Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in recombinant Escherichia coli grown on glucose
|
|
|
- Duane Chase
- 6 years ago
- Views:
Transcription
1 Journal of Biotechnology 58 (1997) Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in recombinant Escherichia coli grown on glucose Henry E. Valentin *, Douglas Dennis Department of Biology, James Madison Uni ersity, Harrisonburg, VA 22807, USA Received 14 March 1997; received in revised form 8 July 1997; accepted 15 July 1997 Abstract A recombinant Escherichia coli strain has been developed that produces poly(3-hydroxybutyrate-co-4-hydroxybutyrate) when grown in complex medium containing glucose. This has been accomplished by introducing into E. coli DH5 separate plasmids harboring the polyhydroxyalkanoate (PHA) biosynthesis genes from Ralstonia eutropha (formerly named Alcaligenes eutrophus) and the succinate degradation genes from Clostridium kluy eri, respectively. Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) levels reached 50% of the cell dry weight and contained up to 2.8 mol.% 4-hydroxybutyrate. The molecular weight of the polymer was Elsevier Science B.V. Keywords: Polyhydroxyalkanoate; Succinate degradation; Ralstonia eutropha; Clostridium kluy eri 1. Introduction * Corresponding author. Present address: Monsanto Corporation, 700 Chesterfield Parkway North, St. Louis, MO 63198, USA. Tel.: ; fax: ; hevale@ccmail.monsanto.com Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) [P(3HB-co-4HB)] is a recently discovered PHA that has attracted considerable interest because of its increased flexibility in thermoplastic applications (Kunioka et al., 1989; Doi et al., 1990a,b, Saito and Doi, 1994). Generally, P(3HB-co-4HB) is produced by feeding precursors such as 4-hydroxybutyrate, 1,4-butanediol or -butyrolactone to wild type or mutant strains of Ralstonia eutropha. More recently, Doi has described a method whereby -butyrolactone can be fed to Alcaligenes latus to obtain P(3HB-co-4HB) levels that are 60% of the cell dry weight and which contain 7 12 mol.% 4-hydroxybutyrate (4HB) monomers (Soejima and Doi, 1996). Normally the molar levels of 4HB in the copolymer are relatively low, but recently R. eutropha mutants have been isolated which were able to accumulate a 4HB homopolyester. When these mutants were supplemented with the R. eutropha PHA synthase gene the 4HB homopolyester was accumulated to /97/$ Elsevier Science B.V. All rights reserved. PII S (97)
2 34 H.E. Valentin, D. Dennis / Journal of Biotechnology 58 (1997) levels of 30% of the cell dry weight (Steinbüchel et al., 1994). Studies have indicated that the pathway for 4HB metabolism in R. eutropha occurs by conversion to succinic semialdehyde via a 4-hydroxybutyrate dehydrogenase and subsequent conversion of succinic semialdehyde to succinate by a succinic semialdehyde dehydrogenase (Valentin et al., 1995). The metabolic link to P(3HB-co-4HB) formation was not elucidated in detail, but the presence of high activities of 3HB-dehydrogenase, an enzyme of the intracellular PHA degradation pathway, in crude extracts of 4HB-induced R. eutropha led the authors to suggest that 4-hydroxybutyryl-CoA (the immediate precursor before polymerization) might be supplied by the action of a succinyl-coa:acetoacetate CoA transferase, or an undefined thiokinase; and that 3-hydroxybutyryl-CoA is supplied by the concerted action of the TCA-cycle, gluconeogenesis and the PHA biosynthesis enzymes. Recently a similar pathway, involved in the cofermentation of succinic acid and ethanol in Clostridium kluy eri has been described (Söhling and Gottschalk, 1996) in which 4-hydroxybutyryl- CoA is an intermediate. In this pathway, CoA is transferred to succinate via the action of an enzyme having succinyl-coa transferase activities. Succinyl-CoA is then reduced to succinic semialdehyde and CoA via a succinic semialdehyde dehydrogenase. Succinic semialdehyde is reduced to 4HB (via a 4-hydroxybutyrate dehydrogenase) which is then activated to 4-hydroxybutyryl-CoA by an enzyme possessing 4-hydroxybutyryl CoA:CoA transferase activity. The genes for this pathway have been located on a 7.5 kb DNA fragment that includes genes for 4-hydroxybutyrate dehydrogenase (4hbD), succinic semialdehyde dehydrogenase (sucd), succinyl-coa:coa transferase (cat1), a membrane protein of unknown function (orfy) and 4-hydroxybutyryl- CoA:CoA transferase (designated as orfz in original paper). GenBank accession number L Furthermore, three of the proteins, 4HBD, SUCD and CAT1 were found to be expressed in Escherichia coli. It is already well established that the PHA biosynthesis operon from R. eutropha is sufficiently expressed in E. coli to mediate PHB production to levels as high as 70 80% of the cell dry weight (Schubert et al., 1988; Slater et al., 1988; Peoples and Sinskey, 1989). Therefore, introduction of the sucd, 4hbD and orfz genes from C. kluy eri into a recombinant E. coli strain containing the PHA biosynthesis genes could facilitate the production of 4-hydroxybutyryl-CoA for incorporation into P(3HB-co-4HB) (Fig. 1). Moreover, this incorporation would not depend on immediate precursors of 4-hydroxybutyrate, but should occur simply from abstraction of succinate or succinyl-coa from the citric acid cycle when the bacterial strain is grown on glucose. This paper describes experiments which suggest that this is the case. Fig. 1. Proposed pathway to poly(3hb-co-4hb) synthesis in recombinant E. coli harboring plasmids carrying the PHA synthesis operon from R. eutropha and the succinate utilization pathway from C. kluy eri. Enzymes: 1, 3-ketothiolase; 2, acetoacetyl-coa reductase; 3, PHA synthase, 4, succinic semialdehyde dehydrogenase; 5, 4-hydroxybutyrate dehydrogenase; 6, 4-hydroxybutyrate-CoA:CoA transferase.
3 H.E. Valentin, D. Dennis / Journal of Biotechnology 58 (1997) Materials and methods 2.1. Bacterial strains and plasmids E. coli DH5 F- 80dlacZ M15 (laczyaargf)u169 deor reca1 enda1 phoa hsdr17 (r K, m K+ ) supe44 -thi-1 gyra96 rela1 was used as the host (Gibco-BRL, Bethesda, MD). The plasmid pjm9238 is a runaway replication vector containing the genes for the poly(3-hydroxybutyrate) biosynthesis pathway from R. eutropha and has been previously described (Kidwell et al., 1995) Plasmid construction pcks carries the genes for the succinate degradation pathway from C. kluy eri and is a derivative of pck3 which has previously been described (Söhling and Gottschalk, 1996). To make pcks, the plasmid pck3 was subjected to partial EcoRI digestion and a 9.4 kb fragment was excised from a gel and ligated. This plasmid, pck3a contained the genes for the succinate degradation pathway minus a 1.1 kb EcoRI fragment which encodes a putative membrane spanning protein of unknown function and the amino terminal region of cat1 (succinyl-coa:coa transferase). The insert that remained was removed by digesting with ApaI and SacI and was cloned back into pbluescript II-SK (Stratagene, La Jolla, CA) such that the lacz promoter promoted transcription in the same direction as the three intact genes on the insert (4hbD, sucd, orfz) Cell growth Bacterial strains used in these experiments were cultured overnight in mm sterile tubes containing 3 ml of LB and the appropriate antibiotics (chloramphenicol at 25 g ml 1 for pjm9238, ampicillin at 100 g ml 1 for pcks). A 1 ml volume of the overnight culture was used to inoculate 50 ml of the same media in 250-ml baffled shake flasks. The cultures were grown at 30 C until they reached an optical density of 0.7 (600 nm), at which time glucose was added to a final concentration of 1% (w/v) and the cultures were shifted to an orbital incubator that was at 37 C to induce pjm9238 into runaway replication. Cells were harvested after 48 h of incubation at 37 C PHA analysis Cells were harvested by centrifugation at 3500 rpm (Heraeus Varafuge F) followed by suspension of the pellet in 0.9% (w/v) saline and recentrifugation to obtain a washed pellet. The washed pellet was frozen and dried by lyophilization. Intracellular polyhydroxyalkanoate was characterized using a previously described modification (Slater et al., 1992) of the gas chromatographic method developed by Braunegg et al. (1978). P(3HB) (Sigma Chemicals) and -butyrolactone were used as standards Extraction of the polymer The polymer was extracted by shaking the dried bacterial cell pellet overnight at 37 C in chloroform, removing the bacterial debris by filtration of the chloroform/bacterial suspension through Whatman paper and precipitating the polyester from the filtered solution by the addition of 10 volumes of ethanol. The precipitated polymer was redissolved in chloroform, reprecipitated with 10 volumes of methanol, filtered onto a Whatman paper filter and allowed to dry. The polymer was redissolved in deuterochloroform or chloroform and used for nuclear magnetic resonance and gel permeation chromatography analysis, respectively H and 13 C{ 1 H} NMR analyses Nuclear magnetic resonance studies were carried out using a Bruker AC/200 spectrometer. The 1 H spectrum (200 MHz) was obtained at 22 C on a solution of polymer that was dissolved at a concentration of 20 mg ml 1 in deuterochloroform. Pulses were taken using a 45 pulse width, 2.46 s acquisition time, 3333 Hz spectral width, 16 k data points and 64 accumulations. Chemical shifts were measured relative to CHCl 3 (d=7.24 ppm). The 13 C{ 1 H} spectra (50 MHz) were taken
4 36 H.E. Valentin, D. Dennis / Journal of Biotechnology 58 (1997) Table 1 Poly(3HB-co-4HB) accumulation in recombinant E. coli Plasmids IPTG (mm) Mg cells Mol.% 3HB Mol.% 4HB %P(3HB-co-4HB) pjm pjm9238, pcks pjm9238, pcks at 22 C on a polymer solution dissolved in deuterochloroform at 50 mg ml 1. The spectra were obtained using Waltz decoupling, 30 pulses, 1 s relaxation delay, 12 khz spectral width, 32 k data points and accumulations. Chemical shifts were measured relative to CDCl 3 (d=77.0 ppm) Gel permeation chromatography Molecular weight analyses were carried out employing gel permeation chromatography as previously described by Koizumi et al. (1995). 3. Results and discussion 3.1. Gas chromatographic analysis of PHA produced in E. coli (pjm9238, pcks) E. coli (pjm9238) and E. coli (pjm9238, pcks) were cultured in LB containing glucose to promote PHA synthesis and provide substrates for glycolysis and the TCA cycle. The cells were collected 48 h after induction of PHA accumulation, dried and analyzed for the PHA polyesters (Table 1). Even though an extremely large amount of dried cell pellet from the E. coli (pjm9238) culture was analyzed (84 mg), no 4HB monomers could be detected as constituents of the polymer. The overall level of PHA (52% of cell dry weight) was consistent with previous experiments using this vector and host (Kidwell et al., 1995). When E. coli (pjm9238, pcks) was analyzed (carrying the PHA biosynthesis genes and the succinate degradation pathway genes), very small peaks could be seen on the chromatogram (data no shown) that corresponded exactly to the retention times of the 4HB standard. These were taken to be 4HB. The final level of 4HB in the polymer was 1.5 mol.%. To determine if the succinate degradation genetic pathway on pcks might be under control of the lac promoter upstream of the genes, the experiment was also carried out in the presence of IPTG (added at the time of temperature upshift) to induce the lac promoter. The level of 4HB in the polyester was virtually unchanged (1.3 mol.%), indicating that the lac promoter probably does not play a role in the expression of the succinate degradation genes. When the original plasmid, pck3 (which contains the succinate degradation pathway of C. kluy eri in the opposite orientation from the lac promoter) was used in a similar experiment, only 0.2 mol.% 4HB was found in the polymer. When the DNA insert in pck3 (containing the entire succinate degradation pathway) was reversed, thereby placing the lac promoter colinear with the succinate degradation genes, the level of 4HB was 0.3 mol.% (data not shown). These results suggest that the presence of an intact succinyl-coa:coa transferase gene does not mediate a measurable increase in 4HB levels, as might be expected, but appears to result in a decrease in 4HB levels (in comparison to E. coli (pjm9238, pcks). However, this cannot be considered strong evidence since succinyl-coa:coa transferase activity was not measured and the contribution of the orfy gene product is unknown NMR analysis of the purified polymer Because the level of 4HB in the polymer was quite low, confirmation of its presence was sought by nuclear magnetic resonance spectrometry of the purified polymer. For NMR analysis a larger culture (500 ml) of E. coli (pjm9238, pcks) was grown under conditions described in Section 2 except that the culture contained 2% glucose. The
5 H.E. Valentin, D. Dennis / Journal of Biotechnology 58 (1997) Fig H NMR (A) and 13 C{ 1 H} NMR (B) of purified polymer. culture yielded 439 mg of cell dry mass from which 115 mg of P(3HB-co-4HB) was extracted. This polymer was subjected to both 1 H and 13 C{ 1 H} NMR spectroscopy (Fig. 2). Both spectrographs displayed a pattern of signals virtually identical to spectra of P(3HB-co-4HB) that have been previously published (Doi et al., 1988; Steinbüchel et al., 1994). Signals 6, 7 and 8 represent the resonances of the corresponding 4HB-methylene groups (Fig. 2A). The 13 C{ 1 H} NMR signals of the 4HB carbonyl carbon (signal 5) and the methylene carbons (signals 6, 7 and 8) can be seen in Fig. 2B at 171.9, 30.8, 24.0 and 63.7 ppm, respectively, Using the integration data from the 1 H NMR, we were able to accurately determine the composition of the polymer, which was 97.2 mol.% 3HB and 2.8 mol.% 4HB. As in the gas chromatographic analysis no other signals were seen that could be interpreted to be additional polyhydroxyalkanoates resident in the polymer. It is important to note that the 4HB produced is done so, in spite of the deletion of the succinyl-coa:coa transferase gene which is necessary for succinate degradation in C. kluy eri. This is almost certainly due to the high flux of intracellular succinyl-coa and its availability, through the TCA cycle.
6 38 H.E. Valentin, D. Dennis / Journal of Biotechnology 58 (1997) Molecular weight of P(3HB-co-4HB) from E. coli (pjm9238, pcks) To determine if the addition of 4HB units into the polymer in E. coli might have detrimental effects on the molecular weight, a portion of the extracted polymer isolated for the NMR experiment was subjected to gel permeation chromatography. The estimated molecular weight of this polymer was Previous analyses of poly(3-hydroxybutyrate) extracted from recombinant E. coli (pjm9238) had shown that the molecular weight of the polymer usually was in the range of 1.0 to (Kidwell et al., 1995; unpublished data) so it does not appear that the incorporation of 4HB into the polymer at these levels causes any significant decrease in the molecular weight. Based on these and the other experiments above, we surmise that it is possible to produce a high molecular weight polymer of P(3HB-co-4HB) in recombinant systems. Acknowledgements We thank Dr Gerhard Gottschalk and Dr Brigitte Söhling for providing the plasmid pck3. We also thank Dr Jun Wu at Monsanto Corporation for the GPC analysis of the polymer, Tom Gallaher for NMR analyses and Andy Davis for his able technical work. A portion of this work was supported by the Alexander von Humboldt Foundation. References Braunegg, G., Sonnleitner, B., Lafferty, R.M., A rapid gas chromatographic method for the determination of poly- -hydroxybutyric acid in microbial biomass. Eur. J. Microbiol. Biotechnol. 6, Doi, Y., Kunioka, M., Nakamura, Y., Soga, K., Nuclear magnetic resonance studies on unusual bacterial copolyesters of 3-hydroxybutyrate and 4-hydroxybutyrate. Macromolecules 21, Doi, Y., Janesawa, Y., Kunioka, M., Saito, T., 1990a. Biodegradation of microbial copolyesters: poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and poly(3-hydroxybutyrate-co-4-hydroxybutyrate). Macromolecules 23, Doi, Y., Segawa, A., Kunioka, M., 1990b. Biosynthesis and characterization of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in A. eutrophus. Int. J. Biol. Macromol. 12, Kidwell, J., Valentin, H.E., Dennis, D., Regulated expression of the A. eutrophus pha biosynthesis genes in E. coli. Appl. Env. Microbiol. 61, Koizumi, F., Hideki, A., Doi, Y., Molecular weight of poly(3-hydroxybutyrate) during biological polymerization in A. eutrophus. J.M.S. Pure Appl. Chem. A32, Kunioka, M., Kawaguchi, Y., Doi, Y., Production of biodegradable copolyesters of 3-hydroxybutyrate and 4- hydroxybutyrate by A. eutrophus. Appl. Mirobiol. Biotechnol. 30, Peoples, O.P., Sinskey, A.J., Poly- -hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbc). J. Biol. Chem. 264, Saito, Y., Doi, Y., Microbial synthesis and properties of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in Comamonas acido orans. Int. J. Biol. Macromol. 16, Schubert, P., Steinbüchel, A., Schlegel, H., Cloning of the A. eutrophus genes for synthesis of poly- -hydroxybutyric acid (PHB) and synthesis of PHB in E. coli. J. Bacteriol. 170, Slater, S.C., Voige, W.H., Dennis, D., Cloning and expression in E. coli of the A. eutrophus H16 poly- -hydroxybutyrate biosynthetic pathway. J. Bacteriol. 170, Slater, S., Gallaher, T., Dennis, D., Production of poly(3-hydroxybutyrateco-3-hydroxyvalerate) in a recombinant E. coli strain. Appl. Env. Microbiol. 58, Soejima, T., Doi, Y., Production of poly(3-hydroxybutyrate-co-4 hydroxybutyrate) by A. latus from sucrose and -butyrolactone. International Symposium on Bacteriol Polyhydroxyalkanoates. Davos, Switzerland. Söhling, B., Gottschalk, G., Molecular analysis of the anaerobic succinate degradation pathway in C. kluy eri. J. Bact. 178, Steinbüchel, A., Valentin, H.E., Schönebaum, A., Application of recombinant gene technology for production of polyhydroxyalkanoic acids: Biosynthesis of poly(4-hydroxybutyric acid) homopolyester. J. Env. Pol. Deg. 2, Valentin, H.E., Zingmann, G., Schonebaum, A., Steinbüchel, A., Metabolic pathway for biosynthesis of poly(3- hydroxybutyrate-co-4-hydroxybutyrate) from 4-hydroxybutyrate by A. eutrophus. Eur. J. Biochem. 227,
A Hydrogenophaga pseudoflava strain was able to synthesize poly(3-hydroxybutyric acid-co-4-hydroxybutyric
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Apr. 1999, p. 1570 1577 Vol. 65, No. 4 0099-2240/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Production of Poly(3-Hydroxybutyric
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Apr. 1999, p. 1570 1577 Vol. 65, No. 4 0099-2240/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Production of Poly(3-Hydroxybutyric
Production of Poly(3-Hydroxybutyrate) by Fed-Batch Culture of Filamentation-Suppressed Recombinant Escherichia coli
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 1997, p. 4765 4769 Vol. 63, No. 12 0099-2240/97/$04.00 0 Copyright 1997, American Society for Microbiology Production of Poly(3-Hydroxybutyrate) by Fed-Batch
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 1997, p. 4765 4769 Vol. 63, No. 12 0099-2240/97/$04.00 0 Copyright 1997, American Society for Microbiology Production of Poly(3-Hydroxybutyrate) by Fed-Batch
High-Level Production of Poly(3-Hydroxybutyrate-co-3- Hydroxyvalerate) by Fed-Batch Culture of Recombinant Escherichia coli
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Oct. 1999, p. 4363 4368 Vol. 65, No. 10 0099-2240/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. High-Level Production of Poly(3-Hydroxybutyrate-co-3-
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Oct. 1999, p. 4363 4368 Vol. 65, No. 10 0099-2240/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. High-Level Production of Poly(3-Hydroxybutyrate-co-3-
Longan Shang,, Seong Chun Yim, Hyun Gyu Park,* and Ho Nam Chang*
 140 Biotechnol. Prog. 2004, 20, 140 144 Sequential Feeding of Glucose and Valerate in a Fed-Batch Culture of Ralstonia eutropha for Production of Poly(hydroxybutyrate-co-hydroxyvalerate) with High 3-Hydroxyvalerate
140 Biotechnol. Prog. 2004, 20, 140 144 Sequential Feeding of Glucose and Valerate in a Fed-Batch Culture of Ralstonia eutropha for Production of Poly(hydroxybutyrate-co-hydroxyvalerate) with High 3-Hydroxyvalerate
Hyperproduction of poly(4-hydroxybutyrate) from glucose by recombinant Escherichia coli
 Zhou et al. Microbial Cell Factories 2012, 11:54 RESEARCH Open Access Hyperproduction of poly(4-hydroxybutyrate) from glucose by recombinant Escherichia coli Xiao-Yun Zhou 1, Xiao-Xi Yuan 1, Zhen-Yu Shi
Zhou et al. Microbial Cell Factories 2012, 11:54 RESEARCH Open Access Hyperproduction of poly(4-hydroxybutyrate) from glucose by recombinant Escherichia coli Xiao-Yun Zhou 1, Xiao-Xi Yuan 1, Zhen-Yu Shi
on September 5, 2018 by guest
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 1998, p. 4897 4903 Vol. 64, No. 12 0099-2240/98/$04.00 0 Copyright 1998, American Society for Microbiology. All Rights Reserved. Cloning of the Alcaligenes
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 1998, p. 4897 4903 Vol. 64, No. 12 0099-2240/98/$04.00 0 Copyright 1998, American Society for Microbiology. All Rights Reserved. Cloning of the Alcaligenes
on March 8, 2019 by guest
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 1998, p. 4897 4903 Vol. 64, No. 12 0099-2240/98/$04.00 0 Copyright 1998, American Society for Microbiology. All Rights Reserved. Cloning of the Alcaligenes
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 1998, p. 4897 4903 Vol. 64, No. 12 0099-2240/98/$04.00 0 Copyright 1998, American Society for Microbiology. All Rights Reserved. Cloning of the Alcaligenes
Elisabeth Linton, Dr. Sridhar Viamajala, Dr. Ronald C. Sims UTAH STATE UNIVERSITY Biological and Irrigation Engineering
 THE PRODUCTION OF POLYHYDROXYALKANOATES USING THE WASTE STREAM OF AN ANAEROBIC DIGESTER Elisabeth Linton, Dr. Sridhar Viamajala, Dr. Ronald C. Sims UTAH STATE UNIVERSITY Biological and Irrigation Engineering
THE PRODUCTION OF POLYHYDROXYALKANOATES USING THE WASTE STREAM OF AN ANAEROBIC DIGESTER Elisabeth Linton, Dr. Sridhar Viamajala, Dr. Ronald C. Sims UTAH STATE UNIVERSITY Biological and Irrigation Engineering
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20
 pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
pgm-t Cloning Kit Cat. # : GVT202 Size : 20 Reactions Store at -20 1 Kit Contents Contents pgm-t Cloning Kit pgm-t Vector (50 ng/μl) 20 μl T4 DNA Ligase (3 U/μl) 20 μl 10X T4 DNA Ligation Buffer 30 μl
Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) produced by Comamonas acidovorans
 Polymer 42 (2001) 3455±3461 www.elsevier.nl/locate/polymer Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) produced by Comamonas acidovorans H. Mitomo a, *, W.-C. Hsieh a, K. Nishiwaki a, K. Kasuya a, Y.
Polymer 42 (2001) 3455±3461 www.elsevier.nl/locate/polymer Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) produced by Comamonas acidovorans H. Mitomo a, *, W.-C. Hsieh a, K. Nishiwaki a, K. Kasuya a, Y.
Continuous production of poly-3-hydroxybutyrate by Ralstonia eutropha in a two-stage culture system
 Journal of Biotechnology 88 (2001) 59 65 www.elsevier.com/locate/jbiotec Continuous production of poly-3-hydroxybutyrate by Ralstonia eutropha in a two-stage culture system Guocheng Du a, Jian Chen a,
Journal of Biotechnology 88 (2001) 59 65 www.elsevier.com/locate/jbiotec Continuous production of poly-3-hydroxybutyrate by Ralstonia eutropha in a two-stage culture system Guocheng Du a, Jian Chen a,
pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases For research use only Cat. # : GVT202 Size : 20 Reactions
 pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases Cat. # : GVT202 Size : 20 Reactions Store at -20 For research use only 1 pgm-t Cloning Kit Cat. No.: GVT202 Kit Contents
pgm-t Cloning Kit For direct cloning of PCR products generated by Taq DNA polymerases Cat. # : GVT202 Size : 20 Reactions Store at -20 For research use only 1 pgm-t Cloning Kit Cat. No.: GVT202 Kit Contents
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Step 1: Digest vector with Reason for Step 1. Step 2: Digest T4 genomic DNA with Reason for Step 2: Step 3: Reason for Step 3:
 Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
Biol/Chem 475 Spring 2007 Study Problems for Quiz 2 Quiz 2 (~50 pts) is scheduled for Monday May 14 It will cover all handouts and lab exercises to date except the handout/worksheet (yet to be distributed)
GeNei TM Transformation Teaching Kit Manual
 Teaching Kit Manual Cat No. New Cat No. KT07 107385 KT07A 106220 Revision No.: 00060505 CONTENTS Page No. Objective 3 Principle 3 Kit Description 6 Materials Provided 7 Procedure 9 Observation & Interpretation
Teaching Kit Manual Cat No. New Cat No. KT07 107385 KT07A 106220 Revision No.: 00060505 CONTENTS Page No. Objective 3 Principle 3 Kit Description 6 Materials Provided 7 Procedure 9 Observation & Interpretation
Recovery and Characterization of Poly(3-Hydroxybutyric Acid) Synthesized in Alcaligenes eutrophus and Recombinant Escherichia coli
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Jan., 1995, p. 34 39 Vol. 61, No. 1 0099-2240/95/$04.00 0 Copyright 1995, American Society for Microbiology Recovery and Characterization of Poly(3-Hydroxybutyric
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Jan., 1995, p. 34 39 Vol. 61, No. 1 0099-2240/95/$04.00 0 Copyright 1995, American Society for Microbiology Recovery and Characterization of Poly(3-Hydroxybutyric
proportion of 3-hydroxyoctanoate [3HO] units) from a wide range of unrelated carbon sources.
![proportion of 3-hydroxyoctanoate [3HO] units) from a wide range of unrelated carbon sources. proportion of 3-hydroxyoctanoate [3HO] units) from a wide range of unrelated carbon sources.](/thumbs/88/114959340.jpg) APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Nov. 1990, p. 3354-3359 0099-2240/90/113354-06$02.00/0 Copyright C 1990, American Society for Microbiology Vol. 56, No. 11 Accumulation of a Polyhydroxyalkanoate
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Nov. 1990, p. 3354-3359 0099-2240/90/113354-06$02.00/0 Copyright C 1990, American Society for Microbiology Vol. 56, No. 11 Accumulation of a Polyhydroxyalkanoate
ITS Sequencing in Millepora. 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector
 Page 1 of 5 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector 1. Digest 1 µg of pbluescript with Eco RI 2. Following digestion, add 0.1 volumes of 3M sodium acetate (ph
Page 1 of 5 10/09 Subcloning DNA Fragments into pbluescript Preparation of pbluescript Vector 1. Digest 1 µg of pbluescript with Eco RI 2. Following digestion, add 0.1 volumes of 3M sodium acetate (ph
Reading Lecture 3: 24-25, 45, Lecture 4: 66-71, Lecture 3. Vectors. Definition Properties Types. Transformation
 Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
Lecture 3 Reading Lecture 3: 24-25, 45, 55-66 Lecture 4: 66-71, 75-79 Vectors Definition Properties Types Transformation 56 VECTORS- Definition Vectors are carriers of a DNA fragment of interest Insert
MOK. Media Optimization Kit
 MOK Media Optimization Kit The Media Optimization Kit determines the best medium formulation for maximizing accumulation of recombinant proteins expressed in E. coli, utilizing a series of Athena s superior
MOK Media Optimization Kit The Media Optimization Kit determines the best medium formulation for maximizing accumulation of recombinant proteins expressed in E. coli, utilizing a series of Athena s superior
Continuous production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate): Effects of C/N ratio and dilution rate on HB/HV ratio
 Korean J. Chem. Eng., 26(2), 411-416 (2009) SHORT COMMUNICATION Continuous production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate): Effects of C/N ratio and dilution rate on HB/HV ratio Shwu-Tzy Wu,
Korean J. Chem. Eng., 26(2), 411-416 (2009) SHORT COMMUNICATION Continuous production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate): Effects of C/N ratio and dilution rate on HB/HV ratio Shwu-Tzy Wu,
HiPer Transformation Teaching Kit
 HiPer Transformation Teaching Kit Product Code: HTBM017 Number of experiments that can be performed: 10 Duration of Experiment: 4 days Day 1- Preparation of media and revival of E. coli Host Day 2- Inoculation
HiPer Transformation Teaching Kit Product Code: HTBM017 Number of experiments that can be performed: 10 Duration of Experiment: 4 days Day 1- Preparation of media and revival of E. coli Host Day 2- Inoculation
TIANpure Mini Plasmid Kit
 TIANpure Mini Plasmid Kit For purification of molecular biology grade DNA www.tiangen.com PP080822 TIANpure Mini Plasmid Kit Kit Contents Storage Contents RNaseA (10 mg/ml) Buffer BL Buffer P1 Buffer P2
TIANpure Mini Plasmid Kit For purification of molecular biology grade DNA www.tiangen.com PP080822 TIANpure Mini Plasmid Kit Kit Contents Storage Contents RNaseA (10 mg/ml) Buffer BL Buffer P1 Buffer P2
Hetero-Stagger PCR Cloning Kit
 Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
Product Name: Code No: Size: DynaExpress Hetero-Stagger PCR Cloning Kit DS150 20 reactions Kit Components: Box 1 (-20 ) phst-1 Vector, linearized Annealing Buffer Ligase Mixture phst Forward Sequence Primer
New Platform for Cytochrome P450 Reaction Combining in Situ Immobilization on Biopolymer
 Supporting Information New Platform for Cytochrome P450 Reaction Combining in Situ Immobilization on Biopolymer Jae Hyung Lee, Dong Heon Nam, Sahng Ha Lee, Jong Hyun Park, Si Jae Park, Seung Hwan Lee,
Supporting Information New Platform for Cytochrome P450 Reaction Combining in Situ Immobilization on Biopolymer Jae Hyung Lee, Dong Heon Nam, Sahng Ha Lee, Jong Hyun Park, Si Jae Park, Seung Hwan Lee,
Supplementary Material. Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological
 Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Supplementary Material Increased heterocyst frequency by patn disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density Applied Microbiology
Title. CitationBIORESOURCE TECHNOLOGY, 102(3): Issue Date Doc URL. Type. File Information.
 Title Effect of feeding regimens on polyhydroxybutyrate pr Author(s)Hafuka, Akira; Sakaida, Kenji; Satoh, Hisashi; Takah CitationBIORESOURCE TECHNOLOGY, 102(3): 3551-3553 Issue Date 2011-02 Doc URL http://hdl.handle.net/2115/44977
Title Effect of feeding regimens on polyhydroxybutyrate pr Author(s)Hafuka, Akira; Sakaida, Kenji; Satoh, Hisashi; Takah CitationBIORESOURCE TECHNOLOGY, 102(3): 3551-3553 Issue Date 2011-02 Doc URL http://hdl.handle.net/2115/44977
Transformation (The method of CaCl 2 )
 PROTOCOLS E. coli Transformation (The method of CaCl 2 ) Ligation PRODUCTION competent cells of E. COLI. (Rubidium cells). Gel DNA Recovery Kit Electroporation Digestiones Plasmid purification (The method
PROTOCOLS E. coli Transformation (The method of CaCl 2 ) Ligation PRODUCTION competent cells of E. COLI. (Rubidium cells). Gel DNA Recovery Kit Electroporation Digestiones Plasmid purification (The method
Molecular Biology. Manufactured by. Online Ordering Available
 Molecular Biology Manufactured by Online Ordering Available Molecular Biology Table of Contents 1 Bacterial Culture Media Powder 2 Bacterial Culture Agar Plates 2 Bacterial Culture Agar Heat and Pour Format
Molecular Biology Manufactured by Online Ordering Available Molecular Biology Table of Contents 1 Bacterial Culture Media Powder 2 Bacterial Culture Agar Plates 2 Bacterial Culture Agar Heat and Pour Format
TIANpure Midi Plasmid Kit
 TIANpure Midi Plasmid Kit For purification of molecular biology grade DNA www.tiangen.com Kit Contents Storage TIANpure Midi Plasmid Kit Contents RNaseA (10 mg/ml) Buffer BL Buffer P1 Buffer P2 Buffer
TIANpure Midi Plasmid Kit For purification of molecular biology grade DNA www.tiangen.com Kit Contents Storage TIANpure Midi Plasmid Kit Contents RNaseA (10 mg/ml) Buffer BL Buffer P1 Buffer P2 Buffer
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Handbook of Biodegradable Polymers ISBN:
 Title: Handbook of Biodegradable Polymers Author: C. Bastoli ISBN: 1-85957-389-4 Contents 1 Biodegradability of Polymers Mechanisms and Evaluation Methods 1.1 Introduction 1.2 Background 1.3 Defining Biodegradability
Title: Handbook of Biodegradable Polymers Author: C. Bastoli ISBN: 1-85957-389-4 Contents 1 Biodegradability of Polymers Mechanisms and Evaluation Methods 1.1 Introduction 1.2 Background 1.3 Defining Biodegradability
Molecular Biology. Manufactured by. Online Ordering Available
 Molecular Biology Manufactured by Online Ordering Available Molecular Biology Table of Contents 1 Bacterial Culture Media Powder 2 Bacterial Growth Culture Media 2 Bacterial Culture Media Broth Bottles
Molecular Biology Manufactured by Online Ordering Available Molecular Biology Table of Contents 1 Bacterial Culture Media Powder 2 Bacterial Growth Culture Media 2 Bacterial Culture Media Broth Bottles
Figure 1. Map of cloning vector pgem T-Easy (bacterial plasmid DNA)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 6: Ligation & Bacterial Transformation (Bring your text and laptop to class if you wish to work on your assignment during
Growth, Purification, and Characterization of P450 cam
 1. Cell growth without isotopic labeling Growth, Purification, and Characterization of P450 cam Growth medium Per liter (all components are previously sterilized by either autoclave or filtration): 5 M9
1. Cell growth without isotopic labeling Growth, Purification, and Characterization of P450 cam Growth medium Per liter (all components are previously sterilized by either autoclave or filtration): 5 M9
Figure S1. Biosynthetic pathway of GDP-PerNAc. Mass spectrum of purified GDP-PerNAc. Nature Protocols: doi: /nprot
 Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Synthesis of GDP-PerNAc In E. coli O157, the biosynthesis of GDP- -N-acetyl-D-perosamine (GDP-PerNAc) involves three enzymes (Fig. S1). GDP-D-Mannose is converted by GDP-mannose-4,6-dehydratase (GMD) into
Plasmid Midiprep Purification Kit
 Plasmid Midiprep Purification Kit Cat. # : DP01MD/ DP01MD-100 Size : 20/100 Reactions Store at RT For research use only 1 Description: The Plasmid Midiprep Purification Kit provides simple rapid protocol
Plasmid Midiprep Purification Kit Cat. # : DP01MD/ DP01MD-100 Size : 20/100 Reactions Store at RT For research use only 1 Description: The Plasmid Midiprep Purification Kit provides simple rapid protocol
The Role of genes PA PA2005 in the Metabolism of. D- 3- Hydroxybutyrate in Pseudomonas aeruginosa PAO1. Joshua R. Harris
 The Role of genes PA2003 - PA2005 in the Metabolism of D- 3- Hydroxybutyrate in Pseudomonas aeruginosa PAO1 by Joshua R. Harris Candidate for Bachelor of Science Chemistry Department and Biotechnology
The Role of genes PA2003 - PA2005 in the Metabolism of D- 3- Hydroxybutyrate in Pseudomonas aeruginosa PAO1 by Joshua R. Harris Candidate for Bachelor of Science Chemistry Department and Biotechnology
Production of polyhydroxyalkanoates by Ralstonia eutropha from volatile fatty acids
 Korean J. Chem. Eng., 30(12), 2223-2227 (2013) DOI: 10.1007/s11814-013-0190-9 INVITED REVIEW PAPER INVITED REVIEW PAPER pissn: 0256-1115 eissn: 1975-7220 Production of polyhydroxyalkanoates by Ralstonia
Korean J. Chem. Eng., 30(12), 2223-2227 (2013) DOI: 10.1007/s11814-013-0190-9 INVITED REVIEW PAPER INVITED REVIEW PAPER pissn: 0256-1115 eissn: 1975-7220 Production of polyhydroxyalkanoates by Ralstonia
Chapter 4. Recombinant DNA Technology
 Chapter 4 Recombinant DNA Technology 5. Plasmid Cloning Vectors Plasmid Plasmids Self replicating Double-stranded Mostly circular DNA ( 500 kb) Linear : Streptomyces, Borrelia burgdorferi Replicon
Chapter 4 Recombinant DNA Technology 5. Plasmid Cloning Vectors Plasmid Plasmids Self replicating Double-stranded Mostly circular DNA ( 500 kb) Linear : Streptomyces, Borrelia burgdorferi Replicon
Confirming the Phenotypes of E. coli Strains
 Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
HiPer Plasmid DNA Cloning Teaching Kit
 HiPer Plasmid DNA Cloning Teaching Kit Product Code: HTBM022 Number of experiments that can be performed: 5 Duration of Experiment: 4 days Day 1- Preparation of media and revival of E. coli Host Day2-
HiPer Plasmid DNA Cloning Teaching Kit Product Code: HTBM022 Number of experiments that can be performed: 5 Duration of Experiment: 4 days Day 1- Preparation of media and revival of E. coli Host Day2-
CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution
 CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution Cat. Nos. C400EL10, C400CH10, and CIS40025 Available exclusively thru Lucigen. lucigen.com/epibio
CopyCutter EPI400 Electrocompetent E. coli CopyCutter EPI400 Chemically Competent E. coli CopyCutter Induction Solution Cat. Nos. C400EL10, C400CH10, and CIS40025 Available exclusively thru Lucigen. lucigen.com/epibio
Cat # Box1 Box2. DH5a Competent E. coli cells CCK-20 (20 rxns) 40 µl 40 µl 50 µl x 20 tubes. Choo-Choo Cloning TM Enzyme Mix
 Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
Molecular Cloning Laboratories User Manual Version 3.3 Product name: Choo-Choo Cloning Kits Cat #: CCK-10, CCK-20, CCK-096, CCK-384 Description: Choo-Choo Cloning is a highly efficient directional PCR
Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive
 Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive to ionizing radiation. However, why these mutants are
Designing and creating your gene knockout Background The rada gene was identified as a gene, that when mutated, caused cells to become hypersensitive to ionizing radiation. However, why these mutants are
The current polymer materials in common use are nearly all
 A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme Seiichi Taguchi a,1, Miwa Yamada a, Ken ichiro Matsumoto a, Kenji Tajima a, Yasuharu Satoh a, Masanobu Munekata a, Katsuhiro
A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme Seiichi Taguchi a,1, Miwa Yamada a, Ken ichiro Matsumoto a, Kenji Tajima a, Yasuharu Satoh a, Masanobu Munekata a, Katsuhiro
Presto Mini Plasmid Kit
 Instruction Manual Ver. 03.06.17 For Research Use Only Presto Mini Plasmid Kit PDH004 (4 Preparation Sample Kit) PDH100 (100 Preparation Kit) PDH300 (300 Preparation Kit) Advantages Sample: 1-7 ml of cultured
Instruction Manual Ver. 03.06.17 For Research Use Only Presto Mini Plasmid Kit PDH004 (4 Preparation Sample Kit) PDH100 (100 Preparation Kit) PDH300 (300 Preparation Kit) Advantages Sample: 1-7 ml of cultured
HE Swift Cloning Kit
 HE Swift Cloning Kit For high-efficient cloning of PCR products either blunt or sticky-end Kit Contents Contents VTT-BB05 phe Vector (35 ng/µl) 20 µl T4 DNA Ligase (3 U/µl) 20 µl 2 Reaction Buffer 100
HE Swift Cloning Kit For high-efficient cloning of PCR products either blunt or sticky-end Kit Contents Contents VTT-BB05 phe Vector (35 ng/µl) 20 µl T4 DNA Ligase (3 U/µl) 20 µl 2 Reaction Buffer 100
Transforming Bacteria
 Properties of E. coli Strains for Subcloning Common laboratory strains of E. coli, like JM109, DH5α, and XL-1 Blue, are different from their wildtype counterparts. These strains carry some mutations designed
Properties of E. coli Strains for Subcloning Common laboratory strains of E. coli, like JM109, DH5α, and XL-1 Blue, are different from their wildtype counterparts. These strains carry some mutations designed
Poly (3-hydroxybutyrate) production from glycerol by marine microorganisms. Abstract. 1. Introduction
 573 KKU Res. J. 2012; 17(4):573-579 http : //resjournal.kku.ac.th Poly (3-hydroxybutyrate) production from glycerol by marine microorganisms Sathita Phol-in 1, Donlaya Kamkalong 1, Adisak Jaturapiree 2
573 KKU Res. J. 2012; 17(4):573-579 http : //resjournal.kku.ac.th Poly (3-hydroxybutyrate) production from glycerol by marine microorganisms Sathita Phol-in 1, Donlaya Kamkalong 1, Adisak Jaturapiree 2
Vectors for Gene Cloning: Plasmids and Bacteriophages
 Vectors for Gene Cloning: Plasmids and Bacteriophages DNA molecule must be able to replicate within the host cell to be able to act as a vector for gene cloning, so that numerous copies of the recombinant
Vectors for Gene Cloning: Plasmids and Bacteriophages DNA molecule must be able to replicate within the host cell to be able to act as a vector for gene cloning, so that numerous copies of the recombinant
ml recombinant E. coli cultures (at a density of A 600 units per ml)
 for purification of plasmid DNA, from bacterial cultures Cat. No. 1 754 777 (50 purifications) Cat. No. 1 754 785 (50 purifications) Principle Alkaline lysis releases plasmid DNA from bacteria and RNase
for purification of plasmid DNA, from bacterial cultures Cat. No. 1 754 777 (50 purifications) Cat. No. 1 754 785 (50 purifications) Principle Alkaline lysis releases plasmid DNA from bacteria and RNase
Optimization of Protein Expression in an E. coli System using Thermo Scientific MaxQ 8000 Refrigerated Stackable Shakers
 APPLICATION NOTE Optimization of Protein Expression in an E. coli System using Thermo Scientific MaxQ 8000 Refrigerated Stackable Shakers Authors: Mark Schofield, Research Scientist II, Thermo Fisher Scientific
APPLICATION NOTE Optimization of Protein Expression in an E. coli System using Thermo Scientific MaxQ 8000 Refrigerated Stackable Shakers Authors: Mark Schofield, Research Scientist II, Thermo Fisher Scientific
PRODUCT INFORMATION. Composition of SOC medium supplied :
 Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Product Name : Competent Cell BL21(DE3)pLysS Code No. : DS260 Size : 100 μl 10 Competency : > 5 10 7 cfu/μg (puc19) Supplied product : SOC medium, 1 ml 10 This product is for research use only Description
Data Sheet Quick PCR Cloning Kit
 Data Sheet Quick PCR Cloning Kit 6044 Cornerstone Ct. West, Ste. E DESCRIPTION: The Quick PCR Cloning Kit is a simple and highly efficient method to insert any gene or DNA fragment into a vector, without
Data Sheet Quick PCR Cloning Kit 6044 Cornerstone Ct. West, Ste. E DESCRIPTION: The Quick PCR Cloning Kit is a simple and highly efficient method to insert any gene or DNA fragment into a vector, without
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli
 TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300102, EC300110, EC300150, and C300C105 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300102, EC300110, EC300150, and C300C105 Available exclusively thru Lucigen. lucigen.com/epibio www.lucigen.com
psg5 Vector INSTRUCTION MANUAL Catalog # Revision A For In Vitro Use Only
 psg5 Vector INSTRUCTION MANUAL Catalog #216201 Revision A For In Vitro Use Only 216201-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other warranties
psg5 Vector INSTRUCTION MANUAL Catalog #216201 Revision A For In Vitro Use Only 216201-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other warranties
E ect of Vitreoscilla hemoglobin biosynthesis in Escherichia coli on production of poly(l-hydroxybutyrate) and fermentative parameters
 FEMS Microbiology Letters 214 (2002) 223^227 www.fems-microbiology.org E ect of Vitreoscilla hemoglobin biosynthesis in Escherichia coli on production of poly(l-hydroxybutyrate) and fermentative parameters
FEMS Microbiology Letters 214 (2002) 223^227 www.fems-microbiology.org E ect of Vitreoscilla hemoglobin biosynthesis in Escherichia coli on production of poly(l-hydroxybutyrate) and fermentative parameters
Supplemental materials
 1 2 Supplemental materials 3 4 The terminal oxidase cbb 3 functions in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense 5 Running title: Oxygen respiration and magnetite
1 2 Supplemental materials 3 4 The terminal oxidase cbb 3 functions in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense 5 Running title: Oxygen respiration and magnetite
Overnight Express TM Autoinduction
 About the System Overnight Express Autoinduction System 1 71300-3 71300-4 Overnight Express Autoinduction System 2 71366-3 71366-4 Description The Overnight Express Autoinduction 1 and 2 are designed for
About the System Overnight Express Autoinduction System 1 71300-3 71300-4 Overnight Express Autoinduction System 2 71366-3 71366-4 Description The Overnight Express Autoinduction 1 and 2 are designed for
EFFECT OF GLUCOSE ON PHB PRODUCTION USING Alcaligenes eutrophus DSM 545 AND TISTR 1095
 EFFECT OF GLUCOSE ON PHB PRODUCTION USING Alcaligenes eutrophus DSM AND TISTR Jaruwan Marudkla, Chiravoot Pechyen, and Sarote Sirisansaneeyakul,* Department of Biotechnology, Kasetsart University, Bangkok,,
EFFECT OF GLUCOSE ON PHB PRODUCTION USING Alcaligenes eutrophus DSM AND TISTR Jaruwan Marudkla, Chiravoot Pechyen, and Sarote Sirisansaneeyakul,* Department of Biotechnology, Kasetsart University, Bangkok,,
Detection of bacterial aggregation by flow cytometry
 Detection of bacterial aggregation by flow cytometry Jack Coleman 1 and Melis McHenry 2 1 Enzo Life Sciences, Farmingdale NY, USA, 2 Beckman Coulter Inc., Miami FL, USA PROTEOSTAT Aggresome Detection Kit
Detection of bacterial aggregation by flow cytometry Jack Coleman 1 and Melis McHenry 2 1 Enzo Life Sciences, Farmingdale NY, USA, 2 Beckman Coulter Inc., Miami FL, USA PROTEOSTAT Aggresome Detection Kit
Subcloning 1. Overview 2. Method
 Subcloning 1. Overview: 1.1. Digest plasmids for vector + insert 1.2. CIP treat vector 1.3. Gel purify vector + insert 1.4. Ligate 1.5. Transform bacteria with ligation reactions 1.6. Qiagen or Eppendorf
Subcloning 1. Overview: 1.1. Digest plasmids for vector + insert 1.2. CIP treat vector 1.3. Gel purify vector + insert 1.4. Ligate 1.5. Transform bacteria with ligation reactions 1.6. Qiagen or Eppendorf
Aggregation using Ag43
 Project overview Project overview Aggregation using Ag43 Ag43 EASY DIFFICULT Project overview The best way to produce Bio-plastic Project overview Aggregation Module Plastic Producing Module Bio capsule
Project overview Project overview Aggregation using Ag43 Ag43 EASY DIFFICULT Project overview The best way to produce Bio-plastic Project overview Aggregation Module Plastic Producing Module Bio capsule
Roles of Multiple Acetoacetyl Coenzyme A Reductases in Polyhydroxybutyrate Biosynthesis in Ralstonia eutropha H16
 JOURNAL OF BACTERIOLOGY, Oct. 2010, p. 5319 5328 Vol. 192, No. 20 0021-9193/10/$12.00 doi:10.1128/jb.00207-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Roles of Multiple Acetoacetyl
JOURNAL OF BACTERIOLOGY, Oct. 2010, p. 5319 5328 Vol. 192, No. 20 0021-9193/10/$12.00 doi:10.1128/jb.00207-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Roles of Multiple Acetoacetyl
Nuclear Magnetic Resonance Studies of Poly(3-Hydroxybutyrate) and Polyphosphate Metabolism in Alcaligenes eutrophus
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Nov. 1989, p. 2932-2938 0099-2240/89/112932-07$02.00/0 Copyright 1989, American Society for Microbiology Vol. 55, No. 11 Nuclear Magnetic Resonance Studies of Poly(3-Hydroxybutyrate)
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Nov. 1989, p. 2932-2938 0099-2240/89/112932-07$02.00/0 Copyright 1989, American Society for Microbiology Vol. 55, No. 11 Nuclear Magnetic Resonance Studies of Poly(3-Hydroxybutyrate)
Regulation of enzyme synthesis
 Regulation of enzyme synthesis The lac operon is an example of an inducible operon - it is normally off, but when a molecule called an inducer is present, the operon turns on. The trp operon is an example
Regulation of enzyme synthesis The lac operon is an example of an inducible operon - it is normally off, but when a molecule called an inducer is present, the operon turns on. The trp operon is an example
Table of Contents. IX. Application example XII. Related products... 11
 Table of Contents I. Description...2 II. Components... 3 III. Vector map... 4 IV. Form... 5 V. Purity... 5 VI. Storage... 5 VII. Protocol... 6 VIII. Multicloning site of pcold I-IV (SP4) DNAs... 7 IX.
Table of Contents I. Description...2 II. Components... 3 III. Vector map... 4 IV. Form... 5 V. Purity... 5 VI. Storage... 5 VII. Protocol... 6 VIII. Multicloning site of pcold I-IV (SP4) DNAs... 7 IX.
462 BCH. Biotechnology & Genetic engineering. (Practical)
 462 BCH Biotechnology & Genetic engineering (Practical) Nora Aljebrin Office: Building 5, 3 rd floor, 5T304 E.mail: naljebrin@ksu.edu.sa All lectures and lab sheets are available on my website: Fac.ksu.edu.sa\naljebrin
462 BCH Biotechnology & Genetic engineering (Practical) Nora Aljebrin Office: Building 5, 3 rd floor, 5T304 E.mail: naljebrin@ksu.edu.sa All lectures and lab sheets are available on my website: Fac.ksu.edu.sa\naljebrin
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending)
 peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
peco TM -T7-nGST, Eco cloning Kit User Manual (Patent pending) Cloning PCR products for E Coli expression of N-term GST-tagged protein Cat# Contents Amounts Application IC-1004 peco-t7-ngst vector built-in
Plasmid Midiprep Plus Purification Kit. Cat. # : DP01MD-P10/ DP01MD-P50 Size : 10/50 Reactions Store at RT For research use only
 Plasmid Midiprep Plus Purification Kit Cat. # : DP01MD-P10/ DP01MD-P50 Size : 10/50 Reactions Store at RT For research use only 1 Description: The Plasmid Midiprep Plus Purification Kit provides simple
Plasmid Midiprep Plus Purification Kit Cat. # : DP01MD-P10/ DP01MD-P50 Size : 10/50 Reactions Store at RT For research use only 1 Description: The Plasmid Midiprep Plus Purification Kit provides simple
molecular microbiology
 molecular microbiology 030618dl Sold exclusively through Manufactured by molecular microbiology 1 Culture Media Powder 2 Prepared Culture Media 2 Prepared Culture Media (Broth Bottles and Flasks) 3 Antibiotics
molecular microbiology 030618dl Sold exclusively through Manufactured by molecular microbiology 1 Culture Media Powder 2 Prepared Culture Media 2 Prepared Culture Media (Broth Bottles and Flasks) 3 Antibiotics
Amgen Laboratory Series. Tabs C and E
 Amgen Laboratory Series Tabs C and E Chapter 2A Goals Describe the characteristics of plasmids Explain how plasmids are used in cloning a gene Describe the function of restriction enzymes Explain how to
Amgen Laboratory Series Tabs C and E Chapter 2A Goals Describe the characteristics of plasmids Explain how plasmids are used in cloning a gene Describe the function of restriction enzymes Explain how to
PLNT2530 (2018) Unit 6b Sequence Libraries
 PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
PLNT2530 (2018) Unit 6b Sequence Libraries Molecular Biotechnology (Ch 4) Analysis of Genes and Genomes (Ch 5) Unless otherwise cited or referenced, all content of this presenataion is licensed under the
Understanding Gene Function and Control in Lignin Formation In Wood
 Understanding Gene Function and Control in Lignin Formation In Wood Vincent L. Chiang North Carolina State University Raleigh, NC Tremendous effort has been devoted to developing genetically engineered
Understanding Gene Function and Control in Lignin Formation In Wood Vincent L. Chiang North Carolina State University Raleigh, NC Tremendous effort has been devoted to developing genetically engineered
Transformation of DNA in competent E. coil
 Transformation of DNA in competent E. coil Reagents: SOC medium (1L) (a) 20g tryptone, 5g yeast extract, 0.5g NaCl in 950ml dh 2 O. (b) 250mM KCl: 1.86 KCl in 100ml dh 2 O. Add 10ml of solution (b) to
Transformation of DNA in competent E. coil Reagents: SOC medium (1L) (a) 20g tryptone, 5g yeast extract, 0.5g NaCl in 950ml dh 2 O. (b) 250mM KCl: 1.86 KCl in 100ml dh 2 O. Add 10ml of solution (b) to
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
5/24/11. Agenda. Agenda. Identification and sequencing of VHH against ErbB1, ErbB2 and ErbB3 Experimental section Course Immunobiology
 Agenda Identification and sequencing of VHH against ErbB1, ErbB2 and ErbB3 Experimental section Course Immunobiology Camelid heavy chain antibodies and VHH Structure Principle library construction Phage
Agenda Identification and sequencing of VHH against ErbB1, ErbB2 and ErbB3 Experimental section Course Immunobiology Camelid heavy chain antibodies and VHH Structure Principle library construction Phage
Construction of a Mutant pbr322 Using Site-Directed Mutagenesis to Investigate the Exclusion Effects of pbr322 During Co-transformation with puc19
 Construction of a Mutant pbr322 Using Site-Directed Mutagenesis to Investigate the Exclusion Effects of pbr322 During Co-transformation with puc19 IVANA KOMLJENOVIC Department of Microbiology and Immunology,
Construction of a Mutant pbr322 Using Site-Directed Mutagenesis to Investigate the Exclusion Effects of pbr322 During Co-transformation with puc19 IVANA KOMLJENOVIC Department of Microbiology and Immunology,
N-terminal dual protein functionalization by strainpromoted alkyne nitrone cycloaddition
 Electronic Supplementary Material (ESI) for rganic & Biomolecular Chemistry -terminal dual protein functionalization by strainpromoted alkyne nitrone cycloaddition Rinske P. Temming, a Loek Eggermont,
Electronic Supplementary Material (ESI) for rganic & Biomolecular Chemistry -terminal dual protein functionalization by strainpromoted alkyne nitrone cycloaddition Rinske P. Temming, a Loek Eggermont,
Bacterial genetic exchange:transformation
 Experiment 11 Laboratory to Biology III Diversity of Microorganisms / Wintersemester / page 1 Experiment Advisor Reading Objectives Background Literature www. Links Bacterial genetic exchange:transformation
Experiment 11 Laboratory to Biology III Diversity of Microorganisms / Wintersemester / page 1 Experiment Advisor Reading Objectives Background Literature www. Links Bacterial genetic exchange:transformation
Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein Dronpa. Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Optically Controlled Reversible Protein Hydrogels Based on Photoswitchable Fluorescent Protein
Molecular and functional analysis of the poly-β-hydroxybutyrate biosynthesis operon
 Molecular and functional analysis of the poly-β-hydroxybutyrate biosynthesis operon of Pseudomonas sp BJ-1 S.W. Zhu, Z.Y. Fang, H.Y. Jiang and B.J. Cheng School of Life Science, Anhui Agricultural University,
Molecular and functional analysis of the poly-β-hydroxybutyrate biosynthesis operon of Pseudomonas sp BJ-1 S.W. Zhu, Z.Y. Fang, H.Y. Jiang and B.J. Cheng School of Life Science, Anhui Agricultural University,
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli
 TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300105, EC300110, EC300150, and C300C105 Connect with Epicentre on our blog (epicentral.blogspot.com),
TransforMax EPI300 Electrocompetent E. coli TransforMax EPI300 Chemically Competent E. coli Cat. Nos. EC300105, EC300110, EC300150, and C300C105 Connect with Epicentre on our blog (epicentral.blogspot.com),
StrataPrep Plasmid Miniprep Kit
 StrataPrep Plasmid Miniprep Kit INSTRUCTION MANUAL Catalog #400761 and #400763 Revision A For In Vitro Use Only 400761-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this
StrataPrep Plasmid Miniprep Kit INSTRUCTION MANUAL Catalog #400761 and #400763 Revision A For In Vitro Use Only 400761-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this
A Discovery Laboratory Investigating Bacterial Gene Regulation
 Chapter 8 A Discovery Laboratory Investigating Bacterial Gene Regulation Robert Moss Wofford College 429 N. Church Street Spartanburg, SC 29307 mosssre@wofford.edu Bob Moss is an Associate Professor of
Chapter 8 A Discovery Laboratory Investigating Bacterial Gene Regulation Robert Moss Wofford College 429 N. Church Street Spartanburg, SC 29307 mosssre@wofford.edu Bob Moss is an Associate Professor of
Transformation of Escherichia coli With a Chimeric Plasmid
 Transformation of Escherichia coli With a Chimeric Plasmid Now that we have generated recombinant molecules, we must next amplify them by inserting them into an acceptable host so that they may be analyzed
Transformation of Escherichia coli With a Chimeric Plasmid Now that we have generated recombinant molecules, we must next amplify them by inserting them into an acceptable host so that they may be analyzed
DNA Cloning with Cloning Vectors
 Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
Cloning Vectors A M I R A A. T. A L - H O S A R Y L E C T U R E R O F I N F E C T I O U S D I S E A S E S F A C U L T Y O F V E T. M E D I C I N E A S S I U T U N I V E R S I T Y - E G Y P T DNA Cloning
NZYGene Synthesis kit
 Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
Kit components Component Concentration Amount NZYGene Synthesis kit Catalogue number: MB33901, 10 reactions GS DNA Polymerase 1U/ μl 30 μl Reaction Buffer for GS DNA Polymerase 10 150 μl dntp mix 2 mm
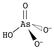 ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
ph and redox potential (pe) are the most important factors controlling arsenic speciation. Phosphate (A) and arsenate (B) speciation are shown as a function of ph for the (V) oxidation states. H 3 PO 4
Protein isotopic enrichment for NMR studies
 Protein isotopic enrichment for NMR studies Protein NMR studies ARTGKYVDES sequence structure Structure of protein- ligand, protein-protein complexes Binding of molecules (perturbation mapping) Dynamic
Protein isotopic enrichment for NMR studies Protein NMR studies ARTGKYVDES sequence structure Structure of protein- ligand, protein-protein complexes Binding of molecules (perturbation mapping) Dynamic
BL21(DE3) expression competent cell pack DS265. Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube)
 Product Name : Code No. : Kit Component : BL21(DE3) expression competent cell pack DS265 Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube) transfromation efficiency: 5 10
Product Name : Code No. : Kit Component : BL21(DE3) expression competent cell pack DS265 Component Code No. Contents Competent cell BL21(DE3) DS250 5 tubes (100 μl/tube) transfromation efficiency: 5 10
Purification and Characterization of a DNA Plasmid Part A CHEM 4581: Biochemistry Laboratory I Version: January 18, 2008
 Purification and Characterization of a DNA Plasmid Part A CHEM 4581: Biochemistry Laboratory I Version: January 18, 2008 INTRODUCTION DNA Plasmids. A plasmid is a small double-stranded, circular DNA molecule
Purification and Characterization of a DNA Plasmid Part A CHEM 4581: Biochemistry Laboratory I Version: January 18, 2008 INTRODUCTION DNA Plasmids. A plasmid is a small double-stranded, circular DNA molecule
Received 19 July 2001/Accepted 3 October 2001
 JOURNAL OF BACTERIOLOGY, Jan. 2002, p. 59 66 Vol. 184, No. 1 0021-9193/02/$04.00 0 DOI: 10.1128/JB.184.1.59 66.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. The Ralstonia
JOURNAL OF BACTERIOLOGY, Jan. 2002, p. 59 66 Vol. 184, No. 1 0021-9193/02/$04.00 0 DOI: 10.1128/JB.184.1.59 66.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. The Ralstonia
I-Blue Midi Plasmid Kit. I-Blue Midi Plasmid Kit. (Endotoxin Free) IBI SCIENTIFIC. Instruction Manual Ver For Research Use Only.
 Instruction Manual Ver. 05.11.17 For Research Use Only I-Blue Midi Plasmid Kit & I-Blue Midi Plasmid Kit (Endotoxin Free) IB47180, IB47190 (2 Preparation Sample Kit) IB47181, IB47191 (25 Preparation Kit)
Instruction Manual Ver. 05.11.17 For Research Use Only I-Blue Midi Plasmid Kit & I-Blue Midi Plasmid Kit (Endotoxin Free) IB47180, IB47190 (2 Preparation Sample Kit) IB47181, IB47191 (25 Preparation Kit)
Plasmid DNA Isolation Column Kit Instruction Manual Catalog No. SA-40012: 50 reactions SA-40011: 100 reactions
 Plasmid DNA Isolation Column Kit Instruction Manual Catalog No. SA-40012: 50 reactions SA-40011: 100 reactions Maxim Biotech, Inc. 780 Dubuque Avenue, So. San Francisco, CA 94080, U.S.A. Tel: (800) 989-6296
Plasmid DNA Isolation Column Kit Instruction Manual Catalog No. SA-40012: 50 reactions SA-40011: 100 reactions Maxim Biotech, Inc. 780 Dubuque Avenue, So. San Francisco, CA 94080, U.S.A. Tel: (800) 989-6296
Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost
 Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen University,
Expression and Purification of the Thermus thermophilus Argonaute Protein Daan C. Swarts *, Matthijs M. Jore and John van der Oost Department of Agrotechnology and Food Sciences, Wageningen University,
TightRegulation, Modulation, and High-Level Expression byvectors ContainingtheArabinose Promoter
 TightRegulation, Modulation, and High-Level Expression byvectors ContainingtheArabinose Promoter L M Guzman et al. (1995) Journal of Bacteriology 177: 4121-4130 Outline 1. Introduction 2. Objective 3.
TightRegulation, Modulation, and High-Level Expression byvectors ContainingtheArabinose Promoter L M Guzman et al. (1995) Journal of Bacteriology 177: 4121-4130 Outline 1. Introduction 2. Objective 3.
