Initiation of Replication. This section will begin with a brief discussion of chromosomal DNA sequences that sponsor the initiation of replication.
|
|
|
- Emily Anthony
- 6 years ago
- Views:
Transcription
1 Initiation of Replication This section will begin with a brief discussion of chromosomal DNA sequences that sponsor the initiation of replication. Replicons and origins of replication The unit of DNA replication is called the replicon. It is at the level of the replicon that DNA replication is regulated. Consider that DNA replication must be coordinated with the cell cycle, which demands a strict alteration of DNA replication and mitosis. Thus each replicon must be replicated at the appropriate time, and must be replicated only once per cell cycle (i.e. once per S phase). Replicons have a unique origin of replication (ori), where replication initiates. (Ori's are sometimes called replicators, but we will use the term ori, or origin.) Origin sequences bind initiator proteins that promote the formation of replication forks. A replicon may also have a discrete terminus, where replication stops. Replicons may be unidirectional or bidirectional, depending on whether one or two replication forks advance from the origin. In the case of cellular chromosomes (prokaryotic and eukaryotic), replication occurs bidirectionally. Simple origins (those from bacterial chromosomes, viral chromosomes, and chromosomes of lower eukaryotes such as budding yeast) can be identified genetically as cis-acting sequences that sponsor the replication (extrachromosomal maintenance) of DNA molecules in which they are inserted. These simple origins are modular in nature, and consist of a series of short sequence blocks spaced over a region of bp. In budding yeast, for example, replication origins have been identified as autonomously replicating sequences (ARS). ARS's were discovered by fragmenting the yeast genome by restriction endonuclease digestion, and inserting the fragments into plasmids containing a selectable marker (e.g. nutritional marker or antibiotic resistance) (Fig. 59). These plasmids were then used to transform yeast, and cells capable of forming colonies contained plasmids that were capable of replicating in yeast cells; i.e. the DNA fragment inserted into the plasmid contained an ARS. Some ARS sequences have been confirmed as bona fide origins of replication. In contrast, complex oris (those found in the chromosomes of multicellular eukaryotes) usually cannot be identified in this way. That is, they do not behave as ARS's, probably because their function requires chromatin structure. Thus, relatively few oris from higher organisms have been 68
2 identified, and relatively little is known of their structure. Most encompass several kilobases of DNA and it has been difficult to identify critical sequences within them. That is, multiple sequence elements over a large stretch of DNA contribute to origin function, and these sequences seem to be redundant. On the other hand, in some early embryos (Drosophila and Xenopus) origins appear to require little or no sequence specificity, perhaps to allow for a very rapid S phase. Key to understanding how cells regulate DNA synthesis is understanding the nature of DNA replication origins. Replication origins play two important roles in DNA replication: They provide discrete starting points where replication begins. This ensures that each time the DNA must be replicated, all the necessary components of the replication machinery are brought to the correct location for assembly. They provide a means to regulate when initiation events occur. This ensures that the DNA is replicated only once per cell division; that is, a chromosomal replication origin is used once and only once per cell cycle. Consider the E. coli chromosome. The genome exists as a single large circular molecule (1N = 4.2 x 10 6 bp). It constitutes a single replicon, as it contains only one origin of replication (oric). Replication initiates at the origin and proceeds bidirectionally (theta mode replication). Two replication forks leave the single origin in opposite directions and proceed at about the same rate until they meet. Replication of the E. coli chromosome is relatively rapid: 4,200,000 bp/100,000 bp per min = 42 minutes needed to replicate the entire genome. In contrast, eukaryotic genomes are very much larger and fork movement is considerably slower (500-4,000 bp/min vs. 50,000 bp/min in E. coli). To compensate, eukaryotic genomes are divided into several (linear) chromosomes, each of which is divided into a large number of relatively small replicons. In eukaryotes, one origin of replication occurs every kb (compare with the 4,200 kb E. coli genome) (Fig. 60). Replication proceeds bidirectionally from each origin, until it encounters a replication fork from an adjacent origin (or reaches the end of the DNA molecule). Consider the mouse genome (1N = 2 x 10 9 bp; 2N = 4 x 10 9 bp). Even though it is about a 1,000 times larger than the E. coli genome, the fact that it is divided into 25,000 replicons of average 69
3 size 150 kb allows for rapid duplication. Just over an hour would be required if all replicons fired at the same time. But in fact, eukaryotic genomes typically require 6-8 hours to be replicated, because not all origins fire at once. Replicons containing active genes tend to be replicated early, while others are replicated later. (This implies additional regulation, which is not yet understood.) Obviously, events which occur at the origin of replication are of central importance for replication and its control. There are many questions: How is replication initiated? How is initiation coordinated with the cell cycle? How is initiation restricted to once per S phase? How is the replication of thousands of origins coordinated? How do viruses escape these controls? At present, we have only partial answers to these questions, and of course we know more about prokaryotic systems than we do about eukaryotic systems. But again, despite the differences between pro- and eukaryotes, there are elements of mechanism that are common to both. Here we will focus on origin structure and how replication is initiated. Characteristics of simple origins What do replication origins look like? Origins of DNA replication can be divided into two general groups: simple origins such as E. coli oric and those found in simple eukaryotic genomes (viruses and yeast), and those found in complex genomes of plants and metazoa (flies, frogs and mammals). Simple origins have a modular anatomy composed of unique DNA sequence motifs, at least one of which interacts with soluble proteins (Fig. 61). They usually consist of at least four motifs/elements. The organization of these elements relative to each other is similar in most simple origins (DePamphilis, M. (1996) Origins of DNA replication. In DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press. pp ) 1. The origin recognition element (ORE). An ORE is a DNA binding site for one or more origin recognition proteins (ORPs) that are required for initiation of DNA replication. Binding is sequence specific in simple oris, although other factors (such as chromatin structure) can influence binding. Origin recognition proteins serve at least two functions: They initiate DNA unwinding using either their own helicase activity (some viral proteins), by recruiting a helicase, or by distorting the helix. 70
4 They bind other replication proteins and recruit them to the origin. Therefore, binding of the ORE by ORPs is the key step that determines where replication will begin. 2. The DNA unwinding element (DUE). DNA unwinding begins at an easily unwound DNA sequence called the DUE. A DUE is defined by cis-acting mutations that both increase the stability of the double helix and reduce DNA replication efficiency. DUE's are usually A/T rich but lack any other sequence specificity. Any easily unwound DNA sequences can substitute for a bona fide DUE. Therefore, it is unlikely that a specific replication protein interacts with the DUE. Rather, binding of the ORE by ORP results in unwinding at the DUE. In support of this idea, spacing between the ORE and DUE is critical for unwinding. The DUE corresponds to the origin of bidirectional replication (OBR). This site is defined by the transition between continuous and discontinuous DNA synthesis on each template. Thus, the DUE is the entry site for the replication machinery. 3. The A/T element This element is critical for origin function, although what it does is not entirely clear. It should not be confused with the DUE, which is usually also A/T rich. A/T elements characteristically have a "bent" structure (bent DNA), which is favored by the appearance of an unbroken stretch of A-T base pairs. DNA bending may contort the origin and facilitate protein binding to the ORE and unwinding of the DUE (or both). The ORE, the DUE and the A/T rich element are essential and comprise the origin core. 4. Auxiliary elements Auxiliary elements are not essential, but stimulate replication 2 to 1000-fold. typically bind transcription factors. Aux elements How do transcription factors stimulate origin activity? This is not entirely clear. But it is likely that the transcription factors recruit proteins capable of remodeling chromatin structure; thus they may help to open chromatin and allow replication factors (such as an ORP) to access the DNA. 71
5 Initiation- Assembly of replication forks Replication forks are assembled at origins of replication. The components needed to build the replication fork include proteins that recognize the origin (origin recognition proteins), proteins required for assembly but not for subsequent DNA synthesis, and components of the fork replication machinery. Initiation at oric in E. coli. Using plasmids containing oric, an in vitro system utilizing purified proteins has been established. This has lead to significant advances in our understanding of initiation. The goal of the first step in initiation is to open up the duplex, and to recruit replication proteins (in particular, helicase). The product is a prepriming complex. The proteins required for formation of the prepriming complex include DnaA, DnaB, DnaC, and SSB. The reaction is favored by template supercoiling, which facilitates strand separation. OriC contains 3 x 13 bp repeats (the DUE) and 4 x 9 bp repeats (R1-4; the ORE). DnaA acts as an origin recognition protein (initiator protein). It recognizes its cognate origin (oric) in a sequence specific manner. This is the foundation for all subsequent events (Fig. 62). DnaA is both a sequence-specific dsdna binding protein and an ssdna binding protein. It is also an ATPase. In the presence of ATP, DnaA-ATP first acts as a dsdna binding protein and binds cooperatively to the ORE until a complex consisting of DnaA monomers is formed. OriC ( bp of DNA) is wrapped around the DnaA complex. This distorts the helix, which promotes melting at the DUE. This step is facilitated by supercoiling. DNA-ATP then interacts with ssdna at the DUE to stabilize the open conformation. In the next step, the hexameric helicase (DnaB) is recruited and loaded onto the ssdna. Recruiting is accomplished by DnaA which interacts with a helicase loading factor, DnaC. DnaC also interacts with DnaB in the presence of ATP. Six molecules of DnaC-ATP bind six molecules of DnaB. Two such DnaB:DnaC complexes are recruited to the origin by the ability of DnaC to bind DnaA. The presence of ssdna and/or interaction with DnaA triggers ATP hydrolysis by DnaC. The free DnaC then leaves DnaB loaded onto the lagging strand template. Again, two DnaB helicases are 72
6 loaded at oric, one for each fork. At this point, the prepriming complex is formed. It consists of stable replication protein complexes assembled on the ori. The next step in initiation is priming. The helicase, with the aid of SSB, begins to expand the replication bubble. The DnaG primase is recruited next, it forms a mobile complex with DnaB (the primosome) (Fig. 63). After the primosome is formed, primase recognizes the template and synthesizes an RNA primer NT long. This provides the 3'-OH terminus that is recognized by Pol III holoenzyme. The interaction between DnaB and the τ subunit no doubt assists in recruiting Pol III. β clamps are loaded on the primer termini by the γ complex clamp loader. Polymerase loading is the final step in the assembly of the replisome at the replication fork. But replicative synthesis cannot proceed until the complex is further remodeled. Recent evidence indicates that this final remodeling step is ATP hydrolysis by DnaA-ATP. ATP hydrolysis is triggered by the presence of the β clamp at the primer terminus. ATP hydrolysis inactivates and releases DnaA from the origin, which allows elongation to proceed. This provides a mechanism to ensure that replication does not occur until all components are assembled. In addition, since DnaA cannot bind oric unless it is complexed with ATP, this mechanism also prevents re-replication. References: Baker, TA, and Wickner, S.H. (1992) Genetics and enzymology of DNA replication in Escherichia coli. Annual Review of Genetics 26: Katayama, T., Kubota, T., Kurokawa, K., Crooke, E., and Sekimizu, K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94: Kurokawa, K., Nishida, S., Emoto, A., Sekimizu, K., and Katayama, T. (1999) The EMBO Journal 18: Lee, D.G., and Bell, S.P. (2000) ATPase switches controlling DNA replication initiation. Current Opinion in Cell Biology 12: Initiation at eukaryotic chromosomal origins Much less is known about initiation at eukaryotic chromosomal origins. However, a general picture is beginning to emerge. Most of the recent information has come from genetic and biochemical studies using budding yeast (Saccharomyces cerevisiae), fission yeast 73
7 (Schizosaccharomyces pombe), Drosophila melanogaster, the African toad (Xenopus laevis), mouse and human cells. In budding yeast, ARS sequences contain elements which function similarly to those of other simple origins. All ARS's contain an A element, which is characterized by a conserved, 11-bp ARS consensus sequence (an ORE). The A element binds the origin recognition complex (ORC; see below). Three B elements have also been defined. B1, which is adjacent to A, facilitates ORC binding. B2 appears to be a DUE. In the ARS1 ori, B3 is a binding site for transcription factor Abf1, and thus it probably functions as an auxiliary element. The organization of complex metazoan oris is not clear. It is known that they bind ORC, but exactly how is not known, as they do not contain an ARS consensus sequence, or any other obviously conserved sequence. Chromatin structure also plays an important role in origin recognition. Nevertheless, the sequence of events appears to be very similar in yeast and higher eukaryotes. We can say this because the proteins involved are highly conserved and in many cases are interchangeable between organisms. The initiation process is tied to the cell cycle through the action of cyclin dependent kinases (CDKs) which either inhibit or stimulate complex formation by phosphorylating specific proteins, depending on cell cycle timing. The CDKs are a family of related protein kinases that require association with an activating cyclin protein for catalytic function. Cyclin expression/accumulation is regulated in a cell cycle-dependent manner. For example, G1/S-cyclins are active at the end of G1; S-cyclins are active during S phase, M-cyclins promote mitosis, and G1-cyclins allow passage through G1. The exact targets of the CDKs within the initiation complex are not yet known. Origins are recognized by a group of proteins known as the origin recognition complex (ORC). ORC is a complex of six different proteins (ORC 1-6), which binds the ORE in the presence of ATP (Fig. 64). Several of the ORC proteins are ATPases, and the complex has both dsdna and ssdna binding activity. ORC-ATP binds dsdna at the origin. Interestingly, ssdna appears to stimulate ORC-ATPase activity in a length-dependent manner. Thus unwinding at the origin may result in the loss of ORC dsdna binding activity. ORC appears to be associated with the origin through through G2-M-G1, and may be released in S. 74
8 In G1, Cdc6 enters the complex. Cdc6 binding to the origin requires ORC. Cdc6 is also binds ATP and is active as a Cdc6-ATP complex. Cdt1 (not shown on the Fig. 64 diagram) forms a complex with Cdc6. Current data suggests that the Cdc6:Cdt1 complex is the helicase-loader. The Mcm proteins (Mcm 2-7) enter next. Mcm proteins interact with Cdc6 and are not bound to origins until Cdc6 is present. A subset of the Mcm complex, Mcm 4, 6 and 7 (two copies of each), is a weak hexameric helicase in vitro. The other Mcm proteins, particularly Mcm 2, may inhibit helicase activity. Later, at the time of initiation, the helicase could be activated by removal of Mcm 2, 3, and 5. Presumably the active Mcm helicase functions in local unwinding at the origin, and also acts to separate parental strands during replication fork progression. A complex with Mcm (helicase) at the nascent fork constitutes the pre-initiation complex. The transition to replication occurs at the G1/S boundary and involves removal of Cdc6 and Cdt1. CDK phosphorylation of Cdc6 results in its degradation (yeast) or export from the nucleus (mammalian cells). A novel protein called geminin (found in metazoans; not shown) inhibits and probably removes Cdt1. Once Cdc6 and Cdt1 are removed, Cdc45 protein associates with the origin. Recrutment of Cdc45 requires ORC and MCM proteins. Cdc45 also interacts with RPA (SSB), DNA Pol α:primase, and DNA Pol δ/ε. Cdc45 appears to be part of the replisome. So it may not only recruit the polymerase:primase, it may also tether it to the helicase (Mcm:Cdc45:Polα: primase; a mobile primosome?). With the arrival of the primase and polymerases, all components needed for replication are present. But additional proteins that were necessary for initiation but inhibitory to further growth of the fork must first be released. This remodeling step may be mediated in part by Cdk7-Dbf4 kinase (also known as DDK). This kinase is known to phosphorylate Mcm proteins. It is possible that this releases Mcm 2, 3, 5, which could activate the Mcm 4,6,7 helicase. Additional unwinding generates additional ssdna, which stimulates the ORC ATPase. hydrolysis inactivates the ORP function of ORC, and allow its release from the origin. ATP Note that there are several steps that prevent re-replication (re-initiation): CDK-mediated prevention of re-replication. CDK phosphorylation inactivates Cdc6 (clamp loader). This allows complexes established in G1 to transition toward replication, but prevents 75
9 formation of new pre-priming complexes through S, G2, and M. CDKs also phosphorylate ORC and MCM components, although the functional consequences are less clear. Geminin-mediated prevention of re-replication. Geminin appears after cells enter S-phase and levels fall during mitosis. This protein inhibits Cdt1 (part of the clamp loader) and the consequences of this are similar to CDK phosphorylation of Cdc6. ORC-mediated prevention of re-initiation. ORC binds the origin as a nucleotide complex (ORC- ATP). Unwinding generates ssdna, which stimulates of ORC ATPase, which inactivates ORC as an ORP. Although there are differences in detail, what is striking about this mechanism is its general similarity to initiation at E. coli oric: both mechanisms employ a series of discrete steps involving rounds of recruitment and remodeling to produce an active replication fork on which primases and DNA polymerases can be loaded. Most notably, ATP is used as a switch to separate and regulate recruitment and remodeling. (Fig. 65). In each mechanism, protein + ATP is required for an early step, and later ATP hydrolysis results in a switch in protein activity (e.g. DnaA, ORC, DnaC, Cdc6). Previously, we saw a similar sequence of events used during clamp loading by the γ complex and RFC. References: Baker, T.A., and Bell, S.P. (1998) Polymerases and the replisome: Machines within machines. Cell 92: Kelly, T.J., and Brown, G.W. (2000) Regulation of chromosomal replication. Annual Review of Biochemistry 69: Lee, D.G., and Bell, S.P. (2000) ATPase switches controlling DNA replication initiation. Current Opinion in Cell Biology 12: Bell, S.P. and Dutta, A. (2002) DNA replication in eukaryotic cells (2002) Annual Review of Biochemistry 71:
The Size and Packaging of Genomes
 DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Replication The Size and Packaging of Genomes Vary greatly in size Ø Smallest viruses- 4 or 5 genes Ø Escherichia coli- 4,288 genes Ø Human cell- 20,000 to 25,000 genes E. coli 4 million base pairs
DNA Replication II Biochemistry 302. January 25, 2006
 DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 January 25, 2006 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
Chapter 11 DNA Replication and Recombination
 Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
Chapter 11 DNA Replication and Recombination Copyright Copyright 2009 Pearson 2009 Pearson Education, Education, Inc. Inc. 11.1 DNA is reproduced by Semiconservative Replication The complementarity of
DNA Replication II Biochemistry 302. Bob Kelm January 26, 2005
 DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302 Bob Kelm January 26, 2005 Following in Dad s footsteps Original A. Kornberg E. coli DNA Pol I is a lousy replicative enzyme. 400 molecules/cell but ~2 replication forks/cell
DNA Replication II Biochemistry 302. Bob Kelm January 28, 2004
 DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
DNA Replication II Biochemistry 302 Bob Kelm January 28, 2004 Conceptual model for proofreading based on kinetic considerations Fig. 24.44 stalling transient melting exonuclease site occupancy Following
III. Detailed Examination of the Mechanism of Replication A. Initiation B. Priming C. Elongation D. Proofreading and Termination
 Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
Outline for Replication I. General Features of Replication A. Semi-Conservative B. Starts at Origin C. Bidirectional D. Semi-Discontinuous II. Proteins and Enzymes of Replication III. Detailed Examination
DNA Replication in Prokaryotes and Eukaryotes
 DNA Replication in Prokaryotes and Eukaryotes 1. Overall mechanism 2. Roles of Polymerases & other proteins 3. More mechanism: Initiation and Termination 4. Mitochondrial DNA replication DNA replication
DNA Replication in Prokaryotes and Eukaryotes 1. Overall mechanism 2. Roles of Polymerases & other proteins 3. More mechanism: Initiation and Termination 4. Mitochondrial DNA replication DNA replication
 The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
The replication of DNA Kornberg 1957 Meselson and Stahl 1958 Cairns 1963 Okazaki 1968 DNA Replication The driving force for DNA synthesis. The addition of a nucleotide to a growing polynucleotide
Molecular Biology (2)
 Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
Molecular Biology (2) DNA replication Mamoun Ahram, PhD Second semester, 2018-2019 Resources This lecture Cooper, pp. 191-207 2 Some basic information The entire DNA content of the cell is known as genome.
The replication forks Summarising what we know:
 When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
When does replication occur? MBLG1001 lecture 10 Replication the once in a lifetime event! Full blown replication only occurs once, just before cell division BUT the DNA template is constantly being repaired.
The flow of Genetic information
 The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
The flow of Genetic information http://highered.mcgrawhill.com/sites/0072507470/student_view0/chapter3/animation dna_replication quiz_1_.html 1 DNA Replication DNA is a double-helical molecule Watson and
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory
 Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Molecular Biology: General Theory Author: Dr Darshana Morar Licensed under a Creative Commons Attribution license. DNA REPLICATION DNA replication is the process of duplicating the DNA sequence in the
Requirements for the Genetic Material
 Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
Requirements for the Genetic Material 1. Replication Reproduced and transmitted faithfully from cell to cell-generation to generation. 2. Information Storage Biologically useful information in a stable
DNA REPLICATION. Third Stage. Lec. 12 DNA Replication. Lecture No.: 12. A. Watson & Crick (1952) C. Cairns (1963) autoradiographic experiment
 Lec. 12 DNA Replication A. Watson & Crick (1952) Proposed a model where hydrogen bonds break, the two strands separate, and DNA synthesis occurs semi-conservatively in the same net direction. While a straightforward
Lec. 12 DNA Replication A. Watson & Crick (1952) Proposed a model where hydrogen bonds break, the two strands separate, and DNA synthesis occurs semi-conservatively in the same net direction. While a straightforward
Questions from chapters in the textbook that are relevant for the final exam
 Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
Questions from chapters in the textbook that are relevant for the final exam Chapter 9 Replication of DNA Question 1. Name the two substrates for DNA synthesis. Explain why each is necessary for DNA synthesis.
CSIR UGC NET, GATE (ENGINEERING), GATE (Science), IIT-JAM, UGC NET, TIFR, IISc, NIMCET, JEST etc. JNU CEEB SAMPLE THEORY
 JNU CEEB SAMPLE THEORY REPLICATION TYPE OF DNA REPLICATION REPLICATION IN PROKARYOTES REPLICATION IN EUKARYOTES For IIT-JAM, JNU, GATE, NET, NIMCET and Other Entrance Exams 1-C-8, Sheela Chowdhary Road,
JNU CEEB SAMPLE THEORY REPLICATION TYPE OF DNA REPLICATION REPLICATION IN PROKARYOTES REPLICATION IN EUKARYOTES For IIT-JAM, JNU, GATE, NET, NIMCET and Other Entrance Exams 1-C-8, Sheela Chowdhary Road,
Replication. Obaidur Rahman
 Replication Obaidur Rahman DIRCTION OF DNA SYNTHESIS How many reactions can a DNA polymerase catalyze? So how many reactions can it catalyze? So 4 is one answer, right, 1 for each nucleotide. But what
Replication Obaidur Rahman DIRCTION OF DNA SYNTHESIS How many reactions can a DNA polymerase catalyze? So how many reactions can it catalyze? So 4 is one answer, right, 1 for each nucleotide. But what
Replication of DNA and Chromosomes
 Chapter 9. Replication of DNA and Chromosomes 1. Semiconservative Replication 2. DNA Polymerases and DNA Synthesis In Vitro 3. The Complex Replication Apparatus 4. Unique Aspects of Eukaryotic Chromosome
Chapter 9. Replication of DNA and Chromosomes 1. Semiconservative Replication 2. DNA Polymerases and DNA Synthesis In Vitro 3. The Complex Replication Apparatus 4. Unique Aspects of Eukaryotic Chromosome
DNA Metabolism. I. DNA Replication. A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding
 DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
DNA Metabolism I. DNA Replication A. Template concept: 1. How can you make a copy of a molecule? 2. Complementary Hydrogen bonding B. DNA replication follows a set of fundamental rules 1. Semiconservative
Replication of DNA Virus Genomes. Lecture 7 Virology W3310/4310 Spring 2013
 Replication of DNA Virus Genomes Lecture 7 Virology W3310/4310 Spring 2013 It s all about Initiation Problems faced by DNA replication machinery Viruses must replicate their genomes to make new progeny
Replication of DNA Virus Genomes Lecture 7 Virology W3310/4310 Spring 2013 It s all about Initiation Problems faced by DNA replication machinery Viruses must replicate their genomes to make new progeny
Zoo-342 Molecular biology Lecture 2. DNA replication
 Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
Zoo-342 Molecular biology Lecture 2 DNA replication DNA replication DNA replication is the process in which one doubled-stranded DNA molecule is used to create two double-stranded molecules with identical
DNA Replication I Biochemistry 302. Bob Kelm January 24, 2005
 DNA Replication I Biochemistry 302 Bob Kelm January 24, 2005 Watson Crick prediction: Each stand of parent DNA serves as a template for synthesis of a new complementary daughter strand Fig. 4.12 Proof
DNA Replication I Biochemistry 302 Bob Kelm January 24, 2005 Watson Crick prediction: Each stand of parent DNA serves as a template for synthesis of a new complementary daughter strand Fig. 4.12 Proof
Tutorial Week #9 Page 1 of 11
 Tutorial Week #9 Page 1 of 11 Tutorial Week #9 DNA Replication Before the tutorial: Read ECB Chapter 6 p195-207, and review your lecture notes Read this tutorial and create a table of definitions and functions
Tutorial Week #9 Page 1 of 11 Tutorial Week #9 DNA Replication Before the tutorial: Read ECB Chapter 6 p195-207, and review your lecture notes Read this tutorial and create a table of definitions and functions
Gene Expression- Protein Synthesis
 Gene Expression- Protein Synthesis Protein synthesis is the key to expression of biological information - Structural Protein: bones, cartilage - Contractile Proteins: myosin, actin - Enzymes - Transport
Gene Expression- Protein Synthesis Protein synthesis is the key to expression of biological information - Structural Protein: bones, cartilage - Contractile Proteins: myosin, actin - Enzymes - Transport
Prokaryotic cells divide by pinching in two. Fig. 10-CO, p.240
 Prokaryotic cells divide by pinching in two Fig. 10-CO, p.240 Learning Objectives 1. What Is the Flow of Genetic Information in the Cell? 2. What Are the General Considerations in the Replication of DNA?
Prokaryotic cells divide by pinching in two Fig. 10-CO, p.240 Learning Objectives 1. What Is the Flow of Genetic Information in the Cell? 2. What Are the General Considerations in the Replication of DNA?
Welcome to Class 18! Lecture 18: Outline and Objectives. Replication is semiconservative! Replication: DNA DNA! Introductory Biochemistry!
 Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Lecture 18: Outline and Objectives Welcome to Class 18! Introductory Biochemistry! l DNA Replication! l DNA polymerase! l the enzymatic reaction! l proofreading and accuracy! l DNA synthesis! l origins
Chapter 3: Duplicating the DNA- Replication
 3. Basic Genetics Plant Molecular Biology Chapter 3: Duplicating the DNA- Replication Double helix separation New strand synthesis Plant Biotechnology Lecture 2 1 I've missed more than 9000 shots in my
3. Basic Genetics Plant Molecular Biology Chapter 3: Duplicating the DNA- Replication Double helix separation New strand synthesis Plant Biotechnology Lecture 2 1 I've missed more than 9000 shots in my
Genetic Information: DNA replication
 Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Genetic Information: DNA replication Umut Fahrioglu, PhD MSc DNA Replication Replication of DNA is vital to the transmission of genomes and the genes they contain from one cell generation to the other.
Enzymes used in DNA Replication
 Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
Enzymes used in DNA Replication This document holds the enzymes used in DNA replication, their pictorial representation and functioning. DNA polymerase: DNA polymerase is the chief enzyme of DNA replication.
Biologia Genômica. 2º Semestre, Replicação de DNA em Bactérias e no Núcleo Eucariótico. Prof. Marcos Túlio
 Biologia Genômica 2º Semestre, 2017 Replicação de DNA em Bactérias e no Núcleo Eucariótico Prof. Marcos Túlio mtoliveria@fcav.unesp.br Faculdade de Ciências Agrárias e Veterinárias de Jaboticabal Instituto
Biologia Genômica 2º Semestre, 2017 Replicação de DNA em Bactérias e no Núcleo Eucariótico Prof. Marcos Túlio mtoliveria@fcav.unesp.br Faculdade de Ciências Agrárias e Veterinárias de Jaboticabal Instituto
Fidelity of DNA polymerase
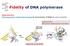 Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
Fidelity of DNA polymerase Shape selectivity: DNA polymerase's conformational change for determination of fidelity for each nucleotide Induced fit: Structure determines function Matched nucleotide Fidelity
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 11 DNA Replication
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 11 DNA Replication 2 3 4 5 6 7 8 9 Are You Getting It?? Which characteristics will be part of semi-conservative replication? (multiple answers) a) The
BS GENOMES. DNA replication and repair
 BS2009 - GENOMES DNA replication and repair REPLICATION GENERAL PRINCIPLES START Must be ready Must know where to start FINISH Must all finish Must ensure that each piece of DNA is replicated only once
BS2009 - GENOMES DNA replication and repair REPLICATION GENERAL PRINCIPLES START Must be ready Must know where to start FINISH Must all finish Must ensure that each piece of DNA is replicated only once
IDTutorial: DNA Replication
 IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
IDTutorial: DNA Replication Introduction In their report announcing the structure of the DNA molecule, Watson and Crick (Nature, 171: 737-738, 1953) observe, It has not escaped our notice that the specific
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI REPLICATION - ENZYMES.
 ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
ARUNAI ACADEMY FOR PG TRB-BOTANY DHARMAPURI.9500244679 REPLICATION - ENZYMES DNA HELICASE Sparation of two strands- DNA helicase enzyme functions Unwinds DNA. DNA double helix by breaking the hydrogen
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function. Gary Peter
 Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
Plant Molecular and Cellular Biology Lecture 4: E. coli DNA Replicase Structure & Function Gary Peter Learning Objectives 1. List and explain the mechanisms by which E. coli DNA is replicated 2. Describe
DNA REPLICATION. Anna Onofri Liceo «I.Versari»
 DNA REPLICATION Anna Onofri Liceo «I.Versari» Learning objectives 1. Understand the basic rules governing DNA replication 2. Understand the function of key proteins involved in a generalised replication
DNA REPLICATION Anna Onofri Liceo «I.Versari» Learning objectives 1. Understand the basic rules governing DNA replication 2. Understand the function of key proteins involved in a generalised replication
DNA replication. - proteins for initiation of replication; - proteins for polymerization of nucleotides.
 DNA replication Replication represents the duplication of the genetic information encoded in DNA that is the crucial step in the reproduction of living organisms and the growth of multicellular organisms.
DNA replication Replication represents the duplication of the genetic information encoded in DNA that is the crucial step in the reproduction of living organisms and the growth of multicellular organisms.
BCMB Chapters 34 & 35 DNA Replication and Repair
 BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
BCMB Chapters 34 & 35 DNA Replication and Repair
 BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
BCMB 3100 - Chapters 34 & 35 DNA Replication and Repair Semi-conservative DNA replication DNA polymerase DNA replication Replication fork; Okazaki fragments Sanger method for DNA sequencing DNA repair
DNA Replication: Prokaryotes and Yeast
 DNA Replication: Prokaryotes and Yeast Derek J Hoelz, Indiana University School of Medicine, Indianapolis, IN, USA Robert J Hickey, Indiana University School of Medicine, Indianapolis, IN, USA Linda H
DNA Replication: Prokaryotes and Yeast Derek J Hoelz, Indiana University School of Medicine, Indianapolis, IN, USA Robert J Hickey, Indiana University School of Medicine, Indianapolis, IN, USA Linda H
DNA Replication in Eukaryotes
 OpenStax-CNX module: m44517 1 DNA Replication in Eukaryotes OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
OpenStax-CNX module: m44517 1 DNA Replication in Eukaryotes OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section,
BIO 311C Spring Lecture 34 Friday 23 Apr.
 BIO 311C Spring 2010 1 Lecture 34 Friday 23 Apr. Summary of DNA Replication in Prokaryotes origin of replication initial double helix origin of replication new growing polynucleotide chains Circular molecule
BIO 311C Spring 2010 1 Lecture 34 Friday 23 Apr. Summary of DNA Replication in Prokaryotes origin of replication initial double helix origin of replication new growing polynucleotide chains Circular molecule
DNA Structure. DNA: The Genetic Material. Chapter 14
 DNA: The Genetic Material Chapter 14 DNA Structure DNA is a nucleic acid. The building blocks of DNA are nucleotides, each composed of: a 5-carbon sugar called deoxyribose a phosphate group (PO 4 ) a nitrogenous
DNA: The Genetic Material Chapter 14 DNA Structure DNA is a nucleic acid. The building blocks of DNA are nucleotides, each composed of: a 5-carbon sugar called deoxyribose a phosphate group (PO 4 ) a nitrogenous
Nucleic Acid Structure:
 Nucleic Acid Structure: Purine and Pyrimidine nucleotides can be combined to form nucleic acids: 1. Deoxyribonucliec acid (DNA) is composed of deoxyribonucleosides of! Adenine! Guanine! Cytosine! Thymine
Nucleic Acid Structure: Purine and Pyrimidine nucleotides can be combined to form nucleic acids: 1. Deoxyribonucliec acid (DNA) is composed of deoxyribonucleosides of! Adenine! Guanine! Cytosine! Thymine
Cyclin- Dependent Protein Kinase. Function
 Complex Cyclin Cyclin- Dependent rotein Kinase Function promotes passage through restriction G1-CDK Complex Cyclin D CDK4 or CDK6 point in late G1 G1/S-CDK Complex Cyclin E CDK2 commits the cell to DA
Complex Cyclin Cyclin- Dependent rotein Kinase Function promotes passage through restriction G1-CDK Complex Cyclin D CDK4 or CDK6 point in late G1 G1/S-CDK Complex Cyclin E CDK2 commits the cell to DA
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA
 RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
RNA Expression of the information in a gene generally involves production of an RNA molecule transcribed from a DNA template. RNA differs from DNA that it has a hydroxyl group at the 2 position of the
Viral DNA replication
 Viral DNA replication Lecture 8 Biology 3310/4310 Virology Spring 2017 The more the merrier --ANONYMOUS Viral DNA genomes must be replicated to make new progeny Parvovirus Retrovirus Poliovirus VII Hepatitis
Viral DNA replication Lecture 8 Biology 3310/4310 Virology Spring 2017 The more the merrier --ANONYMOUS Viral DNA genomes must be replicated to make new progeny Parvovirus Retrovirus Poliovirus VII Hepatitis
DNA, RNA, Replication and Transcription
 Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College DNA, RNA, Replication and Transcription The metabolic processes described earlier (glycolysis, cellular respiration, photophosphorylation,
Harriet Wilson, Lecture Notes Bio. Sci. 4 - Microbiology Sierra College DNA, RNA, Replication and Transcription The metabolic processes described earlier (glycolysis, cellular respiration, photophosphorylation,
Bacterial DNA replication
 Bacterial DNA replication Summary: What problems do these proteins solve? Tyr OH attacks PO4 and forms a covalent intermediate Structural changes in the protein open the gap by 20 Å! 1 Summary: What problems
Bacterial DNA replication Summary: What problems do these proteins solve? Tyr OH attacks PO4 and forms a covalent intermediate Structural changes in the protein open the gap by 20 Å! 1 Summary: What problems
Storage and Expression of Genetic Information
 Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Storage and Expression of Genetic Information 29. DNA structure, Replication and Repair ->Ch 25. DNA metabolism 30. RNA Structure, Synthesis and Processing ->Ch 26. RNA metabolism 31. Protein Synthesis
Please sign below if you wish to have your grades posted by the last five digits of your SSN
 BIO 226R EXAM II (Sample) PRINT YOUR NAME SSN Please sign below if you wish to have your grades posted by the last five digits of your SSN Signature BIO 226R Exam II has 6 pages, and 27 questions. There
BIO 226R EXAM II (Sample) PRINT YOUR NAME SSN Please sign below if you wish to have your grades posted by the last five digits of your SSN Signature BIO 226R Exam II has 6 pages, and 27 questions. There
Prokaryotic Physiology. March 3, 2017
 1. (10 pts) Explain the replication of both strands of DNA in prokaryotes. At a minimum explain the direction of synthesis, synthesis of the leading and lagging strand, separation of the strands and the
1. (10 pts) Explain the replication of both strands of DNA in prokaryotes. At a minimum explain the direction of synthesis, synthesis of the leading and lagging strand, separation of the strands and the
DNA Replication and Repair
 DN Replication and Repair http://hyperphysics.phy-astr.gsu.edu/hbase/organic/imgorg/cendog.gif DN Replication genetic information is passed on to the next generation semi-conservative Parent molecule with
DN Replication and Repair http://hyperphysics.phy-astr.gsu.edu/hbase/organic/imgorg/cendog.gif DN Replication genetic information is passed on to the next generation semi-conservative Parent molecule with
Fig. 16-7a. 5 end Hydrogen bond 3 end. 1 nm. 3.4 nm nm
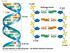 Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Fig. 16-7a end Hydrogen bond end 1 nm 3.4 nm 0.34 nm (a) Key features of DNA structure end (b) Partial chemical structure end Fig. 16-8 Adenine (A) Thymine (T) Guanine (G) Cytosine (C) Concept 16.2: Many
Proposed Models of DNA Replication. Conservative Model. Semi-Conservative Model. Dispersive model
 5.2 DNA Replication Cell Cycle Life cycle of a cell Cells can reproduce Daughter cells receive an exact copy of DNA from parent cell DNA replication happens during the S phase Proposed Models of DNA Replication
5.2 DNA Replication Cell Cycle Life cycle of a cell Cells can reproduce Daughter cells receive an exact copy of DNA from parent cell DNA replication happens during the S phase Proposed Models of DNA Replication
DNA Transcription. Dr Aliwaini
 DNA Transcription 1 DNA Transcription-Introduction The synthesis of an RNA molecule from DNA is called Transcription. All eukaryotic cells have five major classes of RNA: ribosomal RNA (rrna), messenger
DNA Transcription 1 DNA Transcription-Introduction The synthesis of an RNA molecule from DNA is called Transcription. All eukaryotic cells have five major classes of RNA: ribosomal RNA (rrna), messenger
DNA replication: Enzymes link the aligned nucleotides by phosphodiester bonds to form a continuous strand.
 DNA replication: Copying genetic information for transmission to the next generation Occurs in S phase of cell cycle Process of DNA duplicating itself Begins with the unwinding of the double helix to expose
DNA replication: Copying genetic information for transmission to the next generation Occurs in S phase of cell cycle Process of DNA duplicating itself Begins with the unwinding of the double helix to expose
DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA
 DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
DNA Replication DNA vs. RNA DNA: deoxyribonucleic acid (double stranded) RNA: ribonucleic acid (single stranded) Both found in most bacterial and eukaryotic cells RNA molecule can assume different structures
3.A.1 DNA and RNA: Structure and Replication
 3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
3.A.1 DNA and RNA: Structure and Replication Each DNA polymer is made of Nucleotides (monomer) which are made of: a) Phosphate group: Negatively charged and polar b) Sugar: deoxyribose- a 5 carbon sugar
Eukaryotic DNA Replication Mechanism:
 Eukaryotic DNA Replication Mechanism: Eukaryotic cells are endowed with a very complicated but sophisticated regulation of cell division. Embryonic cells go through first seven to eight cell divisions
Eukaryotic DNA Replication Mechanism: Eukaryotic cells are endowed with a very complicated but sophisticated regulation of cell division. Embryonic cells go through first seven to eight cell divisions
2/6/18. file:///users/jlc/ Downloads/ DNAi_replicatio n_vo2-lg.mov. 3x10 13 cells (Bianconi et al. Ann Hum Biol, 2013) DNA Replication in vivo
 file:///users/jlc/ Downloads/ DNAi_replicatio n_vo2-lg.mov 3x10 13 cells (Bianconi et al. Ann Hum Biol, 2013) 10 12 replicating cells (Cairns: Genetics (2006)174,1069). Lose about 1/3 of these per day
file:///users/jlc/ Downloads/ DNAi_replicatio n_vo2-lg.mov 3x10 13 cells (Bianconi et al. Ann Hum Biol, 2013) 10 12 replicating cells (Cairns: Genetics (2006)174,1069). Lose about 1/3 of these per day
There are four phases: M = mitosis is when chromosomes become condensed and separate and the cell divides: visible changes. ~ 1 hr.
 The cell cycle in eukaryotes Jason Kahn, Biochem 465 Spring 2006 (Mostly from Alberts et al., Molecular Biology of the Cell, 2002). The book is available for free on line at http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mboc4
The cell cycle in eukaryotes Jason Kahn, Biochem 465 Spring 2006 (Mostly from Alberts et al., Molecular Biology of the Cell, 2002). The book is available for free on line at http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mboc4
Chapter Twelve: DNA Replication and Recombination
 This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
This is a document I found online that is based off of the fourth version of your book. Not everything will apply to the upcoming exam so you ll have to pick out what you thing is important and applicable.
You Should Be Able To
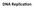 DNA Replica,on You Should Be Able To 1. Describe the func9on of: DNA POL1, DNA POL3, Sliding Clamp, SSBPs, Ligase, Topoisomerase, Helicase, Primase 2. Describe DNA synthesis on the leading and lagging
DNA Replica,on You Should Be Able To 1. Describe the func9on of: DNA POL1, DNA POL3, Sliding Clamp, SSBPs, Ligase, Topoisomerase, Helicase, Primase 2. Describe DNA synthesis on the leading and lagging
DNA ORGANIZATION AND REPLICATION
 DNA ORGANIZATION AND REPLICATION THE CENTRAL DOGMA DNA Replication Transcription Translation STRUCTURAL ORGANIZATION OF DNA DNA is present in the nucleus as CHROMATIN. The basic unit of chromatin is NUCLEOSOME
DNA ORGANIZATION AND REPLICATION THE CENTRAL DOGMA DNA Replication Transcription Translation STRUCTURAL ORGANIZATION OF DNA DNA is present in the nucleus as CHROMATIN. The basic unit of chromatin is NUCLEOSOME
DNA REPLICATION. DNA structure. Semiconservative replication. DNA structure. Origin of replication. Replication bubbles and forks.
 DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
DNA REPLICATION 5 4 Phosphate 3 DNA structure Nitrogenous base 1 Deoxyribose 2 Nucleotide DNA strand = DNA polynucleotide 2004 Biology Olympiad Preparation Program 2 2004 Biology Olympiad Preparation Program
Chapter 9. Topics - Genetics - Flow of Genetics - Regulation - Mutation - Recombination
 Chapter 9 Topics - Genetics - Flow of Genetics - Regulation - Mutation - Recombination 1 Genetics Genome Chromosome Gene Protein Genotype Phenotype 2 Terms and concepts gene Fundamental unit of heredity
Chapter 9 Topics - Genetics - Flow of Genetics - Regulation - Mutation - Recombination 1 Genetics Genome Chromosome Gene Protein Genotype Phenotype 2 Terms and concepts gene Fundamental unit of heredity
DNA Replication III & DNA Damage/Modification Biochemistry 302. January 27, 2006
 DNA Replication III & DNA Damage/Modification Biochemistry 302 January 27, 2006 Bacterial replication organized into a membrane-bound replication factory Two replisomes do not travel away from one another
DNA Replication III & DNA Damage/Modification Biochemistry 302 January 27, 2006 Bacterial replication organized into a membrane-bound replication factory Two replisomes do not travel away from one another
Chapter 12. DNA Replication and Recombination
 Chapter 12 DNA Replication and Recombination I. DNA replication Three possible modes of replication A. Conservative entire original molecule maintained B. Semiconservative one strand is template for new
Chapter 12 DNA Replication and Recombination I. DNA replication Three possible modes of replication A. Conservative entire original molecule maintained B. Semiconservative one strand is template for new
DNA REPLICATION IN EUKARYOTIC CELLS
 Annu. Rev. Biochem. 2002. 71:333 74 DOI: 10.1146/annurev.biochem.71.110601.135425 Copyright 2002 by Annual Reviews. All rights reserved First published as a Review in Advance on February 12, 2002 DNA REPLICATION
Annu. Rev. Biochem. 2002. 71:333 74 DOI: 10.1146/annurev.biochem.71.110601.135425 Copyright 2002 by Annual Reviews. All rights reserved First published as a Review in Advance on February 12, 2002 DNA REPLICATION
RNA synthesis/transcription I Biochemistry 302. February 6, 2004 Bob Kelm
 RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
RNA synthesis/transcription I Biochemistry 302 February 6, 2004 Bob Kelm Overview of RNA classes Messenger RNA (mrna) Encodes protein Relatively short half-life ( 3 min in E. coli, 30 min in eukaryotic
Hershey & Chase Avery, MacLeod, & McCarty DNA: The Genetic Material
 DA: The Genetic Material Chapter 14 Griffith s experiment with Streptococcus pneumoniae Live S strain cells killed the mice Live R strain cells did not kill the mice eat-killed S strain cells did not kill
DA: The Genetic Material Chapter 14 Griffith s experiment with Streptococcus pneumoniae Live S strain cells killed the mice Live R strain cells did not kill the mice eat-killed S strain cells did not kill
The Genetic Material. The Genetic Material. The Genetic Material. DNA: The Genetic Material. Chapter 14
 DNA: Chapter 14 Frederick Griffith, 1928 studied Streptococcus pneumoniae, a pathogenic bacterium causing pneumonia there are 2 strains of Streptococcus: - S strain is virulent - R strain is nonvirulent
DNA: Chapter 14 Frederick Griffith, 1928 studied Streptococcus pneumoniae, a pathogenic bacterium causing pneumonia there are 2 strains of Streptococcus: - S strain is virulent - R strain is nonvirulent
DNA replication. DNA replication. replication model. replication fork. chapter 6
 DN chapter 6 DN two complementary s bases joined by hydrogen bonds separation of s each - template determines order of nucleotides in duplicate parent DN s separate two identical daughter s model dispersive
DN chapter 6 DN two complementary s bases joined by hydrogen bonds separation of s each - template determines order of nucleotides in duplicate parent DN s separate two identical daughter s model dispersive
The Structure of DNA
 The Structure of DNA Questions to Ponder 1) How is the genetic info copied? 2) How does DNA store the genetic information? 3) How is the genetic info passed from generation to generation? The Structure
The Structure of DNA Questions to Ponder 1) How is the genetic info copied? 2) How does DNA store the genetic information? 3) How is the genetic info passed from generation to generation? The Structure
Molecular Biology, Lecture 3 DNA Replication
 Molecular Biology, Lecture 3 DNA Replication We will continue talking about DNA replication. We have previously t discussed the structure of DNA. DNA replication is the copying of the whole DNA content
Molecular Biology, Lecture 3 DNA Replication We will continue talking about DNA replication. We have previously t discussed the structure of DNA. DNA replication is the copying of the whole DNA content
DNA metabolism. DNA Replication DNA Repair DNA Recombination
 DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
DNA metabolism DNA Replication DNA Repair DNA Recombination Chutima Talabnin Ph.D. School of Biochemistry,Institute of Science, Suranaree University of Technology Central Dogma or Flow of genetic information
Tala Saleh. Tamer Barakat ... Anas Abu. Humaidan
 7 Tala Saleh Tamer Barakat... Anas Abu. Humaidan Some Information in this lecture may not be mentioned by the Dr. as thoroughly as this sheet. But they cannot be overlooked for a better understanding,
7 Tala Saleh Tamer Barakat... Anas Abu. Humaidan Some Information in this lecture may not be mentioned by the Dr. as thoroughly as this sheet. But they cannot be overlooked for a better understanding,
Mechanisms of Transcription. School of Life Science Shandong University
 Mechanisms of Transcription School of Life Science Shandong University Ch 12: Mechanisms of Transcription 1. RNA polymerase and the transcription cycle 2. The transcription cycle in bacteria 3. Transcription
Mechanisms of Transcription School of Life Science Shandong University Ch 12: Mechanisms of Transcription 1. RNA polymerase and the transcription cycle 2. The transcription cycle in bacteria 3. Transcription
M1 - Biochemistry. Nucleic Acid Structure II/Transcription I
 M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
M1 - Biochemistry Nucleic Acid Structure II/Transcription I PH Ratz, PhD (Resources: Lehninger et al., 5th ed., Chapters 8, 24 & 26) 1 Nucleic Acid Structure II/Transcription I Learning Objectives: 1.
DNA: The Genetic Material. Chapter 14
 DNA: The Genetic Material Chapter 14 The Genetic Material Frederick Griffith, 1928 studied Streptococcus pneumoniae, a pathogenic bacterium causing pneumonia there are 2 strains of Streptococcus: - S strain
DNA: The Genetic Material Chapter 14 The Genetic Material Frederick Griffith, 1928 studied Streptococcus pneumoniae, a pathogenic bacterium causing pneumonia there are 2 strains of Streptococcus: - S strain
Molecular Biology Midterm Exam 2
 Molecular Biology Midterm Exam 2 1. The experiments by Frank Stahl and Matthew Messelson demonstrated that DNA strands separate during DNA replication. They showed that DNA replication is what kind of
Molecular Biology Midterm Exam 2 1. The experiments by Frank Stahl and Matthew Messelson demonstrated that DNA strands separate during DNA replication. They showed that DNA replication is what kind of
MCB 110 Spring 2017 Exam 1 SIX PAGES
 MCB 110 Spring 2017 Exam 1 SIX PAGES NAME: SID Number: Question Maximum Points Your Points I 28 II 32 III 32 IV 30 V 28 150 PLEASE WRITE your NAME or SID number on each page. This exam must be written
MCB 110 Spring 2017 Exam 1 SIX PAGES NAME: SID Number: Question Maximum Points Your Points I 28 II 32 III 32 IV 30 V 28 150 PLEASE WRITE your NAME or SID number on each page. This exam must be written
Prokaryotic cells divide by pinching in two. Fig. 10-CO, p.240
 Prokaryotic cells divide by pinching in two Fig. 10-CO, p.240 Learning Objectives 1. What Is the Flow of Genetic Information in the Cell? 2. What Are the General Considerations in the Replication of DNA?
Prokaryotic cells divide by pinching in two Fig. 10-CO, p.240 Learning Objectives 1. What Is the Flow of Genetic Information in the Cell? 2. What Are the General Considerations in the Replication of DNA?
1 of 42 9/30/2010 3:17 PM
 1 of 42 9/30/2010 3:17 PM When you have read Chapter 13, you should be able to: State what is meant by the topological problem and explain how DNA topoisomerases solve this problem Describe the key experiment
1 of 42 9/30/2010 3:17 PM When you have read Chapter 13, you should be able to: State what is meant by the topological problem and explain how DNA topoisomerases solve this problem Describe the key experiment
1. True or False? At the DNA level, recombination is initiated by a single stranded break in a DNA molecule.
 1. True or False? At the DNA level, recombination is initiated by a single stranded break in a DNA molecule. 2. True or False? Dideoxy sequencing is a chain initiation method of DNA sequencing. 3. True
1. True or False? At the DNA level, recombination is initiated by a single stranded break in a DNA molecule. 2. True or False? Dideoxy sequencing is a chain initiation method of DNA sequencing. 3. True
DNA Replication. The Organization of DNA. Recall:
 Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
Recall: The Organization of DNA DNA Replication Chromosomal form appears only during mitosis, and is used in karyotypes. folded back upon itself (chromosomes) coiled around itself (chromatin) wrapped around
DNA is the genetic material. DNA structure. Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test
 DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
DNA is the genetic material Chapter 7: DNA Replication, Transcription & Translation; Mutations & Ames test Dr. Amy Rogers Bio 139 General Microbiology Hereditary information is carried by DNA Griffith/Avery
Moayyad Al-shafei. Mohammad Tarabeih. Dr Ma'mon Ahram. 1 P a g e
 3 Moayyad Al-shafei Mohammad Tarabeih Dr Ma'mon Ahram 1 P a g e In this sheet, we are going to discuss 2 main topics: 1- The advantages of restriction endonucleases. 2- DNA replication. Before we start
3 Moayyad Al-shafei Mohammad Tarabeih Dr Ma'mon Ahram 1 P a g e In this sheet, we are going to discuss 2 main topics: 1- The advantages of restriction endonucleases. 2- DNA replication. Before we start
DNA Transcription. Visualizing Transcription. The Transcription Process
 DNA Transcription By: Suzanne Clancy, Ph.D. 2008 Nature Education Citation: Clancy, S. (2008) DNA transcription. Nature Education 1(1) If DNA is a book, then how is it read? Learn more about the DNA transcription
DNA Transcription By: Suzanne Clancy, Ph.D. 2008 Nature Education Citation: Clancy, S. (2008) DNA transcription. Nature Education 1(1) If DNA is a book, then how is it read? Learn more about the DNA transcription
DNA replication. DNA replication. replication model. replication fork. chapter 6
 DN chapter 6 DN two complementary s bases joined by hydrogen bonds separation of s each - template determines order of nucleotides in duplicate parent DN s separate two identical daughter s model dispersive
DN chapter 6 DN two complementary s bases joined by hydrogen bonds separation of s each - template determines order of nucleotides in duplicate parent DN s separate two identical daughter s model dispersive
DNA Model Building and Replica3on
 DNA Model Building and Replica3on DNA Replication S phase Origins of replication in E. coli and eukaryotes (a) Origin of replication in an E. coli cell Origin of replication Bacterial chromosome Doublestranded
DNA Model Building and Replica3on DNA Replication S phase Origins of replication in E. coli and eukaryotes (a) Origin of replication in an E. coli cell Origin of replication Bacterial chromosome Doublestranded
DNA: Structure & Replication
 DNA Form & Function DNA: Structure & Replication Understanding DNA replication and the resulting transmission of genetic information from cell to cell, and generation to generation lays the groundwork
DNA Form & Function DNA: Structure & Replication Understanding DNA replication and the resulting transmission of genetic information from cell to cell, and generation to generation lays the groundwork
MIDTERM I NAME: Student ID Number: I 32 II 33 III 24 IV 30 V
 MIDTERM I NAME: Student ID Number: Question Maximum Points Your Points I 32 II 33 III 24 IV 30 V 31 150 Please write your name/student ID number on each of the following five pages. This exam must be written
MIDTERM I NAME: Student ID Number: Question Maximum Points Your Points I 32 II 33 III 24 IV 30 V 31 150 Please write your name/student ID number on each of the following five pages. This exam must be written
Student name ID # Second Mid Term Exam, Biology 2020, Spring 2002 Scores Total
 Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Second Mid Term Exam, Biology 2020, Spring 2002 Scores 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Total 1 1. Matching (7 pts). Each answer is used exactly once Helicase
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site.
 Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Replication. Replication fork
 Replication In their seminal paper on the DNA double helix published in Nature on April 25 th, 1953, Watson and Crick wrote: It has not escaped our notice that the specific pairing we have postulated immediately
Replication In their seminal paper on the DNA double helix published in Nature on April 25 th, 1953, Watson and Crick wrote: It has not escaped our notice that the specific pairing we have postulated immediately
Eukaryotic Transcription
 Eukaryotic Transcription I. Differences between eukaryotic versus prokaryotic transcription. II. (core vs holoenzyme): RNA polymerase II - Promotor elements. - General Pol II transcription factors (GTF).
Eukaryotic Transcription I. Differences between eukaryotic versus prokaryotic transcription. II. (core vs holoenzyme): RNA polymerase II - Promotor elements. - General Pol II transcription factors (GTF).
