RNP purification, components and activity.
|
|
|
- Hugo Wilkinson
- 6 years ago
- Views:
Transcription
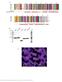
1 Supplementary Figure 1 RNP purification, components and activity. (a) Intein-mediated RNP production and purification. The Ll.LtrB intron RNA (red) (Exons E1 and E2 in green) and associated intron encoded protein (LtrA in yellow) were co-expressed with nisin induction. The RNP was captured and purified on chitin resin column through the chitin binding domain (CBD) that is fused to the LtrA C-terminus, released by DTT cleavage of the intein (I), and further purified by sucrose gradient sedimentation. (b) Product purity. LtrB intron RNA (902 nt) and associated LtrA protein (~70 kd) were analyzed, respectively, on a 1.2% denaturing formaldehyde agarose gel (left, lane 1, R) and on 12% denaturing SDS-PAGE gels (right, lanes 1, P), stained with Coomassie (blue) and silver (brown). Silver staining also revealed a higher molecular weight product, likely corresponding to intron RNA which stains with silver, not seen in a purified LtrA control (lanes 2). (c) RNA analysis. RNA in purified spliced RNPs (lanes 3 and 5) or extracted from the RNP (lane 4) were assayed by primer extension and the cdna analyzed on an 8% urea- PAGE gel. RNPs were either wildtype (wt) or a splicing-defective mutant of the catalytic triad (Triad). A primer, specific to the 5' end of the
2 intron (black arrow and P), was used to reveal precursor (pre-mrna) and spliced intron, from which 68-nt and 41-nt cdna products, respectively, were generated. Lane 1 corresponds to total RNA isolated from cells expressing the group II intron RNP and lane 2 to no RNA. The purified RNP resulted in predominantly a 41-nt band with a minor amount of precursor (lane 3), indicating that most of the pre-mrna had been removed. In contrast, the splicing defective Triad RNP yielded a primer extension product that corresponded exclusively to the intron-containing precursor (lane 5). (d) Activity assays with target DNA. Purified RNPs incubated with target DNA (T, 52nt) were assayed for DNA binding (left) and reverse splicing activity (right) on a 4% native- PAGE and a 10% urea-page gel, respectively. Lanes 1-3 contain no RNP, wild-type RNP and RNP from the catalytic triad mutant, respectively. Only the wt RNP, and not the catalytic mutant Triad, yielded shifted target DNA (left, black triangles). Likewise, only the wt RNP was active in reverse splicing (right) on the target DNA (IS=integration site, CS=cleavage site), producing a high molecular-weight integration product (black triangle) and the corresponding reverse splicing product (RSP) and cleavage product (CP).
3 Supplementary Figure 2 Negative-staining EM and random conical tilt reconstruction of the Ll.LtrB group II intron RNP. (a, b) One pair of electron micrographs of an untilted (a) and tilted (b) negatively stained specimen of the group II intron RNP complex. Four tilt pair particles are labeled with colored boxes, one color labeling one molecule. (c) Two-dimensional class averages of the particle images picked from micrographs of untilted specimen. Two major populated class averages are marked in cyan and lavender respectively. (d) The top row shows the RCT 3D reconstruction volumes from the tilted particle images corresponding to the two major class averages shown in (c). The bottom row shows 3D reconstructions obtained by refining the dataset merged from all the untilted and tilted particle images against the two RCT reconstruction volumes, respectively.
4
5 Supplementary Figure 3 Cryo-EM of the Ll.LtrB group II intron RNP and LtrA-depleted intron. (a) A representative raw micrograph of frozen-hydrated intron RNP particles generated from the 300 kv Titan Krios microscope. Few typical particles are identified by green square boxes. (b) Computed power spectrum of the micrograph in panel A. (c) Outline of the image processing steps to obtain the 3.8 Å resolution cryo-em map of the group II intron RNP complex and the 4.5 Å resolution map of the LtrA-depleted intron RNA. Reference-free 2D classification of 608,019 particle images generated ~200 2D class averages of the group II intron RNP comprising good particles free of ice contaminants and other junks. A 3D classification of the selected particles into four classes yielded three classes with strong LtrA protein occupancy that were merged as a total particle dataset of 450,296 particles and one class lacking density for the LtrA protein as a dataset of 102,522 particles. The first round of 3D auto-refinement of the merged dataset yielded a reconstruction at 4.2 Å resolution. A soft mask (shown in light grey semitransparency) around the most rigid portion of the RNP complex was used for further refinement to generate a final 3D reconstruction at 3.8 Å resolution. Local resolution maps of the 3D reconstructions without and with a soft mask during the refinement are shown on the right of the 4.2-Å and 3.8-Å resolution maps, respectively. The volumes are sliced to show the central portion of the local resolution distribution. A refinement of the LtrA-depleted intron dataset yielded a reconstruction at 4.5 Å resolution. The Euler angle distributions of particle images used for the intron RNP and the LtrA-depleted intron RNA reconstructions are shown next to corresponding EM maps. (d) Corrected gold-standard FSC curves of the final 3D reconstructions of RNP or LtrA-depleted intron RNA with or without a soft mask, respectively, after post-processing in RELION 1.3.
6
7 Supplementary Figure 4 Structure comparisons of the group IIA intron in the LtrA-bound state with that in the LtrA-depleted state and the group IIB intron. (a) Comparison of group IIA intron structure in LtrA-bound state (color coding of RNA domains are according to Fig. 1b) and IEP-depleted state (dark slate gray) shows a RMSD value of 2.4 Å. Regions corresponding to helices Id1 and Id3ii encompassing EBS2 and EBS1, respectively, are enlarged on upper left and right corners. (b) Comparison between tertiary structures of individual domains of the group IIA intron (this study) and group IIB intron (PDB ID: 4R0D). Domains of the group IIA intron are color coded and labeled as in Fig. 2, whereas that of the group IIB intron are shown as light grey color. Differences are displayed for each domain between the two introns. The β-β' interaction site, which is unique to group IIA intron, is boxed in red in the upper left panel. (c) Difference in relative positions of RNA helices of domain I, Id(iv) and domain IV with respect to domain V (red) between the group IIA and group IIB introns. The relative movements in group IIA intron helices are indicated by black arrows. RNA helices from group IIA and group IIB introns are shown in matching colors, except that the IIB intron helices are shown as semitransparent tubes. (d) Fourier shell correlation between atomic coordinates with the corresponding EM densities. RNP, intron-ltra complex; and DV, domain V of the intron RNA.
8
9 Supplementary Figure 5 Overall fitting of the initial homology model and final model of LtrA domains into the cryo-em map. (a) The fingers-palm domain initial homology model (left panel) and final model (middle panel) docked into the corresponding cryo-em density. Superposition of initial and final models (right panel) shows an RMSD value of 1.9 Å. (b) The thumb domain initial homology model (left panel) and final model (middle panel) docked into the corresponding cryo-em density. Superposition of initial and final models (right panel) shows an RMSD value of 1.6 Å. (c) The EN domain initial homology model (left panel) and final model after flipping α-helix (middle panel) docked into the corresponding cryo-em density. It should be noted that the α-helix corresponds to the DBD but was segmented with the EN domain. Superimposition of the aligned segments of the initial homology model and final EN model (right panel) shows an RMSD value of 1.7 Å. Density corresponding to the loop that connects the two-β strands was disordered and hence not modeled. (d) and (e), Overall fittings of NTD and DBD into the corresponding cryo-em masses. The 3.8 Å resolution cryo-em map (shown as semitransparent densities in matching colors) was used to model fingers-palm (a), thumb (b) and NTD (d). The dockings of EN (c) and DBD (e) domains were performed in the 5 Å resolution map (corresponding densities shown as semitransparent mass).
10
11 Supplementary Figure 6 Structure-based sequence alignment of the RT and EN domains. Structure based sequence alignments are performed using PROMALS3D 45. (a) Structure based sequence alignment of the RT fingers-palm domains of LtrA and Tribolium castaneum TERT (PDB: 3DU5). Secondary structures are shown with arrows for β-strands and bars for α helices. Identical residues are shaded in grey and indicated underneath with asterisks. Residues involved in TPRT in TERT and corresponding residues in LtrA are highlighted in different colors. Red, catalytic residues; Turquoise, dntp binding pocket; Green, DNA primer grip; Purple, RNA template interaction. (b) Alignment of the RT thumb domains of LtrA and Saccharomyces cerevisiae Prp8 (PDB: 3ZEF). α-helices are shown as bars, identical residues are shaded in grey and indicated underneath with asterisks. (c) Structure based sequence alignment of the EN domains from LtrA and the putative HNH endonuclease Gmet_0936 protein from Geobacter metallireducens GS-15 (PDB: 4H9D), HNH homing endonuclease I-HmuI (PDB: 1U3E), and E7 toxin (PDB: 1MZ8). Secondary structures are shown with arrows for β-strands and bars for α helices. The typical HNH motif residues are in red and four cysteine residues conserved between LtrA and Geobacter HNH endonuclease and proposed to be in additional metal ion coordination are in magenta. (d) Structure based sequence alignment of RT fingers-palm and thumb domains from LtrA, TERT, Prp8, and HIV-1 RT (PDB: 2HMI). Conserved sequence blocks RT1-RT7 in the fingerspalm domains (rectangular boxes) and the three parallel helices from Prp8 (H1-H3, boxes are in red, orange and green, as in panel b) in thumb domains in LtrA, TERT and HIV-1 RT, are defined using HIV-1 RT and Prp8 as references, respectively. Sequences in α-helices and β-strands are in red and blue, respectively. Insertions between RT sequence blocks, 2a, 4a, 5a, and 7a, and the thumb insertion ti are shown and labeled in red. Sequences of IFD-like motifs, are shaded in grey. Conserved aspartic acid residues at the RT active site are marked by red arrows. Motifs 1, 2, A, B, C, D, E (tan letters) in Tribolium castaneum TERT are in dark red boxes. Sequences that are not structurally resolved are boxed with dashed grey lines.
12
13 Supplementary Figure 7 Comparison of the fingers palm domain of LtrA with that of TERT and Prp8. (a) Docking of fingers-palm domain of TERT (PDB code: 3DU6; left panel) and Prp8 (PDB code: 3JB9; right panel) into the cryo-em density corresponding to the LtrA fingers-palm domain. (b) Structure superimposition of LtrA fingers-palm domains with corresponding domains from TERT (left panel) and Prp8 (right panel) show the RMSD values of 2.0 Å and 2.7 Å, respectively. (c) Rigid-body docking of TERT (left panel) and Prp8 (right panel) fingers domain onto group IIA intron shows a significant steric clash between Prp8 and intron domain Id(iii)a.
14 Supplementary Figure 8 Comparison of the thumb domain of LtrA with those of Prp8 and TERT. (a) Docking of the thumb domain of Prp8 (PDB code: 3JB9; right panel) and TERT (PDB code: 3DU6; left panel) into the cryo-em density corresponding to LtrA s thumb domain. (b) Structure superimposition of the LtrA thumb domain with corresponding domains from Prp8 (left panel) and TERT (right panel) show the RMSD values of 1.5 Å and 2.8 Å, respectively.
15 Supplementary Table 1 RMSD (Å) between LtrA RT figures/palm and thumb domains with those in TERT and Prp8 TERT Prp8 LtrA (Fingers-Palm) LtrA (Thumb)
16 Supplementary Note: Similarities of LtrA subdomains with Prp8 and TERT The docking of the atomic coordinates of the fingers-palm domains of Prp8 32 and TERT 29 into the corresponding cryo-em density of LtrA shows an overall similarity with both structures (Supplementary Fig. 7a). However, the RT domain of Prp8 is significantly larger due to the presence of several insertions in Prp8 RT, such as 4a, 5a and 7a (Fig. 3c; Supplementary Fig. 6d) that could not be accommodated by the cryo-em density (Supplementary Fig. 7a, right panel). Nevertheless, the conserved structural segments of the fingers-palm domain of LtrA superimpose well with corresponding segments from both TERT and Prp8 (Supplementary Fig. 7b; Supplementary Videos 3, 4) with RMSD values of 2.0 and 2.7 Å, respectively. Insertions 4a, 5a and 7a in the finger/palm domain of Prp8 would make a significant steric clash with the d(iii)a helix within D1 of the intron RNA (Supplementary Fig. 7c, right panel). Furthermore, both LtrA and TERT carry two conserved aspartate residues in the RT5 segment (Fig. 3c) that are involved in reverse transcription, whereas Prp8 carries only one aspartate in the homologous segment and the second aspartate is replaced by an arginine, which makes Prp8 an inactive reverse transcriptase. These observations suggest that the fingers-palm domain of TERT is most closely related to that in LtrA (Fig. 3b; Supplementary Fig. 6a). As described above, LtrA is also functionally related to the Prp8 RT of the spliceosome 3,25-28,37,38. Accordingly, we find that the thumb of LtrA possesses three parallel α- helices that more closely resemble the thumb domain of Prp8 (Figs. 3b-d; Supplementary Figs. 6b, 6d; Supplementary Video 6) than the thumb domain of TERT. Whereas coordinates of the Prp8 thumb domain 32 could be well docked into the cryo EM mass corresponding to the LtrA thumb domain (Supplementary Fig 8a, left panel), the thumb domain of TERT is much larger (Supplementary Fig. 8a, right panel) and would make a significant steric clash with the intron RNA. Our model of the thumb domain of LtrA superimposes well with coordinates of the Prp8 thumb domain (Supplementary Fig. 8b, left panel; Supplementary Video 6) with an RMSD of 1.5Å, whereas the thumb domain of LtrA only partially superimposes with the thumb domain of TERT with an RMSD of 2.8 Å (Supplementary Fig. 8b, right panel; Supplementary Table 2). The above observations further support our finding that the RT domain of LtrA carries a TERTlike fingers-palm domain and a Prp8-like thumb domain.
Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a
 Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a HNH-state 1 in PDB 4OO8, in which the distance from the C atom of the HNH catalytic residue 840 to the
Supplementary Figure 1 Two distinct conformational states of the HNH domain in crystal structures. a HNH-state 1 in PDB 4OO8, in which the distance from the C atom of the HNH catalytic residue 840 to the
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
SUPPLEMENTARY INFORMATION Conserved arginines on the rim of Hfq catalyze base pair formation and exchange Subrata Panja and Sarah A. Woodson T.C. Jenkins Department of Biophysics, Johns Hopkins University,
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
 X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction Tomohisa Shimasaki 1, Hiromi Yoshida 2, Shigehiro Kamitori 2 & Koji
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
has only one nucleotide, U20, between G19 and A21, while trna Glu CUC has two
 SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
SPPLEMENTRY INFORMTION doi:1.138/nature9411 Supplementary Discussion The structural characteristics of trn Gln G in comparison to trn Glu 16 in trn Gln G is directed towards the G19 56 pair, or the outer
PrimePCR Assay Validation Report
 Gene Information Gene Name SRY (sex determining region Y)-box 6 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SOX6 Human This gene encodes a member of the
Gene Information Gene Name SRY (sex determining region Y)-box 6 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SOX6 Human This gene encodes a member of the
A) B) Ladder. Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical
 A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
A) B) Ladder C) r4 r4 Nt- -Ct 78 kda Supplementary Figure 1. Recombinant calpain 14 purification analysis. A) The general domain structure of classical calpains is shown. The protease core consists of
Enhancers mutations that make the original mutant phenotype more extreme. Suppressors mutations that make the original mutant phenotype less extreme
 Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Interactomics and Proteomics 1. Interactomics The field of interactomics is concerned with interactions between genes or proteins. They can be genetic interactions, in which two genes are involved in the
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
3.1.4 DNA Microarray Technology
 3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
3.1.4 DNA Microarray Technology Scientists have discovered that one of the differences between healthy and cancer is which genes are turned on in each. Scientists can compare the gene expression patterns
HiPer RT-PCR Teaching Kit
 HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit
 Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Application Note 13 RNA Sample Preparation Non-Organic-Based Isolation of Mammalian microrna using Norgen s microrna Purification Kit B. Lam, PhD 1, P. Roberts, MSc 1 Y. Haj-Ahmad, M.Sc., Ph.D 1,2 1 Norgen
Hmwk # 8 : DNA-Binding Proteins : Part II
 The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
The purpose of this exercise is : Hmwk # 8 : DNA-Binding Proteins : Part II 1). to examine the case of a tandem head-to-tail homodimer binding to DNA 2). to view a Zn finger motif 3). to consider the case
All Rights Reserved. U.S. Patents 6,471,520B1; 5,498,190; 5,916, North Market Street, Suite CC130A, Milwaukee, WI 53202
 Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D
 Supplementary Information for Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D Federico Forneris a,b, B. Tom Burnley a,b,c and Piet Gros a * a Crystal
Supplementary Information for Ensemble refinement shows conformational flexibility in crystal structures of human complement factor D Federico Forneris a,b, B. Tom Burnley a,b,c and Piet Gros a * a Crystal
Student Learning Outcomes (SLOS)
 Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
Student Learning Outcomes (SLOS) KNOWLEDGE AND LEARNING SKILLS USE OF KNOWLEDGE AND LEARNING SKILLS - how to use Annhyb to save and manage sequences - how to use BLAST to compare sequences - how to get
CSE : Computational Issues in Molecular Biology. Lecture 19. Spring 2004
 CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
CSE 397-497: Computational Issues in Molecular Biology Lecture 19 Spring 2004-1- Protein structure Primary structure of protein is determined by number and order of amino acids within polypeptide chain.
CHAPTER 9 DNA Technologies
 CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
CHAPTER 9 DNA Technologies Recombinant DNA Artificially created DNA that combines sequences that do not occur together in the nature Basis of much of the modern molecular biology Molecular cloning of genes
Nature Structural & Molecular Biology: doi: /nsmb.2548
 Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Supplementary Figure 1. Structure of GltPhout. (a) Stereo view of a slice through a single GltPhout protomer shown in stick representation along with 2Fo-Fc and anomalous difference electron maps. The
Nature Structural & Molecular Biology: doi: /nsmb.3175
 Supplementary Figure 1 Medium- and low-fret groups probably representing varying positions on the DNA show similar changes between H3 and CENP-A nucleosomes with and without CENP-C CD, as does the high-fret
Supplementary Figure 1 Medium- and low-fret groups probably representing varying positions on the DNA show similar changes between H3 and CENP-A nucleosomes with and without CENP-C CD, as does the high-fret
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11726 Supplementary Figure 1 SRP RNA constructs used in single molecule experiments. a, Secondary structure of wildtype E. coli SRP RNA, which forms an elongated hairpin structure capped
doi:10.1038/nature11726 Supplementary Figure 1 SRP RNA constructs used in single molecule experiments. a, Secondary structure of wildtype E. coli SRP RNA, which forms an elongated hairpin structure capped
Transcription. DNA to RNA
 Transcription from DNA to RNA The Central Dogma of Molecular Biology replication DNA RNA Protein transcription translation Why call it transcription and translation? transcription is such a direct copy
Transcription from DNA to RNA The Central Dogma of Molecular Biology replication DNA RNA Protein transcription translation Why call it transcription and translation? transcription is such a direct copy
Protein Characterization/ Purification. Dr. Kevin Ahern
 Protein Characterization/ Purification Dr. Kevin Ahern Protein Purification Applications of Biochemistry Knowledge Protein Purification Applications of Biochemistry Knowledge Opening Cells Protein Purification
Protein Characterization/ Purification Dr. Kevin Ahern Protein Purification Applications of Biochemistry Knowledge Protein Purification Applications of Biochemistry Knowledge Opening Cells Protein Purification
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin)
Protein Synthesis Notes
 Protein Synthesis Notes Protein Synthesis: Overview Transcription: synthesis of mrna under the direction of DNA. Translation: actual synthesis of a polypeptide under the direction of mrna. Transcription
Protein Synthesis Notes Protein Synthesis: Overview Transcription: synthesis of mrna under the direction of DNA. Translation: actual synthesis of a polypeptide under the direction of mrna. Transcription
Chapter Fundamental Molecular Genetic Mechanisms
 Chapter 5-1 - Fundamental Molecular Genetic Mechanisms 5.1 Structure of Nucleic Acids 5.2 Transcription of Protein-Coding Genes and Formation of Functional mrna 5.3 The Decoding of mrna by trnas 5.4 Stepwise
Chapter 5-1 - Fundamental Molecular Genetic Mechanisms 5.1 Structure of Nucleic Acids 5.2 Transcription of Protein-Coding Genes and Formation of Functional mrna 5.3 The Decoding of mrna by trnas 5.4 Stepwise
Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Wang et al.
 Architecture of the Saccharomyces cerevisiae NuA/TIP0 complex 9 10 Wang et al. 1 9 10 11 1 1 Supplementary Figure 1 Preparation and EM analysis of the endogenous NuA complex. a, SDS-PAGE analysis of the
Architecture of the Saccharomyces cerevisiae NuA/TIP0 complex 9 10 Wang et al. 1 9 10 11 1 1 Supplementary Figure 1 Preparation and EM analysis of the endogenous NuA complex. a, SDS-PAGE analysis of the
INTRODUCTION TO REVERSE TRANSCRIPTION PCR (RT-PCR) ABCF 2016 BecA-ILRI Hub, Nairobi 21 st September 2016 Roger Pelle Principal Scientist
 INTRODUCTION TO REVERSE TRANSCRIPTION PCR (RT-PCR) ABCF 2016 BecA-ILRI Hub, Nairobi 21 st September 2016 Roger Pelle Principal Scientist Objective of PCR To provide a solution to one of the most pressing
INTRODUCTION TO REVERSE TRANSCRIPTION PCR (RT-PCR) ABCF 2016 BecA-ILRI Hub, Nairobi 21 st September 2016 Roger Pelle Principal Scientist Objective of PCR To provide a solution to one of the most pressing
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Nature Structural and Molecular Biology: doi: /nsmb.2937
 Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2F o F c electron density map of CtIP-NTD. (a) Multiple sequence alignment,
Supplementary Figure 1 Multiple sequence alignment of the CtIP N-terminal domain, purified CtIP protein constructs and details of the 2F o F c electron density map of CtIP-NTD. (a) Multiple sequence alignment,
Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2
 Structural evidence for consecutive -like modules in the spliceosomal ATPase Brr2 Lingdi Zhang, Tao Xu, Corina Maeder, Laura-Oana Bud, James Shanks, Jay Nix, Christine Guthrie, Jeffrey A. Pleiss, Rui Zhao
Structural evidence for consecutive -like modules in the spliceosomal ATPase Brr2 Lingdi Zhang, Tao Xu, Corina Maeder, Laura-Oana Bud, James Shanks, Jay Nix, Christine Guthrie, Jeffrey A. Pleiss, Rui Zhao
CGTAAGAGTACGTCCAGCATCGGF ATCATTATCTACATCXXXXXTACCATTCATTCAGATCTCACTATCGCATTCTCATGCAGGTCGTAGCCXS
 5 CGTAAGAGTACGTCCAGCATCGGF ATCATTATCTACATCXXXXXTACCATTCATTCAGATCTCACTATCGCATTCTCATGCAGGTCGTAGCCXS 29 Supplementary Figure 1 Sequence of the DNA primer/template pair used in Figure 1bc. A 23nt primer is
5 CGTAAGAGTACGTCCAGCATCGGF ATCATTATCTACATCXXXXXTACCATTCATTCAGATCTCACTATCGCATTCTCATGCAGGTCGTAGCCXS 29 Supplementary Figure 1 Sequence of the DNA primer/template pair used in Figure 1bc. A 23nt primer is
MMLV Reverse Transcriptase 1st-Strand cdna Synthesis Kit
 MMLV Reverse Transcriptase 1st-Strand cdna Synthesis Kit Cat. No. MM070150 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA265E MMLV Reverse Transcriptase 1st-Strand cdna Synthesis
MMLV Reverse Transcriptase 1st-Strand cdna Synthesis Kit Cat. No. MM070150 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA265E MMLV Reverse Transcriptase 1st-Strand cdna Synthesis
Structural Bioinformatics (C3210) DNA and RNA Structure
 Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Structural Bioinformatics (C3210) DNA and RNA Structure Importance of DNA/RNA 3D Structure Nucleic acids are essential materials found in all living organisms. Their main function is to maintain and transmit
Supplementary Information
 Supplementary Information Minimal mechanistic model of sirna-dependent target RNA slicing by recombinant human Argonaute2 protein Andrea Deerberg*, Sarah Willkomm* and Tobias Restle Institute of Molecular
Supplementary Information Minimal mechanistic model of sirna-dependent target RNA slicing by recombinant human Argonaute2 protein Andrea Deerberg*, Sarah Willkomm* and Tobias Restle Institute of Molecular
RNA does not adopt the classic B-DNA helix conformation when it forms a self-complementary double helix
 Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
Reason: RNA has ribose sugar ring, with a hydroxyl group (OH) If RNA in B-from conformation there would be unfavorable steric contact between the hydroxyl group, base, and phosphate backbone. RNA structure
Sam68 STARR Sam68 QUA1- KH. p(r ) [Å] [Å] TSTAR STAR. Sam68 QUA1-KH and. constructs are
![Sam68 STARR Sam68 QUA1- KH. p(r ) [Å] [Å] TSTAR STAR. Sam68 QUA1-KH and. constructs are Sam68 STARR Sam68 QUA1- KH. p(r ) [Å] [Å] TSTAR STAR. Sam68 QUA1-KH and. constructs are](/thumbs/73/69166612.jpg) a b Sam68 STARR Sam68 QUA1- KH c d e ) p(r p(r ) r [Å] r [Å] Supplementary Figure 1: The QUA2 domain is not involved in i the overall conformation of the STAR domain (a) Overlay of T-STAR QUA1-KH in complex
a b Sam68 STARR Sam68 QUA1- KH c d e ) p(r p(r ) r [Å] r [Å] Supplementary Figure 1: The QUA2 domain is not involved in i the overall conformation of the STAR domain (a) Overlay of T-STAR QUA1-KH in complex
HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION Nikesh Patel *, Simon J. White *, Rebecca F Thompson, Richard Bingham 1, Eva U. Weiß 1, Daniel P. Maskell, Adam Zlotnick 2,
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION Nikesh Patel *, Simon J. White *, Rebecca F Thompson, Richard Bingham 1, Eva U. Weiß 1, Daniel P. Maskell, Adam Zlotnick 2,
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
PrimePCR Assay Validation Report
 Gene Information Gene Name collagen, type IV, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID COL4A1 Human This gene encodes the major type IV alpha
Gene Information Gene Name collagen, type IV, alpha 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID COL4A1 Human This gene encodes the major type IV alpha
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Fatchiyah
 Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
Fatchiyah Email: fatchiya@yahoo.co.id RNAs: mrna trna rrna RNAi DNAs: Protein: genome DNA cdna mikro-makro mono-poly single-multi Analysis: Identification human and animal disease Finger printing Sexing
The Cys 2. His 2. Research Article 451
 Research Article 451 High-resolution structures of variant Zif268 DNA complexes: implications for understanding zinc finger DNA recognition Monicia Elrod-Erickson 1, Timothy E Benson 1 and Carl O Pabo
Research Article 451 High-resolution structures of variant Zif268 DNA complexes: implications for understanding zinc finger DNA recognition Monicia Elrod-Erickson 1, Timothy E Benson 1 and Carl O Pabo
PrimePCR Assay Validation Report
 Gene Information Gene Name laminin, beta 3 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID LAMB3 Human The product encoded by this gene is a laminin that
Gene Information Gene Name laminin, beta 3 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID LAMB3 Human The product encoded by this gene is a laminin that
Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD
 Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD Department of Biopharmacy, Faculty of Pharmacy, Silpakorn University Example of critical checkpoints
Molecular characterization, detection & quantitation of biological products Purin Charoensuksai, PhD Department of Biopharmacy, Faculty of Pharmacy, Silpakorn University Example of critical checkpoints
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Quiz Submissions Quiz 4
 Quiz Submissions Quiz 4 Attempt 1 Written: Nov 1, 2015 17:35 Nov 1, 2015 22:19 Submission View Released: Nov 4, 2015 20:24 Question 1 0 / 1 point Three RNA polymerases synthesize most of the RNA present
Quiz Submissions Quiz 4 Attempt 1 Written: Nov 1, 2015 17:35 Nov 1, 2015 22:19 Submission View Released: Nov 4, 2015 20:24 Question 1 0 / 1 point Three RNA polymerases synthesize most of the RNA present
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Bi 8 Lecture 7. Ellen Rothenberg 26 January Reading: Ch. 3, pp ; panel 3-1
 Bi 8 Lecture 7 PROTEIN STRUCTURE, Functional analysis, and evolution Ellen Rothenberg 26 January 2016 Reading: Ch. 3, pp. 109-134; panel 3-1 (end with free amine) aromatic, hydrophobic small, hydrophilic
Bi 8 Lecture 7 PROTEIN STRUCTURE, Functional analysis, and evolution Ellen Rothenberg 26 January 2016 Reading: Ch. 3, pp. 109-134; panel 3-1 (end with free amine) aromatic, hydrophobic small, hydrophilic
Section 10.3 Outline 10.3 How Is the Base Sequence of a Messenger RNA Molecule Translated into Protein?
 Section 10.3 Outline 10.3 How Is the Base Sequence of a Messenger RNA Molecule Translated into Protein? Messenger RNA Carries Information for Protein Synthesis from the DNA to Ribosomes Ribosomes Consist
Section 10.3 Outline 10.3 How Is the Base Sequence of a Messenger RNA Molecule Translated into Protein? Messenger RNA Carries Information for Protein Synthesis from the DNA to Ribosomes Ribosomes Consist
Reverse transcription-pcr (rt-pcr) Dr. Hani Alhadrami
 Reverse transcription-pcr (rt-pcr) Dr. Hani Alhadrami hanialhadrami@kau.edu.sa www.hanialhadrami.kau.edu.sa Overview Several techniques are available to detect and analyse RNA. Examples of these techniques
Reverse transcription-pcr (rt-pcr) Dr. Hani Alhadrami hanialhadrami@kau.edu.sa www.hanialhadrami.kau.edu.sa Overview Several techniques are available to detect and analyse RNA. Examples of these techniques
MonsterScript Reverse Transcriptase
 MonsterScript Reverse Transcriptase Cat. Nos. MSTA5110, and MSTA5124 Connect with Epicentre on our blog (epicentral.blogspot.com), Facebook (facebook.com/epicentrebio), and Twitter (@EpicentreBio). www.epicentre.com
MonsterScript Reverse Transcriptase Cat. Nos. MSTA5110, and MSTA5124 Connect with Epicentre on our blog (epicentral.blogspot.com), Facebook (facebook.com/epicentrebio), and Twitter (@EpicentreBio). www.epicentre.com
PrimePCR Assay Validation Report
 Gene Information Gene Name transforming growth factor, beta 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID TGFB1 Human This gene encodes a member of the
Gene Information Gene Name transforming growth factor, beta 1 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID TGFB1 Human This gene encodes a member of the
Specificity: Induced Fit
 Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
Specificity: Induced Fit Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) This adaptation is called the induced fit Induced fit allows for tighter binding of the ligand Induced
SSA Signal Search Analysis II
 SSA Signal Search Analysis II SSA other applications - translation In contrast to translation initiation in bacteria, translation initiation in eukaryotes is not guided by a Shine-Dalgarno like motif.
SSA Signal Search Analysis II SSA other applications - translation In contrast to translation initiation in bacteria, translation initiation in eukaryotes is not guided by a Shine-Dalgarno like motif.
Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element
 Molecular Cell Supplemental Information Mechanism of ranscription ermination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element Aneeshkumar G. Arimbasseri and Richard
Molecular Cell Supplemental Information Mechanism of ranscription ermination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element Aneeshkumar G. Arimbasseri and Richard
 Watson BM Gene Capitolo 11 Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. ? Le proteine della trasposizione Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A.
Watson BM Gene Capitolo 11 Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. ? Le proteine della trasposizione Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A.
Genes found in the genome include protein-coding genes and non-coding RNA genes. Which nucleotide is not normally found in non-coding RNA genes?
 Midterm Q Genes found in the genome include protein-coding genes and non-coding RNA genes Which nucleotide is not normally found in non-coding RNA genes? G T 3 A 4 C 5 U 00% Midterm Q Which of the following
Midterm Q Genes found in the genome include protein-coding genes and non-coding RNA genes Which nucleotide is not normally found in non-coding RNA genes? G T 3 A 4 C 5 U 00% Midterm Q Which of the following
Nature Methods: doi: /nmeth Supplementary Figure 1. Retention of RNA with LabelX.
 Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Supplementary Figure 1 Retention of RNA with LabelX. (a) Epi-fluorescence image of single molecule FISH (smfish) against GAPDH on HeLa cells expanded without LabelX treatment. (b) Epi-fluorescence image
Roche Molecular Biochemicals Technical Note No. LC 9/2000
 Roche Molecular Biochemicals Technical Note No. LC 9/2000 LightCycler Optimization Strategy Introduction Purpose of this Note Table of Contents The LightCycler system provides different detection formats
Roche Molecular Biochemicals Technical Note No. LC 9/2000 LightCycler Optimization Strategy Introduction Purpose of this Note Table of Contents The LightCycler system provides different detection formats
DNA - The Double Helix
 DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including cell reproduction,
DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including cell reproduction,
Zool 3200: Cell Biology Exam 3 3/6/15
 Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Name: Trask Zool 3200: Cell Biology Exam 3 3/6/15 Answer each of the following questions in the space provided; circle the correct answer or answers for each multiple choice question and circle either
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Methods of Biomaterials Testing Lesson 3-5. Biochemical Methods - Molecular Biology -
 Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
Methods of Biomaterials Testing Lesson 3-5 Biochemical Methods - Molecular Biology - Chromosomes in the Cell Nucleus DNA in the Chromosome Deoxyribonucleic Acid (DNA) DNA has double-helix structure The
BETA STRAND Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna
 Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna E-mail: a.hochkoeppler@unibo.it C-ter NH and CO groups: right, left, right (plane of the slide)
Prof. Alejandro Hochkoeppler Department of Pharmaceutical Sciences and Biotechnology University of Bologna E-mail: a.hochkoeppler@unibo.it C-ter NH and CO groups: right, left, right (plane of the slide)
DNA - The Double Helix
 Name Date Period DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including
Name Date Period DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including
Supplementary Figure 1: Two modes of low concentration of BsSMC on a DNA (a) Protein staining (left) and fluorescent imaging of Cy3 (right) confirm
 Supplementary Figure 1: Two modes of low concentration of BsSMC on a DNA (a) Protein staining (left) and fluorescent imaging of Cy3 (right) confirm that BsSMC was labeled with Cy3 NHS-Ester. In each panel,
Supplementary Figure 1: Two modes of low concentration of BsSMC on a DNA (a) Protein staining (left) and fluorescent imaging of Cy3 (right) confirm that BsSMC was labeled with Cy3 NHS-Ester. In each panel,
Introduction to Microarray Analysis
 Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Introduction to Microarray Analysis Methods Course: Gene Expression Data Analysis -Day One Rainer Spang Microarrays Highly parallel measurement devices for gene expression levels 1. How does the microarray
Review of Old Information: What is the monomer and polymer of: Macromolecule Monomer Polymer Carbohydrate Lipid Protein
 Section 1.8 Question of the Day: Name: Review of Old Information: What is the monomer and polymer of: Macromolecule Monomer Polymer Carbohydrate Lipid Protein New Information: One of the most important
Section 1.8 Question of the Day: Name: Review of Old Information: What is the monomer and polymer of: Macromolecule Monomer Polymer Carbohydrate Lipid Protein New Information: One of the most important
2910 GENES & DEVELOPMENT
 A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the
A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the
SuperScript IV Reverse Transcriptase as a better alternative to AMV-based enzymes
 WHITE PAPER SuperScript IV Reverse Transcriptase SuperScript IV Reverse Transcriptase as a better alternative to AMV-based enzymes Abstract Reverse transcriptases (RTs) from avian myeloblastosis virus
WHITE PAPER SuperScript IV Reverse Transcriptase SuperScript IV Reverse Transcriptase as a better alternative to AMV-based enzymes Abstract Reverse transcriptases (RTs) from avian myeloblastosis virus
DNA - The Double Helix
 DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including cell reproduction,
DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including cell reproduction,
CRISPR/Cas9 Genome Editing: Transfection Methods
 CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
CRISPR/ Genome Editing: Transfection Methods For over 20 years Mirus Bio has developed and manufactured high performance transfection products and technologies. That expertise is now being applied to the
8/21/2014. From Gene to Protein
 From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
From Gene to Protein Chapter 17 Objectives Describe the contributions made by Garrod, Beadle, and Tatum to our understanding of the relationship between genes and enzymes Briefly explain how information
Description...1 Components...1 Storage... 1 Technical Information...1 Protocol...2 Examples using the kit...4 Troubleshooting...
 QuickClean II Gel Extraction Kit Cat. No. L00418 Technical Manual No. TM0594 Version: 03042011 I II III IV V VI VII VIII Description......1 Components.....1 Storage.... 1 Technical Information....1 Protocol.....2
QuickClean II Gel Extraction Kit Cat. No. L00418 Technical Manual No. TM0594 Version: 03042011 I II III IV V VI VII VIII Description......1 Components.....1 Storage.... 1 Technical Information....1 Protocol.....2
CHAPTER 20 DNA TECHNOLOGY AND GENOMICS. Section A: DNA Cloning
 Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA.
 Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
Summary of Supplemental Information Figure S1: NUN preparation yields nascent, unadenylated RNA with a different profile from Total RNA. Figure S2: rrna removal procedure is effective for clearing out
PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%)
 1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
1 AFFINITY HIS-TAG PURIFICATION PROCEDURE FOR USE NICKEL NTA Magnetic Agarose Beads (5%) DESCRIPTION Nickel NTA Magnetic Agarose Beads are products that allow rapid and easy small-scale purification of
NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses)
 NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses) Consist of chemically linked sequences of nucleotides Nitrogenous base Pentose-
NUCLEIC ACIDS Genetic material of all known organisms DNA: deoxyribonucleic acid RNA: ribonucleic acid (e.g., some viruses) Consist of chemically linked sequences of nucleotides Nitrogenous base Pentose-
AmpliScribe T7-Flash Transcription Kit
 AmpliScribe T7-Flash Transcription Kit Cat. Nos. ASF3257 and ASF3507 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA191E AmpliScribe T7-Flash Transcription Kit 12/2016 1 1. Introduction
AmpliScribe T7-Flash Transcription Kit Cat. Nos. ASF3257 and ASF3507 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com MA191E AmpliScribe T7-Flash Transcription Kit 12/2016 1 1. Introduction
Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words).
 1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
1 Quiz1 Q1 2011 Alpha-helices, beta-sheets and U-turns within a protein are stabilized by (hint: two words) Value Correct Answer 1 noncovalent interactions 100% Equals hydrogen bonds (100%) Equals H-bonds
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Viral RNAi suppressor reversibly binds sirna to. outcompete Dicer and RISC via multiple-turnover
 Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death
 SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
SUPPLEMENTAL DATA Transcriptional Regulation of Pro-apoptotic Protein Kinase C-delta: Implications for Oxidative Stress-induced Neuronal Cell Death Huajun Jin 1, Arthi Kanthasamy 1, Vellareddy Anantharam
Molecular Cell Biology - Problem Drill 11: Recombinant DNA
 Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Molecular Cell Biology - Problem Drill 11: Recombinant DNA Question No. 1 of 10 1. Which of the following statements about the sources of DNA used for molecular cloning is correct? Question #1 (A) cdna
Chromatin Structure. a basic discussion of protein-nucleic acid binding
 Chromatin Structure 1 Chromatin DNA packaging g First a basic discussion of protein-nucleic acid binding Questions to answer: How do proteins bind DNA / RNA? How do proteins recognize a specific nucleic
Chromatin Structure 1 Chromatin DNA packaging g First a basic discussion of protein-nucleic acid binding Questions to answer: How do proteins bind DNA / RNA? How do proteins recognize a specific nucleic
Transcription is the first step of gene expression, in which a particular segment of DNA is copied into RNA by the enzyme, RNA polymerase.
 Transcription in Bacteria Transcription in Bacteria Transcription in Bacteria Transcription is the first step of gene expression, in which a particular segment of DNA is copied into RNA by the enzyme,
Transcription in Bacteria Transcription in Bacteria Transcription in Bacteria Transcription is the first step of gene expression, in which a particular segment of DNA is copied into RNA by the enzyme,
Biotechnology. Explorer Program. Serious About Science Education 5/17/09 1
 Biotechnology Explorer Program Serious About Science Education 5/17/09 1 Chromosome 8: PCR TM PCR Workshop Kirk Brown,, Tracy High School; Tracy, Ca Stan Hitomi,, Monte Vista High School; Danville, CA
Biotechnology Explorer Program Serious About Science Education 5/17/09 1 Chromosome 8: PCR TM PCR Workshop Kirk Brown,, Tracy High School; Tracy, Ca Stan Hitomi,, Monte Vista High School; Danville, CA
MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE?
 MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE? Lesson Plan: Title Introduction to the Genome Browser: what is a gene? JOYCE STAMM Objectives Demonstrate basic skills in using the UCSC Genome
MODULE 1: INTRODUCTION TO THE GENOME BROWSER: WHAT IS A GENE? Lesson Plan: Title Introduction to the Genome Browser: what is a gene? JOYCE STAMM Objectives Demonstrate basic skills in using the UCSC Genome
XactEdit Cas9 Nuclease with NLS User Manual
 XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
XactEdit Cas9 Nuclease with NLS User Manual An RNA-guided recombinant endonuclease for efficient targeted DNA cleavage Catalog Numbers CE1000-50K, CE1000-50, CE1000-250, CE1001-250, CE1001-1000 Table of
mmu-mir-200a-3p Real-time RT-PCR Detection and U6 Calibration Kit User Manual MyBioSource.com Catalog # MBS826230
 mmu-mir-200a-3p Real-time RT-PCR Detection and U6 Calibration Kit User Manual Catalog # MBS826230 For the detection and quantification of mirnas mmu-mir-200a-3p normalized by U6 snrna using Real-time RT-PCR
mmu-mir-200a-3p Real-time RT-PCR Detection and U6 Calibration Kit User Manual Catalog # MBS826230 For the detection and quantification of mirnas mmu-mir-200a-3p normalized by U6 snrna using Real-time RT-PCR
Preparing Samples for Analysis of Small RNA
 Preparing Samples for Analysis of Small RNA Topics 3 Introduction 4 Kit Contents and Equipment Checklist 6 Isolate Small RNA by Denaturing PAGE Gel 9 Ligate 5' RNA Adapters 12 Ligate 3' RNA Adapters 15
Preparing Samples for Analysis of Small RNA Topics 3 Introduction 4 Kit Contents and Equipment Checklist 6 Isolate Small RNA by Denaturing PAGE Gel 9 Ligate 5' RNA Adapters 12 Ligate 3' RNA Adapters 15
Spring 2006 Biochemistry 302 Exam 1
 1 Name Spring 2006 Biochemistry 302 Exam 1 Directions: This exam has 36 questions/problems totaling 90 points. Check to make sure you have all six pages. Some questions have multiple parts so read each
1 Name Spring 2006 Biochemistry 302 Exam 1 Directions: This exam has 36 questions/problems totaling 90 points. Check to make sure you have all six pages. Some questions have multiple parts so read each
Supplementary Figure 1. Pressure dependence of the self-cleavage reaction of the modified hairpin ribozyme. Time evolution of the modified hairpin
 Supplementary Figure 1. Pressure dependence of the self-cleavage reaction of the modified hairpin ribozyme. Time evolution of the modified hairpin ribozyme s fraction cleaved for the reaction in the presence
Supplementary Figure 1. Pressure dependence of the self-cleavage reaction of the modified hairpin ribozyme. Time evolution of the modified hairpin ribozyme s fraction cleaved for the reaction in the presence
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
Learning Objectives. Define RNA interference. Define basic terminology. Describe molecular mechanism. Define VSP and relevance
 Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
Learning Objectives Define RNA interference Define basic terminology Describe molecular mechanism Define VSP and relevance Describe role of RNAi in antigenic variation A Nobel Way to Regulate Gene Expression
DNA - The Double Helix
 DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including cell reproduction,
DNA - The Double Helix Recall that the nucleus is a small spherical, dense body in a cell. It is often called the "control center" because it controls all the activities of the cell including cell reproduction,
Review of Protein (one or more polypeptide) A polypeptide is a long chain of..
 Gene expression Review of Protein (one or more polypeptide) A polypeptide is a long chain of.. In a protein, the sequence of amino acid determines its which determines the protein s A protein with an enzymatic
Gene expression Review of Protein (one or more polypeptide) A polypeptide is a long chain of.. In a protein, the sequence of amino acid determines its which determines the protein s A protein with an enzymatic
