Candidate region (0.74 Mb) ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC
|
|
|
- Britton Ryan
- 6 years ago
- Views:
Transcription
1 A idm-3 idm-3 B Physical distance (Mb) C Chr.3 Recom. Rate (%) ATG Candidate region (.74 Mb) n=4 TAA D idm-3 G3T(E4) G4A(W988) WT idm-3 ATC TCT GGG ACT CAT GAG CAG GAG GCT AGC ATC TCT GGG ACT CAT TAG CAG GAG GCT AGC WT AGT TTA GTG GAA ACT TGG ACT GAA GGT TTT AGT TTA GTG GAA ACT TGA ACT GAA GGT TTT
2 E X Col- F X idm- F Supplemental Figure. IDM prevents transcriptional gene silencing. (A) LUC luminescence image showing that the IDM mutations cause silencing of the 3S-LUC reporter gene. Seedlings grown in MS plates were imaged after being sprayed with luciferase substrate. (B) Rough mapping of the mutation. Shown are the molecular markers and their positions on chromosome III around the region. About 4 mutant plants from the progenies of a cross between (in Col- ) and Ler were used to define the position of. The recombination rate at these markers is indicated. (C) Schematic diagram showing the positions of the point mutations at the IDM locus. Black rectangles represent exons. (D) DNA sequence alignments between WT and mutated IDM. Point mutations in the mutants are highlighted in red. (E) Allelism analyses of the mutant. Seedlings grown in MS plates were imaged after being sprayed with luciferase substrate.
3 A AtSN HaeIII Solo-LTR AluI No digest control B drd- nrpe-9 C Physical distance (Mb) Chr Recom. Rate (%) drd- n=63 Physical distance (Mb) Chr. Candidate region (.9 Mb) nrpe D Recom. Rate (%) ATG Candidate region (.3 Mb) n=96 drd- G33A (R74H) TAA DRD ATG nrpe-9 G779A (W638) TAA NRPE
4 E drd- drd- + DRD-Flag drd- drd- + DRD-Flag F nrpe-9 nrpe-9 + NRPE-Flag nrpe-9 nrpe-9 + NRPE-Flag suvh- rdr- Supplemental Figure. Identification of RdDM pathway components as IDM suppressors and effects of their mutations on transcriptional gene silencing of the reporter genes. (A) Analysis of DNA methylation levels at the AtSNI and Solo-LTR loci by chop-pcr. Genomic DNA from and the mutants of IDM suppressors was digested by the DNA methylation-sensitive restriction enzyme HaeIII or AluI and then subjected to semi-quantitative PCR. Undigested samples were used as controls. (B) Effects of drd- and nrpe-9 mutations on 3S-LUC expression. (C) Rough mapping of the drd- and nrpe-9 mutations. (D) Schematic diagram showing the positions of the point mutations at the DRD and NRPE loci. Black rectangles represent exons. (E) Complementation assay showing that introduction of the prodrd::drd-flag or pronrpe::nrpe-flag transgene into drd- or nrpe-9 respectively restored 3S-LUC silencing. (F) Effects of rdr- and suvh- mutations on 3S-LUC expression. The 3S-LUC reporter gene was introduced into suvh- and rdr- mutant plants by crossing the suvh- and rdr- mutants with the plants in background.
5 A morc6- morc6-6 B Physical distance (Mb) Chr. Recom. Rate (%) n=96 Candidate region (.84 Mb) WT morc6-4 TCC AAT GCT ACC TCA CAT AAA TGG GCT TTT GGA TCC AAT GCT ACC TCA CAT AAA TGA GCT TTT GGA C morc6-3 (GK-99B6) morc6-6 (G37A) ATG () TAG (6) ATG9 morc6-4 (G48A) morc6- (G46A) D morc6-4 morc6- (W8) (W3) 36 morc6-6 (G43E) MORC6 morc6-3
6 E WT nrpe-9 morc6-4 nrpe-9 + NRPE-Flag morc6-4 + MORC6-Flag morc6-4 morc6-4 + MORC6-Flag Supplemental Figure 3. Map-based cloning of morc6-4. (A) The morc6- and morc6-6 mutations cause partial release of the LUC reporter genes. Seedlings grown in MS plates were imaged after being sprayed with luciferase substrate. (B) Rough mapping of the morc6-4 mutation. Shown are the molecular markers and their positions on chromosome I around the morc6-4 region. Ninety-six mutant plants from the progenies of a cross between morc6-4 (in Col-) and ros4/idm-3 (in C4) were used to define the position of morc6-4. The recombination rate at these markers is indicated. A point mutation (G/A) was identified in MORC6. (C) Schematic diagram showing the positions of the point mutations and T- DNA insertion at the MORC6 locus. Black rectangles represent exons. Gray rectangles represent UTRs. (D) Schematic diagram of MORC6 protein domain structure. MORC6 contains a GHKL domain, a S ATPase domain and a putative Coiled-Coil (CC) domain. The point mutations are in the GHKL domain and S domain. (E) Complementation assay showing that introduction of the promorc6::morc6-flag transgene into morc6-4 restored 3S-LUC silencing.
7 A Physical distance (Mb) Chr Recom. Rate (%) n=93 B Candidate region (4. Mb) C ATG () TAA (93) AT4G346 suvh9- (salk_4833) suvh9- (C66T) D suvh9- (Q36) SUVH9
8 E suvh9- suvh9-+suvh9-flag suvh9- suvh9-+suvh9-flag Supplemental Figure 4. Map-based cloning of suvh9-. (A) Rough mapping of the suvh9- mutation. Shown are the molecular markers and their positions on chromosome IV around the suvh9- region. Ninety-three mutant plants from the progenies of a cross between suvh9- (in Col-) and ros4/idm-3 (in C4) were used to define the position of suvh9-. The recombination rate at these markers is indicated. (B) Genome sequencing of suvh9-. A point mutation (G/A) was identified in SUVH9. (C) Schematic diagram showing the positions of the point mutation and T-DNA insertion at the SUVH9 locus. Black rectangles represent exons. (D) Schematic diagram of SUVH9 protein domain structure. SUVH9 contains a SRA domain, a preset domain and a SET domain. The point mutation is in the SRA domain. (E) Complementation assay showing introduction of the prosuvh9::suvh9- Flag transgene into suvh9- restored 3S-LUC silencing.
9 A B. nluc + cluc;. nluc + SUVH9-cLUC; 3. MORC-nLUC + cluc; 4. MORC-nLUC + SUVH9-cLUC;. MORC-nLUC + cluc; 6. MORC-nLUC + SUVH9-cLUC; 7. IDN-nLUC + cluc; 8. IDN-nLUC + SUVH9-cLUC. SD-TL SD-TLH AD/BD SUVH9/IDN IDN/SUVH9 SUVH9/SWI3B SWI3B/SUVH9 C MORC6/SUVH nluc + cluc. nluc + SUVH9-cLUC 3. SWI3A-nLUC + cluc 4. SWI3A-nLUC + SUVH9-cLUC. SWI3B-nLUC + cluc 6. SWI3B-nLUC + SUVH9-cLUC 7. SWI3C-nLUC + cluc 8. SWI3C-nLUC + SUVH9-cLUC 9. SWI3D-nLUC + cluc. SWI3D-nLUC + SUVH9-cLUC Supplemental Figure. SUVH9 interacts with SWI/SNF chromatin-remodeling proteins. (A) Detection of the interactions of SUVH9 with MORC, MORC and IDN by split-luc assays. (B) Detection of the interactions of SUVH9 with IDN and SWI3B by Y--H assays. (C) Detection of the interactions of SUVH9 with SWI/SNF chromatin-remodeling proteins by split-luc assays. nluc, C- terminal region of firefly luciferase; cluc, N-terminal region of firefly luciferase. Three independent experiments were done with similar results obtained. Red indicates positive protein interaction.
10 nluc + cluc. nluc + SUVH-cLUC 3. IDN-nLUC + cluc 4. IDN-nLUC+ SUVH-cLUC. MORC-nLUC + cluc 6. MORC-nLUC + SUVH-cLUC 7. MORC-nLUC + cluc 8. MORC-nLUC + SUVH-cLUC 9. MORC6-nLUC + cluc. MORC6-nLUC + SUVH-cLUC. SWI3A-nLUC + cluc. SWI3A-nLUC + SUVH-cLUC 3. SWI3B-nLUC + cluc 4. SWI3B-nLUC + SUVH-cLUC. SWI3C-nLUC + cluc 6. SWI3C-nLUC + SUVH-cLUC 7. SWI3D-nLUC + cluc 8. SWI3D-nLUC + SUVH-cLUC Supplemental Figure 6. Detection of the interactions between SUVH and other related proteins by split-luc assays. Interactions of SUVH with IDN, MORC family proteins and SWI/SNF chromatin-remodeling proteins as demonstrated by firefly luciferase complementation imaging assays in Nicotiana benthamiana leaves. nluc, C-terminal region of firefly luciferase; cluc, N-terminal region of firefly luciferase. Three independent experiments were done with similar results obtained. Red indicates positive protein interaction.
11 A Relative transcript level TE84 TE46 TE TE497 8 Solo-LTR COPIA B Relative transcript level SDC At3g769 At4g At4g394 C Relative transcript level At3g769 At4g469 At4g969 At4g
12 D 4 3 morc6-3 Genes DNA methylation level log (morc6/col-) Log (fold change of normalized reads) E DNA methylation level log (suvhsuvh9/col-) suvhsuvh9 Genes Log (fold change of normalized reads) Supplemental Figure 7. Effects of morc6-4 on RNA transcript level and effects of suvhsuvh9 and morc6-3 on RNA transcript level as determined by RNA-seq. (A) Up-regulated TEs in morc6-3 and morc6-4. (B) Up-regulated genes in morc6-4. (C) Real-time PCR was performed to confirm the RNA-seq results for genes. ACTIN 7 was used as internal control. Standard errors were calculated from three biological replicates, P <.. (D) DNA methylation levels for up-regulated genes in morc6-3. (E) DNA methylation levels for up-regulated genes in suvhsuvh9. Differentially expressed genes were identified when log (fold changes of normalized reads) or - and P value<..
13 Supplemental Table. Mass-spectrometric analysis of SUVH9 co-purifying proteins. Supplemental Table. Differentially expressed genes in suvhsuvh9 and morc6-3. Supplemental Table 3. Differentially expressed TEs in suvhsuvh9 and morc6-3. Supplemental Table 4. Primers used in this study. Supplemental Table. Mass-spectrometric analysis of SUVH9 co-purifying proteins. Protein Accession Gene Accession Protein Mass Spectra Unique Peptides IPI977 AT4G346 SUVH IPI94 ATG9 MORC6 749 IPI4938 AT3G4867 IDN 777 IPI763 AT4G3443 SWI3B 873 Supplemental Table 4. Primers used in this study. Primer Name Sequence ('-3') Purpose 3S-BS-F GATGTTTTTGTYGATAGTGGTTTTAAAGATGGAT Bisulfite sequencing for 3S-BS-R CTCTTCATAACCTTATACAATTACTCTCCAA 3S promoter SDC-BS-F GAAAAAGTTGGAATGGGTTTGGAGAGTTTAA Bisulfite sequencing for SDC-BS-R CAACAAACCCTAATATATTTTATATTAAAAC SDC Solo LTR-BS-F GATATAAAGGAATGGTTAGATAATATGYGATT Bisulfite sequencing for Solo LTR-BS-R CRATATAACTCAAAATTTATATTACTCTTAA solo-ltr ATCOPIA8-BS-F TATTTATTTYGTTCATTTGGATTAGTTTT Bisulfite sequencing for ATCOPIA8-BS-R ACRATATCAAAATAATTATCATCATCTTAA COPIA 8 LUC-RT-F CTCCCCTCTCTAAGGAAGTCG Real-time PCR for LUC LUC-RT-R CCAGAATGTAGCCATCCATC NPTII-RT-F AGGTTCCATCTGCCAGGTATCA Real-time PCR for NPT II NPTII-RT-R CCCGGTATCCAGATCCACAA SDC-RT-F AATGTAAGTTGTAAACCATTTGAACGTGACC Real-time PCR for SDC SDC-RT-R CAGGCATCCGTAGAACTCATGAGC Solo LTR-F AACTAACGTCATTACATACACATCTTG Real-time PCR for Solo LTR-R AATTAGGATCTTGTTTGCCAGCTA solo-ltr ATCOPIA8-F AGTCCTTTTGGTTGCTGAACA Real-time PCR for COPIA ATCOPIA8-R CCGGATGTAGCAACATTCACT 8 ACTIN 7-LP TCGTGGTGGTGAGTTTGTTA Real-time PCR, loading ACTIN 7-RP CAGCATCATCACAAGCATCC control SUVH-AD-BamHI-F GCGGGATCCATGAGTACATTGTTACC Amplify SUVH CDS for SUVH-AD-XhoI-R TATCTCGAGGTTGCAGATGGCGAGCT
14 SUVH9-AD-Nde-F AGATTACGCTCATATGATGGGTTCTTCTCAC Amplify SUVH9 CDS for SUVH9-AD-EcoR-R SUVH9-BD-Nde-F SUVH9-BD-EcoR-R CACCCGGGTGGAATTCTTAATTACAAATGGC GGAGGACCTGCATATGATGGGTTCTTCTCAC GGATCCCCGGGAATTCTTAATTACAAATGGC MORC-AD-NdeI-F AGATTACGCTCATATGATGGCGAAAAATTAC Amplify MORC CDS for MORC-AD-EcoRI-R CACCCGGGTGGAATTCCTAAACTTGTTGCATCTCC MORC- AD-NdeI-F AGATTACGCTCATATGATGCCTCCTATGGCG Amplify MORC CDS for MORC- AD-EcoRI-R CACCCGGGTGGAATTCCTAAGCTTGTTGCATC MORC6-AD-in-NdeI-F AGATTACGCTCATATGATGAGTCACGATAG Amplify MORC6 CDS for MORC6-AD-in-EcoRI-R MORC6-BD-EcoRI-F MORC6-BD-SalI-R ACGAGATCTGGTCGACCTACGTATTTACATTTCTTCTG GGCGAATTCATGAGTCACGATAGAAGTG GCCGTCGACCTACGTATTTACATTTC SWI3B-A/BD-NdeI-F GGACATATGATGGCCATGAAAGCTCCCGAT Amplify SWI3B CDS for SWI3B-A/BD-EcoRI-R GGCGAATTCCTAACACTCTATTCTATC SWI3C-BD-NdeI-F GGAGGACCTGCATATGATGCCAGCTTCTGAAG Amplify SWI3C CDS for SWI3C-BD-EcoRI-R GGATCCCCGGGAATTCTTAGTTTAAGCCTAAGC IDN-A/BD-EcoRI-F CCCGAATTCATGGGAAGCACTGTGAT Amplify IDN CDS for IDN-A/BD-BamHI-R ATAGGATCCAGCCATTCCACGCTTGC SUVH-NLUC-BamHI-F CGGTACCCGGGATCCaATGAGTACATTGTTACC Amplify SUVH cdna SUVH-NLUC-SalI- R SUVH-CLUC-BamHI-F SUVH-CLUC-SalI- R ACGAGATCTGGTCGACGTTGCAGATG GCGAGC CGGTACCCGGGATCCaATGAGTACATTGTTACC AGCTCTGCAGGTCGACCTAGTTGCAG ATGGCG for split-luc SUVH9-NLUC-BamHI-F CGGTACCCGGGATCCaATGGGTTCTTCTCAC Amplify SUVH9 cdna SUVH9-NLUC-SalI- R SUVH9-CLUC-BamHI-F SUVH9-CLUC-SalI- R ACGAGATCTGGTCGACATTACAAATGGC CGGTACCCGGGATCCaATGGGTTCTTCTCAC AGCTCTGCAGGTCGACTTAATTACAAATGGC for split-luc MORC-Nluc-KpnI-F CCGGGTACCATGGCGAAAAATTACACAGTC Amplify MORC cdna MORC-Nluc-SalI-R MORC-Cluc-KpnI-F MORC-Cluc-SalI-R CGGGTCGACAACTTGTTGCATCTCCTTCTT CCGGGTACCATGGCGAAAAATTACACAGTC CGGGTCGACCTAAACTTGTTGCATCTCCTT for split-luc MORC-NLUC-BamHI-F CGGTACCCGGGATCCaATGCCTCCTATGGCGA Amplify MORC cdna MORC-NLUC-SalI-R MORC-CLUC-BamHI-F MORC-CLUC-SalI-R ACGAGATCTGGTCGACAGCTTGTTGCATCTCC CGGTACCCGGGATCCaATGCCTCCTATGGCGA AGCTCTGCAGGTCGACCTAAGCTTGTTGCATC for split-luc MORC6-NLUC-BamHI-F CGGTACCCGGGATCCaATGAGTCACGATAGA Amplify MORC6 cdna MORC6-NLUC-SalI-R ACGAGATCTGGTCGACCGTATTTACATTTCTTCTGTGCT CG for split-luc SWI3A-NLUC-BamHI-F CGGTACCCGGGATCCaATGGAAGCCACTGATC Amplify SWI3A cdna for SWI3A-NLUC-SalI-R SWI3A-CLUC-BamHI-F ACGAGATCTGGTCGACTTTCACGTACGTATGAT CGGTACCCGGGATCCaATGGAAGCCACTGATC split-luc
15 SWI3A-CLUC-SalI-R AGCTCTGCAGGTCGACTCATTTCACGTACGTAT SWI3B-NLUC-BamHI-F CGGTACCCGGGATCCaATGGCCATGAAAGCTC Amplify SWI3B cdna for SWI3B-NLUC-SalI-R SWI3B-CLUC-BamHI-F SWI3B-CLUC-SalI-R ACGAGATCTGGTCGACACACTCTATTCTATCT CGGTACCCGGGATCCaATGGCCATGAAAGCTC AGCTCTGCAGGTCGACCTAACACTCTATTCT split-luc SWI3C-NLUC-BamHI- F CGGTACCCGGGATCCaATGCCAGCTTCTGAA Amplify SWI3C cdna for SWI3C-NLUC-SalI-R SWI3C-CLUC-BamHI- F SWI3C-CLUC-SalI-R ACGAGATCTGGTCGACGTTTAAGCCTAAGCC CGGTACCCGGGATCCaATGCCAGCTTCTGAA AGCTCTGCAGGTCGACTTAGTTTAAGCCTAAGC split-luc SWI3D-NLUC-BamHI-F CGGTACCCGGGATCCaATGGAGGAAAAACGAC Amplify SWI3D cdna for SWI3D-NLUC-SalI-R SWI3D-CLUC-BamHI-F SWI3D-CLUC-SalI-R ACGAGATCTGGTCGACAACCGAAGAAACATTG CGGTACCCGGGATCCaATGGAGGAAAAACGAC AGCTCTGCAGGTCGACCTAAACCGAAGAAAC split-luc SDC-3C-fix CAATCGGTTTGTTCGGATTAGG 3C experiments SDC-3C-Ⅰ SDC-3C-Ⅱ SDC-3C-Ⅲ SDC-3C-Ⅳ SDC-3C-Ⅴ SDC-3C-Ⅵ SDC-3C-Ⅶ SoloLTR-3C-fix SoloLTR-3C-Ⅰ SoloLTR-3C-Ⅱ CGATGTGAAAGATCCGCTGAC GCCGCATTACATGGCTCCAA GGGTGTGCTCCAAATGATTG TGGTCTAGGCTACTACCTAG TGCCCATAACAATCGGAC GGTCTTCAGTCTTGTGTTAGG CGAGTTGGAGAATGTAGATTCC ATATTCGAGGGTGACAGCGT GAATCAGTTTGGCAATGTCG AGGACCGCTAGAAGATTGAG TE84-F CCAACTCAAGATGACCTCCA Real-time PCR for TE TE84-R TE947-F TE947-R TE8-F TE8-R TE46-F TE46-R TE487-F TE487-R TE497-F TE497-R CAGTGGTGGTTCCCTTATCT GGTAGAGCAACAATGGAGAG CTTCTTCCTCCTCTCACCTA TTGCCCAAGCATCTGTGATC CCGCCTGCATTTCTGACTAC GAGGTTGAGGGAAGTTGGTT GTGCATCTGCTCTGGAGTTT CATCGCCGAAAGTGACTGTT GCATTACGAAGAGCTAGATGAG CGGTGCAATACGTAAACCAC CCACAATTGATGAGCATCTGAG transcripts 3
Supplemental Figure 1 A
 Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Supplemental Figure 1 A Supplemental Data. Han et al. (2016). Plant Cell 10.1105/tpc.15.00997 BamHI -1616 bp SINE repeats SalI ATG SacI +1653 bp HindIII LB HYG p35s pfwa LUC Tnos RB pfwa-bamhi-f: CGGGATCCCGCCTTTCTCTTCCTCATCTGC
Arabidopsis actin depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB
 Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Supplemental Figure legends Figure S1. (A) (B) (C) (D) Figure S2. Figure S3. (A-E) Figure S4. Figure S5. (A, C, E, G, I) (B, D, F, H, Figure S6.
 Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Figure legends Figure S1. Map-based cloning and complementation testing for ZOP1. (A) ZOP1 was mapped to a ~273-kb interval on Chromosome 1. In the interval, a single-nucleotide G to A substitution
Supplemental Data Supplemental Figure 1.
 Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis
 1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
Supplementary Figure 1A A404 Cells +/- Retinoic Acid
 Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana.
 IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Israel Ausin, Todd C. Mockler, Joanne Chory, and Steven E. Jacobsen Col Ler nrpd1a rdr2 dcl3-1 ago4-1 idn1-1 idn2-1
IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Israel Ausin, Todd C. Mockler, Joanne Chory, and Steven E. Jacobsen Col Ler nrpd1a rdr2 dcl3-1 ago4-1 idn1-1 idn2-1
Supplemental Data. Cui et al. (2012). Plant Cell /tpc a b c d. Stem UBC32 ACTIN
 A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
A Root Stem Leaf Flower Silique Senescence leaf B a b c d UBC32 ACTIN C * Supplemental Figure 1. Expression Pattern and Protein Sequence of UBC32 Homologues in Yeast, Human, and Arabidopsis. (A) Expression
PCR analysis was performed to show the presence and the integrity of the var1csa and var-
 Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH).
 Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
Bisulfite Treatment of DNA Dilute DNA sample to 2µg DNA in 50µl ddh 2 O. Add 5µl of 3N NaOH to DNA sample (final concentration 0.3N NaOH). Incubate in a 37ºC water bath for 30 minutes. To 55µl samples
Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC
 Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR
 Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR 1 The problem We wish to clone a yet unknown gene from a known
Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR 1 The problem We wish to clone a yet unknown gene from a known
PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells
 Supplementary Information for: PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells Ju Hye Jang 1, Hyun Kim 2, Mi Jung Jang 2, Ju Hyun Cho 1,2,* 1 Research Institute
Supplementary Information for: PGRP negatively regulates NOD-mediated cytokine production in rainbow trout liver cells Ju Hye Jang 1, Hyun Kim 2, Mi Jung Jang 2, Ju Hyun Cho 1,2,* 1 Research Institute
Supplemental Data. mir156-regulated SPL Transcription. Factors Define an Endogenous Flowering. Pathway in Arabidopsis thaliana
 Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Table S1. Bacterial strains (Related to Results and Experimental Procedures)
 Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Table S1. Bacterial strains (Related to Results and Experimental Procedures) Strain number Relevant genotype Source or reference 1045 AB1157 Graham Walker (Donnelly and Walker, 1989) 2458 3084 (MG1655)
Supplemental material
 Supplemental material Diversity of O-antigen repeat-unit structures can account for the substantial sequence variation of Wzx translocases Yaoqin Hong and Peter R. Reeves School of Molecular Bioscience,
Supplemental material Diversity of O-antigen repeat-unit structures can account for the substantial sequence variation of Wzx translocases Yaoqin Hong and Peter R. Reeves School of Molecular Bioscience,
Disease and selection in the human genome 3
 Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Supporting information for Biochemistry, 1995, 34(34), , DOI: /bi00034a013
 Supporting information for Biochemistry, 1995, 34(34), 10807 10815, DOI: 10.1021/bi00034a013 LESNIK 10807-1081 Terms & Conditions Electronic Supporting Information files are available without a subscription
Supporting information for Biochemistry, 1995, 34(34), 10807 10815, DOI: 10.1021/bi00034a013 LESNIK 10807-1081 Terms & Conditions Electronic Supporting Information files are available without a subscription
RPA-AB RPA-C Supplemental Figure S1: SDS-PAGE stained with Coomassie Blue after protein purification.
 RPA-AB RPA-C (a) (b) (c) (d) (e) (f) Supplemental Figure S: SDS-PAGE stained with Coomassie Blue after protein purification. (a) RPA; (b) RPA-AB; (c) RPA-CDE; (d) RPA-CDE core; (e) RPA-DE; and (f) RPA-C
RPA-AB RPA-C (a) (b) (c) (d) (e) (f) Supplemental Figure S: SDS-PAGE stained with Coomassie Blue after protein purification. (a) RPA; (b) RPA-AB; (c) RPA-CDE; (d) RPA-CDE core; (e) RPA-DE; and (f) RPA-C
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Hes6. PPARα. PPARγ HNF4 CD36
 SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
Supplementary Information. Construction of Lasso Peptide Fusion Proteins
 Supplementary Information Construction of Lasso Peptide Fusion Proteins Chuhan Zong 1, Mikhail O. Maksimov 2, A. James Link 2,3 * Departments of 1 Chemistry, 2 Chemical and Biological Engineering, and
Supplementary Information Construction of Lasso Peptide Fusion Proteins Chuhan Zong 1, Mikhail O. Maksimov 2, A. James Link 2,3 * Departments of 1 Chemistry, 2 Chemical and Biological Engineering, and
Supplemental Data. Bennett et al. (2010). Plant Cell /tpc
 BRN1 ---------MSSSNGGVPPGFRFHPTDEELLHYYLKKKISYEKFEMEVIKEVDLNKIEPWDLQDRCKIGSTPQNEWYFFSHKDRKYPTGS 81 BRN2 --------MGSSSNGGVPPGFRFHPTDEELLHYYLKKKISYQKFEMEVIREVDLNKLEPWDLQERCKIGSTPQNEWYFFSHKDRKYPTGS 82 SMB
BRN1 ---------MSSSNGGVPPGFRFHPTDEELLHYYLKKKISYEKFEMEVIKEVDLNKIEPWDLQDRCKIGSTPQNEWYFFSHKDRKYPTGS 81 BRN2 --------MGSSSNGGVPPGFRFHPTDEELLHYYLKKKISYQKFEMEVIREVDLNKLEPWDLQERCKIGSTPQNEWYFFSHKDRKYPTGS 82 SMB
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
6/256 1/256 0/256 1/256 2/256 7/256 10/256. At3g06290 (SAC3B)
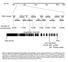 Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Chr.III M 5M 1M 15M 2M 23M BAC clones F22F7 F1A16 F24F17 F24P17 T8E24 F17A9 F21O3 F17A17 F18C1 F2O1 F28L1 F5E6 F3E22 T1B9 MLP3 Number of recombinants 6/256 1/256 /256 1/256 2/256 7/256 1/256 At3g629 (SAC3B)
Supplemental Data. Na Xu et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplemental Figure 1. The weak fluorescence phenotype is not caused by the mutation in At3g60240. (A) A mutation mapped to the gene At3g60240. Map-based cloning strategy was used to map the mutated site
Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the
 Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the portal vein for digesting adult livers, whereas it was
Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the portal vein for digesting adult livers, whereas it was
Supplementary. Table 1: Oligonucleotides and Plasmids. complementary to positions from 77 of the SRα '- GCT CTA GAG AAC TTG AAG TAC AGA CTG C
 Supplementary Table 1: Oligonucleotides and Plasmids 913954 5'- GCT CTA GAG AAC TTG AAG TAC AGA CTG C 913955 5'- CCC AAG CTT ACA GTG TGG CCA TTC TGC TG 223396 5'- CGA CGC GTA CAG TGT GGC CAT TCT GCT G
Supplementary Table 1: Oligonucleotides and Plasmids 913954 5'- GCT CTA GAG AAC TTG AAG TAC AGA CTG C 913955 5'- CCC AAG CTT ACA GTG TGG CCA TTC TGC TG 223396 5'- CGA CGC GTA CAG TGT GGC CAT TCT GCT G
II 0.95 DM2 (RPP1) DM3 (At3g61540) b
 Table S2. F 2 Segregation Ratios at 16 C, Related to Figure 2 Cross n c Phenotype Model e 2 Locus A Locus B Normal F 1 -like Enhanced d Uk-1/Uk-3 149 64 36 49 DM2 (RPP1) DM1 (SSI4) a Bla-1/Hh-0 F 3 111
Table S2. F 2 Segregation Ratios at 16 C, Related to Figure 2 Cross n c Phenotype Model e 2 Locus A Locus B Normal F 1 -like Enhanced d Uk-1/Uk-3 149 64 36 49 DM2 (RPP1) DM1 (SSI4) a Bla-1/Hh-0 F 3 111
(b) Genotyping of SALK_ (AT1g51690) and SALK_ (At1g17720) to find homozygous single mutant plants.
 Supplemental Table 1. Primers used for cloning and RT-PCR. The B-type subunits cdna CDS were amplified using high-fidelity PCR amplification with gene specific primers. The cdna CDS sequences were obtained
Supplemental Table 1. Primers used for cloning and RT-PCR. The B-type subunits cdna CDS were amplified using high-fidelity PCR amplification with gene specific primers. The cdna CDS sequences were obtained
Y-chromosomal haplogroup typing Using SBE reaction
 Schematic of multiplex PCR followed by SBE reaction Multiplex PCR Exo SAP purification SBE reaction 5 A 3 ddatp ddgtp 3 T 5 A G 3 T 5 3 5 G C 5 3 3 C 5 ddttp ddctp 5 T 3 T C 3 A 5 3 A 5 5 C 3 3 G 5 3 G
Schematic of multiplex PCR followed by SBE reaction Multiplex PCR Exo SAP purification SBE reaction 5 A 3 ddatp ddgtp 3 T 5 A G 3 T 5 3 5 G C 5 3 3 C 5 ddttp ddctp 5 T 3 T C 3 A 5 3 A 5 5 C 3 3 G 5 3 G
Cat. # Product Size DS130 DynaExpress TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1
 Product Name: Kit Component TA PCR Cloning Kit (ptakn-2) Cat. # Product Size DS130 TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1 2 Ligation Buffer
Product Name: Kit Component TA PCR Cloning Kit (ptakn-2) Cat. # Product Size DS130 TA PCR Cloning Kit (ptakn-2) 20 reactions Box 1 (-20 ) ptakn-2 Vector, linearized 20 µl (50 ng/µl) 1 2 Ligation Buffer
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified
 Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
Supplementary Figure 1. jmj30-2 and jmj32-1 produce null mutants. (a) Schematic drawing of JMJ30 and JMJ32 genome structure showing regions amplified by primers used for mrna expression analysis. Gray
S156AT168AY175A (AAA) were purified as GST-fusion proteins and incubated with GSTfused
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 Supplemental Materials Supplemental Figure S1 (a) Phenotype of the wild type and grik1-2 grik2-1 plants after 8 days in darkness.
Supplemental Information. Human Senataxin Resolves RNA/DNA Hybrids. Formed at Transcriptional Pause Sites. to Promote Xrn2-Dependent Termination
 Supplemental Information Molecular Cell, Volume 42 Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination Konstantina Skourti-Stathaki, Nicholas
Supplemental Information Molecular Cell, Volume 42 Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination Konstantina Skourti-Stathaki, Nicholas
2
 1 2 3 4 5 6 7 Supplemental Table 1. Magnaporthe oryzae strains generated in this study. Strain background Genotype Strain name Description Guy-11 H1:RFP H1:RFP Strain expressing Histone H1- encoding gene
1 2 3 4 5 6 7 Supplemental Table 1. Magnaporthe oryzae strains generated in this study. Strain background Genotype Strain name Description Guy-11 H1:RFP H1:RFP Strain expressing Histone H1- encoding gene
Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR
 Supplemental Dataset Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. DNA sequence Amino acid sequence WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR Allele 1 CCTGTC------------------GATAGC
Supplemental Dataset Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. DNA sequence Amino acid sequence WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR Allele 1 CCTGTC------------------GATAGC
Supporting Online Information
 Supporting Online Information Isolation of Human Genomic DNA Sequences with Expanded Nucleobase Selectivity Preeti Rathi, Sara Maurer, Grzegorz Kubik and Daniel Summerer* Department of Chemistry and Chemical
Supporting Online Information Isolation of Human Genomic DNA Sequences with Expanded Nucleobase Selectivity Preeti Rathi, Sara Maurer, Grzegorz Kubik and Daniel Summerer* Department of Chemistry and Chemical
Quantitative reverse-transcription PCR. Transcript levels of flgs, flgr, flia and flha were
 1 Supplemental methods 2 3 4 5 6 7 8 9 1 11 12 13 14 15 16 17 18 19 21 22 23 Quantitative reverse-transcription PCR. Transcript levels of flgs, flgr, flia and flha were monitored by quantitative reverse-transcription
1 Supplemental methods 2 3 4 5 6 7 8 9 1 11 12 13 14 15 16 17 18 19 21 22 23 Quantitative reverse-transcription PCR. Transcript levels of flgs, flgr, flia and flha were monitored by quantitative reverse-transcription
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Overexpression Normal expression Overexpression Normal expression. 26 (21.1%) N (%) P-value a N (%)
 SUPPLEMENTARY TABLES Table S1. Alteration of ZNF322A protein expression levels in relation to clinicopathological parameters in 123 Asian and 74 Caucasian lung cancer patients. Asian patients Caucasian
SUPPLEMENTARY TABLES Table S1. Alteration of ZNF322A protein expression levels in relation to clinicopathological parameters in 123 Asian and 74 Caucasian lung cancer patients. Asian patients Caucasian
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
Supporting Information. Copyright Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 2006
 Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Supporting Information for Expanding the Genetic
Supporting Information Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Copyright Wiley-VCH Verlag GmbH & Co. KGaA, 69451 Weinheim, 2006 Supporting Information for Expanding the Genetic
Supplemental Table 1. Primers used for PCR.
 Supplemental Table 1. Primers used for PCR. Gene Type Primer Sequence Genotyping and semi-quantitative RT-PCR F 5 -TTG CCC GAT CAC CAT CTG TA-3 rwa1-1 R 5 -TGT AGC GAT CAA GGC CTG ATC TAA-3 LB 5 -TAG CAT
Supplemental Table 1. Primers used for PCR. Gene Type Primer Sequence Genotyping and semi-quantitative RT-PCR F 5 -TTG CCC GAT CAC CAT CTG TA-3 rwa1-1 R 5 -TGT AGC GAT CAA GGC CTG ATC TAA-3 LB 5 -TAG CAT
Rassf1a -/- Sav1 +/- mice. Supplementary Figures:
 1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
1 Supplementary Figures: Figure S1: Genotyping of Rassf1a and Sav1 mutant mice. A. Rassf1a knockout mice lack exon 1 of the Rassf1 gene. A diagram and typical PCR genotyping of wildtype and Rassf1a -/-
ΔPDD1 x ΔPDD1. ΔPDD1 x wild type. 70 kd Pdd1. Pdd3
 Supplemental Fig. S1 ΔPDD1 x wild type ΔPDD1 x ΔPDD1 70 kd Pdd1 50 kd 37 kd Pdd3 Supplemental Fig. S1. ΔPDD1 strains express no detectable Pdd1 protein. Western blot analysis of whole-protein extracts
Supplemental Fig. S1 ΔPDD1 x wild type ΔPDD1 x ΔPDD1 70 kd Pdd1 50 kd 37 kd Pdd3 Supplemental Fig. S1. ΔPDD1 strains express no detectable Pdd1 protein. Western blot analysis of whole-protein extracts
An engineered tryptophan zipper-type peptide as a molecular recognition scaffold
 SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
SUPPLEMENTARY MATERIAL An engineered tryptophan zipper-type peptide as a molecular recognition scaffold Zihao Cheng and Robert E. Campbell* Supplementary Methods Library construction for FRET-based screening
SUPPLEMENTAL DATA SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL DATA SUPPLEMENTAL FIGURE LEGENDS SUPPLEMENTAL FIGURE S1. Identification of BmCREC. (A) Amino acid sequences of BmCREC show the peptides identified in LC-MS/MS analysis (marked by red letters
SUPPLEMENTAL DATA SUPPLEMENTAL FIGURE LEGENDS SUPPLEMENTAL FIGURE S1. Identification of BmCREC. (A) Amino acid sequences of BmCREC show the peptides identified in LC-MS/MS analysis (marked by red letters
Multiplexing Genome-scale Engineering
 Multiplexing Genome-scale Engineering Harris Wang, Ph.D. Department of Systems Biology Department of Pathology & Cell Biology http://wanglab.c2b2.columbia.edu Rise of Genomics An Expanding Toolbox Esvelt
Multiplexing Genome-scale Engineering Harris Wang, Ph.D. Department of Systems Biology Department of Pathology & Cell Biology http://wanglab.c2b2.columbia.edu Rise of Genomics An Expanding Toolbox Esvelt
Project 07/111 Final Report October 31, Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines
 Project 07/111 Final Report October 31, 2007. Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines Project Leader: Dr Douglas C. Hodgins (519-824-4120 Ex 54758, fax 519-824-5930)
Project 07/111 Final Report October 31, 2007. Project Title: Cloning and expression of porcine complement C3d for enhanced vaccines Project Leader: Dr Douglas C. Hodgins (519-824-4120 Ex 54758, fax 519-824-5930)
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
SUPPLEMENTARY INFORMATION
 Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
Gene replacements and insertions in rice by intron targeting using CRISPR Cas9 Table of Contents Supplementary Figure 1. sgrna-induced targeted mutations in the OsEPSPS gene in rice protoplasts. Supplementary
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
Legends for supplementary figures 1-3
 High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology Sidsel Nag 1,2 *, Marlene D. Dalgaard 3, Poul-Erik
High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology Sidsel Nag 1,2 *, Marlene D. Dalgaard 3, Poul-Erik
Expression of Recombinant Proteins
 Expression of Recombinant Proteins Uses of Cloned Genes sequencing reagents (eg, probes) protein production insufficient natural quantities modify/mutagenesis library screening Expression Vector Features
Expression of Recombinant Proteins Uses of Cloned Genes sequencing reagents (eg, probes) protein production insufficient natural quantities modify/mutagenesis library screening Expression Vector Features
Lacombe et al. Supplemental material ms INS-BR-TR-2
 Supplemental Figure 1: Rabbit α-gla antibodies specifically recognize carboxylated VKD proteins. (A) HEK293 cells were transfected with plasmids encoding GGCX-FLAG, PT- FLAG or tagged proline rich Gla
Supplemental Figure 1: Rabbit α-gla antibodies specifically recognize carboxylated VKD proteins. (A) HEK293 cells were transfected with plasmids encoding GGCX-FLAG, PT- FLAG or tagged proline rich Gla
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11496 Cl. 8 Cl. E93 Rag1 -/- 3H9 + BM Rag1 -/- BM CD CD c-kit c-kit c-kit wt Spleen c-kit B22 B22 IgM IgM IgM Supplementary Figure 1. FACS analysis of single-cell-derived pre-b cell clones.
doi:1.138/nature11496 Cl. 8 Cl. E93 Rag1 -/- 3H9 + BM Rag1 -/- BM CD CD c-kit c-kit c-kit wt Spleen c-kit B22 B22 IgM IgM IgM Supplementary Figure 1. FACS analysis of single-cell-derived pre-b cell clones.
SUPPLEMENTARY INFORMATION
 AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
AS-NMD modulates FLM-dependent thermosensory flowering response in Arabidopsis NATURE PLANTS www.nature.com/natureplants 1 Supplementary Figure 1. Genomic sequence of FLM along with the splice sites. Sequencing
Supporting Information
 Supporting Information Table S1. Oligonucleotide sequences used in this work Oligo DNA A B C D CpG-A CpG-B CpG-C CpG-D Sequence 5 ACA TTC CTA AGT CTG AAA CAT TAC AGC TTG CTA CAC GAG AAG AGC CGC CAT AGT
Supporting Information Table S1. Oligonucleotide sequences used in this work Oligo DNA A B C D CpG-A CpG-B CpG-C CpG-D Sequence 5 ACA TTC CTA AGT CTG AAA CAT TAC AGC TTG CTA CAC GAG AAG AGC CGC CAT AGT
SUPPORTING INFORMATION FILE
 Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules Nan Liu, Mengxi Wang, Wen Deng, Christine S. Schmidt, Weihua Qin, Heinrich Leonhardt and Fabio Spada Department of
Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules Nan Liu, Mengxi Wang, Wen Deng, Christine S. Schmidt, Weihua Qin, Heinrich Leonhardt and Fabio Spada Department of
ORFs and genes. Please sit in row K or forward
 ORFs and genes Please sit in row K or forward https://www.flickr.com/photos/teseum/3231682806/in/photostream/ Question: why do some strains of Vibrio cause cholera and others don t? Methods Mechanisms
ORFs and genes Please sit in row K or forward https://www.flickr.com/photos/teseum/3231682806/in/photostream/ Question: why do some strains of Vibrio cause cholera and others don t? Methods Mechanisms
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Investigation of the Biosynthesis of the Lasso Peptide Chaxapeptin Using an E. coli-based Production System Helena Martin-Gómez, Uwe Linne, Fernando Albericio, Judit Tulla-Puche,*
SUPPORTING INFORMATION Investigation of the Biosynthesis of the Lasso Peptide Chaxapeptin Using an E. coli-based Production System Helena Martin-Gómez, Uwe Linne, Fernando Albericio, Judit Tulla-Puche,*
SUPPLEMENTARY MATERIALS AND METHODS. E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1)
 SUPPLEMENTARY MATERIALS AND METHODS E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1) dinb::kan (lab stock) derivative was used as wild-type. MG1655 alka tag dinb (2) is
SUPPLEMENTARY MATERIALS AND METHODS E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1) dinb::kan (lab stock) derivative was used as wild-type. MG1655 alka tag dinb (2) is
Dierks Supplementary Fig. S1
 Dierks Supplementary Fig. S1 ITK SYK PH TH K42R wt K42R (kinase deficient) R29C E42K Y323F R29C E42K Y323F (reduced phospholipid binding) (enhanced phospholipid binding) (reduced Cbl binding) E42K Y323F
Dierks Supplementary Fig. S1 ITK SYK PH TH K42R wt K42R (kinase deficient) R29C E42K Y323F R29C E42K Y323F (reduced phospholipid binding) (enhanced phospholipid binding) (reduced Cbl binding) E42K Y323F
Supplemental Figure 1.
 Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
Supplemental Data. Charron et al. Dynamic landscapes of four histone modifications during de-etiolation in Arabidopsis. Plant Cell (2009). 10.1105/tpc.109.066845 Supplemental Figure 1. Immunodetection
SUPPLEMENTAL TABLE S1. Additional descriptions of plasmid constructions and the oligonucleotides used Plasmid or Oligonucleotide
 SUPPLEMENTAL TABLE S1. Additional descriptions of plasmid constructions and the oligonucleotides used Plasmid or Oligonucleotide former/ working Description a designation Plasmids pes213a b pes213-tn5
SUPPLEMENTAL TABLE S1. Additional descriptions of plasmid constructions and the oligonucleotides used Plasmid or Oligonucleotide former/ working Description a designation Plasmids pes213a b pes213-tn5
Supplemental Data. Li et al. (2015). Plant Cell /tpc
 Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
Supplemental Data Supplemental Figure 1: Characterization of asr3 T-DNA knockout lines and complementation transgenic lines. (A) The scheme of At2G33550 (ASR3) with gray boxes indicating exons and dash
FAT10 and NUB1L bind the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis
 SUPPLEMENTARY INFORMATION FAT10 and NUB1L bind the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis Neha Rani, Annette Aichem, Gunter Schmidtke, Stefan Kreft, and Marcus Groettrup
SUPPLEMENTARY INFORMATION FAT10 and NUB1L bind the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis Neha Rani, Annette Aichem, Gunter Schmidtke, Stefan Kreft, and Marcus Groettrup
MacBlunt PCR Cloning Kit Manual
 MacBlunt PCR Cloning Kit Manual Shipping and Storage MacBlunt PCR Cloning Kits are shipped on dry ice. Each kit contains a box with cloning reagents and an attached bag with Eco-Blue Competent Cells (optional).
MacBlunt PCR Cloning Kit Manual Shipping and Storage MacBlunt PCR Cloning Kits are shipped on dry ice. Each kit contains a box with cloning reagents and an attached bag with Eco-Blue Competent Cells (optional).
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07182 SUPPLEMENTAL FIGURES AND TABLES Fig. S1. myf5-expressing cells give rise to brown fat depots and skeletal muscle (a) Perirenal BAT from control (cre negative) and myf5-cre:r26r3-yfp
doi: 10.1038/nature07182 SUPPLEMENTAL FIGURES AND TABLES Fig. S1. myf5-expressing cells give rise to brown fat depots and skeletal muscle (a) Perirenal BAT from control (cre negative) and myf5-cre:r26r3-yfp
Supplementary Figures 1-12
 Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
Supplementary Figures 1-12 Supplementary Figure 1. The specificity of anti-abi1 antibody. Total Proteins extracted from the wild type seedlings or abi1-3 null mutant seedlings were used for immunoblotting
SUPPLEMENTARY INFORMATION
 1. RNA/DNA sequences used in this study 2. Height and stiffness measurements on hybridized molecules 3. Stiffness maps at varying concentrations of target DNA 4. Stiffness measurements on RNA/DNA hybrids.
1. RNA/DNA sequences used in this study 2. Height and stiffness measurements on hybridized molecules 3. Stiffness maps at varying concentrations of target DNA 4. Stiffness measurements on RNA/DNA hybrids.
Supplemental Data. Seo et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
Supplemental Figure 1. Protein alignment of ABD1 from other model organisms. The alignment was performed with H. sapiens DCAF8, M. musculus DCAF8 and O. sativa Os10g0544500. The WD40 domains are underlined.
hcd1tg/hj1tg/ ApoE-/- hcd1tg/hj1tg/ ApoE+/+
 ApoE+/+ ApoE-/- ApoE-/- H&E (1x) Supplementary Figure 1. No obvious pathology is observed in the colon of diseased ApoE-/me. Colon samples were fixed in 1% formalin and laid out in Swiss rolls for paraffin
ApoE+/+ ApoE-/- ApoE-/- H&E (1x) Supplementary Figure 1. No obvious pathology is observed in the colon of diseased ApoE-/me. Colon samples were fixed in 1% formalin and laid out in Swiss rolls for paraffin
MCB421 FALL2005 EXAM#3 ANSWERS Page 1 of 12. ANSWER: Both transposon types form small duplications of adjacent host DNA sequences.
 Page 1 of 12 (10pts) 1. There are two mechanisms for transposition used by bacterial transposable elements: replicative (Tn3) and non-replicative (Tn5 and Tn10). Compare and contrast the two mechanisms
Page 1 of 12 (10pts) 1. There are two mechanisms for transposition used by bacterial transposable elements: replicative (Tn3) and non-replicative (Tn5 and Tn10). Compare and contrast the two mechanisms
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supporting Information
 upporting Information hiota et al..73/pnas.159218 I Materials and Methods Yeast trains. Yeast strains used in this study are described in Table 1. TOM22FLAG, a yeast haploid strain for expression of C-terminally
upporting Information hiota et al..73/pnas.159218 I Materials and Methods Yeast trains. Yeast strains used in this study are described in Table 1. TOM22FLAG, a yeast haploid strain for expression of C-terminally
evaluated with UAS CLB eliciting UAS CIT -N Libraries increase in the
 Supplementary Figures Supplementary Figure 1: Promoter scaffold library assemblies. Many ensembless of libraries were evaluated in this work. As a legend, the box outline color in top half of the figure
Supplementary Figures Supplementary Figure 1: Promoter scaffold library assemblies. Many ensembless of libraries were evaluated in this work. As a legend, the box outline color in top half of the figure
Det matematisk-naturvitenskapelige fakultet
 UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam: Friday
UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam: Friday
Lecture 22: Molecular techniques DNA cloning and DNA libraries
 Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
Lecture 22: Molecular techniques DNA cloning and DNA libraries DNA cloning: general strategy -> to prepare large quantities of identical DNA Vector + DNA fragment Recombinant DNA (any piece of DNA derived
Converting rabbit hybridoma into recombinant antibodies with effective transient production in an optimized human expression system
 Converting rabbit hybridoma into recombinant antibodies with effective transient production in an optimized human expression system Dr. Tim Welsink Molecular Biology Transient Gene Expression OUTLINE Short
Converting rabbit hybridoma into recombinant antibodies with effective transient production in an optimized human expression system Dr. Tim Welsink Molecular Biology Transient Gene Expression OUTLINE Short
strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular
![strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular](/thumbs/72/66361212.jpg) Additional file 2 Identification of AOX1 in P. pastoris GS115 with a Mut s phenotype Results and Discussion The HBsAg producing strain was originally identified as a Mut s (methanol utilization slow) strain
Additional file 2 Identification of AOX1 in P. pastoris GS115 with a Mut s phenotype Results and Discussion The HBsAg producing strain was originally identified as a Mut s (methanol utilization slow) strain
Cloning and Expression of a Haloacid Dehalogenase Enzyme. By: Skyler Van Senior Research Advisor: Dr. Anne Roberts
 Cloning and Expression of a Haloacid Dehalogenase Enzyme By: Skyler Van Senior Research Advisor: Dr. Anne Roberts utline The gene being cloned is JHP1130 from Helicobacter pylori (H. pylori) JHP1130 is
Cloning and Expression of a Haloacid Dehalogenase Enzyme By: Skyler Van Senior Research Advisor: Dr. Anne Roberts utline The gene being cloned is JHP1130 from Helicobacter pylori (H. pylori) JHP1130 is
Materials Protein synthesis kit. This kit consists of 24 amino acids, 24 transfer RNAs, four messenger RNAs and one ribosome (see below).
 Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Protein Synthesis Instructions The purpose of today s lab is to: Understand how a cell manufactures proteins from amino acids, using information stored in the genetic code. Assemble models of four very
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for
 Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Fig. S1. Clustering analysis of expression array and ChIP-PCR assay in the ARF3 locus. (A) Typical examples of the transgenic plants used for ChIP-chip and ChIP-PCR assays. The presence of pas1:t7:as1
Supplemental Figure 1. Transcript profiles of Arabidopsis IPMS1 and IPMS2 in different tissues and developmental stages.
 Transcript level (intensity) Supplemental Data. Xing et al. Plant Cell (2017) 10.1105/tpc.17.00186 1600 1400 1200 1000 800 600 400 200 0 IPMS1 IPMS2 Supplemental Figure 1. Transcript profiles of Arabidopsis
Transcript level (intensity) Supplemental Data. Xing et al. Plant Cell (2017) 10.1105/tpc.17.00186 1600 1400 1200 1000 800 600 400 200 0 IPMS1 IPMS2 Supplemental Figure 1. Transcript profiles of Arabidopsis
Supplementary Methods Quantitative RT-PCR. For mrna, total RNA was prepared using TRIzol reagent (Invitrogen) and genomic DNA was eliminated with TURB
 Supplementary Methods Quantitative RT-PCR. For mrna, total RNA was prepared using TRIzol reagent (Invitrogen) and genomic DNA was eliminated with TURBO DNA-free Kit (Ambion). One µg of total RNA was reverse
Supplementary Methods Quantitative RT-PCR. For mrna, total RNA was prepared using TRIzol reagent (Invitrogen) and genomic DNA was eliminated with TURBO DNA-free Kit (Ambion). One µg of total RNA was reverse
Search for and Analysis of Single Nucleotide Polymorphisms (SNPs) in Rice (Oryza sativa, Oryza rufipogon) and Establishment of SNP Markers
 DNA Research 9, 163 171 (2002) Search for and Analysis of Single Nucleotide Polymorphisms (SNPs) in Rice (Oryza sativa, Oryza rufipogon) and Establishment of SNP Markers Shinobu Nasu, Junko Suzuki, Rieko
DNA Research 9, 163 171 (2002) Search for and Analysis of Single Nucleotide Polymorphisms (SNPs) in Rice (Oryza sativa, Oryza rufipogon) and Establishment of SNP Markers Shinobu Nasu, Junko Suzuki, Rieko
Supplemental Figure 1
 Supplemental Figure 1 A LK sls1 lks1-2 F 1 sls1 B LK sls1 lks1-2 F 1 lks1-2 sls1 F 1 lks1-2 sls1 F 2 Col lks1-2 Col+LKS1 Col Col+LKS1 Supplemental Figure 1. Genetic analysis of sls1 mutant. (A) and (B)
Supplemental Figure 1 A LK sls1 lks1-2 F 1 sls1 B LK sls1 lks1-2 F 1 lks1-2 sls1 F 1 lks1-2 sls1 F 2 Col lks1-2 Col+LKS1 Col Col+LKS1 Supplemental Figure 1. Genetic analysis of sls1 mutant. (A) and (B)
Dynamic enhancer-gene body contacts during transcription elongation
 Dynamic enhancer-gene body contacts during transcription elongation Kiwon Lee, Chris C.-S. Hsiung,, Peng Huang, Arjun Raj, *, and Gerd A. Blobel Division of Hematology, The Children s Hospital of Philadelphia,
Dynamic enhancer-gene body contacts during transcription elongation Kiwon Lee, Chris C.-S. Hsiung,, Peng Huang, Arjun Raj, *, and Gerd A. Blobel Division of Hematology, The Children s Hospital of Philadelphia,
Lecture 11: Gene Prediction
 Lecture 11: Gene Prediction Study Chapter 6.11-6.14 1 Gene: A sequence of nucleotides coding for protein Gene Prediction Problem: Determine the beginning and end positions of genes in a genome Where are
Lecture 11: Gene Prediction Study Chapter 6.11-6.14 1 Gene: A sequence of nucleotides coding for protein Gene Prediction Problem: Determine the beginning and end positions of genes in a genome Where are
Supporting Information
 Supporting Information Barderas et al. 10.1073/pnas.0801221105 SI Text: Docking of gastrin to Constructed scfv Models Interactive predocking of the 4-WL-5 motif into the central pocket observed in the
Supporting Information Barderas et al. 10.1073/pnas.0801221105 SI Text: Docking of gastrin to Constructed scfv Models Interactive predocking of the 4-WL-5 motif into the central pocket observed in the
Table S1. Alteration of ZNF322A and FBXW7 protein expression levels in relation to clinicopathological parameters in 135 lung cancer patients.
 SUPPLEMENTARY TABLES Table S1. Alteration of ZNF322A and FBXW7 protein expression levels in relation to clinicopathological parameters in 135 lung cancer patients. ZNF322A Normal Overexpression expression
SUPPLEMENTARY TABLES Table S1. Alteration of ZNF322A and FBXW7 protein expression levels in relation to clinicopathological parameters in 135 lung cancer patients. ZNF322A Normal Overexpression expression
Supplemental Figure 1
 Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
Supplemental Figure 1 A gta2-1 gta2-2 1kb AT4G08350 B Col-0 gta2-1 LP+RP+LBb1.3 C Col-0 gta2-2 LP+RP+LB1 D Col-0 gta2-2 gta2-1 GTA2 TUBLIN E Col0 gta2-1 gta2-2 Supplemental Figure 1. Phenotypic analysis
S4B fluorescence (AU)
 A S4B fluorescence (AU) S4B fluorescence (AU) dsbb csgba csgd dsbb csgba bcsa 5000 * NS NS 4000 * 3000 2000 1000 0 ΔcsgBAΔbcsA ΔcsgDΔdsbBΔbcsA ΔcsgBA ΔdsbBΔcsgBA ΔcsgDΔdsbB B -1000 4000 * * NS 3500 * 3000
A S4B fluorescence (AU) S4B fluorescence (AU) dsbb csgba csgd dsbb csgba bcsa 5000 * NS NS 4000 * 3000 2000 1000 0 ΔcsgBAΔbcsA ΔcsgDΔdsbBΔbcsA ΔcsgBA ΔdsbBΔcsgBA ΔcsgDΔdsbB B -1000 4000 * * NS 3500 * 3000
Supplemental Data. Guo et al. (2015). Plant Cell /tpc
 Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Figure 1. The Mutant exb1-d Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons. (A) Branches were developed in exb1-d but not in wild-type plants. (B) and (C)
Supplemental Data. Lee et al. Plant Cell. (2010) /tpc Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2.
 Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Supplemental Figure 1. Protein and Gene Structures of DWA1 and DWA2. (A) Protein structures of DWA1 and DWA2. WD40 region was determined based on the NCBI conserved domain databases (B, C) Schematic representation
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
