Chromatin immunoprecipitation (ChIP) ES and FACS-sorted GFP+/Flk1+ cells were fixed in 1% formaldehyde and sonicated until fragments of an average
|
|
|
- Helena Rosanna Parrish
- 5 years ago
- Views:
Transcription
1 Chromatin immunoprecipitation (ChIP) ES and FACS-sorted cells were fixed in % formaldehyde and sonicated until fragments of an average size of 5 bp were obtained. Soluble, sheared chromatin was diluted to a concentration of µg/ml in RIPA buffer ( mm Tris-HCl ph 7.5, mm EDTA,.5 mm EGTA, % Triton X-,.% SDS,.% Sodium Deoxycholate, 4 mm NaCl, protease inhibitors). To analyse Ringb binding 3 µg chromatin (3 µl) was added to 5 µl protein G Dynabeads (Invitrogen) saturated with anti-ringb antibody (MBL) or nonspecific IgG (DAKO) and incubated for at least 6 hours at 4 C on a rotating wheel. Nonimmunoprecipitated chromatin ( µl) was kept as input. Immune complexes were washed twice in RIPA buffer, twice in RIPA buffer with 5 mm NaCl, once in RIPA buffer with 5 mm LiCl, and twice in TE. Prior to elution, samples were transferred to a new tube. Elution was done in 5 µl elution buffer ( mm Tris-HCl ph 7.5, 5 mm EDTA, 5 mm NaCl, % SDS, 5 µg/ ml Proteinase K, 5 µg/ml RnaASE A) on a shaking thermal block at 68 C for at least 6 hours. Eluates were transferred to a new tube and beads incubated with additional 5 µl of elution buffer for 5 min. The two eluates were pooled and DNA was purified by phenol:chloroform extraction, precipitated and resuspended in 4-8 µl TE. µl were used in each PCR reaction. To analyse the abundance of modified histones the procedure was modified as follows. Each IP contained µg ( µl) chromatin which was added to 5 µl protein A Dynabeads (Invitrogen) saturated with either anti-h3k4me3 (Abcam), anti-h3k7me3 (Millipore), anti-total H3 (Abcam) or non-specific IgG (DAKO). Different anti-h3k4me3 (Millipore) and anti-h3k7me3 (Millipore) antibodies were used for the domain analysis shown in figure C. The reactions were done in duplicate in a 48- well format. Elution was done as described above or in µl modified elution buffer (5 mm Tris-HCl ph 7.5, mm EDTA, 5 µg/ml Proteinase K, 5 µg/ml RNase A, ph 9.8) by incubation for 3 min at 37 C, 6 hours at 68 C, min at 95 C. The eluate was used directly as template in quantitative PCR reactions (5 µl per sample). For sequential ChIP, µg chromatin was incubated with primary antibody (anti-h3k4me3) cross-linked with dimethyl pimelidate dihydrochloride (DMP, Sigma) to protein A-coated magnetic beads (Invitrogen). Following removal of unbound chromatin, the immunoprecipitated material was eluted in mm Tris-HCl ph 7.5, 5 mm EDTA, 5 mm NaCl, % SDS by incubating 3 min at room temperature and min at 68 C. The eluate was diluted : in RIPA
2 buffer without SDS and split into two aliquots receiving either the second antibody (anti- H3K7me3) or non-specific IgG (DAKO) pre-bound to protein A-coated magnetic beads. Immunoprecipitated DNA was quantitated by real-time PCR using primers amplifying the promoter regions of candidate genes (sequences are available upon request). Hematopoietic assay Assays for hematopoietic colonies were performed essentially as described (Fehling et al, Development. 3; 3:47-47). FACS sorted Flk+ (hemangioblast) Ringa -/- /Ringb fl/fl /Cre-ER T cells were reaggregated in either EB differentiation medium or blast medium containing 8 nm 4-OHT (tamoxifen) or vehicle for -3 days before plating in semi-solid hematopoietic medium. Colony scoring was done after 6 days. Figure S. Changes in gene expression and replication timing of candidate loci upon mesodermal differentiation. (A) Expression of marker (Sox, Oct4, Rex), mesoderm-associated (Bra/T, Flk, Ikaros, Myf5) and neuralassociated (Math, Nkx-, Nkx-9, Sox) genes or a ubiquitously expressed gene (β- Actin), in and FACS-enriched cell samples. Values were normalised to a housekeeping gene (Hmbs) and presented as fold change over positive control samples ( for β-actin, Oct4, Sox, Rex; FACS-sorted cells for Bra/T; FACS sorted cells for Flk; B3 pre-b cells for Ikaros, differentiated CC myoblasts for Myf5; embryonic head (E4.5) for Math, Nkx-; GFP+ FACS-sorted, differentiated Sox-GFP 46C for Sox; embryos (embryonic day 8) for Nkx-9). The average and standard deviation from three experiments are shown. (B) Summary of the replication timing profiles of individual loci during mesodermal commitment, colourcoded to facilitate comparisons as previously described (Azuara et al. Nat Cell Biol. 6; 8:53-538; Perry et al. Cell Cycle. 4; 3:645-65); early replication (peak in G and/or S) in green, middle-early (peak in S) in lime, middle (peak in S and S3) in yellow, middle-late (peak in S3) in orange and late replication (peak in S4 and/or G) in red. *Hem. Prog.: the adult hematopoietic precursor cell line FDCP mix (ref. 8). **Neural prog.: ES-derived neural precursors induced with retinoic acid (ref. 3). : Not analysed. Figure S. Overview of the replication timing assay. (A) Asynchronously dividing cell populations were pulsed with BrdU to label newly replicated
3 DNA. The cells were harvested and stained with propidium iodide (PI), sorted according to DNA content into six fractions representing G, four sequential S-phase stages (S-S4) and G/M, as indicated. As a control for equivalent handling of samples, an equal amount of BrdU-labelled Drosophila genomic DNA is spiked into each cell-cycle fraction. After DNA isolation and sonication, BrdU-labelled DNA is immunoprecipitated using anti-brdu antibodies. Quantitation by real-time PCR of the amount of locus-specific newly replicated DNA in each fraction is used to determine the timing of DNA replication as early (peak abundance in G and S), mid-early (peak in S), middle (peak in S and S3), mid-late (peak in S3) or late (peak in S4 and G/M). (B) Results showing equivalent recovery of spiked DNA (Drosophila Gbe gene), and early (α Globin locus) and late (X4 repeat) replicating loci are provided for reference. Figure S3. Analysis of PRC levels and function in Flk+ hemangioblast cells. (A) Quantitative RT-PCR analysis of the mrna levels of PRC components Mel8, Bmi, Ringa and Ringb during mesodermal differentiation of ES cells. Values were normalised to a housekeeping gene (Hmbs) and presented as fold change over. The average and standard deviation from three experiments are shown. (B) Western blot analysis of Ringb levels in sequential three-fold dilutions of whole cell extracts from ES and sorted cell populations. Anti-Tubulin antibody was used as a loading control. (C) FACS analysis of dissociated Ringa -/- /Ringb fl/fl /CreER T embryoid bodies stained with PE-conjugated anti-flk antibody, where the gate show a typical Flk- positive population. The profile is representative of several experiments (>5). (D) Western blot analysis of Ringb levels in whole cell extracts from untreated (-Tam) and tamoxifen treated (+Tam) FACS-sorted Flk+ Ringa -/- /Ringb fl/fl /CreER T cells. Anti-Lamin antibody was used as a loading control. (E) Quantitative RT-PCR analysis of the Ringb gene, hematopoietic Ringb target genes (Scl, Runx, Fli) or non-ringb target genes (Gata) in Ringa -/- / Ringb fl/fl /Cre-ER T (upper) or EB-derived Flk+ hemangioblast cells (lower) three days after addition of Tamoxifen (Tam) to delete Ringb (black bars), or in untreated controls (white bars). Values were normalized to a control gene (Hmbs) and expressed as fold change relative to untreated. The average and standard deviation from two-three experiments are shown. (F) Hematopoietic colony assay was performed in triplicate for untreated (-) and tamoxifen treated (+) samples after culturing FACS-sorted Flk+ Ringa -/- /Ringb fl/fl /CreER T cells for a single day in differentiation medium (with or without tamoxifen), before replating in semisolid hematopoietic medium. Asterisks indicate
4 significantly different numbers of colonies in treated compared to untreated samples (p<.5, student s t-test).
5 A mrna level relative to control/undifferentiated β-actin Sox Oct Rex Bra/T Ikaros Myf Flk Control Math Nkx- Nkx-9 Sox GFP-/Flk- GFP-/Flk- GFP-/Flk- Control B Ctrl αglobin X4 ES Rex Sox Meso Bry Flk Ikaros Myf5 Neural Math Sox Nkx- Nkx-9 GFP-/Flk- GFP+/Flk- GFP+/Flk+ Hem. prog.* Neural prog.** G-S Early S Mid-early S-S3 Middle S3 Mid-late S4-G/M Late Figure S
6 A B Asynchronously growing cells 5 BrdU pulse to label newly replicated DNA 4 3 Quality control (Drosophila Gbe) PI staining and cell cycle sorting according to DNA content Relative cell number G S S S3 S4 G/M PI intensity (DNA content) Spike in Drosophila DNA Sonication and anti-brdu IP Newly replicated DNA (% of total) G S S S3 S4 G/M Early control (α-globin) Late control (X4) Real time PCR Cell cycle fraction Figure S
7 A C D B mrna relative to Ringb Ringb Mel8 GFP-/Flk Ringa Bmi GFP-/Flk- GFP-/Flk- GFP+/Flk- GFP+/Flk+ E mrna level (fold change relative to -Tam) PE-Flk 33 % Fl Flk+ (hemangioblast) Ringb F Lamin Hematopoietic colonies (per 5 cells) -Tam +Tam Day 4 * Tam +Tam Tubulin RingB Scl Runx Fli Gata Figure S3
1. Cross-linking and cell harvesting
 ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
ChIP is a powerful tool that allows the specific matching of proteins or histone modifications to regions of the genome. Chromatin is isolated and antibodies to the antigen of interest are used to determine
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript;
 Supplemental Methods Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript; Bio-Rad, Hercules, CA, USA) and standard RT-PCR experiments were carried out using the 2X GoTaq
Supplemental Methods Quantitative and non-quantitative RT-PCR. cdna was generated from 500ng RNA (iscript; Bio-Rad, Hercules, CA, USA) and standard RT-PCR experiments were carried out using the 2X GoTaq
Supplementary Information: Materials and Methods. Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered
 Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplementary Information: Materials and Methods Immunoblot and immunoprecipitation. Cells were washed in phosphate buffered saline (PBS) and lysed in TNN lysis buffer (50mM Tris at ph 8.0, 120mM NaCl
Supplemental Data. Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes
 Cell, Volume 135 Supplemental Data Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes Andrzej T. Wierzbicki, Jeremy R. Haag, and
Cell, Volume 135 Supplemental Data Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes Andrzej T. Wierzbicki, Jeremy R. Haag, and
SUPPLEMENTARY INFORMATION. LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans
 SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
SUPPLEMENTARY INFORMATION LIN-28 co-transcriptionally binds primary let-7 to regulate mirna maturation in C. elegans Priscilla M. Van Wynsberghe 1, Zoya S. Kai 1, Katlin B. Massirer 2-4, Victoria H. Burton
Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg
![Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg Product Datasheet. Histone H4 [Dimethyl Lys20] Antibody NB SS. Unit Size: mg](/thumbs/88/115416970.jpg) Product Datasheet Histone H4 [Dimethyl Lys20] Antibody NB21-2089SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related
Product Datasheet Histone H4 [Dimethyl Lys20] Antibody NB21-2089SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related
Myers Lab ChIP-seq Protocol v Modified January 10, 2014
 Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
Myers Lab ChIP-seq Protocol V011014 1 Contact information: Dr. Florencia Pauli Behn HudsonAlpha Institute for Biotechnology 601 Genome Way Huntsville, AL 35806 Telephone: 256-327-5229 Email: fpauli@hudsonalpha.org
ChIP protocol Chromatin fragmentation using the Covaris S2 sonicator by Ethan Ford (version 12/1/11) X- link Cells
 ChIP protocol Chromatin fragmentation using the Covaris S2 sonicator by Ethan Ford (version 12/1/11) X- link Cells 1. Grow six 15 cm plates of HeLa cells to 90% confluency. 2. Remove media and add wash
ChIP protocol Chromatin fragmentation using the Covaris S2 sonicator by Ethan Ford (version 12/1/11) X- link Cells 1. Grow six 15 cm plates of HeLa cells to 90% confluency. 2. Remove media and add wash
Supplementary Information
 Supplementary Information Table of contents: Pages: Supplementary Methods 2-3 Supplementary Figure 1 4 Supplementary Figure 2 5 Supplementary Figure 3 6 Supplementary Figure 4 7 Supplementary Figure 5
Supplementary Information Table of contents: Pages: Supplementary Methods 2-3 Supplementary Figure 1 4 Supplementary Figure 2 5 Supplementary Figure 3 6 Supplementary Figure 4 7 Supplementary Figure 5
ChIPmentation CeMM v1.14 (September 2016)
 ChIPmentation CeMM v1.14 (September 2016) This protocol works well for efficient histone modification and transcription factor antibodies. For less efficient antibodies sonication different sonication-,
ChIPmentation CeMM v1.14 (September 2016) This protocol works well for efficient histone modification and transcription factor antibodies. For less efficient antibodies sonication different sonication-,
Transcriptional regulation of BRCA1 expression by a metabolic switch: Di, Fernandez, De Siervi, Longo, and Gardner. H3K4Me3
 ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
ChIP H3K4Me3 enrichment.25.2.15.1.5 H3K4Me3 H3K4Me3 ctrl H3K4Me3 + E2 NS + E2 1. kb kb +82 kb Figure S1. Estrogen promotes entry of MCF-7 into the cell cycle but does not significantly change activation-associated
HiChIP Protocol Mumbach et al. (CHANG), p. 1 Chang Lab, Stanford University. HiChIP Protocol
 HiChIP Protocol Mumbach et al. (CHANG), p. 1 HiChIP Protocol Citation: Mumbach et. al., HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nature Methods (2016). Cell Crosslinking
HiChIP Protocol Mumbach et al. (CHANG), p. 1 HiChIP Protocol Citation: Mumbach et. al., HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nature Methods (2016). Cell Crosslinking
Chromatin Immunoprecipitation Approaches to Determine Co-transcriptional Nature of Splicing
 Chapter 23 Chromatin Immunoprecipitation Approaches to Determine Co-transcriptional Nature of Splicing Nicole I. Bieberstein, Korinna Straube, and Karla M. Neugebauer Abstract Chromatin immunoprecipitation
Chapter 23 Chromatin Immunoprecipitation Approaches to Determine Co-transcriptional Nature of Splicing Nicole I. Bieberstein, Korinna Straube, and Karla M. Neugebauer Abstract Chromatin immunoprecipitation
Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry
 Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Extended experimental procedure Nascent Chromatin Capture. The NCC protocol is designed for SILAC-based massspectrometry analysis of nascent versus mature chromatin composition (1). It requires a large
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
Supplemental material THE JOURNAL OF CELL BIOLOGY Taylor et al., http://www.jcb.org/cgi/content/full/jcb.201403021/dc1 Figure S1. Representative images of Cav 1a -YFP mutants with and without LMB treatment.
IMMUNOPRECIPITATION (IP)
 1 IMMUNOPRECIPITATION (IP) Overview and Technical Tips 2 CONTENTS 3 7 8 9 12 13 17 18 19 20 Introduction Factors Influencing IP General Protocol Modifications Of IP Protocols Troubleshooting Contact Us
1 IMMUNOPRECIPITATION (IP) Overview and Technical Tips 2 CONTENTS 3 7 8 9 12 13 17 18 19 20 Introduction Factors Influencing IP General Protocol Modifications Of IP Protocols Troubleshooting Contact Us
ChIP-seq Protocol for RNA-Binding Proteins
 ChIP-seq: Cells were grown according to the approved ENCODE cell culture protocols. Cells were fixed in 1% formaldehyde and resuspended in lysis buffer. Chromatin was sheared to 100-500 bp using a Branson
ChIP-seq: Cells were grown according to the approved ENCODE cell culture protocols. Cells were fixed in 1% formaldehyde and resuspended in lysis buffer. Chromatin was sheared to 100-500 bp using a Branson
ChIP-Adembeads (cat #04242/04342)
 ChIP-Adem-Kit (cat # 04243/04343) ChIP-Adembeads (cat #04242/04342) Instruction manual for magnetic Chromatin Immunoprecipitation Ademtech SA Bioparc BioGalien 27, allée Charles Darwin 33600 PESSAC France
ChIP-Adem-Kit (cat # 04243/04343) ChIP-Adembeads (cat #04242/04342) Instruction manual for magnetic Chromatin Immunoprecipitation Ademtech SA Bioparc BioGalien 27, allée Charles Darwin 33600 PESSAC France
Chromatin Immunoprecipitation (ChIP)
 de Lange Lab Chromatin Immunoprecipitation (ChIP) Required Solutions IP Wash A 0.1% SDS 1% Triton X-100 2 mm EDTA ph 8.0 20 mm Tris-HCl ph 8.0 150 mm NaCl 1 mm PMSF 1 µg/ml Leupeptin 1 µg/ml Aprotinin
de Lange Lab Chromatin Immunoprecipitation (ChIP) Required Solutions IP Wash A 0.1% SDS 1% Triton X-100 2 mm EDTA ph 8.0 20 mm Tris-HCl ph 8.0 150 mm NaCl 1 mm PMSF 1 µg/ml Leupeptin 1 µg/ml Aprotinin
The preparation of native chromatin from cultured human cells.
 Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Native chromatin immunoprecipitation protocol The preparation of native chromatin from cultured human cells. All solutions need to be ice cold. Sucrose containing solutions must be made up fresh on the
Product Datasheet. Histone H3 [Trimethyl Lys4] Antibody NB SS. Unit Size: mg
![Product Datasheet. Histone H3 [Trimethyl Lys4] Antibody NB SS. Unit Size: mg Product Datasheet. Histone H3 [Trimethyl Lys4] Antibody NB SS. Unit Size: mg](/thumbs/94/118559095.jpg) Product Datasheet Histone H3 [Trimethyl Lys4] Antibody NB21-1023SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Reviews: 1 Publications: 3
Product Datasheet Histone H3 [Trimethyl Lys4] Antibody NB21-1023SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Reviews: 1 Publications: 3
Supplementary Figure 1. HiChIP provides confident 1D factor binding information.
 Supplementary Figure 1 HiChIP provides confident 1D factor binding information. a, Reads supporting contacts called using the Mango pipeline 19 for GM12878 Smc1a HiChIP and GM12878 CTCF Advanced ChIA-PET
Supplementary Figure 1 HiChIP provides confident 1D factor binding information. a, Reads supporting contacts called using the Mango pipeline 19 for GM12878 Smc1a HiChIP and GM12878 CTCF Advanced ChIA-PET
RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the
 Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Supplementary Methods RT-PCR and real-time PCR analysis RNA was isolated using NucleoSpin RNA II (Macherey-Nagel, Bethlehem, PA) according to the manufacturer s protocol and quantified by measuring the
Chromatin Immunoprecipitation (ChIPs) Protocol for Tissues(Farnham Lab)
 Chromatin Immunoprecipitation (ChIPs) Protocol for Tissues(Farnham Lab) This protocol is based upon protocols from Mark Biggin, Dave Allis and Richard Treisman plus a fair amount of trial and error. We
Chromatin Immunoprecipitation (ChIPs) Protocol for Tissues(Farnham Lab) This protocol is based upon protocols from Mark Biggin, Dave Allis and Richard Treisman plus a fair amount of trial and error. We
Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon Song, Richard M. Amasino, Bosl Noh, and Yoo-Sun Noh
 Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Developmental Cell, Volume 22 Supplemental Information Control of Seed Germination by Light-Induced Histone Arginine Demethylation Activity Jung-Nam Cho, Jee-Youn Ryu, Young-Min Jeong, Jihye Park, Ji-Joon
Supplementary Data. Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells
 Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Data Supplementary Methods Three-step protocol for spontaneous differentiation of mouse induced pluripotent stem (embryonic stem) cells Mouse induced pluripotent stem cells (ipscs) were cultured
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Ren Lab ENCODE in situ HiC Protocol for Tissue
 Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
Ren Lab ENCODE in situ HiC Protocol for Tissue Pulverization, Crosslinking of Tissue Note: Ensure the samples are kept frozen on dry ice throughout pulverization. 1. Pour liquid nitrogen into a mortar
ChIP-chip protocol adapted for the mod-encode project
 ChIP-chip protocol adapted for the mod-encode project Version 1.2 : August 2007 Nicolas Nègre, Xiaochun Ni, Sergey Lavrov, Giacomo Cavalli and Kevin P. White University of Chicago, Department of Human
ChIP-chip protocol adapted for the mod-encode project Version 1.2 : August 2007 Nicolas Nègre, Xiaochun Ni, Sergey Lavrov, Giacomo Cavalli and Kevin P. White University of Chicago, Department of Human
Product Datasheet. Histone H3 [Trimethyl Lys9] Antibody NB Unit Size: 0.05 mg
![Product Datasheet. Histone H3 [Trimethyl Lys9] Antibody NB Unit Size: 0.05 mg Product Datasheet. Histone H3 [Trimethyl Lys9] Antibody NB Unit Size: 0.05 mg](/thumbs/95/122552588.jpg) Product Datasheet Histone H3 [Trimethyl Lys9] Antibody NB21-1073 Unit Size: 0.05 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Publications: 2 Protocols, Publications,
Product Datasheet Histone H3 [Trimethyl Lys9] Antibody NB21-1073 Unit Size: 0.05 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Publications: 2 Protocols, Publications,
Chromatin Immunoprecipitation (ChIP) Assay*
 Chromatin Immunoprecipitation (ChIP) Assay* Chromatin Immunoprecipitation (ChIP) assays are used to evaluate the association of proteins with specific DNA regions. The technique involves crosslinking of
Chromatin Immunoprecipitation (ChIP) Assay* Chromatin Immunoprecipitation (ChIP) assays are used to evaluate the association of proteins with specific DNA regions. The technique involves crosslinking of
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured
 Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
Supplementary Figure 1. Soft fibrin gels promote growth and organized mesodermal differentiation. Representative images of single OGTR1 ESCs cultured in 90-Pa 3D fibrin gels for 5 days in the presence
SUPPLEMENTAL MATERIALS. Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary
 SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
ChIP Capture Sequencing (ChIP-CapSeq): A Protocol for Targeted Sequencing of Immunoprecipitated DNA
 Sequencing Technical Note 2016 ChIP Capture Sequencing (): A Protocol for Targeted Sequencing of Immunoprecipitated DNA Jim Blackburn, PhD Ira Deveson Tim Mercer, PhD Garvan Institute of Medical Research,
Sequencing Technical Note 2016 ChIP Capture Sequencing (): A Protocol for Targeted Sequencing of Immunoprecipitated DNA Jim Blackburn, PhD Ira Deveson Tim Mercer, PhD Garvan Institute of Medical Research,
Product Datasheet. Histone H3 [ac Lys4] Antibody NB SS. Unit Size: mg
![Product Datasheet. Histone H3 [ac Lys4] Antibody NB SS. Unit Size: mg Product Datasheet. Histone H3 [ac Lys4] Antibody NB SS. Unit Size: mg](/thumbs/88/115978493.jpg) Product Datasheet Histone H3 [ac Lys4] Antibody NB21-1024SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Reviews: 1 Publications: 2 Protocols,
Product Datasheet Histone H3 [ac Lys4] Antibody NB21-1024SS Unit Size: 0.025 mg Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Reviews: 1 Publications: 2 Protocols,
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplemental Data. LMO4 Controls the Balance between Excitatory. and Inhibitory Spinal V2 Interneurons
 Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Neuron, Volume 61 Supplemental Data LMO4 Controls the Balance between Excitatory and Inhibitory Spinal V2 Interneurons Kaumudi Joshi, Seunghee Lee, Bora Lee, Jae W. Lee, and Soo-Kyung Lee Supplemental
Like use other ChIP kits, before handle ChIP assay please choose a good antibody suitable for precipitation the crosslinked protein / DNA complexes.
 ChIP Assay Kit Cat:RK20100 Like use other ChIP kits, before handle ChIP assay please choose a good antibody suitable for precipitation the crosslinked protein / DNA complexes. Manufactured by Global Headquarters
ChIP Assay Kit Cat:RK20100 Like use other ChIP kits, before handle ChIP assay please choose a good antibody suitable for precipitation the crosslinked protein / DNA complexes. Manufactured by Global Headquarters
Supporting Information. SI Material and Methods
 Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
Supporting Information SI Material and Methods RNA extraction and qrt-pcr RNA extractions were done using the classical phenol-chloroform method. Total RNA samples were treated with Turbo DNA-free (Ambion)
Luban Lab ChIP-seq protocol
 Luban Lab ChIP-seq protocol (updated by Anetta Nowosielska, December 2016) University of Massachusetts Medical School Program in Molecular Medicine 373 Plantation Street, Biotech 2 suite 319 Worcester,
Luban Lab ChIP-seq protocol (updated by Anetta Nowosielska, December 2016) University of Massachusetts Medical School Program in Molecular Medicine 373 Plantation Street, Biotech 2 suite 319 Worcester,
Cell extracts and western blotting RNA isolation and real-time PCR Chromatin immunoprecipitation (ChIP)
 Cell extracts and western blotting Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. 1 Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose
Cell extracts and western blotting Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with lysis buffer. 1 Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose
Protocol Immunprecipitation
 1 Protocol Immunprecipitation IP Juni 08, S.L. Before starting an IP on should be certain, that one can clearly detect the Protein to precipitate in a standard Western Blot with the lysis conditions used
1 Protocol Immunprecipitation IP Juni 08, S.L. Before starting an IP on should be certain, that one can clearly detect the Protein to precipitate in a standard Western Blot with the lysis conditions used
Chromatin Immunoprecipitation Assay for Early Zebrafish Embryos (Prot59)
 Chromatin Immunoprecipitation Assay for Early Zebrafish Embryos (Prot59) Leif C. Lindeman Leif C. Lindeman 1, Philippe Collas 1 1 Stem Cell Epigenetics Laboratory (Collas lab), Institute of Basic Medical
Chromatin Immunoprecipitation Assay for Early Zebrafish Embryos (Prot59) Leif C. Lindeman Leif C. Lindeman 1, Philippe Collas 1 1 Stem Cell Epigenetics Laboratory (Collas lab), Institute of Basic Medical
1. Collect worms in a 50ml tube. Spin and wait until worms are collected at the bottom. Transfer sample to a 15ml tube and wash with M9 until clean.
 Worm Collection 1. Collect worms in a 50ml tube. Spin and wait until worms are collected at the bottom. Transfer sample to a 15ml tube and wash with M9 until clean. 2. Transfer sample to a 50ml conical.
Worm Collection 1. Collect worms in a 50ml tube. Spin and wait until worms are collected at the bottom. Transfer sample to a 15ml tube and wash with M9 until clean. 2. Transfer sample to a 50ml conical.
EPIGENTEK. EpiQuik Chromatin Immunoprecipitation Kit. Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Chromatin Immunoprecipitation Kit Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Chromatin Immunoprecipitation Kit is suitable for combining the specificity of
EpiQuik Chromatin Immunoprecipitation Kit Base Catalog # P-2002 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Chromatin Immunoprecipitation Kit is suitable for combining the specificity of
EPIGENTEK. EpiQuik Tissue Chromatin Immunoprecipitation Kit. Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
EPIGENTEK. EpiQuik Methylated DNA Immunoprecipitation Kit. Base Catalog # P-2019 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Methylated DNA Immunoprecipitation Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik MeDIP Kit can be used for immunoprecipitating the methylated DNA from a broad range
EpiQuik Methylated DNA Immunoprecipitation Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik MeDIP Kit can be used for immunoprecipitating the methylated DNA from a broad range
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Kit Components. 1M NaHCO 3, Catalog # Lot # One vial containing 500µl of 1M NaHCO 3.
 Certificate of Analysis 48 Barn Road Lake Placid, NY 12946 Technical Support: T: 800 548-7853 F: 518 523-4513 email: techserv@upstate.com Sales Department: T: 800 233-3991 F: 781 890-7738 Licensing Dept.:
Certificate of Analysis 48 Barn Road Lake Placid, NY 12946 Technical Support: T: 800 548-7853 F: 518 523-4513 email: techserv@upstate.com Sales Department: T: 800 233-3991 F: 781 890-7738 Licensing Dept.:
Zebrafish ChIP-array protocol: version 1 August 5 th 2005
 Zebrafish ChIP-array protocol: version 1 August 5 th 2005 PROCEDURE I. Fixation of embryos 1. Dechorionate embryos (Pronase treatment) 2. Fix embryos, 15 mins in 1.85% formaldehyde in Embryo medium, RT,
Zebrafish ChIP-array protocol: version 1 August 5 th 2005 PROCEDURE I. Fixation of embryos 1. Dechorionate embryos (Pronase treatment) 2. Fix embryos, 15 mins in 1.85% formaldehyde in Embryo medium, RT,
Tissue Acetyl-Histone H4 ChIP Kit
 Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Tissue Acetyl-Histone H4 ChIP Kit Catalog Number KA0672 48 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle
Supplementary Figure 1. RAD51 and RAD51 paralogs are enriched spontaneously onto
 Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
Supplementary Figure legends Supplementary Figure 1. and paralogs are enriched spontaneously onto the S-phase chromatin during DN replication. () Chromatin fractionation was carried out as described in
ChIP DNA Clean & Concentrator Catalog Nos. D5201 & D5205
 Page 0 INSTRUCTION MANUAL ChIP DNA Clean & Concentrator Catalog Nos. D5201 & D5205 Highlights Quick (2 minute) recovery of ultra-pure DNA from chromatin immunoprecipitation (ChIP) assays, cell lysates,
Page 0 INSTRUCTION MANUAL ChIP DNA Clean & Concentrator Catalog Nos. D5201 & D5205 Highlights Quick (2 minute) recovery of ultra-pure DNA from chromatin immunoprecipitation (ChIP) assays, cell lysates,
rev. 05/31/12 Box 2 contains the following buffers/reagents:
 Store at RT, 4 C, and 20 C #9002 SimpleChIP Enzymatic Chromatin IP Kit (Agarose Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 05/31/12 Orders
Store at RT, 4 C, and 20 C #9002 SimpleChIP Enzymatic Chromatin IP Kit (Agarose Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 05/31/12 Orders
EZ-Zyme Chromatin Prep Kit
 Instruction Manual for EZ-Zyme Chromatin Prep Kit Catalog # 17 375 Sufficient reagents for 22 enzymatic chromatin preparations per kit. Contents Page I. INTRODUCTION 2 II. EZ-Zyme KIT COMPONENTS 3 A. Provided
Instruction Manual for EZ-Zyme Chromatin Prep Kit Catalog # 17 375 Sufficient reagents for 22 enzymatic chromatin preparations per kit. Contents Page I. INTRODUCTION 2 II. EZ-Zyme KIT COMPONENTS 3 A. Provided
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Fold activation. Development Supplementary information. δ51+s/p5 Nes+S/P5 Nes+BS/BP5
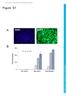 Figure S A DAPI EGFP B 50 0 5 20 Fold activation 00 50 0 δ5+s/p5 Nes+S/P5 Nes+BS/BP5 C Anti-ZIC2 Anti-H2 Anti-OTX2 Anti-H2 + ZIC2 + OTX2 50 40 30 20 0 + ZIC2 40 30 20 0 +OTX2 0 0 0.2 0.4 0.6 0.8.2.4.6.8
Figure S A DAPI EGFP B 50 0 5 20 Fold activation 00 50 0 δ5+s/p5 Nes+S/P5 Nes+BS/BP5 C Anti-ZIC2 Anti-H2 Anti-OTX2 Anti-H2 + ZIC2 + OTX2 50 40 30 20 0 + ZIC2 40 30 20 0 +OTX2 0 0 0.2 0.4 0.6 0.8.2.4.6.8
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Human/Mouse/Rat) *Colorimetric*
 Catalog # Kit Size SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Human/Mouse/Rat) *Colorimetric* AS-55550 One 96-well strip plate This kit is optimized to detect human/mouse/rat alpha-synuclein
Catalog # Kit Size SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Human/Mouse/Rat) *Colorimetric* AS-55550 One 96-well strip plate This kit is optimized to detect human/mouse/rat alpha-synuclein
ChIP-Adem-Kit AutoMag Solution (Cat #04643) Instruction manual for automation protocol
 ChIP-Adem-Kit AutoMag Solution (Cat #04643) Instruction manual for automation protocol ADEMTECH SA Bioparc BioGalien 27, allée Charles Darwin 33600 Pessac France Tel: +33557020201 Fax +3355702020 Visit
ChIP-Adem-Kit AutoMag Solution (Cat #04643) Instruction manual for automation protocol ADEMTECH SA Bioparc BioGalien 27, allée Charles Darwin 33600 Pessac France Tel: +33557020201 Fax +3355702020 Visit
ab ChIP Kit Magnetic One-Step
 ab156907 ChIP Kit Magnetic One-Step Instructions for Use For selective enrichment of a chromatin fraction containing specific DNA sequences in a high throughput format using chromatin isolated from various
ab156907 ChIP Kit Magnetic One-Step Instructions for Use For selective enrichment of a chromatin fraction containing specific DNA sequences in a high throughput format using chromatin isolated from various
Chromatin Immunoprecipitation (ChIP) Assay (PROT11)
 Chromatin Immunoprecipitation (ChIP) Assay (PROT11) Antigone Kouskouti & Irene Kyrmizi Institute of Molecular Biology and Biotechnology FORTH 1527 Vassilika Vouton 711 10, Heraklion, Crete, Greece Email
Chromatin Immunoprecipitation (ChIP) Assay (PROT11) Antigone Kouskouti & Irene Kyrmizi Institute of Molecular Biology and Biotechnology FORTH 1527 Vassilika Vouton 711 10, Heraklion, Crete, Greece Email
Streptavidin Mag Sepharose
 GE Healthcare Life Sciences Data file 28-9921-05 AB Protein sample preparation Streptavidin Mag Sepharose Streptavidin Mag Sepharose (Fig 1) is a magnetic bead for simple and efficient enrichment of target
GE Healthcare Life Sciences Data file 28-9921-05 AB Protein sample preparation Streptavidin Mag Sepharose Streptavidin Mag Sepharose (Fig 1) is a magnetic bead for simple and efficient enrichment of target
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
Anti-CLOCK (Mouse) mab
 Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-CLOCK (Mouse) mab CODE No. D349-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal CLSP4 Mouse IgG1
Page 1 For Research Use Only. Not for use in diagnostic procedures. Anti-CLOCK (Mouse) mab CODE No. D349-3 CLONALITY CLONE ISOTYPE QUANTITY SOURCE IMMUNOGEN FORMURATION STORAGE Monoclonal CLSP4 Mouse IgG1
SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Rat) *Colorimetric*
 SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Rat) *Colorimetric* Revision number: 1.3 Last updated: 1/15/18 Catalog # AS-55550-R Kit Size One 96-well strip plate This kit is optimized to detect
SensoLyte Anti-alpha-Synuclein Quantitative ELISA Kit (Rat) *Colorimetric* Revision number: 1.3 Last updated: 1/15/18 Catalog # AS-55550-R Kit Size One 96-well strip plate This kit is optimized to detect
Supporting Information
 Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
Supporting Information Su et al. 10.1073/pnas.1211604110 SI Materials and Methods Cell Culture and Plasmids. Tera-1 and Tera-2 cells (ATCC: HTB- 105/106) were maintained in McCoy s 5A medium with 15% FBS
MeDIP-seq library construction protocol v2 Costello Lab June Notes:
 MeDIP-seq library construction protocol v2 Costello Lab June 2010 Notes: A. For all Qiagen gel extraction steps (Qiaquick and MinElute), melt gel slice at 37º C instead of 50º (see Quail et al 2008 Nature
MeDIP-seq library construction protocol v2 Costello Lab June 2010 Notes: A. For all Qiagen gel extraction steps (Qiaquick and MinElute), melt gel slice at 37º C instead of 50º (see Quail et al 2008 Nature
Supplemental Information. PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock
 Molecular Cell, Volume 49 Supplemental Information PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock Dafne Campigli Di Giammartino, Yongsheng Shi, and James L. Manley Supplemental Information
Molecular Cell, Volume 49 Supplemental Information PARP1 Represses PAP and Inhibits Polyadenylation during Heat Shock Dafne Campigli Di Giammartino, Yongsheng Shi, and James L. Manley Supplemental Information
Cdc42 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Cdc42 Activation Assay Kit Catalog Number: 80701 20 assays 1 Table of Content Product Description 3 Assay
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for combining the
APPLICATION NOTE. Abstract. Introduction
 From minuscule amounts to magnificent results: reliable ChIP-seq data from 1, cells with the True MicroChIP and the MicroPlex Library Preparation kits Abstract Diagenode has developed groundbreaking solutions
From minuscule amounts to magnificent results: reliable ChIP-seq data from 1, cells with the True MicroChIP and the MicroPlex Library Preparation kits Abstract Diagenode has developed groundbreaking solutions
RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu *
 RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu * School of Biomedical Sciences, Thomas Jefferson University, Philadelphia, USA *For correspondence:
RNP-IP (Modified Method)-Getting Majority RNA from RNA Binding Protein in the Cytoplasm Fengzhi Liu * School of Biomedical Sciences, Thomas Jefferson University, Philadelphia, USA *For correspondence:
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of
 Stem Cell Reports, Volume 8 Supplemental Information RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of PolyA + and SmallRNA Transcriptomes and Splicing in Human Pluripotent Stem Cells Riikka
Stem Cell Reports, Volume 8 Supplemental Information RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of PolyA + and SmallRNA Transcriptomes and Splicing in Human Pluripotent Stem Cells Riikka
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
SUPPLEMENTRY INFORMTION DOI:.38/ncb Kdmb locus kb Long isoform Short isoform Long isoform Jmj XX PHD F-box LRR Short isoform XX PHD F-box LRR Target Vector 3xFlag Left H Neo Right H TK LoxP LoxP Kdmb Locus
B. ADM: C. D. Apoptosis: 1.68% 2.99% 1.31% Figure.S1,Li et al. number. invaded cells. HuH7 BxPC-3 DLD-1.
 A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
A. - Figure.S1,Li et al. B. : - + - + - + E-cadherin CK19 α-sma vimentin β -actin C. D. Apoptosis: 1.68% 2.99% 1.31% - : - + - + - + Apoptosis: 48.33% 45.32% 44.59% E. invaded cells number 400 300 200
RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by. 5 AGACACAAACACCAUUGUCACACUCCACAGC; Rand-2 OMe,
 Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
Materials and methods Oligonucleotides and DNA constructs RNA oligonucleotides and 2 -O-methylated oligonucleotides were synthesized by Dharmacon Inc. (Lafayette, CO). The sequences were: 122-2 OMe, 5
EpiSeeker ChIP Kit Histone H4 (acetyl) Tissue
 ab117151 EpiSeeker ChIP Kit Histone H4 (acetyl) Tissue Instructions for Use For successful chromatin immunoprecipitation for acetyl-histone H4 from mammalian tissues This product is for research use only
ab117151 EpiSeeker ChIP Kit Histone H4 (acetyl) Tissue Instructions for Use For successful chromatin immunoprecipitation for acetyl-histone H4 from mammalian tissues This product is for research use only
Supplementary information
 Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Supplementary information Table of Content: Supplementary Results... 2 Supplementary Figure S1: Experimental validation of AP-MS results by coimmunprecipitation Western blot analysis.... 3 Supplementary
Gα 13 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Gα 13 Activation Assay Kit Catalog Number: 80401 20 assays NewEast Biosciences 1 Table of Content Product
rev. 06/18/18 RPL30 MyoD1
 Store at RT, 4 C, and 20 C #9002 SimpleChIP Enzymatic Chromatin IP Kit (Agarose Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 06/18/18 Orders
Store at RT, 4 C, and 20 C #9002 SimpleChIP Enzymatic Chromatin IP Kit (Agarose Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 06/18/18 Orders
Arf6 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Arf6 Activation Assay Kit Catalog Number: 82401 20 assays NewEast Biosciences 1 Table of Content Product
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium
 Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
Apoptosis assay: Apoptotic cells were identified by Annexin V-Alexa Fluor 488 and Propidium Iodide (Invitrogen, Carlsbad, CA) staining. Briefly, 2x10 5 cells were washed once in cold PBS and resuspended
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10928 Materials and Methods 1. Plant material and growth conditions. All plant lines used were in Col-0 background unless otherwise specified. pif4-101 mutant
SUPPLEMENTARY INFORMATION doi:10.1038/nature10928 Materials and Methods 1. Plant material and growth conditions. All plant lines used were in Col-0 background unless otherwise specified. pif4-101 mutant
Cat. No. MG17PG-1ml XPRESSAFFINITY PROTEIN G- MAGNETIC NANOPARTICLES (MNP) FOR RESEARCH APPLICATIONS
 Cat. No. MG17PG-1ml XPRESSAFFINITY PROTEIN G- MAGNETIC NANOPARTICLES (MNP) FOR RESEARCH APPLICATIONS Product Description: MagGenome s Protein G-MNP provides a fast and convenient method for affinity based
Cat. No. MG17PG-1ml XPRESSAFFINITY PROTEIN G- MAGNETIC NANOPARTICLES (MNP) FOR RESEARCH APPLICATIONS Product Description: MagGenome s Protein G-MNP provides a fast and convenient method for affinity based
Protein A Agarose Immunoprecipitation Kit
 Protein A Agarose Immunoprecipitation Kit Catalog Number KA0568 20 Reactions Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Protein A Agarose Immunoprecipitation Kit Catalog Number KA0568 20 Reactions Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
Supplemental Materials and Methods
 Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
Supplemental Materials and Methods Co-immunoprecipitation (Co-IP) assay Cells were lysed with NETN buffer (20 mm Tris-HCl, ph 8.0, 0 mm NaCl, 1 mm EDT, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate,
SUPPLEMENTARY INFORMATION
 (Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
(Supplementary Methods and Materials) GST pull-down assay GST-fusion proteins Fe65 365-533, and Fe65 538-700 were expressed in BL21 bacterial cells and purified with glutathione-agarose beads (Sigma).
Rab5 Activation Assay Kit
 A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
A helping hand for your research Product Manual Configuration-specific Monoclonal Antibody Based Rab5 Activation Assay Kit Catalog Number: 83701 20 assays 24 Whitewoods Lane 1 Table of Content Product
HPV E6 oncoprotein targets histone methyltransferases for modulating specific. Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu,
 1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
1 HPV E oncoprotein targets histone methyltransferases for modulating specific gene transcription 3 5 Chih-Hung Hsu, Kai-Lin Peng, Hua-Ci Jhang, Chia-Hui Lin, Shwu-Yuan Wu, Cheng-Ming Chiang, Sheng-Chung
For gel-shift assays, 2 ul ivtt synthesized protein (Promega) was incubated at room temperature
 Supplementary Material and Methods EMSA For gel-shift assays, 2 ul ivtt synthesized protein (Promega) was incubated at room temperature for 30 min in a 15 ul volume containing 15 mm Tris-HCl (ph 7.5),
Supplementary Material and Methods EMSA For gel-shift assays, 2 ul ivtt synthesized protein (Promega) was incubated at room temperature for 30 min in a 15 ul volume containing 15 mm Tris-HCl (ph 7.5),
ChromaFlash One-Step Magnetic ChIP Kit
 ChromaFlash One-Step Magnetic ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE Uses: The ChromaFlash One-Step Magnetic ChIP Kit is suitable for selective enrichment of a chromatin
ChromaFlash One-Step Magnetic ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE Uses: The ChromaFlash One-Step Magnetic ChIP Kit is suitable for selective enrichment of a chromatin
#9003. SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) n 1 Kit. (30 Immunoprecipitations) and 20 C. Store at 4 C, RT
 Store at 4 C, RT and 20 C #9003 SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 06/15/18 Orders
Store at 4 C, RT and 20 C #9003 SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) 3 n 1 Kit (30 Immunoprecipitations) For Research Use Only. Not For Use In Diagnostic Procedures. rev. 06/15/18 Orders
Supplementary Figure Legends
 Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
Supplementary Figure Legends Figure S1 gene targeting strategy for disruption of chicken gene, related to Figure 1 (f)-(i). (a) The locus and the targeting constructs showing HpaI restriction sites. The
SUPPLEMENTAL MATERIALS SIRTUIN 1 PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF2 ACTIVATION
 SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
SUPPLEMENTAL MATERIALS SIRTUIN PROMOTES HYPEROXIA-INDUCED LUNG EPITHELIAL DEATH INDEPENDENT OF NRF ACTIVATION Haranatha R. Potteti*, Subbiah Rajasekaran*, Senthilkumar B. Rajamohan*, Chandramohan R. Tamatam,
IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only
 IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only Introduction The IgG TrueBlot for mouse, rabbit, or goat-derived antibodies represents unique series of respective
IgG TrueBlot Protocol for Mouse, Rabbit or Goatderived Antibodies - For Research Use Only Introduction The IgG TrueBlot for mouse, rabbit, or goat-derived antibodies represents unique series of respective
Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition
 SUPPLEMENTARY INFORMATION Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition Mari-Liis Visnapuu 1 and Eric C. Greene 1 1 Department of Biochemistry &
SUPPLEMENTARY INFORMATION Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition Mari-Liis Visnapuu 1 and Eric C. Greene 1 1 Department of Biochemistry &
