The Translational Genomics Core at Partners Personalized Medicine: Facilitating the Transition of Research towards Personalized Medicine
|
|
|
- Cuthbert Jones
- 6 years ago
- Views:
Transcription
1 The Translational Genomics Core at Partners Personalized Medicine: Facilitating the Transition of Research towards Personalized Medicine The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Published Version Accessed Citable Link Terms of Use Blau, Ashley, Alison Brown, Lisa Mahanta, and Sami S. Amr The Translational Genomics Core at Partners Personalized Medicine: Facilitating the Transition of Research towards Personalized Medicine. Journal of Personalized Medicine 6 (1): 10. doi: /jpm doi: /jpm January 19, :04:33 PM EST This article was downloaded from Harvard University's DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at (Article begins on next page)
2 Journal of Personalized Medicine Article The Translational Genomics Core at Partners Personalized Medicine: Facilitating the Transition of Research towards Personalized Medicine Ashley Blau 1, Alison Brown 1, Lisa Mahanta 1 and Sami S. Amr 1,2, * 1 Partners HealthCare Personalized Medicine, 65 Landsdowne Street, Cambridge, MA 02139, USA; ablau@partners.org (A.B.); abrown13@partners.org (A.B.); lmahanta@partners.org (L.M.) 2 Department of Pathology, Brigham and Women s Hospital and Harvard Medical School, Boston, MA 02115, USA * Correspondence: samr@partners.org; Tel.: ; Fax: Academic Editor: Scott T. Weiss Received: 21 October 2015; Accepted: 18 February 2016; Published: 26 February 2016 Abstract: The Translational Genomics Core (TGC) at Partners Personalized Medicine (PPM) serves as a fee-for-service core laboratory for Partners Healthcare researchers, providing access to technology platforms and analysis pipelines for genomic, transcriptomic, and epigenomic research projects. The interaction of the TGC with various components of PPM provides it with a unique infrastructure that allows for greater IT and bioinformatics opportunities, such as sample tracking and data analysis. The following article describes some of the unique opportunities available to an academic research core operating within PPM, such the ability to develop analysis pipelines with a dedicated bioinformatics team and maintain a flexible Laboratory Information Management System (LIMS) with the support of an internal IT team, as well as the operational challenges encountered to respond to emerging technologies, diverse investigator needs, and high staff turnover. In addition, the implementation and operational role of the TGC in the Partners Biobank genotyping project of over 25,000 samples is presented as an example of core activities working with other components of PPM. Keywords: translational genomics core; academic core facility; genomics services; genotyping; seamless workflows 1. Introduction Technological and analytical advancements in genomics and decreasing cost of sequencing and array-based platforms in recent years are a key catalyst in the exponential growth of translational research initiatives and broader integration of clinical genetic testing. Most notably, next-generation sequencing (NGS) technologies have enhanced the capabilities of researchers and clinical laboratories to identify and characterize markers of disease in pursuit of personalized medicine. More recently developed technologies, such as NGS, as well as established platforms, such as array-based technologies, require significant laboratory infrastructure and operational expertise to maximize their utility, and therefore, are often aggregated in core facilities at academic medical institutions. In its commitment to advancing medical genomics research and its integration into clinical practice, Partners Healthcare Personalized Medicine (PPM) established the Translational Genomics Core (TGC) as part of its center to provide access and expertise of leading technologies to academic clinical and translational researchers at Partners Healthcare. The TGC offers end-to-end technical and analytical expertise and services across several leading platforms that support genomic, transcriptomic and epigenomic studies (Table 1). All platforms and J. Pers. Med. 2016, 6, 10; doi: /jpm
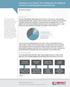
3 J. Pers. Med. 2016, 6, 10 2 of 10 assays performed at the TGC are validated and optimized on automation instruments to provide reproducible, high quality, and high-throughput sample processing and data generation. In addition, the core is responsible for the evaluation and development of novel platforms and assays to further research programs at Partners Healthcare. Alongside its role as a core facility to researchers, the TGC is integrated into the initiatives and operations of other components within Partners Personalized Medicine (described in An overview of Partners Personalized Medicine at Partners Healthcare in this issue). As a result of these interactions, the TGC enjoys a synergistic relationship with these different components that have translated into unique opportunities for the center and Partners researchers. The following describes the role and operations of the TGC within the center and across the Partners Healthcare research community. Table 1. Platforms and services at the Translational Genomics Core. Technology Platforms Next Generation Sequencing (NGS) Array-based platforms Services Genomic analysis Transcriptome analysis Methylation analysis Illumina HiSeq 2500 instrument Illumina MiSeq instrument Illumina iscan System (Infinium BeadChip) Affymetrix GeneChip Scanner Next generation sequencing: Whole genome sequencing (WGS) Whole exome sequencing (WES) Targeted gene panel sequencing (hybrid capture method or amplicon enrichment method) Metagenomic analysis: 16S rrna gene microbial profiling Array based genotyping: Standard or custom arrays (500,000 5,000,000 SNPs) Next generation sequencing: Whole transcriptome sequencing (RNAseq) 3 differential gene expression (DGEseq) Small RNA and mirna analysis (Small RNAseq) Array based expression profiling Whole transcript expression and analysis 3 expression profiling (polya transcript profiling) mirna profiling Next generation sequencing: Reduced Representation Bisulfite Sequencing (RRBS) Whole-Genome Bisulfite Sequencing (WGBS) Targeted Bisulfite Sequencing Array-based methylation analysis Bisulfite converted CpG locus genotyping (~485,000 CpG targets) 2. Supporting Partners Investigators Over the past year, the Translational Genomics Core has served approximately 75 investigators to help them achieve their research goals through access to genomics platforms and services in Table 1, providing end-to-end processing and analysis of research samples. These services are offered to Partners Healthcare researchers with institutional, grant, or commercial funding on a first-come first-serve basis. More than 11,000 samples were submitted to the TGC for processing across the various platforms offered. Of all samples types submitted, approximately 75% were DNA, and the rest were RNA, PCR product, or pre-made libraries. The majority of samples received for processing were for genotyping and library construction and NGS sequencing services. Of the almost 5000 samples
4 J. Pers. Med. 2016, 6, 10 3 of 10 processed on the Infinium platform for either genotyping or methylation this year, about half were for the Partners Biobank on Illumina s Multi-Ethnic Consortium Genotyping Array (MEGArray) chip. J. Pers. Med. A 2016, breakdown 6, 10 of the types of samples submitted for the various services can be viewed below (Figure 1). 3 of 9 (a) (b) Figure 1. Figure Percentage 1. Percentage distribution distribution of of samples submitted by (a) type of of sample submitted and and (b) processing (b) processing platform; platform; represents represents 11,155 11,155 samples from 75 investigators submitted submitted over aover one year a one year period (September 2014 September 2015). period (September 2014 September 2015). The Translational Genomics Core (TGC) pays close attention to researcher focus and technology The Translational Genomics Core (TGC) pays close attention to researcher focus and needs. To make sure researcher needs are met, the TGC is continuously discussing new methods with technology investigators needs. To andmake companies sure and researcher developingneeds new technologies are met, the in the TGC lab. The is continuously disease areas studied discussing by new methods Partners with investigators are and vast companies so many different and developing methods and new platforms technologies are requiredin tothe support lab. The this disease areas studied diverse range by Partners of work. Ainvestigators list of large research are vast areas studied so many by Partners different investigators methods that and have platforms been are required supported to support by the this TGC diverse can be range found inof Table work. 2, and A list a breakdown of large of research the manyareas librarystudied construction by Partners services utilized by investigators can be viewed in Figure 2. investigators that have been supported by the TGC can be found in Table 2, and a breakdown of the many library construction services utilized by investigators can be viewed in Figure 2. Table 2. Representative list of 10 disease areas and omics platforms for translational/clinical research projects studied by 75 Partners investigators over the past year illustrating broad range of clinical Table 2. research Representative activities supported list of by10 thedisease TGC. areas and omics platforms for translational/clinical research projects studied by 75 Partners investigators over the past year illustrating broad range of Disease Areas Platform clinical research activities supported by the TGC. Asthma Expression Analysis, Methlyation Chronic obstructive Disease pulmonary Areas disease (COPD) RNASeq Platform Rheumatoid arthritis (RA) Targeted Gene Panel Asthma Thoracic Cancer ExomeExpression Sequencing, Expression Analysis, Analysis Methlyation Chronic obstructive pulmonary HIV Integration disease (COPD) Amplicon Sequencing RNASeq Coronary artery disease (CHD) Exome Sequencing Rheumatoid arthritis (RA) Targeted Gene Panel Parkinson s/alzheimer s Disease Amplicon Sequencing Thoracic Myopathies Cancer Exome Targeted Sequencing, Gene Panel Expression Analysis Hypohidrotic HIV Integration ectodermal dysplasia 3 DGE Amplicon RNASeq Sequencing Diet effects on gut microflora Coronary artery disease (CHD) 16S Microbiome rrna Sequencing Exome Parkinson s/alzheimer s Disease Amplicon Sequencing Myopathies Targeted Gene Panel Hypohidrotic ectodermal dysplasia 3 DGE RNASeq Diet effects on gut microflora 16S Microbiome rrna Sequencing
5 Coronary artery disease (CHD) Exome Sequencing Parkinson s/alzheimer s Disease Amplicon Sequencing Myopathies Targeted Gene Panel Hypohidrotic ectodermal dysplasia 3 DGE RNASeq Diet effects on gut microflora 16S Microbiome rrna Sequencing J. Pers. Med. 2016, 6, 10 4 of 10 Figure 2. Breakdown of volume of samples submitted for various library construction methods; Figure 2. Breakdown of volume of samples submitted for various library construction methods; represents 3441 samples submitted from 20 unique investigators over a one year period (September represents 2014 September 3441 samples 2015). submitted from 20 unique investigators over a one year period (September 2014 September 2015). 3. Integration of Core Operations through LIMS At the foundation of Partners Personalized Medicine is the operations driven by information technology (IT). The same (Laboratory Information Management) LIM systems, which is a software based management system to support any and all laboratory operations, are utilized across departments, allowing for end to end project management. These LIMS and the IT infrastructure are described in depth in The Information Technology Infrastructure for the Translational Genomics Core and the Partners Biobank at Partners Personalized Medicine in this issue. For research projects two main LIMS are utilized, STARLIMS and GIGPAD. STARLIMS is used to manage sample processing workflows in the lab. It also serves as a sample inventory tracking system capturing the physical location of samples within the lab. If investigators have their own external samples to submit for processing, they can place orders through GIGPAD ( Investigators can upload their sample information directly into the portal and order sample processing and analysis services. The genomic instruments and analysis software in the lab can feed output data directly into this LIMS allowing data to be viewed and downloaded securely by investigators through this platform. GIGPAD is also used to bill for rendered services. GIGPAD interfaces with STARLIMS, so updates in the processing workflow made by the lab in STARLIMS are automatically updated in GIGPAD for the investigator to view. The same IT team supports all departments at Partners Personalized Medicine. One of the roles of the IT team is to manage both GIGPAD and STARLIMS. Having the same team manage both LIMS helps maintain the integrity of data being shared between the two platforms. This unification of support also helps streamline troubleshooting for the ongoing clinical and research processes within Partners Personalized Medicine as well as and external investigators. Members from the lab and IT teams meet weekly to provide updates, feedback on functionality, and bring any issues to discuss. This ongoing dialogue helps keep operations running smoothly. The Partners Biobank houses banked samples (plasma, serum, and DNA) collected from research-consented patients from Partners Healthcare hospitals, which are available to Partners researchers or distribution to Partners HealthCare investigators with appropriate approval from the Partners Institutional Review board (IRB) (described in The Partners HealthCare Biobank at Partners Personalized Medicine article in this issue). Whether samples were obtained through the Partners Biobank or were from an investigator s private cohort, the Biobank can provide samples processing services, such as extraction, quantitation, and plating. The Biobank is proficient at preparing samples to meet required specifications for various downstream processing platforms offered by the Translational Genomics Core. Many investigators take advantage of this end to end workflow, and will submit samples to the Biobank for preparation to be processed by the TGC. The same lab manager
6 J. Pers. Med. 2016, 6, 10 5 of 10 and project manager oversee projects in both the Biobank and the TGC. This integration between departments lends to a smooth transition of projects from sample processing to the genomic analysis stage. The same LIMs and PPM staff track projects from the time the initial request is made, through sample preparation in the Biobank and genomic analysis in the Translational Genomics Core, until the return of data. If analysis is requested, the project will be tracked through the Bioinformatics team until analyzed data is released to the investigator, through a secure File Transfer Protocol (FTP) on the Partners Healthcare network for Partners Healthcare researchers, or through an external hard drive. 4. Providing Bioinformatics (BioFx) Support on Core Areas of Expertise Processing and analysis of sequencing and genotyping data requires complex bioinformatics tools and pipelines to provide reliable results that can be accurately interpreted in the context of the study. Working closely with the bioinformatics team at PPM, the TGC is able to provide bioinformatics services and expertise to translational and clinical researchers. Given the enormous time and effort required to build bioinformatics tools and gain analytical expertise, the TGC has aligned its bioinformatics development goals and services with that of different components of the center. For example, leveraging the LMM s expertise in sequencing analysis, clinical grade variant calling and comprehensive annotation for whole genome/exome and targeted sequencing projects is offered to researchers through the core. For researchers with limited expertise in the development of bioinformatics tools, this has led to confidence in analytical methods and efficient utilization of genomic sequencing data. Although many bioinformatics requests have been encountered, we have limited our services to focus on those that are more common to investigators at Partners Healthcare In addition, we have focused our in-house bioinformatics development efforts to support the technology platforms most utilized by the majority of translational and clinical investigators including: (1) variant calling and annotation for targeted sequencing panels, exomes, and whole genomes, (2) differential expression through RNAseq, (3) allele calling on genotyping arrays, and (4) differential expression on microarray platforms. To this end, our strategy is to partner with research groups with a vested interest in a given assay or platform. This allows both groups to share resources and expertise in building an end-to-end solution for novel techniques and platforms. These partnerships often arise with repeat external users of center resources who want to develop a new experiment protocol that requires special equipment and expertise for the wet bench portion and is an area of interest in the research community. We are able to contribute to this partnership by providing the development of the wet bench process and access to the specialized equipment. One such example, is the development of a robust whole transcriptome sequencing analysis pipeline through a collaboration between the TGC and a Partners investigator with training and expertise in bioinformatics analysis. Through this interaction, the core provided access and technical expertise in library construction from RNA samples and sequencing of libraries on the NGS platform. The investigator, in turn, developed an analysis pipeline using the Tuxedo Suite tools (Broad Institute) that includes alignment of reads to a reference genome, measures transcript abundance, and performs differential expression analysis and visualization. Through several rounds of validation experiments and analyses, we optimized our wet-bench and analysis workflow to include quality control (QC) measures such as synthetic oligo spike-ins and fastq QC analysis which help inform on the quality of samples and their processing in the lab, and then set pass thresholds to avoid the use of inaccurate data. A copy of this pipeline was transferred to PPM servers and is maintained and offered to other researchers through our bioinformatics group, providing a laboratory processing workflow with QC checks at multiple steps and access to a validated transcriptome sequencing analysis pipelines. Similar initiatives to co-develop bioinformatics tools for platforms and techniques that support personalized medicine research are underway, including metagenomics 16S rrna gene microbial profiling and small RNA/miRNA profiling and discovery from serum and plasma samples.
7 J. Pers. Med. 2016, 6, 10 6 of Supporting Biobank Genotyping Initiative A major goal of the Partners Personalized Medicine Biobank is the genotyping of Biobank samples using the MEGArray chip on Illumina s Infinium platform. To complete objectives of this genotyping initiative, the TGC is required to meet a throughput of samples per month to be run on this chip. Genotyping hardware and infrastructure already existed in the TGC for genotyping small projects with a low sample number. The increase in volume and throughput required by the Biobank genotyping project meant that we required additional resources, equipment, modification to existing equipment and a sample handling and wet lab processing LIMS. The Illumina Laboratory Information Management System (LIMS) is a proprietary system used to connect sample ID s in sample plates with all reagent barcodes and to collect technician and instrument ID s and timestamps for complete documentation of the wet lab process from start to finish. Manual collection of this information with a hand-held barcode scanner was possible but slowed the process dramatically and was liable to introduce error. This made it necessary to integrate a robotic manipulator arm (RoMa) to the Tecan EVo liquid handling system (Tecan) for all the post PCR sample handling and manipulation. The RoMa integrates two barcode scanners necessary for capturing sample, reagent, and chip identity throughout the process. The capacity of our scanner was doubled by the introduction of a second scanner with a twister arm to allow walkaway continuous scanning of chips into the LIMS. These improvements allow processing of 384 samples per week by one technician, with throughput doubling with two technicians on a staggered schedule. On completion of scanning the chips, data (.idat) is collected in the Illumina LIMS and a previously generated cluster file (.egt file) from Illumina GenomeStudio software is applied to it along with a SNP manifest file specific for the version of the chip used (.bpm). The cluster file assigns an allele call to each sample per SNP based upon the intensity of the fluorescent dye found at a position on the chip during scanning. The ID of the SNP is based on the position of the call on the chip using the manifest (.bpm) as a reference. This produces the genotype call files (GTC) that are used to create smaller binary PED files. PEDs from each sample are merged together and then curated to remove failing SNPs, which are SNPs having a call rate <0.95% across all samples, or SNPs that are not mapped to the genome. At this point the Biobank IDs are re-associated with the data as well as a gender designation. The genotyped gender is compared to the assigned gender as a QC check. Quality control plots are generated to allow identification of failing samples; those that are lower than 97% success across all targeted positions or show an intensity signal lower than the intensity of the background noise. An overview of the genotyping workflow is shown in Figure 3 which highlights the sample wet bench processing, integration of the Illumina LIMS and our homebrew GIGPAD LIMS, as well as initial bioinformatics analyses that offer basic genotyping calls and quality control check based on SNP intensities and/or call rates. Additional information on the bioinformatics workflow is described in Tsai et al. [1]. For the TGC, the utility of this integration between the LIMS and Biofx processes is in its ability to perform cluster file generation using multiple batches run on the same array, thereby enhancing allele calling, particularly to rare SNPs, and lower the SNP failure rate. In addition, the generation of sample QC plots for multiple batches simultaneously, which occurs at the snpqc_info step in Figure 3 and is described in Tsai et al. [1], allows the laboratory team to more easily identify failed samples and batch specific trends relating to sample quality or chip lot quality due to manufacturer error.
8 J. Pers. Med. 2016, 6, 10 6 of 9 J. Pers. Med. 2016, 6, 10 7 of 10 samples per week by one technician, with throughput doubling with two technicians on a staggered schedule. Figure Figure 3. Summary 3. of of Biobank genotyping workflow (taken from fromtsai Tsai et al. et[1]). al. [1]). 6. Staffing On and completion Operations of scanning Challenges the chips, at an Academic data (.idat) Core is collected in the Illumina LIMS and a previously generated cluster file (.egt file) from Illumina GenomeStudio software is applied to it The along field with of a SNP genomics manifest is file a rapidly specific changing for the version fieldof with the chip theused introduction (.bpm). The ofcluster new techniques file assigns and technologies an allele on call ato regular each sample basis allowing per SNP genomics based upon tothe be intensity carried out of the faster fluorescent and cheaper dye found than previous at a iterations. position Generally, on the chip core during labs are scanning. forcedthe by budgetary ID of the SNP constraints is based on to the recruit position inexperienced of the call on graduates the and train chip them using on the the manifest job. Once (.bpm) hired, as a retention reference. then This becomes produces an the issue, genotype as technical call files (GTC) staff with that growing are experience used to have create the smaller option binary of moving PED files. to higher PEDs from paying each similar sample roles are in merged surrounding together industry and then labs. curated to remove failing SNPs, which are SNPs having a call rate <0.95% across all samples, or SNPs Balancing staffing levels with demand is tricky as volume of incoming work is highly variable with that are not mapped to the genome. At this point the Biobank IDs are re-associated with the data as little long term planning and communication between the researcher and the core lab. High turnaround well as a gender designation. The genotyped gender is compared to the assigned gender as a QC of staff check. and Quality constantly control changing plots are services generated requires to allow close identification management of failing to ensure samples; the laboratory those that remains are in GLP lower (Good than Laboratory 97% success Practice) across all compliance targeted positions [2]. This or show is an an important intensity signal quality lower measure than the for our customers, intensity especially of the background those who noise. mayan beoverview workingof towards the genotyping FDA approval workflow as is shown part ofin their Figure research. 3 Whilewhich success highlights of a core the facility sample can wet bebench measured processing, by financial integration success, of the inillumina an academic LIMS environment and our another homebrew measure GIGPAD of success LIMS, isas through well as initial publication, bioinformatics either analyses directly that byoffer thebasic coregenotyping facility orcalls through recognition and quality of the control corecheck within based publications SNP intensities by our and/or customers, call rates. or the Additional numberinformation of grants that on the a core bioinformatics workflow is described in Tsai et al. [1]. For the TGC, the utility of this integration facility supports. Following below is a more detailed discussion on how we deal with each of these between the LIMS and Biofx processes is in its ability to perform cluster file generation using challenges with the aim of moving forward within the world of genomics and providing a high quality multiple batches run on the same array, thereby enhancing allele calling, particularly to rare SNPs, cost effective service to our customers. Our facility s pool of customers includes a large number of translational and clinical researchers whose needs are wide ranging in the field of genomics. As well as carrying out internal review of new technologies and techniques ourselves, we are often approached by customers with novel methods or instrumentation needs, looking for a service lab to support them. The effort required to bring on a
9 J. Pers. Med. 2016, 6, 10 8 of 10 new technique is high, so detailed consideration may be required. Technique development demands resources be redirected from income generating production work for a period of time while manual testing, optimization, automation of high volume tests, quality control, procedure documentation and analysis of data is carried out. Other points to consider include availability of staff to carry out development, acceptable timeline for development of the test for the requesting customer, the level of demand for a new test, whether proposed cost to customer will be acceptable and if there are cheaper alternative services available to researchers in other institutional facilities. Introduction of new instrumentation is also high risk, due to the potentially high purchase price. Caution may be warranted to allow time to monitor how well that technology performs in other facilities. If possible, alternative systems should be reviewed to ensure the correct decision is made. Proof of concept testing is required, either with the vendor of the instrumentation or with a known collaborator/colleague. One way for a core facility to mitigate the burden of the instrument purchase price is to partner with a PI or consortium who hold a grant to cover the purchase of a high value instrument and offer to house and operate the instrumentation for a reduced fee, while allowing other customers samples to be run on that instrument in downtime. At the opposite end of the spectrum, the decision to stop offering a technique or use of piece of instrumentation, sometimes when there is still low demand for it, is made to offset unnecessary expenditure of resources, labor, and costs associated with maintaining that platform or process. The largest three items in our annual budget are supplies, maintenance contracts and salaries. Supply costs can be negotiated with vendors for best prices, but they also vary with volume. The costs of supplies are covered directly by charges to our customers. Maintenance contracts are more static but can also be negotiated with cheaper contracts arising by locking down coverage over multiple years. Salaries are defined on an institutional level depending on qualifications and experience to ensure parity over staff at a particular level within the institution. This makes it difficult to compete in salary for outstanding new hires. Our current staff includes a Director, a Laboratory Manager, a Project Manager and five junior staff. All junior staff are hired with no experience and go through lengthy training on the job, so in order to make this worthwhile we ask for two years as a minimum commitment. There can be hesitancy in accepting a position within a core facility as there is an incorrect perception of the scope of work carried out by junior staff in a fee-for service lab. Prior to working in our lab, the perception is that it would be mundane, boring and repetitive, but in reality all staff are cross trained on all our services through genotyping, next-generation sequencing and gene expression and so are kept busy and challenged. In addition, during the interviewing process we highlight the broad range of cutting edge clinical and translational research that we are involved in to off-set the negative perception of working in a core laboratory. This has attracted junior technicians who are exploring career paths or considering graduate or professional education, who felt the scope and breadth of work done at the TGC will be helpful for their respective goals. However, because of the cutting edge nature of the techniques we use, we do find we have an issue with staff retention. The experience gained by our junior staff is sought after by many of the surrounding private institutions and we find that we lose staff after two years due to more competitive salaries offered by those institutions. We aim to attract high quality junior staff and prevent loss of exceptional staff by promotion based on ability, and by giving ownership of interesting and varied projects. This allows them to gain skills in project design, customer service, time management, research and development, as well as management of more junior staff. We also allow time for the staff to take advantage of the professional development training classes offered by our institution. Another thing that we promote is to have our senior management engage with our junior staff and provide recognition for good quality work. We hope that these benefits provided to our staff are giving them experience and education that will hold them in good stead to be successful as they move into their next role, whether that is in our lab or out of it. The aim of Good Laboratory Practice (GLP) of the food and drug administration (FDA) is to provide consistently high quality technical work and data over time and over different technical
10 J. Pers. Med. 2016, 6, 10 9 of 10 staff. There are many areas covered by GLP including equipment management, training records, standard operating procedures, materials quality, data management and quality processes as detailed in 21 CFR 58 [2]. The challenge for any lab is the vast amount of organization and documentation to establish and maintain GLP. It requires buy in and input from all members of staff, so they must all receive the necessary GLP training. It is important that all staff see how following GLP regulations can improve the quality of data we generate for our customers. The biggest challenge we face is the amount of time we can dedicate to the implementation of GLP. Due to our small staff size and the large range of available services, we have limited resources to devote to this as the bulk of our effort is put towards the production of data to our customers. We have initiated GLP compliance in our lab for certain core services such as next generation sequencing and Infinium array genotyping, and are in the process of working through each of our services and understand that this process may take several years. A published study into University GLP compliance showed that the average years to compliance was four years, ranging from 10 months to seven years [3]. This study identifies several challenges to GLP compliance related to the management of GLP programs, quality assurance training and implementation for technicians, and inadequate documentation of all processes within a GLP workflow [3]. One of the measures of success and progress of an academic lab is the production of journal articles. This measure is also true in the case of a Core facility; we monitor publications from our institution on a regular basis to aid in measuring the usage of our facility. In the past, it has been common to find that our facility has not been acknowledged, so we remind our customers on data delivery that an acknowledgement is very much appreciated. This process is becoming more common amongst Core facilities and has been recognized as an issue at several national conferences (NERG and ABRF) and highlighted in an editorial in BioTechniques [4]. However, an alternative measure of success has been to identify the number of grants or grant dollars supported by our laboratories. Our figures for the academic year show that we have supported over 60 grants totaling over $165 million. 7. Conclusion Our operational experiences highlight common considerations encountered at academic research core such as evolving with the advancement of genomic and transcriptomic technologies and assays, and responding to the diverse needs of investigators at an academic and clinical research institution. As highlighted in this article, several key components have allowed us to thrive in this environment, including: (1) building a core set of genomics and bioinformatics services developed through partnerships and collaborations with a clinical lab or researchers within the institution, (2) having a broad and flexible LIMS system that can handle the tracking and management of variety of sample and library types and can easily adapt and integrate new technologies, and (3) cross training of junior staff across multiple platforms and techniques to engage current staff and limit the impact of staff turnover on productivity. As many academic research cores are fee-for-service with limited funds for development and validation of new workflows and implementing regulatory compliance for current ones, the decision to pursue these initiatives must be weighed against the impact it will have on current production and productivity, and the perceived added benefit of that endeavor. These will not always be clear to research cores, which emphasizes the importance of building relationships with clinical and research labs within the institution. The integrative nature of all the components of Partners Personalized Medicine situates the Translational Genomics Core in a unique position among academic core laboratories. This is reinforced by a seamless integration of IT infrastructure and bioinformatics pipelines to allow easy communication, management and tracking of samples and data for both external researchers and PPM research initiatives. The Biobank genotyping initiative, which involves Partners Biobank samples, LIMS infrastructure supporting both the Biobank and the TGC, as well bioinformatics pipelines for quality check steps and analysis, highlights the seamless integration of different components for an efficient
11 J. Pers. Med. 2016, 6, of 10 workflow at the TGC. As such, the TGC is able to utilize its interactions within the center to offer end-to-end genomic analyses services to academic and clinical researchers, while ensuring that onboarding of novel platforms and assays can rapidly occur to help meet the goals of the center. Acknowledgments: The Translational Genomics Core at Partners Personalized Medicine is a not-for-profit fee-for-service core facility at Partners HealthCare. Author Contributions: Ashley Blau drafted sections 1 4 including tables and graphs, Alison Brown drafted sections 6 7, and all authors contributed to revision and editing of the manuscript. Conflicts of Interest: The authors are paid employees of Partners Personalized Medicine, otherwise the authors declare no conflict of interest. References 1. Tsai, E.A.; Peirce, J.; Embree, K.; Brown, A.; Boutin, N.T.; Amr, S.S.; Weiss, S.T.; Lebo, M.S. Bioinformatics Infrastructure for the Partners Biobank Initiative. In Poster session presented at the meeting of the American Society of Human Genetics, Baltimore, MD, USA, 6 10 October CFR Code of Federal Regulations Title 21 of the FDA (updated August 2015). Good Laboratory Practice for Nonclinical Laboratory. Available online: scripts/cdrh/cfdocs/ cfcfr/cfrsearch.cfm?cfrpart=58 (accessed on 20 October 2015). 3. Hancock, S. Meeting the Challenges of Implementing Good Laboratory Practices compliance in a university setting. Qual. Assur. J. 2002, 6, [CrossRef] 4. Letter from the Editor. A lack of attribution. BioTechniques, 2014, 57, p Available online: (accessed on 13 October 2015) by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (
Genomic solutions for complex disease
 Genomic solutions for complex disease Power your with our genomic solutions Access a breadth of applications. Gain a depth of insights. To enhance their understanding of complex disease, researchers are
Genomic solutions for complex disease Power your with our genomic solutions Access a breadth of applications. Gain a depth of insights. To enhance their understanding of complex disease, researchers are
G E N OM I C S S E RV I C ES
 GENOMICS SERVICES ABOUT T H E N E W YOR K G E NOM E C E N T E R NYGC is an independent non-profit implementing advanced genomic research to improve diagnosis and treatment of serious diseases. Through
GENOMICS SERVICES ABOUT T H E N E W YOR K G E NOM E C E N T E R NYGC is an independent non-profit implementing advanced genomic research to improve diagnosis and treatment of serious diseases. Through
Illumina s Suite of Targeted Resequencing Solutions
 Illumina s Suite of Targeted Resequencing Solutions Colin Baron Sr. Product Manager Sequencing Applications 2011 Illumina, Inc. All rights reserved. Illumina, illuminadx, Solexa, Making Sense Out of Life,
Illumina s Suite of Targeted Resequencing Solutions Colin Baron Sr. Product Manager Sequencing Applications 2011 Illumina, Inc. All rights reserved. Illumina, illuminadx, Solexa, Making Sense Out of Life,
Pioneering Clinical Omics
 Pioneering Clinical Omics Clinical Genomics Strand NGS An analysis tool for data generated by cutting-edge Next Generation Sequencing(NGS) instruments. Strand NGS enables read alignment and analysis of
Pioneering Clinical Omics Clinical Genomics Strand NGS An analysis tool for data generated by cutting-edge Next Generation Sequencing(NGS) instruments. Strand NGS enables read alignment and analysis of
resequencing storage SNP ncrna metagenomics private trio de novo exome ncrna RNA DNA bioinformatics RNA-seq comparative genomics
 RNA Sequencing T TM variation genetics validation SNP ncrna metagenomics private trio de novo exome mendelian ChIP-seq RNA DNA bioinformatics custom target high-throughput resequencing storage ncrna comparative
RNA Sequencing T TM variation genetics validation SNP ncrna metagenomics private trio de novo exome mendelian ChIP-seq RNA DNA bioinformatics custom target high-throughput resequencing storage ncrna comparative
GENOMICS WORKFLOW SOLUTIONS THAT GO WHERE THE SCIENCE LEADS. Genomics Solutions Portfolio
 GENOMICS WORKFLOW SOLUTIONS THAT GO WHERE THE SCIENCE LEADS Genomics Solutions Portfolio WORKFLOW SOLUTIONS FROM EXTRACTION TO ANALYSIS Application-based answers for every step of your workflow Scientists
GENOMICS WORKFLOW SOLUTIONS THAT GO WHERE THE SCIENCE LEADS Genomics Solutions Portfolio WORKFLOW SOLUTIONS FROM EXTRACTION TO ANALYSIS Application-based answers for every step of your workflow Scientists
The first and only fully-integrated microarray instrument for hands-free array processing
 The first and only fully-integrated microarray instrument for hands-free array processing GeneTitan Instrument Transform your lab with a GeneTitan Instrument and experience the unparalleled power of streamlining
The first and only fully-integrated microarray instrument for hands-free array processing GeneTitan Instrument Transform your lab with a GeneTitan Instrument and experience the unparalleled power of streamlining
GENOMICS WORKFLOW SOLUTIONS THAT GO WHERE THE SCIENCE LEADS. Genomics Solutions Portfolio
 GENOMICS WORKFLOW SOLUTIONS THAT GO WHERE THE SCIENCE LEADS Genomics Solutions Portfolio WORKFLOW SOLUTIONS FROM EXTRACTION TO ANALYSIS Application-based answers for every step of your workflow Scientists
GENOMICS WORKFLOW SOLUTIONS THAT GO WHERE THE SCIENCE LEADS Genomics Solutions Portfolio WORKFLOW SOLUTIONS FROM EXTRACTION TO ANALYSIS Application-based answers for every step of your workflow Scientists
WELCOME. Norma J. Nowak, PhD Executive Director, NY State Center of Excellence in Bioinformatics and Life Sciences (CBLS)
 WELCOME Norma J. Nowak, PhD Executive Director, NY State Center of Excellence in Bioinformatics and Life Sciences (CBLS) Director, UB Genomics and Bioinformatics Core (GBC) o o o o o o o o o o o o Grow
WELCOME Norma J. Nowak, PhD Executive Director, NY State Center of Excellence in Bioinformatics and Life Sciences (CBLS) Director, UB Genomics and Bioinformatics Core (GBC) o o o o o o o o o o o o Grow
DNA. bioinformatics. genomics. personalized. variation NGS. trio. custom. assembly gene. tumor-normal. de novo. structural variation indel.
 DNA Sequencing T TM variation DNA amplicon mendelian trio genomics NGS bioinformatics tumor-normal custom SNP resequencing target validation de novo prediction personalized comparative genomics exome private
DNA Sequencing T TM variation DNA amplicon mendelian trio genomics NGS bioinformatics tumor-normal custom SNP resequencing target validation de novo prediction personalized comparative genomics exome private
Bioinformatics Advice on Experimental Design
 Bioinformatics Advice on Experimental Design Where do I start? Please refer to the following guide to better plan your experiments for good statistical analysis, best suited for your research needs. Statistics
Bioinformatics Advice on Experimental Design Where do I start? Please refer to the following guide to better plan your experiments for good statistical analysis, best suited for your research needs. Statistics
DNA. bioinformatics. epigenetics methylation structural variation. custom. assembly. gene. tumor-normal. mendelian. BS-seq. prediction.
 Epigenomics T TM activation SNP target ncrna validation metagenomics genetics private RRBS-seq de novo trio RIP-seq exome mendelian comparative genomics DNA NGS ChIP-seq bioinformatics assembly tumor-normal
Epigenomics T TM activation SNP target ncrna validation metagenomics genetics private RRBS-seq de novo trio RIP-seq exome mendelian comparative genomics DNA NGS ChIP-seq bioinformatics assembly tumor-normal
Quality Control of Next Generation Sequence Data
 Quality Control of Next Generation Sequence Data January 17, 2018 Kane Tse, Assistant Bioinformatics Coordinator Canada s Michael Smith Genome Sciences Centre BC Cancer Agency Canada s Michael Smith Genome
Quality Control of Next Generation Sequence Data January 17, 2018 Kane Tse, Assistant Bioinformatics Coordinator Canada s Michael Smith Genome Sciences Centre BC Cancer Agency Canada s Michael Smith Genome
The Expanded Illumina Sequencing Portfolio New Sample Prep Solutions and Workflow
 The Expanded Illumina Sequencing Portfolio New Sample Prep Solutions and Workflow Marcus Hausch, Ph.D. 2010 Illumina, Inc. All rights reserved. Illumina, illuminadx, Solexa, Making Sense Out of Life, Oligator,
The Expanded Illumina Sequencing Portfolio New Sample Prep Solutions and Workflow Marcus Hausch, Ph.D. 2010 Illumina, Inc. All rights reserved. Illumina, illuminadx, Solexa, Making Sense Out of Life, Oligator,
ILLUMINA SEQUENCING SYSTEMS
 ILLUMINA SEQUENCING SYSTEMS PROVEN QUALITY. TRUSTED SOLUTIONS. Every day, researchers are using Illumina next-generation sequencing (NGS) systems to better understand human health and disease, as well
ILLUMINA SEQUENCING SYSTEMS PROVEN QUALITY. TRUSTED SOLUTIONS. Every day, researchers are using Illumina next-generation sequencing (NGS) systems to better understand human health and disease, as well
Redefine what s possible with the Axiom Genotyping Solution
 Redefine what s possible with the Axiom Genotyping Solution From discovery to translation on a single platform The Axiom Genotyping Solution enables enhanced genotyping studies to accelerate your research
Redefine what s possible with the Axiom Genotyping Solution From discovery to translation on a single platform The Axiom Genotyping Solution enables enhanced genotyping studies to accelerate your research
Introducing QIAseq. Accelerate your NGS performance through Sample to Insight solutions. Sample to Insight
 Introducing QIAseq Accelerate your NGS performance through Sample to Insight solutions Sample to Insight From Sample to Insight let QIAGEN enhance your NGS-based research High-throughput next-generation
Introducing QIAseq Accelerate your NGS performance through Sample to Insight solutions Sample to Insight From Sample to Insight let QIAGEN enhance your NGS-based research High-throughput next-generation
Capabilities & Services
 Capabilities & Services Accelerating Research & Development Table of Contents Introduction to DHMRI 3 Services and Capabilites: Genomics 4 Proteomics & Protein Characterization 5 Metabolomics 6 In Vitro
Capabilities & Services Accelerating Research & Development Table of Contents Introduction to DHMRI 3 Services and Capabilites: Genomics 4 Proteomics & Protein Characterization 5 Metabolomics 6 In Vitro
GenoLogics LIMS Helps EdgeBio Build Services Business
 LIMS Supports ResEarch & clinical NGS services LAB GenoLogics LIMS Helps EdgeBio Build Services Business LIMS Supports Research & Clinical NGS Services LAB GenoLogics LIMS Helps EdgeBio Build Services
LIMS Supports ResEarch & clinical NGS services LAB GenoLogics LIMS Helps EdgeBio Build Services Business LIMS Supports Research & Clinical NGS Services LAB GenoLogics LIMS Helps EdgeBio Build Services
Introducing a Highly Integrated Approach to Translational Research: Biomarker Data Management, Data Integration, and Collaboration
 Introducing a Highly Integrated Approach to Translational Research: Biomarker Data Management, Data Integration, and Collaboration 1 2 Translational Informatics Overview Precision s suite of services is
Introducing a Highly Integrated Approach to Translational Research: Biomarker Data Management, Data Integration, and Collaboration 1 2 Translational Informatics Overview Precision s suite of services is
solid S Y S T E M s e q u e n c i n g See the Difference Discover the Quality Genome
 solid S Y S T E M s e q u e n c i n g See the Difference Discover the Quality Genome See the Difference With a commitment to your peace of mind, Life Technologies provides a portfolio of robust and scalable
solid S Y S T E M s e q u e n c i n g See the Difference Discover the Quality Genome See the Difference With a commitment to your peace of mind, Life Technologies provides a portfolio of robust and scalable
Surely Better Target Enrichment from Sample to Sequencer
 sureselect TARGET ENRICHMENT solutions Surely Better Target Enrichment from Sample to Sequencer Agilent s market leading SureSelect platform provides a complete portfolio of catalog to custom products,
sureselect TARGET ENRICHMENT solutions Surely Better Target Enrichment from Sample to Sequencer Agilent s market leading SureSelect platform provides a complete portfolio of catalog to custom products,
Global Screening Array (GSA)
 Technical overview - Infinium Global Screening Array (GSA) with optional Multi-disease drop in (MD) The Infinium Global Screening Array (GSA) combines a highly optimized, universal genome-wide backbone,
Technical overview - Infinium Global Screening Array (GSA) with optional Multi-disease drop in (MD) The Infinium Global Screening Array (GSA) combines a highly optimized, universal genome-wide backbone,
Reads to Discovery. Visualize Annotate Discover. Small DNA-Seq ChIP-Seq Methyl-Seq. MeDIP-Seq. RNA-Seq. RNA-Seq.
 Reads to Discovery RNA-Seq Small DNA-Seq ChIP-Seq Methyl-Seq RNA-Seq MeDIP-Seq www.strand-ngs.com Analyze Visualize Annotate Discover Data Import Alignment Vendor Platforms: Illumina Ion Torrent Roche
Reads to Discovery RNA-Seq Small DNA-Seq ChIP-Seq Methyl-Seq RNA-Seq MeDIP-Seq www.strand-ngs.com Analyze Visualize Annotate Discover Data Import Alignment Vendor Platforms: Illumina Ion Torrent Roche
2017 HTS-CSRS COMMUNITY PUBLIC WORKSHOP
 2017 HTS-CSRS COMMUNITY PUBLIC WORKSHOP GenomeNext Overview Olympus Platform The Olympus Platform provides a continuous workflow and data management solution from the sequencing instrument through analysis,
2017 HTS-CSRS COMMUNITY PUBLIC WORKSHOP GenomeNext Overview Olympus Platform The Olympus Platform provides a continuous workflow and data management solution from the sequencing instrument through analysis,
The MiniSeq System. Explore the possibilities. Discover demonstrated NGS workflows for molecular biology applications.
 The MiniSeq System. Explore the possibilities. Discover demonstrated NGS workflows for molecular biology applications. Let your work flow with Illumina NGS. The MiniSeq System delivers powerful and cost-effective
The MiniSeq System. Explore the possibilities. Discover demonstrated NGS workflows for molecular biology applications. Let your work flow with Illumina NGS. The MiniSeq System delivers powerful and cost-effective
Welcome to the NGS webinar series
 Welcome to the NGS webinar series Webinar 1 NGS: Introduction to technology, and applications NGS Technology Webinar 2 Targeted NGS for Cancer Research NGS in cancer Webinar 3 NGS: Data analysis for genetic
Welcome to the NGS webinar series Webinar 1 NGS: Introduction to technology, and applications NGS Technology Webinar 2 Targeted NGS for Cancer Research NGS in cancer Webinar 3 NGS: Data analysis for genetic
Intuitive Lab Management from Bench to Boardroom
 Intuitive Lab Management from Bench to Boardroom Clarity LIMS features are bundled into editions. Flags indicate the features contained within each edition. INTRODUCTION As a genomics or mass spec lab,
Intuitive Lab Management from Bench to Boardroom Clarity LIMS features are bundled into editions. Flags indicate the features contained within each edition. INTRODUCTION As a genomics or mass spec lab,
Overcome limitations with RNA-Seq
 Buyer s Guide Simple, customized RNA-Seq workflows Evaluating options for next-generation RNA sequencing Overcome limitations with RNA-Seq Next-generation sequencing (NGS) has revolutionized the study
Buyer s Guide Simple, customized RNA-Seq workflows Evaluating options for next-generation RNA sequencing Overcome limitations with RNA-Seq Next-generation sequencing (NGS) has revolutionized the study
Development of quantitative targeted RNA-seq methodology for use in differential gene expression
 Development of quantitative targeted RNA-seq methodology for use in differential gene expression Dr. Jens Winter, Market Development Group Biological Biological Research Content EMEA QIAGEN Universal Workflows
Development of quantitative targeted RNA-seq methodology for use in differential gene expression Dr. Jens Winter, Market Development Group Biological Biological Research Content EMEA QIAGEN Universal Workflows
Gene Regulation Solutions. Microarrays and Next-Generation Sequencing
 Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
Gene Regulation Solutions Microarrays and Next-Generation Sequencing Gene Regulation Solutions The Microarrays Advantage Microarrays Lead the Industry in: Comprehensive Content SurePrint G3 Human Gene
CAPTURE-BASED APPROACH FOR COMPREHENSIVE DETECTION OF IMPORTANT ALTERATIONS
 CAPTURE-BASE APPROACH FOR COMPREHENSIVE ETECTION OF IMPORTANT ALTERATIONS SEQUENCE MUTATIONS MICROSATELLITE INSTABILITY AMPLIFICATIONS GENOMIC REARRANGEMENTS For Research Use Only. Not for iagnostic Purposes.
CAPTURE-BASE APPROACH FOR COMPREHENSIVE ETECTION OF IMPORTANT ALTERATIONS SEQUENCE MUTATIONS MICROSATELLITE INSTABILITY AMPLIFICATIONS GENOMIC REARRANGEMENTS For Research Use Only. Not for iagnostic Purposes.
Gene expression microarrays and assays. Because your results can t wait
 Gene expression microarrays and assays Because your results can t wait A simple path from data to decision-making The power of expression microarrays Transcriptome-wide analysis can be complex. Matching
Gene expression microarrays and assays Because your results can t wait A simple path from data to decision-making The power of expression microarrays Transcriptome-wide analysis can be complex. Matching
Magnis NGS Prep System. Your lab s automated library preparation solution for next-generation sequencing
 Magnis NGS Prep System Your lab s automated library preparation solution for next-generation sequencing Magnis NGS Prep System Complete system for NGS preparation One of the challenges with implementing
Magnis NGS Prep System Your lab s automated library preparation solution for next-generation sequencing Magnis NGS Prep System Complete system for NGS preparation One of the challenges with implementing
Illumina Custom Capabilities: Amplicons, Enrichment and iselect
 Illumina Custom Capabilities: Amplicons, Enrichment and iselect Daniel Peiffer, PhD Sr. Product Manager DNA Sequencing Applications Tuesday, March 22, 2011 2010 Illumina, Inc. All rights reserved. Illumina,
Illumina Custom Capabilities: Amplicons, Enrichment and iselect Daniel Peiffer, PhD Sr. Product Manager DNA Sequencing Applications Tuesday, March 22, 2011 2010 Illumina, Inc. All rights reserved. Illumina,
Territory Account Manager (MD, NC or TX based)
 Territory Account Manager (MD, NC or TX based) Looking for bilingual Chinese-English young professionals with passion in selling genomics services. The Territory Account Manager's role is to ensure achievement
Territory Account Manager (MD, NC or TX based) Looking for bilingual Chinese-English young professionals with passion in selling genomics services. The Territory Account Manager's role is to ensure achievement
Applications of Next Generation Sequencing in Metagenomics Studies
 Applications of Next Generation Sequencing in Metagenomics Studies Francesca Rizzo, PhD Genomix4life Laboratory of Molecular Medicine and Genomics Department of Medicine and Surgery University of Salerno
Applications of Next Generation Sequencing in Metagenomics Studies Francesca Rizzo, PhD Genomix4life Laboratory of Molecular Medicine and Genomics Department of Medicine and Surgery University of Salerno
Next Generation Oncology Sequencing in your Laboratory. Built by pioneers in cancer genomics and liquid biopsy approaches PROGENEUS
 PROGENEUS Next Generation Oncology Sequencing in your Laboratory Built by pioneers in cancer genomics and liquid biopsy approaches For Research Use Only. Not for iagnostic Purposes. personalgenome.com/progeneus
PROGENEUS Next Generation Oncology Sequencing in your Laboratory Built by pioneers in cancer genomics and liquid biopsy approaches For Research Use Only. Not for iagnostic Purposes. personalgenome.com/progeneus
 www.illumina.com/hiseq www.illumina.com FOR RESEARCH USE ONLY 2012 2014 Illumina, Inc. All rights reserved. Illumina, BaseSpace, cbot, CSPro, Genetic Energy, HiSeq, Nextera, TruSeq, the pumpkin orange
www.illumina.com/hiseq www.illumina.com FOR RESEARCH USE ONLY 2012 2014 Illumina, Inc. All rights reserved. Illumina, BaseSpace, cbot, CSPro, Genetic Energy, HiSeq, Nextera, TruSeq, the pumpkin orange
Illumina ArrayLab Consulting Service. A comprehensive solution to meet your ultra high-throughput needs
 Illumina ArrayLab Consulting Service A comprehensive solution to meet your ultra high-throughput needs High-throughput laboratory setup Capacity planning Hardware and data management Lab management, staffing,
Illumina ArrayLab Consulting Service A comprehensive solution to meet your ultra high-throughput needs High-throughput laboratory setup Capacity planning Hardware and data management Lab management, staffing,
Satellite Education Workshop (SW4): Epigenomics: Design, Implementation and Analysis for RNA-seq and Methyl-seq Experiments
 Satellite Education Workshop (SW4): Epigenomics: Design, Implementation and Analysis for RNA-seq and Methyl-seq Experiments Saturday March 17, 2012 Orlando, Florida Workshop Description: This full day
Satellite Education Workshop (SW4): Epigenomics: Design, Implementation and Analysis for RNA-seq and Methyl-seq Experiments Saturday March 17, 2012 Orlando, Florida Workshop Description: This full day
Almac Diagnostics. NGS Panels: From Patient Selection to CDx. Dr Katarina Wikstrom Head of US Operations Almac Diagnostics
 Almac Diagnostics NGS Panels: From Patient Selection to CDx Dr Katarina Wikstrom Head of US Operations Almac Diagnostics Overview Almac Diagnostics Overview Benefits and Challenges of NGS Panels for Subject
Almac Diagnostics NGS Panels: From Patient Selection to CDx Dr Katarina Wikstrom Head of US Operations Almac Diagnostics Overview Almac Diagnostics Overview Benefits and Challenges of NGS Panels for Subject
Symphony Genomics Workflow Management System Managing, Integrating, and Delivering Your NGS Data and Results
 Symphony Genomics Workflow Management System Managing, Integrating, and Delivering Your NGS Data and Results Precision Genomics for Immuno-Oncology www.personalis.com info@personalis.com 1 855-GENOME4
Symphony Genomics Workflow Management System Managing, Integrating, and Delivering Your NGS Data and Results Precision Genomics for Immuno-Oncology www.personalis.com info@personalis.com 1 855-GENOME4
DNA. Clinical Trials. Research RNA. Custom. Reports CLIA CAP GCP. Tumor Genomic Profiling Services for Clinical Trials
 Tumor Genomic Profiling Services for Clinical Trials Custom Reports DNA RNA Focused Gene Sets Clinical Trials Accuracy and Content Enhanced NGS Sequencing Extended Panel, Exomes, Transcriptomes Research
Tumor Genomic Profiling Services for Clinical Trials Custom Reports DNA RNA Focused Gene Sets Clinical Trials Accuracy and Content Enhanced NGS Sequencing Extended Panel, Exomes, Transcriptomes Research
Next generation sequencing in diagnostic laboratories: opportunities and challenges
 Next generation sequencing in diagnostic laboratories: opportunities and challenges Vitali Sintchenko Marie Bashir Institute for Emerging Infectious Diseases & Biosecurity Declaration No conflict of interest
Next generation sequencing in diagnostic laboratories: opportunities and challenges Vitali Sintchenko Marie Bashir Institute for Emerging Infectious Diseases & Biosecurity Declaration No conflict of interest
Getting high-quality cytogenetic data is a SNP.
 Getting high-quality cytogenetic data is a SNP. SNP data. Increased insight. Cytogenetics is at the forefront of the study of cancer and congenital disorders. And we put you at the forefront of cytogenetics.
Getting high-quality cytogenetic data is a SNP. SNP data. Increased insight. Cytogenetics is at the forefront of the study of cancer and congenital disorders. And we put you at the forefront of cytogenetics.
Next Generation Sequencing of HLA: Challenges and Opportunities in the era of Precision Medicine. Dr. Paul Keown, 2016
 Next Generation Sequencing of HLA: Challenges and Opportunities in the era of Precision Medicine Dr. Paul Keown, 2016 Statement of Conflict & Collaboration Therapeutics collaborations Novartis, Roche,
Next Generation Sequencing of HLA: Challenges and Opportunities in the era of Precision Medicine Dr. Paul Keown, 2016 Statement of Conflict & Collaboration Therapeutics collaborations Novartis, Roche,
Impact of gdna Integrity on the Outcome of DNA Methylation Studies
 Impact of gdna Integrity on the Outcome of DNA Methylation Studies Application Note Nucleic Acid Analysis Authors Emily Putnam, Keith Booher, and Xueguang Sun Zymo Research Corporation, Irvine, CA, USA
Impact of gdna Integrity on the Outcome of DNA Methylation Studies Application Note Nucleic Acid Analysis Authors Emily Putnam, Keith Booher, and Xueguang Sun Zymo Research Corporation, Irvine, CA, USA
Laboratory Management for Clinical and Research NGS Labs
 Laboratory Management for Clinical and Research NGS Labs Introduction Few can argue that Next Generation Sequencing is having a major impact on genomics research. The progress in this field exceeds even
Laboratory Management for Clinical and Research NGS Labs Introduction Few can argue that Next Generation Sequencing is having a major impact on genomics research. The progress in this field exceeds even
User Requirement Specifications
 User Requirement Specifications for the tender (AZ-2017-0155) Implementation of a LIMS system and 3 years license at subsequently mentioned as purchaser Tenderer = Contractor (AN) Page 1 of 10 Inhalt 1
User Requirement Specifications for the tender (AZ-2017-0155) Implementation of a LIMS system and 3 years license at subsequently mentioned as purchaser Tenderer = Contractor (AN) Page 1 of 10 Inhalt 1
The New Genome Analyzer IIx Delivering more data, faster, and easier than ever before. Jeremy Preston, PhD Marketing Manager, Sequencing
 The New Genome Analyzer IIx Delivering more data, faster, and easier than ever before Jeremy Preston, PhD Marketing Manager, Sequencing Illumina Genome Analyzer: a Paradigm Shift 2000x gain in efficiency
The New Genome Analyzer IIx Delivering more data, faster, and easier than ever before Jeremy Preston, PhD Marketing Manager, Sequencing Illumina Genome Analyzer: a Paradigm Shift 2000x gain in efficiency
A Current Profile of Microarray Laboratories: the ABRF Microarray Research Group Survey of Laboratories Using Microarray Technologies
 A Current Prile Microarray Laboratories: the 2002-2003 2003 ABRF Microarray Research Group Survey Laboratories Using Microarray Technologies K.L. Knudtson 1, C. Griffin 2, D.A. Iacobas 3, K. Johnson 4,
A Current Prile Microarray Laboratories: the 2002-2003 2003 ABRF Microarray Research Group Survey Laboratories Using Microarray Technologies K.L. Knudtson 1, C. Griffin 2, D.A. Iacobas 3, K. Johnson 4,
Agrigenomics Genotyping Solutions. Easy, flexible, automated solutions to accelerate your genomic selection and breeding programs
 Agrigenomics Genotyping Solutions Easy, flexible, automated solutions to accelerate your genomic selection and breeding programs Agrigenomics genotyping solutions A single platform for every phase of your
Agrigenomics Genotyping Solutions Easy, flexible, automated solutions to accelerate your genomic selection and breeding programs Agrigenomics genotyping solutions A single platform for every phase of your
Outsource or in-house? The evolving role of analytical services in pharmaceutical manufacturing
 Outsource or in-house? The evolving role of analytical services in pharmaceutical manufacturing By Impact Analytical Introduction About Impact Analytical Impact Analytical is a contract analytical testing
Outsource or in-house? The evolving role of analytical services in pharmaceutical manufacturing By Impact Analytical Introduction About Impact Analytical Impact Analytical is a contract analytical testing
Undergraduate Research in the Brzustowicz Laboratory Human Genetics Institute Department of Genetics Rutgers University
 Undergraduate Research in the Brzustowicz Laboratory Human Genetics Institute Department of Genetics Rutgers University Genetics plays an important role in the development of many of the major psychiatric
Undergraduate Research in the Brzustowicz Laboratory Human Genetics Institute Department of Genetics Rutgers University Genetics plays an important role in the development of many of the major psychiatric
September 12, Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane Room 1061 Rockville, MD 20852
 September 12, 2016 Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane Room 1061 Rockville, MD 20852 RE: Docket No. FDA-2016-D-0971: Draft Guidance for Industry and
September 12, 2016 Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane Room 1061 Rockville, MD 20852 RE: Docket No. FDA-2016-D-0971: Draft Guidance for Industry and
Axiom Biobank Genotyping Solution
 TCCGGCAACTGTA AGTTACATCCAG G T ATCGGCATACCA C AGTTAATACCAG A Axiom Biobank Genotyping Solution The power of discovery is in the design GWAS has evolved why and how? More than 2,000 genetic loci have been
TCCGGCAACTGTA AGTTACATCCAG G T ATCGGCATACCA C AGTTAATACCAG A Axiom Biobank Genotyping Solution The power of discovery is in the design GWAS has evolved why and how? More than 2,000 genetic loci have been
Introduction. Highlights. Prepare Library Sequence Analyze Data
 BaseSpace Sequence Hub Genomics cloud computing expands opportunities for biological discovery, making analysis and storage of next-generation sequencing data accessible to any scientist. Highlights Centralized
BaseSpace Sequence Hub Genomics cloud computing expands opportunities for biological discovery, making analysis and storage of next-generation sequencing data accessible to any scientist. Highlights Centralized
TBRT Meeting April 2018 Scott Weigel Sales Director
 TBRT Meeting April 2018 Scott Weigel Sales Director About Us AgriPlex Genomics was formed in 2014 with the goal of creating a platform for targeted sequencing and genotyping in large numbers of samples.
TBRT Meeting April 2018 Scott Weigel Sales Director About Us AgriPlex Genomics was formed in 2014 with the goal of creating a platform for targeted sequencing and genotyping in large numbers of samples.
Megan Schmidt Vice President of Product Management, CompuGroup Medical
 Thursday April 6, 2017 A01 Molecular and Genetic Testing: How to Position your Laboratory for New Technology Megan Schmidt Vice President of Product Management, CompuGroup Medical DESCRIPTION: Molecular
Thursday April 6, 2017 A01 Molecular and Genetic Testing: How to Position your Laboratory for New Technology Megan Schmidt Vice President of Product Management, CompuGroup Medical DESCRIPTION: Molecular
CM581A2: NEXT GENERATION SEQUENCING PLATFORMS AND LIBRARY GENERATION
 CM581A2: NEXT GENERATION SEQUENCING PLATFORMS AND LIBRARY GENERATION Fall 2015 Instructors: Coordinator: Carol Wilusz, Associate Professor MIP, CMB Instructor: Dan Sloan, Assistant Professor, Biology,
CM581A2: NEXT GENERATION SEQUENCING PLATFORMS AND LIBRARY GENERATION Fall 2015 Instructors: Coordinator: Carol Wilusz, Associate Professor MIP, CMB Instructor: Dan Sloan, Assistant Professor, Biology,
SEQUENCING FROM SAMPLE TO SEQUENCE READY
 SEQUENCING FROM SAMPLE TO SEQUENCE READY ACCESS ARRAY SYSTEM HIGH-QUALITY LIBRARIES NOT ONCE, BUT EVERY TIME n The highest quality amplicons more sensitive, accurate, and specific n Full support for all
SEQUENCING FROM SAMPLE TO SEQUENCE READY ACCESS ARRAY SYSTEM HIGH-QUALITY LIBRARIES NOT ONCE, BUT EVERY TIME n The highest quality amplicons more sensitive, accurate, and specific n Full support for all
High Cross-Platform Genotyping Concordance of Axiom High-Density Microarrays and Eureka Low-Density Targeted NGS Assays
 High Cross-Platform Genotyping Concordance of Axiom High-Density Microarrays and Eureka Low-Density Targeted NGS Assays Ali Pirani and Mohini A Patil ISAG July 2017 The world leader in serving science
High Cross-Platform Genotyping Concordance of Axiom High-Density Microarrays and Eureka Low-Density Targeted NGS Assays Ali Pirani and Mohini A Patil ISAG July 2017 The world leader in serving science
Innovations in Molecular Lab Operation. Joseph M. Campos, PhD, D(ABMM), F(AAM)
 Innovations in Molecular Lab Operation Lessons in the Best Ways to Blend Automation, Lean Workflow, and Paperless Processes Joseph M. Campos, PhD, D(ABMM), F(AAM) Interim Chief, Division of Laboratory
Innovations in Molecular Lab Operation Lessons in the Best Ways to Blend Automation, Lean Workflow, and Paperless Processes Joseph M. Campos, PhD, D(ABMM), F(AAM) Interim Chief, Division of Laboratory
Agilent NGS Solutions : Addressing Today s Challenges
 Agilent NGS Solutions : Addressing Today s Challenges Charmian Cher, Ph.D Director, Global Marketing Programs 1 10 years of Next-Gen Sequencing 2003 Completion of the Human Genome Project 2004 Pyrosequencing
Agilent NGS Solutions : Addressing Today s Challenges Charmian Cher, Ph.D Director, Global Marketing Programs 1 10 years of Next-Gen Sequencing 2003 Completion of the Human Genome Project 2004 Pyrosequencing
Introduction to the MiSeq
 Introduction to the MiSeq 2011 Illumina, Inc. All rights reserved. Illumina, illuminadx, BeadArray, BeadXpress, cbot, CSPro, DASL, Eco, Genetic Energy, GAIIx, Genome Analyzer, GenomeStudio, GoldenGate,
Introduction to the MiSeq 2011 Illumina, Inc. All rights reserved. Illumina, illuminadx, BeadArray, BeadXpress, cbot, CSPro, DASL, Eco, Genetic Energy, GAIIx, Genome Analyzer, GenomeStudio, GoldenGate,
Your Best Data: Teaming QIAGEN Chemistry & Bioinformatics to Drive Samples to Insight
 Your Best Data: Teaming QIAGEN Chemistry & Bioinformatics to Drive Samples to Insight Aysel Heckel Director Clinical Solutions Sales Dr. Anne Arens Field Application Scientist Course on Variant Detection
Your Best Data: Teaming QIAGEN Chemistry & Bioinformatics to Drive Samples to Insight Aysel Heckel Director Clinical Solutions Sales Dr. Anne Arens Field Application Scientist Course on Variant Detection
Lecture #1. Introduction to microarray technology
 Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
Lecture #1 Introduction to microarray technology Outline General purpose Microarray assay concept Basic microarray experimental process cdna/two channel arrays Oligonucleotide arrays Exon arrays Comparing
Blood Pressure and Hypertension Genetics
 Blood Pressure and Hypertension Genetics Yong Huo, M.D. Wei Gao, M.D. Yan Zhang, M.D. Santhi K. Ganesh, M.D. Outline Blood pressure and hypertension in China Update on genetics of blood pressure BP/HTN
Blood Pressure and Hypertension Genetics Yong Huo, M.D. Wei Gao, M.D. Yan Zhang, M.D. Santhi K. Ganesh, M.D. Outline Blood pressure and hypertension in China Update on genetics of blood pressure BP/HTN
Computational Challenges of Medical Genomics
 Talk at the VSC User Workshop Neusiedl am See, 27 February 2012 [cbock@cemm.oeaw.ac.at] http://medical-epigenomics.org (lab) http://www.cemm.oeaw.ac.at (institute) Introducing myself to Vienna s scientific
Talk at the VSC User Workshop Neusiedl am See, 27 February 2012 [cbock@cemm.oeaw.ac.at] http://medical-epigenomics.org (lab) http://www.cemm.oeaw.ac.at (institute) Introducing myself to Vienna s scientific
Surely Better Target Enrichment from Sample to Sequencer and Analysis
 sureselect TARGET ENRIChment solutions Surely Better Target Enrichment from Sample to Sequencer and Analysis Agilent s market leading SureSelect platform provides a complete portfolio of catalog to custom
sureselect TARGET ENRIChment solutions Surely Better Target Enrichment from Sample to Sequencer and Analysis Agilent s market leading SureSelect platform provides a complete portfolio of catalog to custom
Biology 644: Bioinformatics
 Processes Activation Repression Initiation Elongation.... Processes Splicing Editing Degradation Translation.... Transcription Translation DNA Regulators DNA-Binding Transcription Factors Chromatin Remodelers....
Processes Activation Repression Initiation Elongation.... Processes Splicing Editing Degradation Translation.... Transcription Translation DNA Regulators DNA-Binding Transcription Factors Chromatin Remodelers....
Implementation of Automated Sample Quality Control in Whole Exome Sequencing
 Journal of Life Sciences 11 (2017) 261-268 doi: 10.17265/1934-7391/2017.06.001 D DAVID PUBLISHING Implementation of Automated Sample Quality Control in Whole Exome Sequencing Elisa Viering 1, Jana Molitor
Journal of Life Sciences 11 (2017) 261-268 doi: 10.17265/1934-7391/2017.06.001 D DAVID PUBLISHING Implementation of Automated Sample Quality Control in Whole Exome Sequencing Elisa Viering 1, Jana Molitor
Functional DNA Quality Analysis Improves the Accuracy of Next Generation Sequencing from Clinical Specimens
 Functional DNA Quality Analysis Improves the Accuracy of Next Generation Sequencing from Clinical Specimens Overview We have developed a novel QC, the SuraSeq DNA Quantitative Functional Index (QFI ).
Functional DNA Quality Analysis Improves the Accuracy of Next Generation Sequencing from Clinical Specimens Overview We have developed a novel QC, the SuraSeq DNA Quantitative Functional Index (QFI ).
MicroSEQ TM ID Rapid Microbial Identification System:
 MicroSEQ TM ID Rapid Microbial Identification System: the complete solution for reliable genotypic microbial identification 1 The world leader in serving science Rapid molecular methods for pharmaceutical
MicroSEQ TM ID Rapid Microbial Identification System: the complete solution for reliable genotypic microbial identification 1 The world leader in serving science Rapid molecular methods for pharmaceutical
Sample to Insight. Dr. Bhagyashree S. Birla NGS Field Application Scientist
 Dr. Bhagyashree S. Birla NGS Field Application Scientist bhagyashree.birla@qiagen.com NGS spans a broad range of applications DNA Applications Human ID Liquid biopsy Biomarker discovery Inherited and somatic
Dr. Bhagyashree S. Birla NGS Field Application Scientist bhagyashree.birla@qiagen.com NGS spans a broad range of applications DNA Applications Human ID Liquid biopsy Biomarker discovery Inherited and somatic
High Throughput Sequencing the Multi-Tool of Life Sciences. Lutz Froenicke DNA Technologies and Expression Analysis Cores UCD Genome Center
 High Throughput Sequencing the Multi-Tool of Life Sciences Lutz Froenicke DNA Technologies and Expression Analysis Cores UCD Genome Center Complementary Approaches Illumina Still-imaging of clusters (~1000
High Throughput Sequencing the Multi-Tool of Life Sciences Lutz Froenicke DNA Technologies and Expression Analysis Cores UCD Genome Center Complementary Approaches Illumina Still-imaging of clusters (~1000
Centro Nacional de Análisis Genómico. Where are the Bottlenecks of Genome Analysis Today? Teratec. Ecole Polytechnique, Palaiseau, F.
 Centro Nacional de Análisis Genómico Where are the Bottlenecks of Genome Analysis Today? Teratec Ecole Polytechnique, Palaiseau, F Ivo Glynne Gut 29.06.2016 The genomehenge Sequencing capacity >1000 Gbases/day
Centro Nacional de Análisis Genómico Where are the Bottlenecks of Genome Analysis Today? Teratec Ecole Polytechnique, Palaiseau, F Ivo Glynne Gut 29.06.2016 The genomehenge Sequencing capacity >1000 Gbases/day
dbcamplicons pipeline Amplicons
 dbcamplicons pipeline Amplicons Matthew L. Settles Genome Center Bioinformatics Core University of California, Davis settles@ucdavis.edu; bioinformatics.core@ucdavis.edu Microbial community analysis Goal:
dbcamplicons pipeline Amplicons Matthew L. Settles Genome Center Bioinformatics Core University of California, Davis settles@ucdavis.edu; bioinformatics.core@ucdavis.edu Microbial community analysis Goal:
Design a super panel for comprehensive genetic testing
 Design a super panel for comprehensive genetic testing Rong Chen, Ph.D. Assistant Professor Director of Clinical Genome Sequencing Dept. of Genetics and Genomic Sciences Institute for Genomics and Multiscale
Design a super panel for comprehensive genetic testing Rong Chen, Ph.D. Assistant Professor Director of Clinical Genome Sequencing Dept. of Genetics and Genomic Sciences Institute for Genomics and Multiscale
TAKE YOUR NEXT STEP WITH CONFIDENCE
 TAKE YOUR NEXT STEP WITH CONFIDENCE Biomek-Automated Genomic Solutions IT STARTS WITH DEFINING THE RIGHT WORKFLOW SOLUTION FOR YOU Human Health Agricultural Animal Health Environmental Food & Beverage
TAKE YOUR NEXT STEP WITH CONFIDENCE Biomek-Automated Genomic Solutions IT STARTS WITH DEFINING THE RIGHT WORKFLOW SOLUTION FOR YOU Human Health Agricultural Animal Health Environmental Food & Beverage
DNA METHYLATION RESEARCH TOOLS
 SeqCap Epi Enrichment System Revolutionize your epigenomic research DNA METHYLATION RESEARCH TOOLS Methylated DNA The SeqCap Epi System is a set of target enrichment tools for DNA methylation assessment
SeqCap Epi Enrichment System Revolutionize your epigenomic research DNA METHYLATION RESEARCH TOOLS Methylated DNA The SeqCap Epi System is a set of target enrichment tools for DNA methylation assessment
Expression Array System
 Integrated Science for Gene Expression Applied Biosystems Expression Array System Expression Array System SEE MORE GENES The most complete, most sensitive system for whole genome expression analysis. The
Integrated Science for Gene Expression Applied Biosystems Expression Array System Expression Array System SEE MORE GENES The most complete, most sensitive system for whole genome expression analysis. The
Molecular Diagnostics
 Molecular Diagnostics Part II: Regulations, Markets & Companies By Prof. K. K. Jain MD, FRACS, FFPM Jain PharmaBiotech Basel, Switzerland May 2018 A Jain PharmaBiotech Report A U T H O R ' S B I O G R
Molecular Diagnostics Part II: Regulations, Markets & Companies By Prof. K. K. Jain MD, FRACS, FFPM Jain PharmaBiotech Basel, Switzerland May 2018 A Jain PharmaBiotech Report A U T H O R ' S B I O G R
FFPE in your NGS Study
 FFPE in your NGS Study Richard Corbett Canada s Michael Smith Genome Sciences Centre Vancouver, British Columbia Dec 6, 2017 Our mandate is to advance knowledge about cancer and other diseases and to use
FFPE in your NGS Study Richard Corbett Canada s Michael Smith Genome Sciences Centre Vancouver, British Columbia Dec 6, 2017 Our mandate is to advance knowledge about cancer and other diseases and to use
Development and characterization of a high throughput targeted genotypingby-sequencing solution for agricultural genetic applications
 Development and characterization of a high throughput targeted genotypingby-sequencing solution for agricultural genetic applications Michelle Swimley 1, Angela Burrell 1, Prasad Siddavatam 1, Chris Willis
Development and characterization of a high throughput targeted genotypingby-sequencing solution for agricultural genetic applications Michelle Swimley 1, Angela Burrell 1, Prasad Siddavatam 1, Chris Willis
Guidelines Analysis of RNA Quantity and Quality for Next-Generation Sequencing Projects
 Title: Protocol number: Guidelines Analysis of RNA Quantity and Quality for Next-Generation Sequencing Projects GAF S002 Version: Version 4 Date: December 17 th 2015 Author: P. van der Vlies, C.C. van
Title: Protocol number: Guidelines Analysis of RNA Quantity and Quality for Next-Generation Sequencing Projects GAF S002 Version: Version 4 Date: December 17 th 2015 Author: P. van der Vlies, C.C. van
GENE EXPRESSION REAGENTS MARKETS (SAMPLE COPY, NOT FOR RESALE)
 TriMark Publications April 2007 Volume: TMRGER07-0401 GENE EXPRESSION REAGENTS MARKETS (SAMPLE COPY, NOT FOR RESALE) Trends, Industry Participants, Product Overviews and Market Drivers TABLE OF CONTENTS
TriMark Publications April 2007 Volume: TMRGER07-0401 GENE EXPRESSION REAGENTS MARKETS (SAMPLE COPY, NOT FOR RESALE) Trends, Industry Participants, Product Overviews and Market Drivers TABLE OF CONTENTS
LabOptimize LIMS. Laboratory Tracking Platform
 LabOptimize LIMS Laboratory Tracking Platform BioSoft Integrators offers a comprehensive solution to address tracking, process security, analysis and reporting of data with flexibility, scalability and
LabOptimize LIMS Laboratory Tracking Platform BioSoft Integrators offers a comprehensive solution to address tracking, process security, analysis and reporting of data with flexibility, scalability and
Press Release Presse-Information Information de presse
 Press Release Presse-Information Information de presse Contact/Kontakt Dr. Kathrin Rübberdt Tel. ++49 (0) 69 / 75 64-2 77 Fax ++49 (0) 69 / 75 64-2 72 e-mail: presse@dechema.de Januar 2015 Trend Report
Press Release Presse-Information Information de presse Contact/Kontakt Dr. Kathrin Rübberdt Tel. ++49 (0) 69 / 75 64-2 77 Fax ++49 (0) 69 / 75 64-2 72 e-mail: presse@dechema.de Januar 2015 Trend Report
ICH E18: Guideline for Genomic Sampling and Management of Genomic Data. Prepared by the ICH E18 Expert Working Group
 ICH E18: Guideline for Genomic Sampling and Prepared by the ICH E18 Expert Working Group October 2017 International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use
ICH E18: Guideline for Genomic Sampling and Prepared by the ICH E18 Expert Working Group October 2017 International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use
TREE CODE PRODUCT BROCHURE
 TREE CODE PRODUCT BROCHURE Single Molecule, Real-Time (SMRT) Sequencing technology offers: Long read sequencing ~10 Gb with 20 kb average read lengths for WGS ~20 Gb with 40 kb average read length for
TREE CODE PRODUCT BROCHURE Single Molecule, Real-Time (SMRT) Sequencing technology offers: Long read sequencing ~10 Gb with 20 kb average read lengths for WGS ~20 Gb with 40 kb average read length for
High-throughput scale. Desktop simplicity.
 High-throughput scale. Desktop simplicity. NextSeq 500 System. Flexible power. Speed and simplicity for whole-genome, exome, and transcriptome sequencing. Harness the power of next-generation sequencing.
High-throughput scale. Desktop simplicity. NextSeq 500 System. Flexible power. Speed and simplicity for whole-genome, exome, and transcriptome sequencing. Harness the power of next-generation sequencing.
October 6, Below we provide input from our community of health informatics experts regarding select portions of the Draft Guidance.
 Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane, rm. 1061 Rockville, MD 20852 Submitted electronically via http://www.regulations.gov RE: Use of Public Human Genetic
Division of Dockets Management (HFA-305) Food and Drug Administration 5630 Fishers Lane, rm. 1061 Rockville, MD 20852 Submitted electronically via http://www.regulations.gov RE: Use of Public Human Genetic
Total genomic solutions for biobanks. Maximizing the value of your specimens.
 Total genomic solutions for biobanks. Maximizing the value of your specimens. Unlock the true potential of your biological samples. Greater understanding. Increased value. Value-driven biobanking. Now
Total genomic solutions for biobanks. Maximizing the value of your specimens. Unlock the true potential of your biological samples. Greater understanding. Increased value. Value-driven biobanking. Now
Deep Sequencing technologies
 Deep Sequencing technologies Gabriela Salinas 30 October 2017 Transcriptome and Genome Analysis Laboratory http://www.uni-bc.gwdg.de/index.php?id=709 Microarray and Deep-Sequencing Core Facility University
Deep Sequencing technologies Gabriela Salinas 30 October 2017 Transcriptome and Genome Analysis Laboratory http://www.uni-bc.gwdg.de/index.php?id=709 Microarray and Deep-Sequencing Core Facility University
An innovative approach to genetic testing for improved patient care
 An innovative approach to genetic testing for improved patient care Blueprint Genetics Blueprint Genetics is changing diagnostics by providing fast, affordable and comprehensive genetic knowledge Who we
An innovative approach to genetic testing for improved patient care Blueprint Genetics Blueprint Genetics is changing diagnostics by providing fast, affordable and comprehensive genetic knowledge Who we
About Strand NGS. Strand Genomics, Inc All rights reserved.
 About Strand NGS Strand NGS-formerly known as Avadis NGS, is an integrated platform that provides analysis, management and visualization tools for next-generation sequencing data. It supports extensive
About Strand NGS Strand NGS-formerly known as Avadis NGS, is an integrated platform that provides analysis, management and visualization tools for next-generation sequencing data. It supports extensive
Accelerate your NGS performance through Sample to Insight solutions
 Accelerate your NGS performance through Sample to Insight solutions Complete your NGS workflow and save up to 40% Sample to Insight From Sample to Insight let QIAGEN enhance your NGS-based research High-throughput
Accelerate your NGS performance through Sample to Insight solutions Complete your NGS workflow and save up to 40% Sample to Insight From Sample to Insight let QIAGEN enhance your NGS-based research High-throughput
DNA concentration and purity were initially measured by NanoDrop 2000 and verified on Qubit 2.0 Fluorometer.
 DNA Preparation and QC Extraction DNA was extracted from whole blood or flash frozen post-mortem tissue using a DNA mini kit (QIAmp #51104 and QIAmp#51404, respectively) following the manufacturer s recommendations.
DNA Preparation and QC Extraction DNA was extracted from whole blood or flash frozen post-mortem tissue using a DNA mini kit (QIAmp #51104 and QIAmp#51404, respectively) following the manufacturer s recommendations.
