Transposons Tn1696 and Tn21 and Their Integrons In4 and In2 Have Independent Origins
|
|
|
- Reynold Owen
- 6 years ago
- Views:
Transcription
1 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Apr. 2001, p Vol. 45, No /01/$ DOI: /AAC Copyright 2001, American Society for Microbiology. All Rights Reserved. Transposons Tn1696 and Tn21 and Their Integrons In4 and In2 Have Independent Origins SALLY R. PARTRIDGE, 1,2 HEIDI J. BROWN, 1,2 H. W. STOKES, 2 AND RUTH M. HALL 1 * Sydney Laboratory, CSIRO Molecular Science, North Ryde 2113, 1 and Department of Biological Sciences, Macquarie University Sydney, Sydney 2109, 2 Australia Received 12 September 2000/Returned for modification 9 January 2001/Accepted 24 January 2001 The first 13.6 kb of the mercury and multidrug resistance transposon Tn1696, which includes the class 1 integron In4, has been sequenced. In4 is 8.33 kb long and contains the 5 -conserved segment (5 -CS) and 2.24 kb of the 3 -conserved segment (3 -CS) flanking four integrated cassettes. The 3 -CS region is followed by one full copy and an adjacent partial copy of the insertion sequence IS6100 flanked, in inverse orientation, by two short segments (123 and 152 bp) from the outer right-hand end of class 1 integrons. This structure is representative of a distinct group of class 1 integrons that differs from In2, found in Tn21, and other related class 1 integrons. In4 does not include transposition genes but is bounded by characteristic 25-bp inverted repeats and flanked by a direct duplication of 5 bp of the target sequence, indicating that it was inserted by a transpositional mechanism. In4 lies between the resii and resi sites of a backbone mercury resistance transposon which is >99.5% identical to Tn5036. Although Tn21 and Tn1696 are both classified as members of the Tn21 subfamily of the Tn3 transposon family, the backbone mercury resistance transposons are only 79 to 96% identical. Tn21 also contains a region of about 0.7 kb not found in Tn1696. The integrons In2 and In4 carrying the antibiotic resistance genes have been inserted at different locations into distinct ancestral mercury resistance transposons. Thus, Tn21 and Tn1696 have independent histories and origins. Other transposons (Tn1403 and Tn1412) that include a class 1 integron also have independent origins. In all except Tn21, the integron is located within the res region of the backbone transposon. Tn1696 and Tn21 are large transposons that confer resistance to mercuric ions and to more than one antibiotic. Tn1696 was originally found in plasmid R1033 (36), isolated from a Pseudomonas aeruginosa strain (42), and Tn21 was found in plasmid NR1 (R100) (10), isolated from a Shigella flexneri strain (29). Early comparisons revealed similarities (40). One is that the antibiotic resistance determinants are located between the transposition gene (tnpa and tnpr) module and the mercury resistance (mer) module, and despite differences in the restriction maps (10, 18), these two regions form stable heteroduplexes (40). The central regions containing the antibiotic resistance determinants, sul1 (sulfonamide resistance) and aada1 (streptomycin and spectinomycin resistance) in Tn21 and sul1, aacc1 (gentamicin resistance), aada2, and cmla1 (chloramphenicol resistance) in Tn1696, also formed hybrids in which the aacc1 and cmla1 genes appear as single-stranded loops flanking the aada genes (Fig. 1A). The central regions are now known to be class 1 integrons, In2 in Tn21 and In4 in Tn1696 (4, 14, 43), with one gene cassette (aada1) in In2 (19, 43, 45) and four (aacc1, orfe, aada2, and cmla1) in In4 (2, 8, 13, 44, 50). The integrons found in multiply antibiotic-resistant clinical and environmental isolates generally contain one or more integrated cassettes. Most of them have identical integrase genes and belong to the integron class 1. In class 1 integrons, the region located upstream (5 ) of the integrated gene cassettes (5 -conserved segment; 5 -CS) encodes the IntI1 integrase that * Corresponding author. Mailing address: CSIRO Molecular Science, Sydney Laboratory, P.O. Box 184, North Ryde, NSW 1670, Australia. Phone: Fax: ruth.hall@molsci.csiro.au. is responsible for cassette insertion, and in the majority, the region 3 to the cassettes (3 -conserved segment; 3 -CS) includes a sulfonamide resistance determinant, sul1. Integrons of this type are found in many distinct locations (14, 15, 33, 43), including in the plasmids R46 (IncN), R388 (IncW), R751 (IncP), and pvs1 and in the transposons Tn21 and Tn1696, a fact consistent with the notion that class 1 integrons are themselves mobile elements. However, only a single class 1 integron that is an active transposon, namely Tn402, has been found to date. Tn402 (Fig. 2A) contains the complete transposition gene (tni) module consisting of four genes (tnia, B, Q, and R) required for transposition (22, 33) but lacks the 3 -CS and the sul1 gene (33). When several independently located class 1 integrons that include the 3 -CS and the sul1 gene were examined, a variety of structures were found (14). All of these integrons include the 1.36-kb 5 -CS (15, 43) and the 2.0-kb 3 -CS region identified by Stokes and Hall (43). Thereafter, the sequences diverge one by one from a conserved sequence which is now known to also be part of the 3 -CS (14), indicating that a variety of different events have shaped the 3 -CS and the region beyond it. Beyond their divergence points, three of the integrons examined, In0 from pvs1, In2 from Tn21, and In5 from psch884, are clearly related (4). These three integrons include an insertion sequence, IS1326, and part of the 4-kb segment containing the transposition (tni) genes. In In0, In2, and In5, IS1326 appears to have caused deletions of adjacent sequences, leading to the loss of part of the transposition module (Fig. 2B), and these integrons are thus transposition-defective transposon derivatives (4). A further class 1 integron, In31, is closely related to In5, but the IS1326 element has been lost (23). Similarly, In22, the integron carried by Tn5086, which is a close relative of 1263
2 1264 PARTRIDGE ET AL. ANTIMICROB. AGENTS CHEMOTHER. FIG. 1. Relationship between Tn21 and Tn1696. (A) A schematic representation of the heteroduplex formed by Tn21 and Tn1696. The features shown are from Fig. 4C in reference 40, and gene names and distances are those assigned by the authors. (B) Alignment of Tn21 and Tn1696 sequences. Thick lines represent the backbone transposons and narrow lines the backbones of the integrons. Vertical bars indicate the terminal repeats of the transposons (thick) and integrons (thin). Open boxes show insertion sequences and narrow open boxes with adjacent filled boxes (59-base element) indicate the gene cassettes. Lengths of individual regions are shown in kilobases. Tn21, has arisen by the loss of the IS1326 element from the configuration found in In2 or In0 (33). Though members of this group lack some of the tni genes, In0, In2, In5, and In31 are all bounded by inverted repeats of 25 bp (IRi and IRt, at the integrase and tni ends, respectively) that are also found in Tn402 (4, 5, 14, 33), and two (In0 and In2) are flanked by 5-bp direct duplications, suggesting that they moved to their present locations by transposition (5, 14). However, not all of the 3 -CS (sul1)-containing class 1 integrons that have been examined in detail have the general structure outlined above. In both In1 (from R46) and In4 (from Tn1696) a region identical to one end of the insertion sequence IS6100 (26) was found beyond the point at which their sequences diverge from the 3 -CS sequence (14). The fact that In2 and In4 sequences diverge within the 3 -CS is consistent with early heteroduplex studies (40) which show substantial differences in the regions flanking the 5 - and 3 -CS (Fig. 1A). Thus, though Tn21 and Tn1696 are clearly related, the integron inserts appear to differ in structure and to be located at slightly different positions in the backbone transposon (Fig. 1A). Despite the known differences, Tn21 and Tn1696 have generally been regarded as belonging to a single evolutionary pathway in which the insertion of the integron precedes their divergence (3, 12, 27, 41, 50). The sequence of Tn21 has recently been completed (4) and has been compiled (GenBank accession no. AF071413; 25). Here, we have completed the sequence of the first 13.6 kb of Tn1696, which includes all of In4, the tnp module, and part of the mer module of the backbone mercury resistance transposon. Comparison of the two sequences revealed several significant differences between the two transposons, reflecting an independent origin for Tn21 and Tn1696. FIG. 2. Structure of the backbones of class 1 integrons. Each integron is bounded by inverted repeats (vertical bars) designated IRi and IRt (i and t, respectively) and includes the 5 -CS (medium line), which begins with IRi, contains the inti1 gene, and ends at the vertical arrow in the atti1 site (narrow open box). This arrow also marks the position of integrated cassettes. (A) Tn402 (In16) also includes a complete tni module (thick line), with a full set of transposition genes (tnia, B, and Q), a resolvase gene (tnir), and a res site (filled box). Tn402 contains an array of three cassettes, dfrb3-orfd-qace (not shown). (B) In0, In2, In5, and In31 include the 3 -CS (thin line) and only part of the tni module. The 3 -CS contains qace 1, a truncated version of the qace cassette, the sul1 sulfonamide resistance determinant, and orf5. Insertion sequences are shown as open boxes. The cassette arrays (not shown) are as follows: In0, no cassettes; In2, aada1; In 5, aaca(iia); and In31, blap3-aaca4-catb6-orfn-qacg. (C) In4 includes two copies of very short regions of the IRt end of the tni module separated by one complete and one partial ( ) copy of IS6100 (open box). The cassette array is aacc1-orfe-aada2-cmla1.
3 VOL. 45, 2001 Tn21 AND Tn1696 HAVE INDEPENDENT ORIGINS 1265 TABLE 1. Plasmids Plasmid Description Relevant phenotype Reference R1033 IncP1; contains Tn1696 Ap r Cm r Gm r Km r Sp r Su r Tc r Hg r 18 pxs2 a 7.0-kb EcoRI fragment of R1033 in puc9 Ap r Cm r Su r Hg r 18 prmh kb SalI fragment of R1033 in puc18 Ap r Gm r Sm r Sp r 14 prmh kb SphI fragment of pxs2 in puc19 Ap r This work prmh kb SphI fragment of pxs2 in puc19 Ap r This work prmh kb EcoRI fragment of R1033 in puc19 Ap r Cm r Su r This work prmh kb PstI fragment of prmh493 in puc19 Ap r This work a pxs2 was reported to confer Hg but does not contain all of the mer genes. MATERIALS AND METHODS Bacterial strains and plasmids. Escherichia coli JM109 ( lac-proab) supe thi F trad36 proab laci q lacz M15 was used to propagate plasmid DNA. The plasmids used are listed in Table 1. The prmh series of plasmids was constructed by randomly cloning fragments from the appropriately digested parental plasmid (pxs2 or R1033) into either puc18, puc19 (51), or pacyc184 (6) using standard procedures (37). Plasmids containing the appropriate fragments were identified by screening for antibiotic resistance, by restriction mapping, and by sequencing the fragment ends using universal primer. M13 clones containing the SphI fragment from prmh484 in both orientations were also constructed. Bacteria were routinely cultured at 37 C in Luria-Bertani medium or Luria- Bertani agar supplemented as appropriate with ampicillin (100 g ml 1 )or chloramphenicol (25 g ml 1 ). Antibiotics were obtained from Sigma Chemical Co., St. Louis, Mo. DNA isolation and restriction mapping. For large plasmids, plasmid DNA for restriction analysis was isolated using an alkaline lysis method (1). Restriction enzymes were used in accordance with the manufacturers instructions. Fragments were separated by electrophoresis on 1% (wt/vol) agarose gels and visualized by staining with ethidium bromide. An EcoRI digest of bacteriophage SPP1 (Bresatec, Adelaide, Australia) was used for size markers. Plasmid DNA for sequencing was purified using a Magic Minipreps DNA purification system (Promega Corp., Madison Wis.) or Wizard maxiprep kit (Promega). M13 DNA was prepared by the method of Sanger et al. (38). DNA sequencing and analysis. The DNA sequences on both strands of fragments of Tn1696 cloned in M13 or plasmid vectors were determined. Manual DNA sequencing was performed with a Sequenase 2.0 system (46) as recommended by the manufacturer (U.S. Biochemicals, Cleveland, Ohio) using ditp reaction mixtures followed by a 30-min incubation with a 1-mM deoxynucleoside triphosphates mix and terminal deoxynucleotidyl transferase (Boehringer GmbH, Mannheim, Germany). Automated sequencing was performed by SUPAMAC (Sydney University and Royal Prince Alfred Hospital, Sydney, Australia) or by the sequencing facility at the Department of Biological Sciences, Macquarie University Sydney, on an ABI-PRISM 377 sequencer using the Big Dye system. DNA sequences were assembled using Mac Vector 6.5 and AssemblyLIGN (Oxford Molecular Ltd.). GenBank searches were performed using the BLASTN and FastA programs available through WebANGIS (Australian National Genomic Information Service). Programs in the GCG Wisconsin Package, Version 8.1.0, were used via WAG (WebANGIS GCG) to align and analyze DNA sequences. Nucleotide sequence accession number. The new sequence has been added to the previous compilation of Tn1696 (In4) under GenBank accession no. U RESULTS Structure of In4. A restriction map of the region of R1033 that includes Tn1696 is shown in Fig. 3. The integron In4 contains four integrated gene cassettes, and the sequence of these cassettes and much of the backbone of In4 has previously been reported (2, 8, 14, 44, 50). The sequence of the remainder of the right-hand (RH) end of In4 was determined (Fig. 4B) and revealed the presence of one complete copy of the insertion sequence IS6100 (26) followed by a partial copy that contains the last 321 bp of IS6100. The partial copy of IS6100 is not present in the clone pxs2 isolated by Hirsch et al. (18) and was presumably lost by homologous recombination after cloning. This explains the loss of a small PstI fragment noted by Hirsch et al. (18). The IS6100-IS6100 structure is flanked by short segments from the RH outer end of integrons in inverse orientation, and one of these was identified previously (14). These outer-end segments, 123 bp on the left and 152 bp on FIG. 3. Map of Tn1696. The inverted repeats of Tn1696 and In4 are shown as vertical bars. The features of In4 are shown as in Fig. 2C, with each cassette represented as an open box and adjacent filled box (59-base element). Genes are indicated with arrows. Numbered lines show the extent of sequence previously determined by the following: 1, Wohlleben et al. (50); 2, Stokes and Hall (44); 3, Bissonnette et al. (2); 4, Collis and Hall (8); and 5, Hall et al. (14). The line labeled U12338 represents a previous sequence compilation (14), and the regions sequenced during this work are shown by dotted lines. The fragments cloned in various plasmids are also shown. Restriction enzyme sites are as follows: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; P, PstI; S, SalI; Sp, SphI.
4 1266 PARTRIDGE ET AL. ANTIMICROB. AGENTS CHEMOTHER. FIG. 4. Sequence surrounding IRi (A) and IRt (B) of Tn1696. The numbers correspond to those in the extended GenBank entry (accession no. U12338). Thick horizontal arrows indicate the integron ends, IRi and IRt. Five copies of a 19-bp repeat noted by Rådström et al. (33) are indicated by thin arrows below the sequence labeled i1, i2, t1, t2, and t3. The 5-bp direct duplication flanking In4 is shown in bold and is boxed. The three components of the res site are indicated by horizontal bars. The start codons of genes are underlined and indicated by an M, with the name and the direction of the gene indicated above. Positions of stop codons (underlined) are indicated by asterisks. The boxed regions represent IS6100 and IS6100, and the vertical arrow in IS6100 indicates the position corresponding to the first nucleotide of IS6100. Selected restriction enzyme sites are also shown. the right, both end with the 25-bp sequence that is the RH terminal repeat (IRt) of other class 1 integrons. This configuration is also consistent with the structure seen in electron micrographs (40), namely short inverted repeats (0.1 kb) separated by a region of about 1.1 kb. In4 is thus bounded by the same 25-bp inverted repeats that are found in other class 1 integrons (In2, In0, In5, In31, and Tn402), but it does not contain any of the four tni genes required for the transposition of class 1 integrons and related transposons (22, 33). In4 (Fig. 2C) is thus a transposition-defective transposon derivative. Structure of Tn1696 and location of In4. The sequences of Tn1696 adjacent to the outer ends of In4 (Fig. 4) were compared to other DNA sequences present in GenBank. Both sequences contained regions that are related to sequences found between the tnpr and mere genes in Tn501 and Tn21 and were almost identical to a single continuous region of the
5 VOL. 45, 2001 Tn21 AND Tn1696 HAVE INDEPENDENT ORIGINS 1267 TABLE 2. Nucleotide and amino acid differences between Tn5036 and the Tn1696 backbone Position a Nucleotide(s) Location Amino acid(s) Tn5036 Tn1696 (gene) Tn5036 Tn A C tnpa Ser Ala 2360 G C tnpa Ala Gly 2780 G A tnpa Ser Leu 3036 A G tnpr Thr Thr 3078 C T tnpr Gln Gln 3432 A C tnpr Leu Leu 3460 C G tnpr Cys Ser 3647 A AT resii 3861 A T urf2y Val Glu 4416 G A merd Phe Phe 4424 to 4425 CG GC merd ArgVal ArgLys 4584 C T 4682 C T mera Gln Gln a Numbers refer to position in Tn5036 sequence, accession no. Y mercury resistance transposons Tn5036 ( 99.5% identity) isolated from Enterobacter cloacae (accession no. Y09025; 53) and Tn3926 (97% identity) isolated from Yersinia enterocolitica (accession no. X78059; 30). The 3.7-kb region located to the left of In4 extending from IRtnp of Tn1696 includes the tnpa and tnpr genes (Fig. 3; accession no. U12338) and differs from the Tn5036 sequence at only eight positions (Table 2). In R1033, Tn1696 is located between bases and in the RP1 (RP4) sequence (accession no. L27758; 32). The 1,588-bp region to the right of In4 sequenced in this study is continuous with the tnp region located to the left of In4 and includes a short open reading frame known as urf2y, the merd and mere genes, and part of the mera gene. This region also differs from the Tn5036 sequence at very few positions (6 of 1,588). Thus, In4 was inserted into Tn5036 or a close relative to generate Tn1696. In4 is located between the resii and resi regions of the res site, which was identified by comparison to the experimentally defined res sites of Tn21 and Tn1721 (16, 35). In4 is flanked by a direct duplication of 5 bp of the target sequence (boxed in Fig. 4), indicating that it was inserted by a transposition event. In2 and In4 have different locations within distinct but related transposons. Though the Tn1696 sequence flanking In4 is related to sequences found in Tn21, comparison of the sequences of the genes in the Tn21 backbone and in Tn5036 (25) revealed that the levels of identity of the individual genes range from 79 to 96%. TnX, the ancestral mercury resistance transposon for Tn21, also includes a region of 782 bp located between the urf2y and mere genes that replaces a 65-bp segment in Tn5036 or Tn1696, and In2 is located within this extra segment (25). The location in the Tn21 sequence corresponding to the position of the In4 insertion is separated by 378 bp from the position of In2 in Tn21 (Fig. 5). In heteroduplexes formed between Tn21 and Tn1696 DNA, Schmidt and Klopfer- Kaul (40) found single-stranded loops adjacent to the IRi of the integrons, and the lengths of these loops were estimated to be 0.15 kb in Tn21 and 0.5 kb in Tn1696 (Fig. 1A). From the sequence data, it is clear that these loops have been incorrectly assigned and that the longer single-stranded loop is part of Tn21, as shown in Fig. 1B. The presence of single-stranded loops in both transposons can be explained if the heteroduplex between the 0.1- to 0.15-kb region adjacent to the integron insertion point in Tn1696 and the corresponding region of Tn21 is not as stable as that of surrounding regions. In the region between the start codon of the tnpr gene and the left end of In4, only 68 of 104 bases (65%) are identical in Tn1696 and Tn21, and this is sufficiently low to explain both the observed loop in Tn1696 and the length of the loop in Tn21. Variation in the 5 -CS of In2 and In4. The sequence of the 5 -CS has been determined for many different class 1 integrons and little variation has been found. The sequence of In1 found in R46, which was the first complete sequence of the 5 -CS to be reported (accession no. X06046; 15), is used as the baseline for comparisons. Variation is common in the 10 and 35 boxes of the P c promoter (formerly P ant or P1) that lies within the inti1 gene and directs transcription of the cassette-encoded genes (43). Differences in P c vary the strength of this promoter (7, 24). The P c promoter in In1 is of intermediate strength (24), and In2 and In4 contain, respectively, the weak and the strong version of P c (Fig. 6). However, the 5 -CS of In2 and In4 each includes a further characteristic variation. In In2, a second promoter, P2, has been created by the insertion of three G residues between bp 1238 and 1239 of the 5 -CS (Fig. 6). This insertion increases the spacing between preexisting 35 and 10 boxes from 14 bp, which is outside the range for functional promoters, to 17 bp, which is optimal (17). The P2 promoter has been shown to be functional (7, 39) and is about sixfold stronger than P c (weak) (7). P2 is thus responsible for the bulk of transcription in In2 and other integrons that contain this configuration. In4 includes a duplication of 19 bp (50) that is located in the atti1 region (Fig. 6). This duplication has two effects; it duplicates the strong binding site for IntI1 (9) and it creates an in-frame ATG start codon that is used to translate the aacc1 gene found in the first cassette of In4 (50). The resultant AAC(3)-Ia protein contains an extension of 23 amino acids (20). We examined the possibility that the GGG insertion is diagnostic for integrons carried by transposons that are close relatives of Tn21 in that the backbone is the same (TnX) and the integron is in the same position in this backbone. Amongst over 100 sequences of this part of the 5 -CS in the GenBank database, only In2 (Tn21) and 13 additional entries contain the P2 promoter. In all cases P2 is associated with P c (weak). The additional sequences include In8 in Tn2603 (31), the integron in Tn2426 (54), and In10 in Tn4000 (41), which are known to be closely related to Tn21 on the basis of restriction mapping FIG. 5. Alignment of the Tn21 and Tn1696 backbones. Vertical bars represent the terminal inverted repeats of the transposons. The tnp regions are shown as thin lines, the mer regions as thick lines, and res sites as filled boxes. Vertical arrows mark the sites of integron insertions. tnp and mer genes are lettered. urf2m (2M) in the Tn21 backbone is split into urf2 (2) and tnpm (M) by the insertion of In2. urf2y (2Y) of Tn1696 corresponds to the end of urf2m and tnpm.
6 1268 PARTRIDGE ET AL. ANTIMICROB. AGENTS CHEMOTHER. FIG. 6. Variations in the 5 -CS. (A) Schematic representation of the 5 -CS. The complete 5 -CS (thick line) and part of the adjacent cassette (parallel lines) are shown. The atti1 site is boxed. The extent of the inti1 gene and the positions of promoters P int and P c are indicated. Vertical arrows mark the positions of extra residues present in In2 and In4. The P2 promoter is only present in In2. (B) An expanded view of the promoter and atti1 region. The 5 -CS, adjacent cassette, and atti1 site are indicated as in panel A. Filled boxes show the 10 and 35 regions, which are underlined in the relevant sequences. Differences from the standard 5 -CS sequence (In1; accession no. X06046) are shown in bold and bases that are not present in In1 are boxed. The atti1 sequence shown is from In4, with a vertical arrow indicating the boundary between the 5 -CS (uppercase) and the aacc1 cassette (lowercase). The 7-bp core sites found within the IntI1 binding sites are indicated with arrows. DR1 and DR2 are 15-bp direct repeats. The sequence of the 23-amino-acid extension that results from use of the alternative start codon in the duplication is shown above the DNA sequence, and the normal aacc1 start codon within the cassette is shown by an asterisk. and heteroduplex analysis (39). In two further cases, In14 (plasmid R) (47) and an unnumbered integron found in the chromosome of a multidrug-resistant S. flexneri strain (34), the sequence adjacent to the IRi of the integron has been reported (14, 34) and is identical to the Tn21 sequence. The location of the remaining eight integrons (accession no. U17586, X64368, AF156486, AF202975, AF231133, AF255921, AJ009819, and AJ237702) is not currently known. Thus, the presence of the GGG insertion may be indicative of an integron derived from In2 by loss or gain of gene cassettes. However, the integrons in other close relatives of Tn21 (Tn2424, Tn2410, and Tn5056) do not include P2, and this may be indicative of cassette exchange via homologous recombination, with one crossover within the 5 -CS and the second within either a cassette, the 3 -CS, or another homologous region. The 19-bp duplication found in In4 is also present in six database entries (accession no. S68049, X64525, X64369, AF202035, AF207065, and AJ009820) and one further published sequence (48). However, little is known about the location of these integrons. DISCUSSION The integrons In2 and In4 represent separate evolutionary lineages. The In0-In2-In5 group and the In4 group share the 5 -CS and 3 -CS regions and are bounded by the 25-bp repeats IRi and IRt. However, in the first group IS1326 has caused the loss of parts of the 3 -CS and the tni module, whereas in In4, IS6100 is present and has caused the loss of most of the tni region. The sequences of Tn1696 adjacent to the outer ends of In4 clearly demonstrate both that In4 and In2 were inserted into two transposons that are significantly diverged and that the integron In4 is inserted in Tn1696 at a different location from that of In2 in Tn21. Since the sequences of the conserved segments (5 -CS, 3 -CS, and RH outer-end regions) of In2 and In4 are close to identical, it seems highly unlikely that the transposition regions of Tn21 and Tn1696 diverged after the insertion of the integron. Therefore, the integron insertions are independent events. Thus, Tn21 is not ancestral to all transposons classified as belonging to the Tn501-Tn21 subgroup of the Tn3 transposon family as is frequently claimed (e.g., see references 3, 12, 27, 39, 41, and 50), and In2, the integron found in Tn21, is not ancestral to all other integrons. Tn21 is simply one of many cases where an integron has been acquired by a plasmid or transposon. Nonetheless, Tn21 and its close relatives such as Tn2603 (4) appear to be widely disseminated amongst bacterial species and also globally. They have clearly contributed significantly to the spread of the resistance genes they contain. Evidence for the globalization of mercury resistance transposons, including Tn5036, has also been re-
7 VOL. 45, 2001 Tn21 AND Tn1696 HAVE INDEPENDENT ORIGINS 1269 ported (53). Tn5036 is from an E. cloacae strain isolated from the intestine of a toad found near a mercury mine in Russia and the closely related Tn3926 is from a Y. enterocolitica strain found in milk in France. R1033, which contains Tn1696, was recovered from a clinical Pseudomonas strain isolated in Spain prior to 1975 (42). This emphasizes the connection between environmental, food-borne, and clinical bacteria. In addition to the many transposons that are known to be close relatives of Tn21, further class II transposons that do not determine resistance to mercuric ions are known to include an integron. One of these is Tn1403, for which the sequences of the left-hand end of the integron (In28) and of a short region adjacent to the IRi have been determined (49). In28 is located within the resi site, and Tn1403 is unable to resolve cointegrates. As In4 also lies within the res site of Tn5036, it is likely that Tn1696 is unable to resolve the cointegrates formed as transpositional intermediates. Tn1412 is another transposon that contains a class 1 integron. In this case, the complete sequence is known (accession no. L36547), and Tn1412 consists of Tn5563 (52), a class II transposon belonging to the Tn3 subgroup (12), and a complex integron structure here designated In32. In32 also lies within the res site (28), which is located between the tnpa and tnpr genes of the Tn5563 backbone. Thus, transposons in the Tn3 family have acquired integrons on at least four independent occasions to create Tn21, Tn1696, Tn1403, and Tn1412, and these events occurred relatively recently in the evolution of these transposons. The fact that the three integron insertion events that created Tn1696,Tn1403, and Tn1412 occurred within a relatively short segment may reflect the fact that insertion at other sites would inactivate either transposition functions (tnpr and tnpa) or, in the case of Tn1696, the mercury resistance genes (mer). However, an alternate explanation is that the res region is a preferred target for integron insertions. The latter possibility is supported by an examination of the location of all further class 1 integrons where the left-hand boundary (IRi) has been sequenced. In most cases the adjacent gene encodes a member of the resolvase/invertase family (28). Furthermore, the transposon Tn5053, which has a tni module and outer ends closely related to those of class 1 integrons, has been shown to be preferentially inserted into the mrs region of plasmid RP1 (21). The mrs site is the binding site (res site) for a resolvase (ParA) encoded in the par region of RP1 (RP4) (11). Recently, experimental evidence that Tn5053 also preferentially targets res sites in a number of transposons has been reported (28). Insertions occur preferentially at clusters of positions in or near the res site, and the resolvase is essential to this process. Occasionally, Tn5053 insertions occur at some distance from the res site, up to 200 bp. It is possible that the insertion of In2 into TnX to form Tn21 is similar to these rare insertions, though its position, over 350 bp from the res site, is outside the range observed experimentally. As the location of In2 in Tn21 is an exception, in that it lies quite a distance from res, we conclude that Tn1696 is a better examplar of transposons containing integrons. However, the unusual location of In2 permits the conclusion that an integron is unlikely to have been targeted to this position more than once and hence that all of the transposons with an integron in this position are likely to have evolved from a common ancestor. ACKNOWLEDGMENTS We thank Diana Brookes for technical assistance. The project was supported by a grant from the Australian National Health and Medical Research Council. REFERENCES 1. Birnboim, H. C., and J. Doly A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7: Bissonnette, L., S. Champetier, J.-P. Buisson, and P. H. Roy Characterization of the nonenzymatic chloramphenicol resistance (cmla) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173: Bissonnette, L., and P. H. Roy Characterization of In0 of Pseudomonas aeruginosa plasmid pvs1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 174: Brown, H. J., H. W. Stokes, and R. M. Hall The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178: Brown, N. L., T. K. Misra, J. N. Winnie, A. Schmidt, M. Seiff, and S. Silver The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 202: Chang, A. Y. C., and S. N. Cohen Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134: Collis, C. M., and R. M. Hall Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39: Collis, C. M., and R. M. Hall Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174: Collis, C. M., M.-J. Kim, H. W. Stokes, and R. M. Hall Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29: de la Cruz, F., and J. Grinsted Genetic and molecular characterization of Tn21, a multiple resistance transposon from R J. Bacteriol. 151: Eberl, L., C. S. Kristensen, M. Givskov, E. Grohmann, M. Gerlitz, and H. Schwab Analysis of the multimer resolution system encoded by the parcba operon of broad-host-range plasmid RP4. Mol. Microbiol. 12: Grinsted, J., F. de la Cruz, and R. Schmitt The Tn21 subgroup of bacterial transposable elements. Plasmid 24: Hall, R. M., D. E. Brookes, and H. W. Stokes Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5: Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes Integrons found in different locations have identical 5 ends but variable 3 ends. J. Bacteriol. 176: Hall, R. M., and C. Vockler The region of the IncN plasmid R46 coding for resistance to -lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 15: Hall, S. C., and S. E. Halford Specificity of DNA recognition in the nucleoprotein complex for site-specific recombination by Tn21 resolvase. Nucleic Acids Res. 21: Harley, C. B., and R. P. Reynolds Analysis of E. coli promoter sequences. Nucleic Acids Res. 15: Hirsch, P. R., C. L. Wang, and M. J. Woodward Construction of a Tn5 derivative determining resistance to gentamicin and spectinomycin using a fragment cloned from R1033. Gene 48: Hollingshead, S., and D. Vapnek Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13: Hsiang, M. W., T. J. White, and J. E. Davies NH 2 -terminal sequence of the aminoglycoside acetyltransferase (3)-I mediated by plasmid RIP 135. FEBS Lett. 92: Kholodii, G. Y., Z. M. Gorlenko, O. L. Lomovskaya, S. Z. Mindlin, O. V. Yurieva, and V. G. Nikiforov Molecular characterization of an aberrant mercury resistance transposable element from an environmental Acinetobacter strain. Plasmid 30: Kholodii, G. Y., S. Z. Mindlin, I. A. Bass, O. V. Yurieva, S. V. Minakhina, and V. G. Nikiforov Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol. 17: Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J.-M. Frère, and G. M. Rossolini Structure of In31, a bla IMP -containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an un-
8 1270 PARTRIDGE ET AL. ANTIMICROB. AGENTS CHEMOTHER. usual array of gene cassettes. Antimicrob. Agents Chemother. 43: Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142: Liebert, C. A., R. M. Hall, and A. O. Summers Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63: Martin, C., J. Timm, J. Rauzier, R. Gomez-Lus, J. Davies, and B. Gicquel Transposition of an antibiotic resistance element in mycobacteria. Nature 345: Mercier, J., J. Lachapelle, F. Couture, M. Lafond, G. Vézina, M. Boissinot, and R. C. Levesque Structural and functional characterization of tnpi, a recombinase locus in Tn21 and related -lactamase transposons. J. Bacteriol. 172: Minakhina, S., G. Kholodii, S. Mindlin, O. Yurieva, and V. Nikiforov Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33: Nakaya, R., A. Nakamura, and Y. Murata Resistance transfer agents in Shigella. Biochem. Biophys. Res. Commun. 3: Osbourn, S. E. V., A. K. Turner, and J. Grinsted Nucleotide sequence within Tn3926 confirms this as a Tn21-like transposable element and provides evidence for the origin of the mer operon carried by plasmid pklh2. Plasmid 33: Ouellette, M., L. Bissonnette, and P. H. Roy Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 -lactamase gene. Proc. Natl. Acad. Sci. USA 84: Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. Stanisich, and C. M. Thomas Complete nucleotide sequence of Birmingham IncP plasmids. Compilation and comparative analysis. J. Mol. Biol. 239: Rådström, P., O. Sköld, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176: Rajakumar, K., D. Bulach, J. Davies, L. Ambrose, C. Sasakawa, and B. Adler Identification of a chromosomal Shigella flexneri multi-antibiotic resistance locus which shares sequence and organizational similarity with the resistance region of the plasmid NR1. Plasmid 37: Rogowsky, P., S. E. Halford, and R. Schmitt Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 4: Rubens, C. E., W. F. McNeill, and W. E. Farrar, Jr Transposable plasmid deoxyribonucleic acid sequence in Pseudomonas aeruginosa which mediates resistance to gentamicin and four other antimicrobial agents. J. Bacteriol. 139: Sambrook, J., E. F. Fritsch, and T. Maniatis Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. 38. Sanger, F., S. Nicklen, and A. R. Coulson DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: Schmidt, F The role of insertions, deletions, and substitutions in the evolution of R6 related plasmids encoding aminoglycoside transferase ANT- (2 ). Mol. Gen. Genet. 194: Schmidt, F., and I. Klopfer-Kaul Evolutionary relationship between Tn21-like elements and pbp201, a plasmid from Klebsiella pneumoniae mediating resistance to gentamicin and eight other drugs. Mol. Gen. Genet. 197: Schmidt, F. R. J., E. J. Nücken, and R. B. Henschke Structure and function of hot spots providing signals for site-directed specific recombination and gene expression in Tn21 transposons. Mol. Microbiol. 3: Smith, D. I., R. Gomez Lus, M. C. Rubio Calvo, N. Datta, A. E. Jacob, and R. W. Hedges Third type of plasmid conferring gentamicin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 8: Stokes, H. W., and R. M. Hall A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3: Stokes, H. W., and R. M. Hall Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26: Sundström, L., P. Rådström, G. Swedberg, and O. Sköld Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrv and sull and a recombination active locus of Tn21. Mol. Gen. Genet. 213: Tabor, S., and C. C. Richardson DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 84: Tenover, F. C., D. Filpula, K. L. Phillips, and J. L. Plorde Cloning and sequencing of a gene encoding an aminoglycoside 6 -N-acetyltransferase from an R factor of Citrobacter diversus. J. Bacteriol. 170: Tenover, F. C., K. L. Phillips, T. Gilbert, P. Lockhart, P. J. O Hara, and J. J. Plorde Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob. Agents Chemother. 33: Vézina, G., and R. C. Levesque Molecular characterization of the class II multiresistance transposable element Tn1403 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35: Wohlleben, W., W. Arnold, L. Bissonnette, A. Pelletier, A. Tanguay, P. H. Roy, G. C. Gamboa, G. F. Barry, E. Aubert, J. Davies, and S. A. Kagan On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacci) for gentamicin acetyltransferase-3-i(aac(3)-i), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 217: Yanisch-Perron, C., J. Vieira, and J. Messing Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and puc19 vectors. Gene 33: Yeo, C. C., J. M. Tham, S. M. Kwong, S. Yiin, and C. L. Poh Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pra2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165: Yurieva, O., G. Kholodii, L. Minakhin, Z. Gorlenko, E. Kalyaeva, S. Mindlin, and V. Nikiforov Intercontinental spread of promiscuous mercuryresistance transposons in environmental bacteria. Mol. Microbiol. 24: Zühlsdorf, M. T., and B. Wiedemann Functional and physiological characterization of the Tn21 cassette for resistance genes in Tn2426. J. Gen. Microbiol. 139:
Characterization and Movement of the Class 1 Integron Known as Tn2521 and Tn1405
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, May 2002, p. 1288 1294 Vol. 46, No. 5 0066-4804/02/$04.00 0 DOI: 10.1128/AAC.46.5.1288 1294.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved.
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, May 2002, p. 1288 1294 Vol. 46, No. 5 0066-4804/02/$04.00 0 DOI: 10.1128/AAC.46.5.1288 1294.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved.
Detection of gene cassettes in Tn402-like class 1 integrons. Amplification of the gene cassettes in class 1 integrons by PCR using primers in the
 AAC Accepts, published online ahead of print on June 00 Antimicrob. Agents Chemother. doi:./aac.000-0 Copyright 00, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
AAC Accepts, published online ahead of print on June 00 Antimicrob. Agents Chemother. doi:./aac.000-0 Copyright 00, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
TRANSMISSIBLE GENETIC ELEMENTS: PLASMIDS, TRANSPOSONS & INTEGRONS
 TRANSMISSIBLE GENETIC ELEMENTS: PLASMIDS, TRANSPOSONS & INTEGRONS MOBILE GENETIC ELEMENTS 1 PLASMIDS -MOST OFTEN CIRCULAR MOLECULES OF DOUBLE-STRANDED DNA -VARY WIDELY IN SIZE -SELF-TRANSMISSIBLE ELEMENTS
TRANSMISSIBLE GENETIC ELEMENTS: PLASMIDS, TRANSPOSONS & INTEGRONS MOBILE GENETIC ELEMENTS 1 PLASMIDS -MOST OFTEN CIRCULAR MOLECULES OF DOUBLE-STRANDED DNA -VARY WIDELY IN SIZE -SELF-TRANSMISSIBLE ELEMENTS
MicroReview Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination
 Molecular Microbiology (1995) 15(4), 593 600 MicroReview Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination Ruth M. Hall* and Christina M. Collis CSIRO Division
Molecular Microbiology (1995) 15(4), 593 600 MicroReview Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination Ruth M. Hall* and Christina M. Collis CSIRO Division
Resistance gene mobility mechanisms
 Resistance gene mobility mechanisms Sally Partridge sally.partridge@health.nsw.gov.au CPATH 2015 Kuala Lumpur 13 th June 2015 Mobile resistance genes R R gene plasmid chromosome Mobile resistance genes
Resistance gene mobility mechanisms Sally Partridge sally.partridge@health.nsw.gov.au CPATH 2015 Kuala Lumpur 13 th June 2015 Mobile resistance genes R R gene plasmid chromosome Mobile resistance genes
A Hot Spot in Plasmid F for Site-Specific Recombination Mediated by Tn21 Integron Integrase
 JOURNAL OF BACTERIOLOGY, July 1997, p. 4419 4425 Vol. 179, No. 13 0021-9193/97/$04.00 0 Copyright 1997, American Society for Microbiology A Hot Spot in Plasmid F for Site-Specific Recombination Mediated
JOURNAL OF BACTERIOLOGY, July 1997, p. 4419 4425 Vol. 179, No. 13 0021-9193/97/$04.00 0 Copyright 1997, American Society for Microbiology A Hot Spot in Plasmid F for Site-Specific Recombination Mediated
Genetic support of Extended- Spectrum ß-Lactamases
 Genetic support of Extended- Spectrum ß-Lactamases Laurent Poirel Dept of Microbiology (Pr Nordmann) Bicêtre Hospital. South-Paris Medical School. France ESCMID Conference on ESBL 29-31 May 2006 TEM-like
Genetic support of Extended- Spectrum ß-Lactamases Laurent Poirel Dept of Microbiology (Pr Nordmann) Bicêtre Hospital. South-Paris Medical School. France ESCMID Conference on ESBL 29-31 May 2006 TEM-like
Definition of the atti1 site of class 1 integrons
 Microbiology (2000), 146, 2855 2864 Printed in Great Britain Definition of the atti1 site of class 1 integrons Sally R. Partridge, 1,2 Gavin D. Recchia, 1,2 Carol Scaramuzzi, 1,2 Christina M. Collis, 1
Microbiology (2000), 146, 2855 2864 Printed in Great Britain Definition of the atti1 site of class 1 integrons Sally R. Partridge, 1,2 Gavin D. Recchia, 1,2 Carol Scaramuzzi, 1,2 Christina M. Collis, 1
Genetic Adaptation II. Microbial Physiology Module 3
 Genetic Adaptation II Microbial Physiology Module 3 Topics Topic 4: Topic 5: Transposable Elements Exchange of Genetic Material Between Organisms Topic 5a: Protection Against Foreign DNA Aims and Objectives
Genetic Adaptation II Microbial Physiology Module 3 Topics Topic 4: Topic 5: Transposable Elements Exchange of Genetic Material Between Organisms Topic 5a: Protection Against Foreign DNA Aims and Objectives
New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type dihydrofolate reductase
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2006 New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2006 New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type
Mobile gene cassettes and integrons: moving antibiotic resistance genes in Gram-negative bacteria
 Mobile gene cassettes and integrons: moving antibiotic resistance genes in Gram-negative bacteria Ruth M. Hall Novartis Foundation Symposium Edited by Derek J. Chadwick, Gail Cardew Copyright 0 1997 by
Mobile gene cassettes and integrons: moving antibiotic resistance genes in Gram-negative bacteria Ruth M. Hall Novartis Foundation Symposium Edited by Derek J. Chadwick, Gail Cardew Copyright 0 1997 by
Transposon Tn21, Flagship of the Floating Genome
 MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 1999, p. 507 522 Vol. 63, No. 3 1092-2172/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Transposon Tn21, Flagship
MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 1999, p. 507 522 Vol. 63, No. 3 1092-2172/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Transposon Tn21, Flagship
Gene Integration in the Escherichia coli Chromosome Mediated by Tn21 Integrase (Int21)
 JOURNAL OF BACTERIOLOGY, Feb. 1996, p. 894 898 Vol. 178, No. 3 0021-9193/96/$04.00 0 Copyright 1996, American Society for Microbiology Gene Integration in the Escherichia coli Chromosome Mediated by Tn21
JOURNAL OF BACTERIOLOGY, Feb. 1996, p. 894 898 Vol. 178, No. 3 0021-9193/96/$04.00 0 Copyright 1996, American Society for Microbiology Gene Integration in the Escherichia coli Chromosome Mediated by Tn21
Mutagenesis for Studying Gene Function Spring, 2007 Guangyi Wang, Ph.D. POST103B
 Mutagenesis for Studying Gene Function Spring, 2007 Guangyi Wang, Ph.D. POST103B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm Overview of Last Lecture DNA microarray hybridization
Mutagenesis for Studying Gene Function Spring, 2007 Guangyi Wang, Ph.D. POST103B guangyi@hawaii.edu http://www.soest.hawaii.edu/marinefungi/ocn403webpage.htm Overview of Last Lecture DNA microarray hybridization
Tn502 and Tn512 Are res Site Hunters That Provide Evidence of Resolvase-Independent Transposition to Random Sites
 JOURNAL OF BACTERIOLOGY, Apr. 2010, p. 1865 1874 Vol. 192, No. 7 0021-9193/10/$12.00 doi:10.1128/jb.01322-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Tn502 and Tn512 Are
JOURNAL OF BACTERIOLOGY, Apr. 2010, p. 1865 1874 Vol. 192, No. 7 0021-9193/10/$12.00 doi:10.1128/jb.01322-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. Tn502 and Tn512 Are
Site-Specific Deletion and Rearrangement of Integron Insert Genes Catalyzed by the Integron DNA Integrase
 JOURNAL OF BACTERIOLOGY, Mar. 1992, p. 1574-1585 0021-9193/92/051574-12$02.00/0 Copyright @ 1992, American Society for Microbiology Vol. 174, No. 5 Site-Specific Deletion and Rearrangement of Integron
JOURNAL OF BACTERIOLOGY, Mar. 1992, p. 1574-1585 0021-9193/92/051574-12$02.00/0 Copyright @ 1992, American Society for Microbiology Vol. 174, No. 5 Site-Specific Deletion and Rearrangement of Integron
Molecular Biology: Gene cloning
 Molecular Biology: Gene cloning Author: Prof Marinda Oosthuizen Licensed under a Creative Commons Attribution license. CLONING VECTORS The central component of a gene cloning experiment is the vector or
Molecular Biology: Gene cloning Author: Prof Marinda Oosthuizen Licensed under a Creative Commons Attribution license. CLONING VECTORS The central component of a gene cloning experiment is the vector or
Genetic Engineering & Recombinant DNA
 Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Genetic Engineering & Recombinant DNA Chapter 10 Copyright The McGraw-Hill Companies, Inc) Permission required for reproduction or display. Applications of Genetic Engineering Basic science vs. Applied
Bacterial Transposons
 Bacterial Transposons Cornelis, G. Department of Microbiology. University of Louvain. UCL 30.58. B 1200 Brussels. Belgium A. Bacterial Plasmids Bacterial plasmids have been known for more than twenty years
Bacterial Transposons Cornelis, G. Department of Microbiology. University of Louvain. UCL 30.58. B 1200 Brussels. Belgium A. Bacterial Plasmids Bacterial plasmids have been known for more than twenty years
Lac Operon contains three structural genes and is controlled by the lac repressor: (1) LacY protein transports lactose into the cell.
 Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
Regulation of gene expression a. Expression of most genes can be turned off and on, usually by controlling the initiation of transcription. b. Lactose degradation in E. coli (Negative Control) Lac Operon
independent transposition to random sites
 JB Accepts, published online ahead of print on 29 January 2010 J. Bacteriol. doi:10.1128/jb.01322-09 Copyright 2010, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
JB Accepts, published online ahead of print on 29 January 2010 J. Bacteriol. doi:10.1128/jb.01322-09 Copyright 2010, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights
Mercury Resistance Determinants Related to Tn21, Tn1696, and Tn5053 in Enterobacteria from the Preantibiotic Era
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Mar. 2003, p. 1115 1119 Vol. 47, No. 3 0066-4804/03/$08.00 0 DOI: 10.1128/AAC.47.3.1115 1119.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved.
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Mar. 2003, p. 1115 1119 Vol. 47, No. 3 0066-4804/03/$08.00 0 DOI: 10.1128/AAC.47.3.1115 1119.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved.
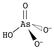 ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
Biological consequences of site specific recombination: integration, excision, deletion
 Biological consequences of site specific recombination: integration, excision, deletion The types of DNA rearrangements promoted by a large number of site specific recombination systems and their physiological
Biological consequences of site specific recombination: integration, excision, deletion The types of DNA rearrangements promoted by a large number of site specific recombination systems and their physiological
GENETICS EXAM 3 FALL a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size.
 Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Student Name: All questions are worth 5 pts. each. GENETICS EXAM 3 FALL 2004 1. a) is a technique that allows you to separate nucleic acids (DNA or RNA) by size. b) Name one of the materials (of the two
Mcbio 316: Exam 3. (10) 2. Compare and contrast operon vs gene fusions.
 Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
Mcbio 316: Exam 3 Name (15) 1. Transposons provide useful tools for genetic analysis. List 5 different uses of transposon insertions. ANSWER: Many answers are possible, however, if multiple items on the
An estimate of the physical distance between two linked markers in Haemophilus influenzae
 J. Biosci., Vol. 13, No. 3, September 1988, pp. 223 228. Printed in India. An estimate of the physical distance between two linked markers in Haemophilus influenzae Ε. Β. SAMIWALA, VASUDHA P. JOSHI and
J. Biosci., Vol. 13, No. 3, September 1988, pp. 223 228. Printed in India. An estimate of the physical distance between two linked markers in Haemophilus influenzae Ε. Β. SAMIWALA, VASUDHA P. JOSHI and
Antibiotic resistance genes located in integrons isolated from Escherichia coli recovered from humans and animals
 University of Wollongong Research Online University of Wollongong Thesis Collection 1954-2016 University of Wollongong Thesis Collections 2009 Antibiotic resistance genes located in integrons isolated
University of Wollongong Research Online University of Wollongong Thesis Collection 1954-2016 University of Wollongong Thesis Collections 2009 Antibiotic resistance genes located in integrons isolated
Molecular Biology Techniques Supporting IBBE
 Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Molecular Biology Techniques Supporting IBBE Jared Cartwright Protein Production Lab Head Contact Details: email jared.cartwright@york.ac.uk Phone 01904 328797 Presentation Aims Gene synthesis Cloning
Antibiotic Resistance: Ampicillin and Gentamicin Bacterial Backbone: pfastbac (Invitrogen)
 G01066 pfbaavmcswtiresmcherrybghpa Plasmid Features: Coordinates Feature 194-348 Tn7L 377-617 SV40pA Complementary 678-818 AAV2 ITR (141bp) 860-955 mcs 956-1542 wtires 1543-2253 mcherry 2268-2481 BgHpA
G01066 pfbaavmcswtiresmcherrybghpa Plasmid Features: Coordinates Feature 194-348 Tn7L 377-617 SV40pA Complementary 678-818 AAV2 ITR (141bp) 860-955 mcs 956-1542 wtires 1543-2253 mcherry 2268-2481 BgHpA
7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions will be posted on the web.
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 4 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
Genetics Lecture Notes Lectures 13 16
 Genetics Lecture Notes 7.03 2005 Lectures 13 16 Lecture 13 Transposable elements Transposons are usually from 10 3 to 10 4 base pairs in length, depending on the transposon type. The key property of transposons
Genetics Lecture Notes 7.03 2005 Lectures 13 16 Lecture 13 Transposable elements Transposons are usually from 10 3 to 10 4 base pairs in length, depending on the transposon type. The key property of transposons
Molecular Genetics Techniques. BIT 220 Chapter 20
 Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Molecular Genetics Techniques BIT 220 Chapter 20 What is Cloning? Recombinant DNA technologies 1. Producing Recombinant DNA molecule Incorporate gene of interest into plasmid (cloning vector) 2. Recombinant
Antibiotic Resistance: Ampicillin and Gentamicin Bacterial Backbone: pfastbac (Invitrogen)
 G01067 pfbaavcagmcswtiresmcherrybghpa Plasmid Features: Coordinates Feature 194-348 Tn7L 377-617 SV40pA Complementary 678-818 AAV2 ITR (141bp) 938-2607 CAG 2600-2685 mcs 2687-3273 wtires 3274-3984 mcherry
G01067 pfbaavcagmcswtiresmcherrybghpa Plasmid Features: Coordinates Feature 194-348 Tn7L 377-617 SV40pA Complementary 678-818 AAV2 ITR (141bp) 938-2607 CAG 2600-2685 mcs 2687-3273 wtires 3274-3984 mcherry
20. . clonal transmission. (conjugation). DNA [1-3]. [4]. [6,7]. DNA. . bacteriophage. clone [7]. DNA [5]. . Integron
![20. . clonal transmission. (conjugation). DNA [1-3]. [4]. [6,7]. DNA. . bacteriophage. clone [7]. DNA [5]. . Integron 20. . clonal transmission. (conjugation). DNA [1-3]. [4]. [6,7]. DNA. . bacteriophage. clone [7]. DNA [5]. . Integron](/thumbs/79/79665377.jpg) 20.. clonal transmission [1-3]. [4]. DNA [5].. Integron. Integron cassette. ( Enterobacteriaceae) integron. 2005. : 05/2/16 : 05/3/14 : (501-759) 588 TEL : 062)220-3259, 3272 FAX : 062)232-2063 E-mail
20.. clonal transmission [1-3]. [4]. DNA [5].. Integron. Integron cassette. ( Enterobacteriaceae) integron. 2005. : 05/2/16 : 05/3/14 : (501-759) 588 TEL : 062)220-3259, 3272 FAX : 062)232-2063 E-mail
MCB 102 University of California, Berkeley August 11 13, Problem Set 8
 MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Regulation of enzyme synthesis
 Regulation of enzyme synthesis The lac operon is an example of an inducible operon - it is normally off, but when a molecule called an inducer is present, the operon turns on. The trp operon is an example
Regulation of enzyme synthesis The lac operon is an example of an inducible operon - it is normally off, but when a molecule called an inducer is present, the operon turns on. The trp operon is an example
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Hetero-Stagger PCR Cloning Kit
 Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
/30/06 Transposons
 411-7 1/30/06 Transposons PP: IV. Transposons A. Classes of transposons B. Mechanisms- 1.replicative (Tn3) 2.conservative (Tn10) C. Genetic Consequences D. Regulation OH: RecA+ RecA Survival UV (100%)
411-7 1/30/06 Transposons PP: IV. Transposons A. Classes of transposons B. Mechanisms- 1.replicative (Tn3) 2.conservative (Tn10) C. Genetic Consequences D. Regulation OH: RecA+ RecA Survival UV (100%)
Antimicrobial Agents and Chemotherapy New Data Letter
 AAC Accepted Manuscript Posted Online 15 August 2016 Antimicrob. Agents Chemother. doi:10.1128/aac.01519-16 Copyright 2016, American Society for Microbiology. All Rights Reserved. 1 Antimicrobial Agents
AAC Accepted Manuscript Posted Online 15 August 2016 Antimicrob. Agents Chemother. doi:10.1128/aac.01519-16 Copyright 2016, American Society for Microbiology. All Rights Reserved. 1 Antimicrobial Agents
CHE.167 Genetics. Bacterial Plasmids
 1 Bacterial Plasmids Open circular form Covalently closed circles (ccc-form) 2 Bacterial plasmids Replication, Maintenance Genes for specific functions Replication - Origin of replication (oriv) - Regulatory
1 Bacterial Plasmids Open circular form Covalently closed circles (ccc-form) 2 Bacterial plasmids Replication, Maintenance Genes for specific functions Replication - Origin of replication (oriv) - Regulatory
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Genetics Time Allowed: 1 hour and 30 minutes Marking Scheme: Total marks available
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Genetics Time Allowed: 1 hour and 30 minutes Marking Scheme: Total marks available
Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe:
 1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
1). (12 Points) Packaging of P22 DNA requires a pac site while packaging of lambda DNA requires a cos site. Briefly describe: 1. The mechanisms used by P22 and lambda to package DNA. P22 uses a headfull
Multiple cloning site between CAG and wild type IRES:
 G0747 pfbaavcagmcs wtiresegfpbghpa Plasmid Features: Coordinates Feature 194-348 Tn7L 376-511 SV40pA Complementary 678-818 AAV2 ITR (141bp) 938-2607 CAG promoter 872-955 mcs 2687-3273 wtires (11th ATG
G0747 pfbaavcagmcs wtiresegfpbghpa Plasmid Features: Coordinates Feature 194-348 Tn7L 376-511 SV40pA Complementary 678-818 AAV2 ITR (141bp) 938-2607 CAG promoter 872-955 mcs 2687-3273 wtires (11th ATG
M. Labbate,* P. Roy Chowdhury, and H. W. Stokes. Department of Chemistry and Biomolecular Sciences, Macquarie University, Sydney, Australia
 JOURNAL OF BACTERIOLOGY, Aug. 2008, p. 5318 5327 Vol. 190, No. 15 0021-9193/08/$08.00 0 doi:10.1128/jb.00199-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. A Class 1 Integron
JOURNAL OF BACTERIOLOGY, Aug. 2008, p. 5318 5327 Vol. 190, No. 15 0021-9193/08/$08.00 0 doi:10.1128/jb.00199-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. A Class 1 Integron
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Xcc strains 8004, ATCC33913, B100 and B Rectangular squares indicate the
 Supplementary information Title: Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Authors: Hui-Zhao Su, Liu Wu, Yan-Hua
Supplementary information Title: Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Authors: Hui-Zhao Su, Liu Wu, Yan-Hua
d. reading a DNA strand and making a complementary messenger RNA
 Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
Biol/ MBios 301 (General Genetics) Spring 2003 Second Midterm Examination A (100 points possible) Key April 1, 2003 10 Multiple Choice Questions-4 pts. each (Choose the best answer) 1. Transcription involves:
DNA Cloning with Cloning Vectors
 Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Lecture Four. Molecular Approaches I: Nucleic Acids
 Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
Lecture Four. Molecular Approaches I: Nucleic Acids I. Recombinant DNA and Gene Cloning Recombinant DNA is DNA that has been created artificially. DNA from two or more sources is incorporated into a single
3 Designing Primers for Site-Directed Mutagenesis
 3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
3 Designing Primers for Site-Directed Mutagenesis 3.1 Learning Objectives During the next two labs you will learn the basics of site-directed mutagenesis: you will design primers for the mutants you designed
B. Incorrect! Ligation is also a necessary step for cloning.
 Genetics - Problem Drill 15: The Techniques in Molecular Genetics No. 1 of 10 1. Which of the following is not part of the normal process of cloning recombinant DNA in bacteria? (A) Restriction endonuclease
Genetics - Problem Drill 15: The Techniques in Molecular Genetics No. 1 of 10 1. Which of the following is not part of the normal process of cloning recombinant DNA in bacteria? (A) Restriction endonuclease
Contextualization and Classification of Natural Genetic Engineering Operators
 Contextualization and Classification of Natural Genetic Engineering Operators James A. Shapiro University of Chicago jsha@uchicago.edu http://shapiro.bsd.uchicago.edu Cells as sophisticated, powerful cognitive
Contextualization and Classification of Natural Genetic Engineering Operators James A. Shapiro University of Chicago jsha@uchicago.edu http://shapiro.bsd.uchicago.edu Cells as sophisticated, powerful cognitive
Determination of Exclusion Effect in Wild Type and Rop Deficient Mutated pbr322 Co-transformations
 Determination of Exclusion Effect in Wild Type and Rop Deficient Mutated pbr322 Co-transformations Suzana Sabaiduc and Andy Lo Microbiology and Immunology University of British Columbia pbr322 is an expression
Determination of Exclusion Effect in Wild Type and Rop Deficient Mutated pbr322 Co-transformations Suzana Sabaiduc and Andy Lo Microbiology and Immunology University of British Columbia pbr322 is an expression
Supporting Information
 Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Supporting Information Gómez-Marín et al. 10.1073/pnas.1505463112 SI Materials and Methods Generation of BAC DK74B2-six2a::GFP-six3a::mCherry-iTol2. BAC clone (number 74B2) from DanioKey zebrafish BAC
Biology 4100 Minor Assignment 1 January 19, 2007
 Biology 4100 Minor Assignment 1 January 19, 2007 This assignment is due in class on February 6, 2007. It is worth 7.5% of your final mark for this course. Your assignment must be typed double-spaced on
Biology 4100 Minor Assignment 1 January 19, 2007 This assignment is due in class on February 6, 2007. It is worth 7.5% of your final mark for this course. Your assignment must be typed double-spaced on
Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for :
 Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Transformation Insertion of DNA of interest Amplification Amplified segment of DNA can be purified from bacteria in sufficient quantity and quality for : DNA Sequence. Understand relatedness of genes and
Genomic Sequencing. Genomic Sequencing. Maj Gen (R) Suhaib Ahmed, HI (M)
 Maj Gen (R) Suhaib Ahmed, HI (M) The process of determining the sequence of an unknown DNA is called sequencing. There are many approaches for DNA sequencing. In the last couple of decades automated Sanger
Maj Gen (R) Suhaib Ahmed, HI (M) The process of determining the sequence of an unknown DNA is called sequencing. There are many approaches for DNA sequencing. In the last couple of decades automated Sanger
Supplementary Material. Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological
 Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Materials and methods. by University of Washington Yeast Resource Center) from several promoters, including
 Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Supporting online material for Elowitz et al. report Materials and methods Strains and plasmids. Plasmids expressing CFP or YFP (wild-type codons, developed by University of Washington Yeast Resource Center)
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Bottom-up genome assembly using the Bacillus subtilis genome vector
 Bottom-up genome assembly using the Bacillus subtilis genome vector Mitsuhiro Itaya, Kyoko Fujita, Azusa Kuroki & Kenji Tsuge Supplementary figures and text: Supplementary Table 1 Primers used to design
Bottom-up genome assembly using the Bacillus subtilis genome vector Mitsuhiro Itaya, Kyoko Fujita, Azusa Kuroki & Kenji Tsuge Supplementary figures and text: Supplementary Table 1 Primers used to design
Supplementary Information
 Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
Supplementary Information Supplementary Figures Figure S1. Study of mgtl translation in vitro. (A) Detection of 5 LR RNA using wild-type and anti-sd (91-95) substituted templates in a transcription-translation
7. (10pts.) The diagram below shows the immunity region of phage l. The genes are:
 7. (10pts.) The diagram below shows the immunity region of phage l. The genes are: ci codes for ci repressor. The ci857 allele is temperature sensitive. cro codes for Cro protein. N codes for the transcription
7. (10pts.) The diagram below shows the immunity region of phage l. The genes are: ci codes for ci repressor. The ci857 allele is temperature sensitive. cro codes for Cro protein. N codes for the transcription
CHAPTER 2A HOW DO YOU BEGIN TO CLONE A GENE? CHAPTER 2A STUDENT GUIDE 2013 Amgen Foundation. All rights reserved.
 CHAPTER 2A HOW DO YOU BEGIN TO CLONE A GENE? 35 INTRODUCTION In the Program Introduction, you learned that the increase in diabetes in the United States has resulted in a great demand for its treatment,
CHAPTER 2A HOW DO YOU BEGIN TO CLONE A GENE? 35 INTRODUCTION In the Program Introduction, you learned that the increase in diabetes in the United States has resulted in a great demand for its treatment,
7.014 Solution Set 4
 7.014 Solution Set 4 Question 1 Shown below is a fragment of the sequence of a hypothetical bacterial gene. This gene encodes production of HWDWN, protein essential for metabolizing sugar yummose. The
7.014 Solution Set 4 Question 1 Shown below is a fragment of the sequence of a hypothetical bacterial gene. This gene encodes production of HWDWN, protein essential for metabolizing sugar yummose. The
Reading Lecture 3: 24-25, 45, Lecture 4: 66-71, Lecture 3. Vectors. Definition Properties Types. Transformation
 Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Characterization of the Class 3 Integron and the Site-Specific Recombination System It Determines
 JOURNAL OF BACTERIOLOGY, June 2002, p. 3017 3026 Vol. 184, No. 11 0021-9193/02/$04.00 0 DOI: 10.1128/JB.184.11.3017 3026.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Characterization
JOURNAL OF BACTERIOLOGY, June 2002, p. 3017 3026 Vol. 184, No. 11 0021-9193/02/$04.00 0 DOI: 10.1128/JB.184.11.3017 3026.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Characterization
Lectures of Dr.Mohammad Alfaham. The Bacterial Genetics
 Lectures of Dr.Mohammad Alfaham The Bacterial Genetics is the total collection of genes carried by a bacterium both on its chromosome and on its extrachromosomal genetic elements (plasmids) A Gene A gene
Lectures of Dr.Mohammad Alfaham The Bacterial Genetics is the total collection of genes carried by a bacterium both on its chromosome and on its extrachromosomal genetic elements (plasmids) A Gene A gene
Construction of shuttle vectors for cloning in Vibrio cholerae and Escherichia coli
 J. Biosci., Vol. 20, Number 3, June 1995, pp 367 376. Printed in India. Construction of shuttle vectors for cloning in Vibrio cholerae and Escherichia coli 1. Introduction DIBYENDU Κ PANDA*, RAJAT BANERJEE
J. Biosci., Vol. 20, Number 3, June 1995, pp 367 376. Printed in India. Construction of shuttle vectors for cloning in Vibrio cholerae and Escherichia coli 1. Introduction DIBYENDU Κ PANDA*, RAJAT BANERJEE
The nucleotide sequences and expression of genes for the beta and epsilon subunits of ATP synthase from rice (Oryza sativa L.)
 Jpn. J. Genet. (1989) 64, pp. 223-229 The nucleotide sequences and expression of genes for the beta and epsilon subunits of ATP synthase from rice (Oryza sativa L.) Yoko NIsxtzAwA1 and Atsushi HIRAI* Graduate
Jpn. J. Genet. (1989) 64, pp. 223-229 The nucleotide sequences and expression of genes for the beta and epsilon subunits of ATP synthase from rice (Oryza sativa L.) Yoko NIsxtzAwA1 and Atsushi HIRAI* Graduate
DNA recombination without ligase: TOPO TA Cloning. topoisomerase
 DNA recombination without ligase: TOPO TA Cloning topoisomerase Cloning strategies, cloning in bacteria other than E.coli Mitesh Shrestha Cloning strategies Cloning in bacteria other than E.coli Convenient
DNA recombination without ligase: TOPO TA Cloning topoisomerase Cloning strategies, cloning in bacteria other than E.coli Mitesh Shrestha Cloning strategies Cloning in bacteria other than E.coli Convenient
Genes specifying degradation of 3-chlorobenzoic acid in plasmids pac27 and pjp4
 Proc. Natl. Acad. Sci. USA Vol. 82, pp. 1638-1642, March 1985 Biochemistry Genes specifying degradation of 3-chlorobenzoic acid in plasmids pac27 and pjp4 (genome organization/gene cloning/gene amplification/genetic
Proc. Natl. Acad. Sci. USA Vol. 82, pp. 1638-1642, March 1985 Biochemistry Genes specifying degradation of 3-chlorobenzoic acid in plasmids pac27 and pjp4 (genome organization/gene cloning/gene amplification/genetic
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Transmission of genetic variation: conjugation. Transmission of genetic variation: conjugation. Transmission of genetic variation: F+ conjugation
 Transmission of genetic variation: conjugation Transmission of genetic variation: conjugation Direct transfer of DNA from one strain to another mediated by fertility factor (F). Best studied in E. coli,
Transmission of genetic variation: conjugation Transmission of genetic variation: conjugation Direct transfer of DNA from one strain to another mediated by fertility factor (F). Best studied in E. coli,
Module Code: BIO00007C
 Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Genetics Time Allowed: 1 Hour 30 Minutes Marking Scheme: Total marks available for
Examination Candidate Number: Desk Number: BSc and MSc Degree Examinations 2018-9 Department : BIOLOGY Title of Exam: Genetics Time Allowed: 1 Hour 30 Minutes Marking Scheme: Total marks available for
7.1 Techniques for Producing and Analyzing DNA. SBI4U Ms. Ho-Lau
 7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
7.1 Techniques for Producing and Analyzing DNA SBI4U Ms. Ho-Lau What is Biotechnology? From Merriam-Webster: the manipulation of living organisms or their components to produce useful usually commercial
Bacterial Genetics. Prof. Dr. Asem Shehabi Faculty of Medicine University of Jordan
 Bacterial Genetics Prof. Dr. Asem Shehabi Faculty of Medicine University of Jordan Bacterial Genes-1 All patterns of growth, metabolism, essential cellular structures, biological characteristics of bacteria
Bacterial Genetics Prof. Dr. Asem Shehabi Faculty of Medicine University of Jordan Bacterial Genes-1 All patterns of growth, metabolism, essential cellular structures, biological characteristics of bacteria
Name Section Problem Set 3
 Name Section 7.013 Problem Set 3 The completed problem must be turned into the wood box outside 68120 by 4:40 pm, Thursday, March 13. Problem sets will not be accepted late. Question 1 Based upon your
Name Section 7.013 Problem Set 3 The completed problem must be turned into the wood box outside 68120 by 4:40 pm, Thursday, March 13. Problem sets will not be accepted late. Question 1 Based upon your
BIO 202 Midterm Exam Winter 2007
 BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
BIO 202 Midterm Exam Winter 2007 Mario Chevrette Lectures 10-14 : Question 1 (1 point) Which of the following statements is incorrect. a) In contrast to prokaryotic DNA, eukaryotic DNA contains many repetitive
Chapter 15 Recombinant DNA and Genetic Engineering. Restriction Enzymes Function as Nature s Pinking Shears
 Chapter 15 Recombinant DNA and Genetic Engineering In this chapter you will learn How restriction enzyme work and why they are essential to DNA technology. About various procedures such as cloning and
Chapter 15 Recombinant DNA and Genetic Engineering In this chapter you will learn How restriction enzyme work and why they are essential to DNA technology. About various procedures such as cloning and
Evolution of Tn22-related Transposons: Isolation of Tn2425, which Harbours IS262
 Journal of General Microbiology (1 985), 131, I 123-1 130. Printed in Great Britain 1123 Evolution of Tn22-related Transposons: Isolation of Tn2425, which Harbours IS262 By JOACHIM F. MEYER, BERTHOLD A.
Journal of General Microbiology (1 985), 131, I 123-1 130. Printed in Great Britain 1123 Evolution of Tn22-related Transposons: Isolation of Tn2425, which Harbours IS262 By JOACHIM F. MEYER, BERTHOLD A.
CHAPTER 20 DNA TECHNOLOGY AND GENOMICS. Section A: DNA Cloning
 Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Section A: DNA Cloning 1. DNA technology makes it possible to clone genes for basic research and commercial applications: an overview 2. Restriction enzymes are used to make recombinant DNA 3. Genes can
Genetic Basis of Variation in Bacteria
 Mechanisms of Infectious Disease Fall 2009 Lecture 2 Jonathan Dworkin, PhD Department of Microbiology jonathan.dworkin@columbia.edu Genetic Basis of Variation in Bacteria I. Organization of genetic material
Mechanisms of Infectious Disease Fall 2009 Lecture 2 Jonathan Dworkin, PhD Department of Microbiology jonathan.dworkin@columbia.edu Genetic Basis of Variation in Bacteria I. Organization of genetic material
The Regulation of Bacterial Gene Expression
 The Regulation of Bacterial Gene Expression Constitutive genes are expressed at a fixed rate Other genes are expressed only as needed Inducible genes Repressible genes Catabolite repression Pre-transcriptional
The Regulation of Bacterial Gene Expression Constitutive genes are expressed at a fixed rate Other genes are expressed only as needed Inducible genes Repressible genes Catabolite repression Pre-transcriptional
Chapter 20 DNA Technology & Genomics. If we can, should we?
 Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
Chapter 20 DNA Technology & Genomics If we can, should we? Biotechnology Genetic manipulation of organisms or their components to make useful products Humans have been doing this for 1,000s of years plant
GENE(S) Carried by transposon
 ANSWER KEY Microbial Genetics BIO 510/610 Fall Quarter 2009 Final Exam 1.) Some Insertion Sequences transpose through a Replicative mechanism of transposition. Other Insertion Sequences utilize a Cut and
ANSWER KEY Microbial Genetics BIO 510/610 Fall Quarter 2009 Final Exam 1.) Some Insertion Sequences transpose through a Replicative mechanism of transposition. Other Insertion Sequences utilize a Cut and
Characteristics of bacterial Plasmid : Size : Conformation : Replication origin of replication : Replication Protein :
 Characteristics of bacterial Plasmid : Size : Conformation : Replication origin of replication : Replication Protein : Definition of Plasmid Plasmids are extrachromosomal circular, double stranded DNA
Characteristics of bacterial Plasmid : Size : Conformation : Replication origin of replication : Replication Protein : Definition of Plasmid Plasmids are extrachromosomal circular, double stranded DNA
FROM EXPERIMENTS IN BACTERIAL GENETICS AND GENE TECHNIQUE
 Uppsala 2001-04-01 REPORT FROM EXPERIMENTS IN BACTERIAL GENETICS AND GENE TECHNIQUE Laboratory assistants: Maria Jönsson Amera Gibreel Students: Contents ASSIGNMENT:... 3 INTRODUCTION:... 3 MATERIAL AND
Uppsala 2001-04-01 REPORT FROM EXPERIMENTS IN BACTERIAL GENETICS AND GENE TECHNIQUE Laboratory assistants: Maria Jönsson Amera Gibreel Students: Contents ASSIGNMENT:... 3 INTRODUCTION:... 3 MATERIAL AND
Mutating Asn-666 to Glu in the O-helix region of the taq DNA polymerase gene
 Research in Pharmaceutical Sciences, April 2010; 5(1): 15-19 Received: Oct 2009 Accepted: Jan 2010 School of Pharmacy & Pharmaceutical Sciences 15 Isfahan University of Medical Sciences Original Article
Research in Pharmaceutical Sciences, April 2010; 5(1): 15-19 Received: Oct 2009 Accepted: Jan 2010 School of Pharmacy & Pharmaceutical Sciences 15 Isfahan University of Medical Sciences Original Article
Selected Techniques Part I
 1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
1 Selected Techniques Part I Gel Electrophoresis Can be both qualitative and quantitative Qualitative About what size is the fragment? How many fragments are present? Is there in insert or not? Quantitative
Motivation From Protein to Gene
 MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
MOLECULAR BIOLOGY 2003-4 Topic B Recombinant DNA -principles and tools Construct a library - what for, how Major techniques +principles Bioinformatics - in brief Chapter 7 (MCB) 1 Motivation From Protein
Genetics and Genomics in Medicine Chapter 3. Questions & Answers
 Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
Genetics and Genomics in Medicine Chapter 3 Multiple Choice Questions Questions & Answers Question 3.1 Which of the following statements, if any, is false? a) Amplifying DNA means making many identical
4/3/2013. DNA Synthesis Replication of Bacterial DNA Replication of Bacterial DNA
 4/3/03 3 4 5 6 7 8 9 0 Chapter 8 Microbial Genetics Terminology Genetics: The study of what genes are, how they carry information, how information is expressed, and how genes are replicated Gene: A segment
4/3/03 3 4 5 6 7 8 9 0 Chapter 8 Microbial Genetics Terminology Genetics: The study of what genes are, how they carry information, how information is expressed, and how genes are replicated Gene: A segment
In other words there are 4.0 x10 5 phosphodiester groups in the basic form to one in the acidic form at ph 7.0.
 In other words there are 4.0 x10 5 phosphodiester groups in the basic form to one in the acidic form at ph 7.0. There are a number of shorthand abbreviations a linear polymer of deoxyribonucleotides. One
In other words there are 4.0 x10 5 phosphodiester groups in the basic form to one in the acidic form at ph 7.0. There are a number of shorthand abbreviations a linear polymer of deoxyribonucleotides. One
MUTANT: A mutant is a strain that has suffered a mutation and exhibits a different phenotype from the parental strain.
 OUTLINE OF GENETICS LECTURE #1 A. TERMS PHENOTYPE: Phenotype refers to the observable properties of an organism, such as morphology, growth rate, ability to grow under different conditions or media. For
OUTLINE OF GENETICS LECTURE #1 A. TERMS PHENOTYPE: Phenotype refers to the observable properties of an organism, such as morphology, growth rate, ability to grow under different conditions or media. For
Chapter 10 Genetic Engineering: A Revolution in Molecular Biology
 Chapter 10 Genetic Engineering: A Revolution in Molecular Biology Genetic Engineering Direct, deliberate modification of an organism s genome bioengineering Biotechnology use of an organism s biochemical
Chapter 10 Genetic Engineering: A Revolution in Molecular Biology Genetic Engineering Direct, deliberate modification of an organism s genome bioengineering Biotechnology use of an organism s biochemical
ET - Recombination. Introduction
 GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
GENERAL & APPLIED GENETICS Geert Van Haute August 2003 ET - Recombination Introduction Homologous recombination is of importance to a variety of cellular processes, including the maintenance of genomic
