAMICS AND INTERACTIONS OF PROTEINS BY NMR SPECTROSCOPY
 STRUCTURE, DYNAMICS AND INTERACTIONS OF PROTEINS BY NMR SPECTROSCOPY Constantin T. Craescu INSERM & Institut Curie - Recherche Orsay, France A SHORT INTRODUCTION TO PROTEIN STRUCTURE Chemical composition
STRUCTURE, DYNAMICS AND INTERACTIONS OF PROTEINS BY NMR SPECTROSCOPY Constantin T. Craescu INSERM & Institut Curie - Recherche Orsay, France A SHORT INTRODUCTION TO PROTEIN STRUCTURE Chemical composition
Nagahama Institute of Bio-Science and Technology. National Institute of Genetics and SOKENDAI Nagahama Institute of Bio-Science and Technology
 A Large-scale Batch-learning Self-organizing Map for Function Prediction of Poorly-characterized Proteins Progressively Accumulating in Sequence Databases Project Representative Toshimichi Ikemura Authors
A Large-scale Batch-learning Self-organizing Map for Function Prediction of Poorly-characterized Proteins Progressively Accumulating in Sequence Databases Project Representative Toshimichi Ikemura Authors
Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL*
 Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL* Site AA Sequence 3x10 6 3x10 6 20x10 6 20x10 6 100x10 6 100x10 6 Post-1 st Post-2 nd Post-1 st Post-2 nd Post-1 st Post-2
Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL* Site AA Sequence 3x10 6 3x10 6 20x10 6 20x10 6 100x10 6 100x10 6 Post-1 st Post-2 nd Post-1 st Post-2 nd Post-1 st Post-2
Textbook Reading Guidelines
 Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: January 16, 2013 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Understanding Bioinformatics by Marketa Zvelebil and Jeremy Baum Last updated: January 16, 2013 Textbook Reading Guidelines Preface: Read the whole preface, and especially: For the students with Life Science
Distributions of Beta Sheets in Proteins with Application to Structure Prediction
 Distributions of Beta Sheets in Proteins with Application to Structure Prediction Ingo Ruczinski Department of Biostatistics Johns Hopkins University Email: ingo@jhu.edu http://biostat.jhsph.edu/ iruczins
Distributions of Beta Sheets in Proteins with Application to Structure Prediction Ingo Ruczinski Department of Biostatistics Johns Hopkins University Email: ingo@jhu.edu http://biostat.jhsph.edu/ iruczins
Introduction to Proteins
 Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Introduction to Proteins Lecture 4 Module I: Molecular Structure & Metabolism Molecular Cell Biology Core Course (GSND5200) Matthew Neiditch - Room E450U ICPH matthew.neiditch@umdnj.edu What is a protein?
Protein Structure Prediction. christian studer , EPFL
 Protein Structure Prediction christian studer 17.11.2004, EPFL Content Definition of the problem Possible approaches DSSP / PSI-BLAST Generalization Results Definition of the problem Massive amounts of
Protein Structure Prediction christian studer 17.11.2004, EPFL Content Definition of the problem Possible approaches DSSP / PSI-BLAST Generalization Results Definition of the problem Massive amounts of
Biochem Fall Sample Exam I Protein Structure. Vasopressin: CYFQNCPRG Oxytocin: CYIQNCPLG
 Biochem Fall 2011 1. Primary Structure and amino acid chemistry Sample Exam I Protein Structure The peptide hormones vasopressin (ADH) and oxytocin each contain only nine amino acids. Vasopressin is an
Biochem Fall 2011 1. Primary Structure and amino acid chemistry Sample Exam I Protein Structure The peptide hormones vasopressin (ADH) and oxytocin each contain only nine amino acids. Vasopressin is an
This practical aims to walk you through the process of text searching DNA and protein databases for sequence entries.
 PRACTICAL 1: BLAST and Sequence Alignment The EBI and NCBI websites, two of the most widely used life science web portals are introduced along with some of the principal databases: the NCBI Protein database,
PRACTICAL 1: BLAST and Sequence Alignment The EBI and NCBI websites, two of the most widely used life science web portals are introduced along with some of the principal databases: the NCBI Protein database,
Supplementary Figures Supplementary Figure 1
 Supplementary Figures Supplementary Figure 1 Supplementary Figure 1 COMSOL simulation demonstrating flow characteristics in hydrodynamic trap structures. (a) Flow field within the hydrodynamic traps before
Supplementary Figures Supplementary Figure 1 Supplementary Figure 1 COMSOL simulation demonstrating flow characteristics in hydrodynamic trap structures. (a) Flow field within the hydrodynamic traps before
Molecular Cell Biology - Problem Drill 06: Genes and Chromosomes
 Molecular Cell Biology - Problem Drill 06: Genes and Chromosomes Question No. 1 of 10 1. Which of the following statements about genes is correct? Question #1 (A) Genes carry the information for protein
Molecular Cell Biology - Problem Drill 06: Genes and Chromosomes Question No. 1 of 10 1. Which of the following statements about genes is correct? Question #1 (A) Genes carry the information for protein
Supplementary Information
 Supplementary Information Site-specific Isopeptide Bridge Tethering of Chimeric gp41 N-terminal Heptad Repeat Helical Trimers for the Treatment of HIV-1 Infection Chao Wang, 1, Xue Li, 1, Fei Yu, 2 Lu
Supplementary Information Site-specific Isopeptide Bridge Tethering of Chimeric gp41 N-terminal Heptad Repeat Helical Trimers for the Treatment of HIV-1 Infection Chao Wang, 1, Xue Li, 1, Fei Yu, 2 Lu
Collagen. 7.88J Protein Folding. Prof. David Gossard October 20, 2003
 Collagen 7.88J Protein Folding Prof. David Gossard October 20, 2003 PDB Acknowledgements The Protein Data Bank (PDB - http://www.pdb.org/) is the single worldwide repository for the processing and distribution
Collagen 7.88J Protein Folding Prof. David Gossard October 20, 2003 PDB Acknowledgements The Protein Data Bank (PDB - http://www.pdb.org/) is the single worldwide repository for the processing and distribution
Homology Modeling of the Chimeric Human Sweet Taste Receptors Using Multi Templates
 2013 International Conference on Food and Agricultural Sciences IPCBEE vol.55 (2013) (2013) IACSIT Press, Singapore DOI: 10.7763/IPCBEE. 2013. V55. 1 Homology Modeling of the Chimeric Human Sweet Taste
2013 International Conference on Food and Agricultural Sciences IPCBEE vol.55 (2013) (2013) IACSIT Press, Singapore DOI: 10.7763/IPCBEE. 2013. V55. 1 Homology Modeling of the Chimeric Human Sweet Taste
Computational Methods for Protein Structure Prediction and Fold Recognition... 1 I. Cymerman, M. Feder, M. PawŁowski, M.A. Kurowski, J.M.
 Contents Computational Methods for Protein Structure Prediction and Fold Recognition........................... 1 I. Cymerman, M. Feder, M. PawŁowski, M.A. Kurowski, J.M. Bujnicki 1 Primary Structure Analysis...................
Contents Computational Methods for Protein Structure Prediction and Fold Recognition........................... 1 I. Cymerman, M. Feder, M. PawŁowski, M.A. Kurowski, J.M. Bujnicki 1 Primary Structure Analysis...................
TERTIARY MOTIF INTERACTIONS ON RNA STRUCTURE
 1 TERTIARY MOTIF INTERACTIONS ON RNA STRUCTURE Bioinformatics Senior Project Wasay Hussain Spring 2009 Overview of RNA 2 The central Dogma of Molecular biology is DNA RNA Proteins The RNA (Ribonucleic
1 TERTIARY MOTIF INTERACTIONS ON RNA STRUCTURE Bioinformatics Senior Project Wasay Hussain Spring 2009 Overview of RNA 2 The central Dogma of Molecular biology is DNA RNA Proteins The RNA (Ribonucleic
Identification and characterization of DNA aptamers specific for
 SUPPLEMENTARY INFORMATION Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein I-Ting Teng 1,, Xiaowei Li 1,, Hamad Ahmad Yadikar #, Zhihui Yang #, Long
SUPPLEMENTARY INFORMATION Identification and characterization of DNA aptamers specific for phosphorylation epitopes of tau protein I-Ting Teng 1,, Xiaowei Li 1,, Hamad Ahmad Yadikar #, Zhihui Yang #, Long
RPA-AB RPA-C Supplemental Figure S1: SDS-PAGE stained with Coomassie Blue after protein purification.
 RPA-AB RPA-C (a) (b) (c) (d) (e) (f) Supplemental Figure S: SDS-PAGE stained with Coomassie Blue after protein purification. (a) RPA; (b) RPA-AB; (c) RPA-CDE; (d) RPA-CDE core; (e) RPA-DE; and (f) RPA-C
RPA-AB RPA-C (a) (b) (c) (d) (e) (f) Supplemental Figure S: SDS-PAGE stained with Coomassie Blue after protein purification. (a) RPA; (b) RPA-AB; (c) RPA-CDE; (d) RPA-CDE core; (e) RPA-DE; and (f) RPA-C
Structural Bioinformatics GENOME 541 Spring 2018
 Molecular composition of a rapidly dividing Escherichia coli cell Structural Bioinformatics GENOME 541 Spring 2018 Lecture 4: Nucleic Acids Frank DiMaio (dimaio@uw.edu) The major biopolymers DNA structure
Molecular composition of a rapidly dividing Escherichia coli cell Structural Bioinformatics GENOME 541 Spring 2018 Lecture 4: Nucleic Acids Frank DiMaio (dimaio@uw.edu) The major biopolymers DNA structure
Supplemental Information Molecular Cell, Volume 41
 Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
Supplemental Information Molecular Cell, Volume 41 Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2 Sutapa Chakrabarti, Uma Jayachandran, Fabien Bonneau, Francesca
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I
 BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
BMB/Bi/Ch 170 Fall 2017 Problem Set 1: Proteins I Please use ray-tracing feature for all the images you are submitting. Use either the Ray button on the right side of the command window in PyMOL or variations
C2H2 zinc finger proteins greatly expand the human regulatory lexicon
 Supplementary Information for: C2H2 zinc finger proteins greatly expand the human regulatory lexicon Hamed S. Najafabadi*,1, Sanie Mnaimneh*,1, Frank W. Schmitges*,1, Michael Garton 1, Kathy N. Lam 2,
Supplementary Information for: C2H2 zinc finger proteins greatly expand the human regulatory lexicon Hamed S. Najafabadi*,1, Sanie Mnaimneh*,1, Frank W. Schmitges*,1, Michael Garton 1, Kathy N. Lam 2,
RNA folding on the 3D triangular lattice. Joel Gillespie, Martin Mayne and Minghui Jiang
 RNA folding on the 3D triangular lattice Joel Gillespie, Martin Mayne and Minghui Jiang University of Waterloo Alan Tsang July 19, 2010 Deoxyribonucleic Acid (DNA) Sequence of bases: Adenine (A) Cytosine
RNA folding on the 3D triangular lattice Joel Gillespie, Martin Mayne and Minghui Jiang University of Waterloo Alan Tsang July 19, 2010 Deoxyribonucleic Acid (DNA) Sequence of bases: Adenine (A) Cytosine
Immune system IgGs. Carla Cortinas, Eva Espigulé, Guillem Lopez-Grado, Margalida Roig, Valentina Salas. Group 2
 Immune system IgGs Carla Cortinas, Eva Espigulé, Guillem Lopez-Grado, Margalida Roig, Valentina Salas Group 2 Index 1. Introduction 1.1. 1.2. 1.3. 1.4. 2. Immunoglobulins IgG formation IgG subclasses Structural
Immune system IgGs Carla Cortinas, Eva Espigulé, Guillem Lopez-Grado, Margalida Roig, Valentina Salas Group 2 Index 1. Introduction 1.1. 1.2. 1.3. 1.4. 2. Immunoglobulins IgG formation IgG subclasses Structural
BIOLOGY 200 Molecular Biology Students registered for the 9:30AM lecture should NOT attend the 4:30PM lecture.
 BIOLOGY 200 Molecular Biology Students registered for the 9:30AM lecture should NOT attend the 4:30PM lecture. Midterm date change! The midterm will be held on October 19th (likely 6-8PM). Contact Kathy
BIOLOGY 200 Molecular Biology Students registered for the 9:30AM lecture should NOT attend the 4:30PM lecture. Midterm date change! The midterm will be held on October 19th (likely 6-8PM). Contact Kathy
Unit 1: DNA and the Genome. Sub-Topic (1.3) Gene Expression
 Unit 1: DNA and the Genome Sub-Topic (1.3) Gene Expression Unit 1: DNA and the Genome Sub-Topic (1.3) Gene Expression On completion of this subtopic I will be able to State the meanings of the terms genotype,
Unit 1: DNA and the Genome Sub-Topic (1.3) Gene Expression Unit 1: DNA and the Genome Sub-Topic (1.3) Gene Expression On completion of this subtopic I will be able to State the meanings of the terms genotype,
Across-proteome modeling of dimer structures for the bottom-up assembly of protein-protein interaction networks
 Maheshwari and Brylinski BMC Bioinformatics (2017) 18:257 DOI 10.1186/s12859-017-1675-z RESEARCH ARTICLE Across-proteome modeling of dimer structures for the bottom-up assembly of protein-protein interaction
Maheshwari and Brylinski BMC Bioinformatics (2017) 18:257 DOI 10.1186/s12859-017-1675-z RESEARCH ARTICLE Across-proteome modeling of dimer structures for the bottom-up assembly of protein-protein interaction
" Rational drug design; a Structural Genomics approach"
 " Rational drug design; a Structural Genomics approach" Meeting the need of time; Dr Sanjan K. Das, PhD. Chief Scientific Officer (CSO), BBUK. Institute of Cancer Therapeutics University of Bradford, UK.
" Rational drug design; a Structural Genomics approach" Meeting the need of time; Dr Sanjan K. Das, PhD. Chief Scientific Officer (CSO), BBUK. Institute of Cancer Therapeutics University of Bradford, UK.
A data-driven protein-structure prediction algorithm
 A data-driven protein-structure prediction algorithm Clinton J. Robinson March 13, 26 David B. Skillicorn School of Computing Queen s University Kingston, Ontario, Canada K7L 3N6 Document prepared March
A data-driven protein-structure prediction algorithm Clinton J. Robinson March 13, 26 David B. Skillicorn School of Computing Queen s University Kingston, Ontario, Canada K7L 3N6 Document prepared March
