Supporting Information for
|
|
|
- Luke Wilkins
- 5 years ago
- Views:
Transcription
1 Supporting Information for CharmeRT: Boosting peptide identifications by chimeric spectra identification and retention time prediction Viktoria Dorfer*,,a, Sergey Maltsev,b, Stephan Winkler a, Karl Mechtler*,b,c. a University of Applied Sciences Upper Austria, Bioinformatics Research Group, Softwarepark 11, 4232 Hagenberg, Austria b Research Institute of Molecular Pathology (IMP), Vienna Biocenter (VBC), Campus-Vienna- Biocenter 1, 1030 Vienna, Austria c IMBA Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna Biocenter (VBC), Dr. Bohr-Gasse 3, 1030 Vienna, Austria These authors contributed equally. * Corresponding authors: Viktoria Dorfer: viktoria.dorfer@fh-hagenberg.at, Phone: Karl Mechtler: karl.mechtler@imp.ac.at, Phone: S-1
2 Table of contents: Table S1 Retention time prediction model Neighboring amino acids Figure S1 Figure S2 Figure S3 Figure S4 Table S2 Figure S5 Figure S6 Figure S7 Figure S8 Figure S9 Figure S10 Table S3 Table S4 Figure S11 Figure S12 Figure S13 Figure S14 Figure S15 Figure S16 Figure S17 Figure S18 All features used in Elutator to validate PSMs and peptides Description of the model used for retention time prediction Description of the impact of neighboring amino acids considered in the RT model Correlation of theoretically calculated hydrophobicity index to the measured retention time for high confident matches (FDR=0.001) of in-house HeLa and TiO2 enriched data sets. Correlation of theoretically calculated hydrophobicity index to the measured retention time for high confident matches (FDR=0.001) of in-house and external HeLa data set Histogram of mass deviations for highly reliable identifications before and after recalibration Longest consecutive series A+B+Y Shared ions between first and second peptides Results for data of O Connell et al. Protein evidence origin Presence of chimeric spectra in data sets with different isolation widths and gradient times Score distributions of MS Amanda scores RNA abundance of HeLa proteins Proportion of second search PSMs for spike-in data Identified PSMs and unique peptides at 1% FDR Mapping grouped proteins identified in first and second searches to RNA HeLa protein expression data Chimeric spectrum example Chimeric spectrum example Chimeric spectrum example Chimeric spectrum example Chimeric spectrum example Chimeric spectrum example Chimeric spectrum example Chimeric spectrum example S-2
3 Elutator features Feature MS Amanda Score Delta Score Delta Cn Retention Time [min] Delta RT [min] Absolute Description The PSM score assigned by the MS Amanda algorithm. Difference of the scores between 1st and 2nd rank matches. Nonzero for 1st rank matches only. Normalized score difference relative to the first best scoring PSM of the spectrum. Zero for 1st rank matches and non-zero for rank 2 and above. Measured peptide retention time. Deviation of the measured retention time (time of spectrum scan) from the predicted. Absolute value of the delta retention time. Delta RT [min] Combined Score % Isolation Interference MH+ [Da] m/z Calibrated Delta m/z [Th] Calibrated Delta Mass [ppm] Peptide Length Combined score of the MS Amanda score and retention time deviation. Fraction of ion current in the isolation width not attributed to the identified precursor. Singly charged mass of the peptide. Measured m/z value. Absolute calibrated deviation of the measured m/z from the theoretical value of the peptide. Calibrated deviation of the measured mass from the theoretical mass of the peptide in ppm. Length of the peptide in residues as a set of binary flags: length <= 6; length = 7; length = 8; length = 9; length = 10; length >= 11 Charge State Precursor charge state, as a set of binary flags: z <= 2; z >= 3 # Missed Cleavages Number of missed cleavages. Log Peptides Matched Log Total Intensity Fraction Matched Intensity [%] Log Total Intensity of Fragments Longest Consecutive Series Y Logarithm of the number of candidates (search space) in the precursor mass window. Logarithm of the total ion current of the fragment spectrum. Fraction of the total ion current of the fragment spectrum that is matched by fragments of the PSM. Similar to Log Total Intensity; peaks corresponding to precursor peaks (including isotopes) are excluded. Length of the longest consecutive matched sequence among the y fragment ion series peaks. S-3
4 Longest Consecutive Series A+B+Y Mean Squared Delta m/z for Fragments Top Y Fragment Delta m/z Second Top Y Fragment Delta m/z Length of the longest consecutive matched sequence confirmed by a, b or y ions. Average of squared mass errors of all fragments in Th, calculated as m i 2, where m i is the mass difference of measured and calculated masses of fragment i, and n is the number of identified fragments. Absolute calibrated deviation of the measured m/z from the theoretical value for the top-intense y fragment. y1 and y2 ions are not considered. Absolute calibrated deviation of the measured m/z from the theoretical value for the second top-intense y fragment. y1 and y2 ions are not considered. n Table S1. All features used in Elutator to validate PSMs and peptides. Retention time prediction model Our retention time prediction model can be fully described as the following non-linear sequence dependent function, which has been described by Krokhin 31, 2006: H = F + newiso(seq, F) + helices1(seq) + helices2(seq) where H is the hydrophobicity, newiso is a function modeling the isoelectric charge, seq is the peptide sequence, helices1 and helices2 are adjustments for short and long helices, and F is defined by: F = sumscale(lengthscale(length) R)) with sumscale being a polynomial function over the argument, lengthscale a polynomial factor dependent on the length of the peptide, length the length of the peptides sequence and R defined as: G R = G + smallness ( length ) undigested(sequence) clusterness(sequence) proline(sequence) with smallness being a correction factor depending on the length of the peptide, undigested a function to handle special positively charged amino acids (L/H/K), clusterness a function for handling clusters of hydrophobic amino acids, decreasing the hydrophobicity, proline a function to handle sequences with >=2 prolines in the peptide sequence, and G defined as: G = basesumofretentioncoefficients(sequence) + C(sequence) with basesumofretentioncoefficients being the sum of all retention time coefficients of all amino residuals of the peptide sequence and C modeling the impact of neighboring amino acids (see below). S-4
5 Interactions between neighboring amino acids We describe the cumulative contribution of neighbor residual s interactions C for peptide sequence s to the hydrophobicity index as summed over all residues i. f(s, i) is defined as 4 C(s) = α k f(s, i) k i k=0 f(s, i) = λ(j i)β(s i )γ(s j ) i 9 j i+9,j i The summation by i, j runs through all amino residuals in the sequence s with a maximal difference of ±9 amino acid positions. For each amino acid pair, we consider two coefficients, β for the amino residual at position i in sequence s and γ for the amino residual j in sequence s, such that the interaction between the amino residuals at positions i and j is described by the product β(s i )γ(s j ). Distance coefficients λ(δ) = λ(j i) account for the contribution of residual pairs with a distance δ between them. All coefficients, including α 0 4, all lambda values, and the values of the lookup tables βand γ are optimized during training of the RT model. Figure S1 Correlation of theoretically calculated hydrophobicity index to the measured retention time for high confident matches (FDR=0.001) of in-house HeLa and TiO2 enriched data sets. 70% of all matches in the TiO2 enriched data set contain one or more phosphorylated sites. Outliers were not removed. S-5
6 Figure S2 Correlation of theoretically calculated hydrophobicity index to the measured retention time for high confident matches (FDR=0.001) of in-house and external HeLa data set. Outliers were not removed. Figure S3 Histogram of mass deviations for highly reliable identifications before and after recalibration, with disabled lock mass. External human dataset has been taken from Michalski et al. 32 S-6
7 y5 y2 b3 a4 Figure S4 Longest Consecutive Series A+B+Y. a4 and b3 ions confirm the sequence, filling gaps between y5 and y2 ions. The length of the Longest Consecutive Series A+B+Y is four in this case. Average fragment ion overlap in % between Dataset replicate first and second peptides second peptides A (1h, 2m/z) B (3h, 2m/z) C (1h, 4m/z) D (3h, 4m/z) E (1h, 8m/z) F (3h, 8m/z) Table S2 Shared ions between first and second peptides. Overlap of fragment ions given in percent between peptides identified in the first and in the second search or between peptides in the second search, when multiple precursors were identified in the second search. S-7
8 Figure S5 Results for data of O Connell et al. 35 Data have been analyzed with Protein Discoverer 1.4.using 10ppm(a)/50 ppm(b) precursor mass tolerance and 0.02Da/0.9Da fragment mass tolerance. a) Label free data acquired at 1.4 m/z isolation width shows a high number of chimeric spectra that can be identified by CharmeRT. b) TMT data has been measured with an isolation window of 0.4 m/z showing a very low number of high confident interfering peptides. Still, the usage of CharmeRT is beneficial in this case as well, as it leads to the identification of more PSMs at the same FDR than the combination of MS Amanda and Percolator. Figure S6 Protein evidence origin. Proteins from a single HeLa run (4m/z, 3h gradient) are investigated and identified peptides are analyzed. The major part of proteins can be confirmed by peptide identifications from both searches, some proteins are only found in one of the two search iterations. Protein inference and grouping has been performed with Proteome Discoverer 1.4. S-8
9 Figure S7 Presence of chimeric spectra in data sets with different isolation widths and gradient times. All spectra having two or more reliably identified precursors are chimeric spectra. As expected, the presence of chimeric spectra rises with increasing isolation width. Figure S8 Proportion of second search PSMs for spike-in data 40. For low spike-in amounts, the proportion of UPS peptides is higher in the second search, as these originate from rare proteins and are therefore more likely to be coeluting peptides. S-9
10 Figure S9 Score distributions of MS Amanda scores for target (blue) and decoy (red) peptides identified in the first (A) or second (B) search. The spectrum quality for co-eluting peptides is lower and the score distributions of target matches of the second search look very similar to decoy matches, so the effect of including auxiliary information used in Elutator is higher. Figure S10 RNA abundance of HeLa proteins. All HeLa proteins are depicted in red, all proteins identified in the first search in light green, all proteins identified in the second search in dark green and proteins solely identified in the second search are purple. S-10
11 Data set Figure Method Ø PSMs first search Ø PSMs second search Ø PSMs added through validation Ø Unique peptides first search Ø Unique peptides overlap Ø Unique peptides second search A 3 CharmeRT B 3 CharmeRT C 3 CharmeRT D 3 CharmeRT E 3 CharmeRT F 3 CharmeRT G 4 Mascot + Percolator G 4 MaxQuant G 4 pparse + Mascot + Percolator G 4 MS Amanda + Percolator G 4 MS Amanda + Elutator (no RT) G 4 CharmeRT H 4 Mascot + Percolator H 4 MaxQuant H 4 pparse + Mascot + Percolator H 4 MS Amanda + Percolator H 4 MS Amanda + Elutator (no RT) H 4 CharmeRT I 2 Mascot + Percolator I 2 MaxQuant I 2 pparse + Mascot + Percolator I 2 MS Amanda + Percolator I 2 MS Amanda + Elutator (no RT) I 2 CharmeRT Table S3: Identified PSMs and unique peptides at 1% FDR (PSM or peptide level) for all Figures presented in the manuscript S-11
12 Total protein groups RNA Expressed Contaminants No RNA Expression Data Zero RNA Expression First search only First + Second searches Table S4: Mapping grouped proteins identified in first and second searches to RNA HeLa protein expression data. Figure S11 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide NANAVMEYEK are given in blue and ions of peptide SNcMDcLDR (with Cs being carbamidomethylated) are given in red. NANAVMEYEK SNcMDcLDR matched ion m/z delta mass [ppm] matched ion m/z delta mass [ppm] [M+2H] ImmL ImmY y y b b y y b b y y y b y y y y y y b y y S-12
13 Figure S12 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide cqaaepqiitgshdttir (with C being carbamidomethylated) are given in blue and ions of peptide QLVAEQVTYQR are given in red. cqaaepqiitgshdttir QLVAEQVTYQR matched ion m/z delta mass [ppm] matched ion m/z delta mass [ppm] y [M+2H] b y y b b y y b b y y y b y y y b y y y b y y y y y y y y y y y b y y S-13
14 Figure S13 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide GTITVSAQELK are given in blue and ions of peptide VMEIVDADEK are given in red. GTITVSAQELK VMEIVDADEK matched ion m/z delta mass [ppm] matched ion m/z delta mass [ppm] ImmIL ImmI y y b b y y b b y y b b y y y b b y y b y y y y y y S-14
15 Figure S14 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide LQELPDAVPHGEMPR are given in blue, ions of peptide YGPLPGPAVPR are given in red, and ions of peptide ELTGEDVLVR are given in green. LQELPDAVPHGEMPR YGPLPGPAVPR ELTGEDVLVR m/z delta matched m/z delta matched m/z mass ion mass ion [ppm] [ppm] matched ion delta mass [ppm] y [M+2H] y b ImmY b y y y b b b y y y b b b y y y y b y y y y y y y y y y y y y y y y S-15
16 Figure S15 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide AISHEHSPSDLEAHFVPLVK are given in blue, ions of peptide NDLSPTTVMSEGAR are given in red, and ions of peptide TPAFAESVTEGDVR are given in green. AISHEHSPSDLEAHFVPLVK NDLSPTTVMSEGAR TPAFAESVTEGDVR m/z delta matched m/z delta matched m/z mass ion mass ion [ppm] [ppm] matched ion delta mass [ppm] [M+3H] [M+2H] [M+2H] b y y y b b b y y y b b b y y y b b b y b y y y b y y y y y b y y y y y y y y y b y b y b b y b b b b b b b y b b b b S-16
17 Figure S16 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide WVGGQHPcFIIAEIGQNHQGDLDVAK (with C being carbamidomethylated) are given in blue and ions of peptide VNLLSFTGSTQVGK are given in red. WVGGQHPcFIIAEIGQNHQGDLDVAK VNLLSFTGSTQVGK matched ion m/z delta mass [ppm] matched ion m/z delta mass [ppm] b b y y b b y y y b y y b y b y y y y b b y y y b y y y b y y y y y y y y y y b y y y y y S-17
18 Figure S17 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide YLEVVLNTLQQASQAQVDK are given in blue and ions of peptide GIDVQQVSLVINYDLPTNR are given in red. YLEVVLNTLQQASQAQVDK GIDVQQVSLVINYDLPTNR matched ion m/z delta mass [ppm] matched ion m/z delta mass [ppm] b y y b b y y b b b y y b b y y b b y y b y y y b y y y y y y y b y y b b y y y y y y y S-18
19 Figure S18 Chimeric spectrum example. Spectrum is part of data set D (HeLa tryptic digest, Q Exactive Hybrid, 3h gradient, 4m/z isolation width).matched ions of peptide GVDEVTIVNILTNR are given in blue and ions of peptide VVIGMDVAASEFFR are given in red. GVDEVTIVNILTNR VVIGMDVAASEFFR matched ion m/z delta mass [ppm] matched ion m/z delta mass [ppm] y ImmF b y y b b y y b b y y b b y y y b b y y b b y y y y b y y y y y y y y S-19
Nature Biotechnology: doi: /nbt Supplementary Figure 1. The workflow of Open-pFind.
 Supplementary Figure 1 The workflow of Open-pFind. The MS data are first preprocessed by pparse, and then the MS/MS data are searched by the open search module. Next, the MS/MS data are re-searched by
Supplementary Figure 1 The workflow of Open-pFind. The MS data are first preprocessed by pparse, and then the MS/MS data are searched by the open search module. Next, the MS/MS data are re-searched by
Cell Signaling Technology
 Cell Signaling Technology PTMScan Direct: Multipathway v2.0 Proteomics Service Group January 14, 2013 PhosphoScan Deliverables Project Overview Methods PTMScan Direct: Multipathway V2.0 (Tables 1,2) Qualitative
Cell Signaling Technology PTMScan Direct: Multipathway v2.0 Proteomics Service Group January 14, 2013 PhosphoScan Deliverables Project Overview Methods PTMScan Direct: Multipathway V2.0 (Tables 1,2) Qualitative
Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs
 Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
Technical Overview Highly Confident Peptide Mapping of Protein Digests Using Agilent LC/Q TOFs Authors Stephen Madden, Crystal Cody, and Jungkap Park Agilent Technologies, Inc. Santa Clara, California,
RockerBox. Filtering massive Mascot search results at the.dat level
 RockerBox Filtering massive Mascot search results at the.dat level Challenges Big experiments High amount of data Large raw and.dat files (> 2GB) How to handle our results?? The 2.2 peptide summary could
RockerBox Filtering massive Mascot search results at the.dat level Challenges Big experiments High amount of data Large raw and.dat files (> 2GB) How to handle our results?? The 2.2 peptide summary could
N- The rank of the specified protein relative to all other proteins in the list of detected proteins.
 PROTEIN SUMMARY file N- The rank of the specified protein relative to all other proteins in the list of detected proteins. Unused (ProtScore) - A measure of the protein confidence for a detected protein,
PROTEIN SUMMARY file N- The rank of the specified protein relative to all other proteins in the list of detected proteins. Unused (ProtScore) - A measure of the protein confidence for a detected protein,
Supporting Information. Scanning Quadrupole Data Independent Acquisition Part A Qualitative and Quantitative Characterization
 S Supporting Information Scanning Quadrupole Data Independent Acquisition Part A Qualitative and Quantitative Characterization M. Arthur Moseley, Christopher J. Hughes 2, Praveen R. Juvvadi 3, Erik J.
S Supporting Information Scanning Quadrupole Data Independent Acquisition Part A Qualitative and Quantitative Characterization M. Arthur Moseley, Christopher J. Hughes 2, Praveen R. Juvvadi 3, Erik J.
Spectral Counting Approaches and PEAKS
 Spectral Counting Approaches and PEAKS INBRE Proteomics Workshop, April 5, 2017 Boris Zybailov Department of Biochemistry and Molecular Biology University of Arkansas for Medical Sciences 1. Introduction
Spectral Counting Approaches and PEAKS INBRE Proteomics Workshop, April 5, 2017 Boris Zybailov Department of Biochemistry and Molecular Biology University of Arkansas for Medical Sciences 1. Introduction
ProteinPilot Report for ProteinPilot Software
 ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Powerful mass spectrometers like
About OMICS Group Conferences
 About OMICS Group OMICS Group International is an amalgamation of Open Access publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of
About OMICS Group OMICS Group International is an amalgamation of Open Access publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of
Peptide and protein identification in mass spectrometry based proteomics. Yafeng Zhu, PhD student Karolinska Institutet, Scilifelab
 Peptide and protein identification in mass spectrometry based proteomics Yafeng Zhu, PhD student Karolinska Institutet, Scilifelab 2017-10-12 Content How is the peptide sequence identified? What is the
Peptide and protein identification in mass spectrometry based proteomics Yafeng Zhu, PhD student Karolinska Institutet, Scilifelab 2017-10-12 Content How is the peptide sequence identified? What is the
Supplementary Tables. Note: Open-pFind is embedded as the default open search workflow of the pfind tool. Nature Biotechnology: doi: /nbt.
 Supplementary Tables Supplementary Table 1. Detailed information for the six datasets used in this study Dataset Mass spectrometer # Raw files # MS2 scans Reference Dong-Ecoli-QE Q Exactive 5 202,452 /
Supplementary Tables Supplementary Table 1. Detailed information for the six datasets used in this study Dataset Mass spectrometer # Raw files # MS2 scans Reference Dong-Ecoli-QE Q Exactive 5 202,452 /
The effect of simulated microgravity on the Brassica napus seedling proteome
 10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
10.1071/FP16378_AC CSIRO 2018 Supplementary Material: Functional Plant Biology, 2018, 45(4), 440 452. Supplementary Material The effect of simulated microgravity on the Brassica napus seedling proteome
Liver Mitochondria Proteomics Employing High-Resolution MS Technology
 Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Liver Mitochondria Proteomics Employing High-Resolution MS Technology Jenny Ho, 1 Loïc Dayon, 2 John Corthésy, 2 Umberto De Marchi, 2 Antonio Núñez, 2 Andreas Wiederkehr, 2 Rosa Viner, 3 Michael Blank,
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO monoclonal antibody. (a) LC-MS analyses of tryptic
 Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
Supplementary Figure 1. Immunoprecipitation of synthetic SUMOm-remnant peptides using UMO 1-7-7 monoclonal antibody. (a) LC-MS analyses of tryptic digest from HEK293 cells spiked with 6 SUMOmremnant peptides
New Approaches to Quantitative Proteomics Analysis
 New Approaches to Quantitative Proteomics Analysis Chris Hodgkins, Market Development Manager, SCIEX ANZ 2 nd November, 2017 Who is SCIEX? Founded by Dr. Barry French & others: University of Toronto Introduced
New Approaches to Quantitative Proteomics Analysis Chris Hodgkins, Market Development Manager, SCIEX ANZ 2 nd November, 2017 Who is SCIEX? Founded by Dr. Barry French & others: University of Toronto Introduced
Next Generation Technology for Reproducible and Precise Proteome Profiling
 7 th Czech Mass Spectrometry Conference April 11th, 2018 Next Generation Technology for Reproducible and Precise Proteome Profiling Lars Kristensen, Ph.D. Application and training specialist The world
7 th Czech Mass Spectrometry Conference April 11th, 2018 Next Generation Technology for Reproducible and Precise Proteome Profiling Lars Kristensen, Ph.D. Application and training specialist The world
Quantification of Isotope Encoded Proteins in 2D Gels
 Quantification of Isotope Encoded Proteins in 2D Gels Using Surface Enhanced Resonance Raman Giselle M. Knudsen 1, Brandon M. Davis 2, Shirshendu K. Deb 1, Yvette Loethen 2, Ravindra Gudihal 1, Pradeep
Quantification of Isotope Encoded Proteins in 2D Gels Using Surface Enhanced Resonance Raman Giselle M. Knudsen 1, Brandon M. Davis 2, Shirshendu K. Deb 1, Yvette Loethen 2, Ravindra Gudihal 1, Pradeep
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
PTM Identification and Localization from MS Proteomics Data
 05.10.17 PTM Identification and Localization from MS Proteomics Data Marc Vaudel Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Bergen, Norway KG Jebsen Center for Diabetes
05.10.17 PTM Identification and Localization from MS Proteomics Data Marc Vaudel Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Bergen, Norway KG Jebsen Center for Diabetes
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University
 Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Quantitation techniques Label-free TMT SILAC Peptide identification and FDR 194 Lecture
Mass Spectrometry and Proteomics - Lecture 6 - Matthias Trost Newcastle University matthias.trost@ncl.ac.uk Previously Quantitation techniques Label-free TMT SILAC Peptide identification and FDR 194 Lecture
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 The mass accuracy of fragment ions is important for peptide recovery in wide-tolerance searches. The same data as in Figure 1B was searched with varying fragment ion tolerances (FIT).
Supplementary Figure 1 The mass accuracy of fragment ions is important for peptide recovery in wide-tolerance searches. The same data as in Figure 1B was searched with varying fragment ion tolerances (FIT).
ProMass HR Applications!
 ProMass HR Applications! ProMass HR Features Ø ProMass HR includes features for high resolution data processing. Ø ProMass HR includes the standard ProMass deconvolution algorithm as well as the full Positive
ProMass HR Applications! ProMass HR Features Ø ProMass HR includes features for high resolution data processing. Ø ProMass HR includes the standard ProMass deconvolution algorithm as well as the full Positive
High Resolution Accurate Mass Peptide Quantitation on Thermo Scientific Q Exactive Mass Spectrometers. The world leader in serving science
 High Resolution Accurate Mass Peptide Quantitation on Thermo Scientific Q Exactive Mass Spectrometers The world leader in serving science Goals Explore the capabilities of High Resolution Accurate Mass
High Resolution Accurate Mass Peptide Quantitation on Thermo Scientific Q Exactive Mass Spectrometers The world leader in serving science Goals Explore the capabilities of High Resolution Accurate Mass
Quantitative Analysis on the Public Protein Prospector Web Site. Introduction
 Quantitative Analysis on the Public Protein Prospector Web Site Peter R. Baker 1, Nicholas J. Agard 2, Alma L. Burlingame 1 and Robert J. Chalkley 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical
Quantitative Analysis on the Public Protein Prospector Web Site Peter R. Baker 1, Nicholas J. Agard 2, Alma L. Burlingame 1 and Robert J. Chalkley 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical
Protein Reports CPTAC Common Data Analysis Pipeline (CDAP)
 Protein Reports CPTAC Common Data Analysis Pipeline (CDAP) v. 4/13/2015 Summary The purpose of this document is to describe the protein reports generated as part of the CPTAC Common Data Analysis Pipeline
Protein Reports CPTAC Common Data Analysis Pipeline (CDAP) v. 4/13/2015 Summary The purpose of this document is to describe the protein reports generated as part of the CPTAC Common Data Analysis Pipeline
Top-Down Proteomics Enables Comparative Analysis of Brain. Proteoforms Between Mouse Strains
 Top-Down Proteomics Enables Comparative Analysis of Brain Proteoforms Between Mouse Strains Roderick G. Davis 1, Hae-Min Park 1, Kyunggon Kim #1, Joseph B. Greer 1, Ryan T. Fellers 1, Richard D. LeDuc
Top-Down Proteomics Enables Comparative Analysis of Brain Proteoforms Between Mouse Strains Roderick G. Davis 1, Hae-Min Park 1, Kyunggon Kim #1, Joseph B. Greer 1, Ryan T. Fellers 1, Richard D. LeDuc
基于质谱的蛋白质药物定性定量分析技术及应用
 基于质谱的蛋白质药物定性定量分析技术及应用 蛋白质组学质谱数据分析 单抗全蛋白测序及鉴定 Bioinformatics Solutions Inc. Waterloo, ON, Canada 上海 中国 2017 1 3 PEAKS Studio GUI Introduction Overview of PEAKS Studio GUI Project View Tasks, running info,
基于质谱的蛋白质药物定性定量分析技术及应用 蛋白质组学质谱数据分析 单抗全蛋白测序及鉴定 Bioinformatics Solutions Inc. Waterloo, ON, Canada 上海 中国 2017 1 3 PEAKS Studio GUI Introduction Overview of PEAKS Studio GUI Project View Tasks, running info,
Spectronaut Pulsar X. Maximize proteome coverage and data completeness by utilizing the power of Hybrid Libraries
 Spectronaut Pulsar X Maximize proteome coverage and data completeness by utilizing the power of Hybrid Libraries More versatility in proteomics research Spectronaut has delivered highest performance in
Spectronaut Pulsar X Maximize proteome coverage and data completeness by utilizing the power of Hybrid Libraries More versatility in proteomics research Spectronaut has delivered highest performance in
Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows
 Supporting Information Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows Ekaterina Stepanova 1, Steven P. Gygi 1, *, Joao A. Paulo 1, * 1 Department of
Supporting Information Filter-based Protein Digestion (FPD): A Detergent-free and Scaffold-based Strategy for TMT workflows Ekaterina Stepanova 1, Steven P. Gygi 1, *, Joao A. Paulo 1, * 1 Department of
Introduction. Benefits of the SWATH Acquisition Workflow for Metabolomics Applications
 SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
SWATH Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics A Data Independent Acquisition Technique Employed on the TripleTOF 6600 System Zuzana
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis
 Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Pushing the Leading Edge in Protein Quantitation: Integrated, Precise, and Reproducible Protein Quantitation Workflow Solutions
 2017 Metabolomics Seminars Pushing the Leading Edge in Protein Quantitation: Integrated, Precise, and Reproducible Protein Quantitation Workflow Solutions The world leader in serving science 2 3 Cancer
2017 Metabolomics Seminars Pushing the Leading Edge in Protein Quantitation: Integrated, Precise, and Reproducible Protein Quantitation Workflow Solutions The world leader in serving science 2 3 Cancer
Important Information for MCP Authors
 Guidelines to Authors for Publication of Manuscripts Describing Development and Application of Targeted Mass Spectrometry Measurements of Peptides and Proteins and Submission Checklist The following Guidelines
Guidelines to Authors for Publication of Manuscripts Describing Development and Application of Targeted Mass Spectrometry Measurements of Peptides and Proteins and Submission Checklist The following Guidelines
Quality Control Measures for Routine, High-Throughput Targeted Protein Quantitation Using Tandem Capillary Column Separation
 Quality Control Measures for Routine, High-Throughput Targeted Protein Quantitation Using Tandem Capillary Column Separation Sebastien Gallien 1, Elodie Duriez 1, Amol Prakash, Scott Peterman, Andreas
Quality Control Measures for Routine, High-Throughput Targeted Protein Quantitation Using Tandem Capillary Column Separation Sebastien Gallien 1, Elodie Duriez 1, Amol Prakash, Scott Peterman, Andreas
Appendix. Table of contents
 Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Appendix Table of contents 1. Appendix figures 2. Legends of Appendix figures 3. References Appendix Figure S1 -Tub STIL (41) DVFFYQADDEHYIPR (55) (64) AVLLDLEPR (72) 1 451aa (1071) YLNENQLSQLSVTR (1084)
Data Pre-processing in Liquid Chromatography-Mass Spectrometry Based Proteomics
 Bioinformatics Advance Access published September 8, 25 The Author (25). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Bioinformatics Advance Access published September 8, 25 The Author (25). Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oxfordjournals.org
Just launched. Operated by. Karel Harant
 Just launched Operated by Karel Harant 1 Rapid development of instrumentation Decline of MALDI-based instruments 2D protein electrophoresis abandoned 80% of proteomic data acquired in last two years (Somebody,
Just launched Operated by Karel Harant 1 Rapid development of instrumentation Decline of MALDI-based instruments 2D protein electrophoresis abandoned 80% of proteomic data acquired in last two years (Somebody,
Supplemental Materials
 Supplemental Materials MSGFDB Parameters For all MSGFDB searches, the following parameters were used: 30 ppm precursor mass tolerance, enable target-decoy search, enzyme specific scoring (for Arg-C, Asp-N,
Supplemental Materials MSGFDB Parameters For all MSGFDB searches, the following parameters were used: 30 ppm precursor mass tolerance, enable target-decoy search, enzyme specific scoring (for Arg-C, Asp-N,
Center for Mass Spectrometry and Proteomics Phone (612) (612)
 Outline Database search types Peptide Mass Fingerprint (PMF) Precursor mass-based Sequence tag Results comparison across programs Manual inspection of results Terminology Mass tolerance MS/MS search FASTA
Outline Database search types Peptide Mass Fingerprint (PMF) Precursor mass-based Sequence tag Results comparison across programs Manual inspection of results Terminology Mass tolerance MS/MS search FASTA
How to view Results with Scaffold. Proteomics Shared Resource
 How to view Results with Scaffold Proteomics Shared Resource Starting out Download Scaffold from http://www.proteomes oftware.com/proteom e_software_prod_sca ffold_download.html Follow installation instructions
How to view Results with Scaffold Proteomics Shared Resource Starting out Download Scaffold from http://www.proteomes oftware.com/proteom e_software_prod_sca ffold_download.html Follow installation instructions
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS
 Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Key questions of proteomics. Bioinformatics 2. Proteomics. Foundation of proteomics. What proteins are there? Protein digestion
 s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture X roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture X roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
Strategies for Quantitative Proteomics. Atelier "Protéomique Quantitative" La Grande Motte, France - June 26, 2007
 Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1.
 GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
GR24 (μm) 0 20 0 20 GST-ShHTL7 anti-gst His-MAX2 His-COI1 PVDF staining Supplementary information, Figure S1A ShHTL7 interacted with MAX2 but not another F-box protein COI1. Pull-down assays using GST-ShHTL7
FACTORS THAT AFFECT PROTEIN IDENTIFICATION BY MASS SPECTROMETRY HAOFEI TIFFANY WANG. (Under the Direction of Ron Orlando) ABSTRACT
 FACTORS THAT AFFECT PROTEIN IDENTIFICATION BY MASS SPECTROMETRY by HAOFEI TIFFANY WANG (Under the Direction of Ron Orlando) ABSTRACT Mass spectrometry combined with database search utilities is a valuable
FACTORS THAT AFFECT PROTEIN IDENTIFICATION BY MASS SPECTROMETRY by HAOFEI TIFFANY WANG (Under the Direction of Ron Orlando) ABSTRACT Mass spectrometry combined with database search utilities is a valuable
Proteomics and some of its Mass Spectrometric Applications
 Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Supplementary Fig. 1. S-1. Supplementary Fig. 2. S-2. Supplementary Fig. 3. S-3. Supplementary Fig. 4. S-4. Supplementary Fig. 5.
 Supplementary Information - An IonStar experimental strategy for MS1 ion current-based quantification using ultra-high-field Orbitrap: reproducible, in-depth and accurate protein measurement in larger
Supplementary Information - An IonStar experimental strategy for MS1 ion current-based quantification using ultra-high-field Orbitrap: reproducible, in-depth and accurate protein measurement in larger
Shotgun Proteomics: How Confident are you in that Identification? or Statistical Evaluation of Shotgun Proteomic Data
 Shotgun Proteomics: How Confident are you in that Identification? or Statistical Evaluation of Shotgun Proteomic Data Ron Orlando Complex Carbohydrate Research Center University of Georgia Athens, GA 30602
Shotgun Proteomics: How Confident are you in that Identification? or Statistical Evaluation of Shotgun Proteomic Data Ron Orlando Complex Carbohydrate Research Center University of Georgia Athens, GA 30602
BIOINFORMATICS ORIGINAL PAPER
 BIOINFORMATICS ORIGINAL PAPER Vol 25 no 22 29, pages 2969 2974 doi:93/bioinformatics/btp5 Data and text mining Improving peptide identification with single-stage mass spectrum peaks Zengyou He and Weichuan
BIOINFORMATICS ORIGINAL PAPER Vol 25 no 22 29, pages 2969 2974 doi:93/bioinformatics/btp5 Data and text mining Improving peptide identification with single-stage mass spectrum peaks Zengyou He and Weichuan
How to view Results with. Proteomics Shared Resource
 How to view Results with Scaffold 3.0 Proteomics Shared Resource An overview This document is intended to walk you through Scaffold version 3.0. This is an introductory guide that goes over the basics
How to view Results with Scaffold 3.0 Proteomics Shared Resource An overview This document is intended to walk you through Scaffold version 3.0. This is an introductory guide that goes over the basics
timstof Pro powered by PASEF and the Evosep One for high speed and sensitive shotgun proteomics
 timstof Pro powered by PASEF and the Evosep One for high speed and sensitive shotgun proteomics The Parallel Accumulation Serial Fragmentation (PASEF) method for trapped ion mobility spectrometry (TIMS)
timstof Pro powered by PASEF and the Evosep One for high speed and sensitive shotgun proteomics The Parallel Accumulation Serial Fragmentation (PASEF) method for trapped ion mobility spectrometry (TIMS)
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Thursday December 20 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Thursday December 20 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Estoril Education Day
 Estoril Education Day -Experimental design in Proteomics October 23rd, 2010 Peter James Note Taking All the Powerpoint slides from the Talks are available for download from: http://www.immun.lth.se/education/
Estoril Education Day -Experimental design in Proteomics October 23rd, 2010 Peter James Note Taking All the Powerpoint slides from the Talks are available for download from: http://www.immun.lth.se/education/
Accelerating Throughput for Targeted Quantitation of Proteins/Peptides in Biological Samples
 Technical Note Accelerating Throughput for Targeted Quantitation of Proteins/Peptides in Biological Samples Biomarker Verification Studies using the QTRAP 5500 Systems and the Reagents Triplex Sahana Mollah,
Technical Note Accelerating Throughput for Targeted Quantitation of Proteins/Peptides in Biological Samples Biomarker Verification Studies using the QTRAP 5500 Systems and the Reagents Triplex Sahana Mollah,
Modification Site Localization Scoring Integrated into a Search Engine
 Modification Site Localization Scoring Integrated into a Search Engine Peter R. Baker 1, Jonathan C. Trinidad 1, Katalin F. Medzihradszky 1, Alma L. Burlingame 1 and Robert J. Chalkley 1 1 Mass Spectrometry
Modification Site Localization Scoring Integrated into a Search Engine Peter R. Baker 1, Jonathan C. Trinidad 1, Katalin F. Medzihradszky 1, Alma L. Burlingame 1 and Robert J. Chalkley 1 1 Mass Spectrometry
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements
 Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Simultaneous Quantitation of a Monoclonal Antibody and Two Proteins in Human Plasma by High Resolution and Accurate Mass Measurements Paul-Gerhard Lassahn 1, Kai Scheffler 2, Myriam Demant 3, Nathanael
Basic protein and peptide science for proteomics. Henrik Johansson
 Basic protein and peptide science for proteomics Henrik Johansson Proteins are the main actors in the cell Membranes Transport and storage Chemical factories DNA Building proteins Structure Proteins mediate
Basic protein and peptide science for proteomics Henrik Johansson Proteins are the main actors in the cell Membranes Transport and storage Chemical factories DNA Building proteins Structure Proteins mediate
Nano LC at 20 nl/min Made Easy: A Splitless Pump Combined with Fingertight UHPLC Nano Column to Boost LC-MS Sensitivity in Proteomics
 Nano LC at 20 nl/min Made Easy: A Splitless Pump Combined with Fingertight UHPLC Nano Column to Boost LC-MS Sensitivity in Proteomics Rieux L 1., De Pra M 1., Köcher T 2., Mechtler K 2., Swart R 1 1 Thermo
Nano LC at 20 nl/min Made Easy: A Splitless Pump Combined with Fingertight UHPLC Nano Column to Boost LC-MS Sensitivity in Proteomics Rieux L 1., De Pra M 1., Köcher T 2., Mechtler K 2., Swart R 1 1 Thermo
Improving Productivity with Applied Biosystems GPS Explorer
 Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B
 Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Practical Tips. : Practical Tips Matrix Science
 Practical Tips : Practical Tips 2006 Matrix Science 1 Peak detection Especially critical for Peptide Mass Fingerprints A tryptic digest of an average protein (30 kda) should produce of the order of 50
Practical Tips : Practical Tips 2006 Matrix Science 1 Peak detection Especially critical for Peptide Mass Fingerprints A tryptic digest of an average protein (30 kda) should produce of the order of 50
Application Note TOF/MS
 Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
MassHunter Profinder: Batch Processing Software for High Quality Feature Extraction of Mass Spectrometry Data
 MassHunter Profinder: Batch Processing Software for High Quality Feature Extraction of Mass Spectrometry Data Technical Overview Introduction LC/MS metabolomics workflows typically involve the following
MassHunter Profinder: Batch Processing Software for High Quality Feature Extraction of Mass Spectrometry Data Technical Overview Introduction LC/MS metabolomics workflows typically involve the following
timstof Innovation with Integrity Powered by PASEF TIMS-QTOF MS
 timstof Powered by PASEF Innovation with Integrity TIMS-QTOF MS timstof The new standard for high speed, high sensitivity shotgun proteomics The timstof Pro with PASEF technology delivers revolutionary
timstof Powered by PASEF Innovation with Integrity TIMS-QTOF MS timstof The new standard for high speed, high sensitivity shotgun proteomics The timstof Pro with PASEF technology delivers revolutionary
Application Note 639. Daniel Lopez-Ferrer 1, Michael Blank 1, Stephan Meding 2, Aran Paulus 1, Romain Huguet 1, Remco Swart 2, Andreas FR Huhmer 1
 Pushing the Limits of Bottom-Up Proteomics with State-Of-The-Art Capillary UHPLC and Orbitrap Mass Spectrometry for Reproducible Quantitation of Proteomes Daniel Lopez-Ferrer 1, Michael Blank 1, Stephan
Pushing the Limits of Bottom-Up Proteomics with State-Of-The-Art Capillary UHPLC and Orbitrap Mass Spectrometry for Reproducible Quantitation of Proteomes Daniel Lopez-Ferrer 1, Michael Blank 1, Stephan
Supplementary Information
 Identifying sources of tick blood meals using unidentified tandem mass spectral libraries Özlem Önder 1, Wenguang Shao 2, Brian Kemps 1, Henry Lam 2,3,*, Dustin Brisson 1,* 1 Department of Biology, University
Identifying sources of tick blood meals using unidentified tandem mass spectral libraries Özlem Önder 1, Wenguang Shao 2, Brian Kemps 1, Henry Lam 2,3,*, Dustin Brisson 1,* 1 Department of Biology, University
Supplementary Figure 1: MS Data Quality (1/2)
 Supplementary Figure 1: MS Data Quality (1/2) a.4.3 Precursor Mass Error Plot Median Absolute Mass Error:.72 ppm b.2.15 Fragment Mass Error Plot Median Absolute Mass Error: 1.95 ppm.2.1.1.5. 6 4 2 2 4
Supplementary Figure 1: MS Data Quality (1/2) a.4.3 Precursor Mass Error Plot Median Absolute Mass Error:.72 ppm b.2.15 Fragment Mass Error Plot Median Absolute Mass Error: 1.95 ppm.2.1.1.5. 6 4 2 2 4
timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics
 timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics The timstof Pro powered by PASEF enables sequencing speed of > 2 Hz, high sensitivity
timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics The timstof Pro powered by PASEF enables sequencing speed of > 2 Hz, high sensitivity
ProteinPilot Software Overview
 ProteinPilot Software Overview High Quality, In-Depth Protein Identification and Protein Expression Analysis Sean L. Seymour and Christie L. Hunter SCIEX, USA As mass spectrometers for quantitative proteomics
ProteinPilot Software Overview High Quality, In-Depth Protein Identification and Protein Expression Analysis Sean L. Seymour and Christie L. Hunter SCIEX, USA As mass spectrometers for quantitative proteomics
Xevo G2-S QTof and TransOmics: A Multi-Omics System for the Differential LC/MS Analysis of Proteins, Metabolites, and Lipids
 Xevo G2-S QTof and TransOmics: A Multi-Omics System for the Differential LC/MS Analysis of Proteins, Metabolites, and Lipids Ian Edwards, Jayne Kirk, and Joanne Williams Waters Corporation, Manchester,
Xevo G2-S QTof and TransOmics: A Multi-Omics System for the Differential LC/MS Analysis of Proteins, Metabolites, and Lipids Ian Edwards, Jayne Kirk, and Joanne Williams Waters Corporation, Manchester,
Spectrum Mill MS Proteomics Workbench. Comprehensive tools for MS proteomics
 Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
An Improved Approach for Glycan Structure Identification from HCD MS/MS Spectra
 Introduction Method Experiments An Improved Approach for Glycan Structure Identification from HCD MS/MS Spectra Weiping Sun, Yi Liu, Gilles Lajoie, Bin Ma and Kaizhong Zhang Department of Computer Science,
Introduction Method Experiments An Improved Approach for Glycan Structure Identification from HCD MS/MS Spectra Weiping Sun, Yi Liu, Gilles Lajoie, Bin Ma and Kaizhong Zhang Department of Computer Science,
Bioinformatics for proteomics
 Purdue-UAB Botanicals Center for Age-Related Disease Bioinformatics for proteomics Stephen Barnes, PhD Purdue-UAB Botanical Center Workshop 2002 Mass Spectrometry Methods in Botanicals Research Objectives
Purdue-UAB Botanicals Center for Age-Related Disease Bioinformatics for proteomics Stephen Barnes, PhD Purdue-UAB Botanical Center Workshop 2002 Mass Spectrometry Methods in Botanicals Research Objectives
Proteomics software at MSI. Pratik Jagtap Minnesota Supercomputing institute
 Proteomics software at MSI. Pratik Jagtap Minnesota Supercomputing institute http://www.mass.msi.umn.edu/ Proteomics software at MSI. proteomics : emerging technology proteomics workflow search algorithms
Proteomics software at MSI. Pratik Jagtap Minnesota Supercomputing institute http://www.mass.msi.umn.edu/ Proteomics software at MSI. proteomics : emerging technology proteomics workflow search algorithms
See More, Much More Clearly
 TRIPLETOF 6600 SYSTEM WITH SWATH ACQUISITION 2.0 See More, Much More Clearly NEXT-GENERATION PROTEOMICS PLATFORM Every Peak. Every Run. Every Time. The Next-generation Proteomics Platform is here: The
TRIPLETOF 6600 SYSTEM WITH SWATH ACQUISITION 2.0 See More, Much More Clearly NEXT-GENERATION PROTEOMICS PLATFORM Every Peak. Every Run. Every Time. The Next-generation Proteomics Platform is here: The
A Complete Workflow Solution for Intact Monoclonal Antibody Characterization Using a New High-Performance Benchtop Quadrupole- Orbitrap LC-MS/MS
 A Complete Workflow Solution for Intact Monoclonal Antibody Characterization Using a New High-Performance Benchtop Quadrupole- Orbitrap LC-MS/MS Zhiqi Hao, 1 Yi Zhang, 1 David Horn, 1 Seema Sharma, 1 Shiaw-Lin
A Complete Workflow Solution for Intact Monoclonal Antibody Characterization Using a New High-Performance Benchtop Quadrupole- Orbitrap LC-MS/MS Zhiqi Hao, 1 Yi Zhang, 1 David Horn, 1 Seema Sharma, 1 Shiaw-Lin
ipep User s Guide Proteomics
 Proteomics ipep User s Guide ipep has been designed to aid in designing sample preparation techniques around the detection of specific sites or motifs in proteins and proteomes. The first application,
Proteomics ipep User s Guide ipep has been designed to aid in designing sample preparation techniques around the detection of specific sites or motifs in proteins and proteomes. The first application,
Thermo Scientific Q Exactive HF Orbitrap LC-MS/MS System. Higher-Quality Data, Faster Than Ever. Speed Productivity Confidence
 Thermo Scientific Q Exactive HF Orbitrap LC-MS/MS System Higher-Quality Data, Faster Than Ever Speed Productivity Confidence Transforming discovery quantitation The Thermo Scientific Q Exactive HF hybrid
Thermo Scientific Q Exactive HF Orbitrap LC-MS/MS System Higher-Quality Data, Faster Than Ever Speed Productivity Confidence Transforming discovery quantitation The Thermo Scientific Q Exactive HF hybrid
Identification of Microprotein-Protein Interactions via APEX Tagging
 Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Supporting Information Identification of Microprotein-Protein Interactions via APEX Tagging Qian Chu, Annie Rathore,, Jolene K. Diedrich,, Cynthia J. Donaldson, John R. Yates III, and Alan Saghatelian
Data Quality Control in Peptide Identification
 Data Quality Control in Peptide Identification Yunping Zhu State Key Laboratory of Proteomics Beijing Proteome Research Center Beijing Institute of Radiation Medicine Beijing, 2010-11-11 Data quality control
Data Quality Control in Peptide Identification Yunping Zhu State Key Laboratory of Proteomics Beijing Proteome Research Center Beijing Institute of Radiation Medicine Beijing, 2010-11-11 Data quality control
ProteinPilot Software for Protein Identification and Expression Analysis
 ProteinPilot Software for Protein Identification and Expression Analysis Providing expert results for non-experts and experts alike ProteinPilot Software Overview New ProteinPilot Software transforms protein
ProteinPilot Software for Protein Identification and Expression Analysis Providing expert results for non-experts and experts alike ProteinPilot Software Overview New ProteinPilot Software transforms protein
Smartube Benchmark of Nano-LC
 Smartube Benchmark of Nano-LC The state of the art Smartube is a high performance chromatographic column product line. It is made for high efficiency, high selectivity microscale separations. Compared
Smartube Benchmark of Nano-LC The state of the art Smartube is a high performance chromatographic column product line. It is made for high efficiency, high selectivity microscale separations. Compared
ENHANCED PROTEIN AND PEPTIDE CHARACTERIZATION
 Agilent MassHunter BioConfirm Software ENHANCED PROTEIN AND PEPTIDE CHARACTERIZATION Spectrum Algorithm Peak Modeling Deconvolution Mass 16951.778 Sequence Name Myoglobin (horse) Target mass 16951.673
Agilent MassHunter BioConfirm Software ENHANCED PROTEIN AND PEPTIDE CHARACTERIZATION Spectrum Algorithm Peak Modeling Deconvolution Mass 16951.778 Sequence Name Myoglobin (horse) Target mass 16951.673
A Unique LC-MS Assay for Host Cell Proteins(HCPs) ) in Biologics
 A Unique LC-MS Assay for Host Cell Proteins(HCPs) ) in Biologics Catalin Doneanu,, Ph.D. Biopharmaceutical Sciences, Waters September 16, 2009 Mass Spec 2009 2009 Waters Corporation Host Cell Proteins
A Unique LC-MS Assay for Host Cell Proteins(HCPs) ) in Biologics Catalin Doneanu,, Ph.D. Biopharmaceutical Sciences, Waters September 16, 2009 Mass Spec 2009 2009 Waters Corporation Host Cell Proteins
Nature Methods: doi: /nmeth Supplementary Figure 1
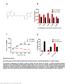 Supplementary Figure 1 Enrichment with anti-biotin antibody significantly increases detection of biotinylated peptides in complex samples. (A) Structure of NHS-biotin (B) Results of spike-in studies. Bar
Supplementary Figure 1 Enrichment with anti-biotin antibody significantly increases detection of biotinylated peptides in complex samples. (A) Structure of NHS-biotin (B) Results of spike-in studies. Bar
Protein Interaction Network of Human Protein Kinase. D2 Revealed by Chemical Cross-linking/Mass. Spectrometry
 SUPPORTING INFORMATION Protein Interaction Network of Human Protein Kinase D2 Revealed by Chemical Cross-linking/Mass Spectrometry Björn Häupl, Christian H. Ihling, and Andrea Sinz * Department of Pharmaceutical
SUPPORTING INFORMATION Protein Interaction Network of Human Protein Kinase D2 Revealed by Chemical Cross-linking/Mass Spectrometry Björn Häupl, Christian H. Ihling, and Andrea Sinz * Department of Pharmaceutical
Supplementary Figure 1. Processing and quality control for recombinant proteins.
 Supplementary Figure 1 Processing and quality control for recombinant proteins. (a) Schematic representation of processing of a recombinant protein library. Recombinant proteins were generated individually.
Supplementary Figure 1 Processing and quality control for recombinant proteins. (a) Schematic representation of processing of a recombinant protein library. Recombinant proteins were generated individually.
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
ChIP-seq and RNA-seq. Farhat Habib
 ChIP-seq and RNA-seq Farhat Habib fhabib@iiserpune.ac.in Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions
ChIP-seq and RNA-seq Farhat Habib fhabib@iiserpune.ac.in Biological Goals Learn how genomes encode the diverse patterns of gene expression that define each cell type and state. Protein-DNA interactions
ApplicationNOTE THE USE OF A NANOLOCKSPRAY ELECTROSPRAY INTERFACE FOR EXACT MASS PROTEOMICS STUDIES
 OVERVIEW This application note describes the implementation of a dual sprayer NanoLockSpray TM source on the Waters Micromass Q-Tof micro TM mass spectrometer This source consists of a dual sprayer arrangement;
OVERVIEW This application note describes the implementation of a dual sprayer NanoLockSpray TM source on the Waters Micromass Q-Tof micro TM mass spectrometer This source consists of a dual sprayer arrangement;
Ensure your Success with Agilent s Biopharma Workflows
 Ensure your Success with Agilent s Biopharma Workflows Steve Madden Software Product Manager Agilent Technologies, Inc. June 5, 2018 Agenda Agilent BioPharma Workflow Platform for LC/MS Intact Proteins
Ensure your Success with Agilent s Biopharma Workflows Steve Madden Software Product Manager Agilent Technologies, Inc. June 5, 2018 Agenda Agilent BioPharma Workflow Platform for LC/MS Intact Proteins
Quantification of Host Cell Protein Impurities Using the Agilent 1290 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System
 Application Note Biotherapeutics Quantification of Host Cell Protein Impurities Using the Agilent 9 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System Authors Linfeng Wu and Yanan Yang
Application Note Biotherapeutics Quantification of Host Cell Protein Impurities Using the Agilent 9 Infinity II LC Coupled with the 6495B Triple Quadrupole LC/MS System Authors Linfeng Wu and Yanan Yang
Protein Grouping, FDR Analysis and Databases.
 Protein Grouping, FDR Analysis and Databases. March 15th 2012 Pratik Jagtap The Minnesota http://www.mass.msi.umn.edu/ Protein Grouping, FDR Analysis and Databases Overview. Protein Grouping : Concept
Protein Grouping, FDR Analysis and Databases. March 15th 2012 Pratik Jagtap The Minnesota http://www.mass.msi.umn.edu/ Protein Grouping, FDR Analysis and Databases Overview. Protein Grouping : Concept
Host Cell Protein Analysis Using Agilent AssayMAP Bravo and 6545XT AdvanceBio LC/Q-TOF
 Application Note Biotherapeutics and Biosimilars Host Cell Protein Analysis Using Agilent AssayMAP Bravo and 6545XT AdvanceBio LC/Q-TOF Authors Linfeng Wu, Shuai Wu and Te-Wei Chu Agilent Technologies,
Application Note Biotherapeutics and Biosimilars Host Cell Protein Analysis Using Agilent AssayMAP Bravo and 6545XT AdvanceBio LC/Q-TOF Authors Linfeng Wu, Shuai Wu and Te-Wei Chu Agilent Technologies,
High-throughput Proteomic Data Analysis. Suh-Yuen Liang ( 梁素雲 ) NRPGM Core Facilities for Proteomics and Glycomics Academia Sinica Dec.
 High-throughput Proteomic Data Analysis Suh-Yuen Liang ( 梁素雲 ) NRPGM Core Facilities for Proteomics and Glycomics Academia Sinica Dec. 9, 2009 High-throughput Proteomic Data Are Information Rich and Dependent
High-throughput Proteomic Data Analysis Suh-Yuen Liang ( 梁素雲 ) NRPGM Core Facilities for Proteomics and Glycomics Academia Sinica Dec. 9, 2009 High-throughput Proteomic Data Are Information Rich and Dependent
Mass Spectrometry Based Proteomics Data Analysis Using GalaxyP
 Mass Spectrometry Based Proteomics Data Analysis Using GalaxyP GCC 2015 GalaxyP Workshop July 6th, 2015 Norwich, UK Presenters: Tim Griffin, Pratik Jagtap and James Johnson Documentation: Kevin Murray,
Mass Spectrometry Based Proteomics Data Analysis Using GalaxyP GCC 2015 GalaxyP Workshop July 6th, 2015 Norwich, UK Presenters: Tim Griffin, Pratik Jagtap and James Johnson Documentation: Kevin Murray,
PEAKS 8 User Manual. PEAKS Team
 PEAKS 8 User Manual PEAKS Team PEAKS 8 User Manual PEAKS Team Publication date 2016 Table of Contents 1. Overview... 1 1. How to Use This Manual... 1 2. What Is PEAKS?... 1 3. What Is New in PEAKS 8?...
PEAKS 8 User Manual PEAKS Team PEAKS 8 User Manual PEAKS Team Publication date 2016 Table of Contents 1. Overview... 1 1. How to Use This Manual... 1 2. What Is PEAKS?... 1 3. What Is New in PEAKS 8?...
Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification
 Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification Protein Expression Analysis using the TripleTOF 5600 System and itraq Reagents Vojtech Tambor 1, Christie
Combination of Isobaric Tagging Reagents and Cysteinyl Peptide Enrichment for In-Depth Quantification Protein Expression Analysis using the TripleTOF 5600 System and itraq Reagents Vojtech Tambor 1, Christie
Automated platform of µlc-ms/ms using SAX trap column for
 Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
