Life Sciences Reporting Summary
|
|
|
- Tamsyn Potter
- 5 years ago
- Views:
Transcription
1 Life Sciences Reporting Summary Corresponding author(s): Vinod Balachandran Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This form is intended for publication with all accepted life science papers and provides structure for consistency and transparency in reporting. Every life science submission will use this form; some list items might not apply to an individual manuscript, but all fields must be completed for clarity. For further information on the points included in this form, see Reporting Life Sciences Research. For further information on Nature Research policies, including our data availability policy, see Authors & Referees and the Editorial Policy Checklist. Experimental design 1. Sample size Describe how sample size was determined. 2. Data exclusions Describe any data exclusions. 3. Replication Describe whether the experimental findings were reliably reproduced. 4. Randomization Describe how samples/organisms/participants were allocated into experimental groups. 5. Blinding Describe whether the investigators were blinded to group allocation during data collection and/or analysis. We were unable to calculate pre-specified effect sizes to optimally determine sample sizes in the two groups, given the extreme rarity of published literature examining tumor specimens in long term survivors of pancreatic cancer. The largest study (prior to ours) to perform whole exome sequencing on long term survivors had a sample size of 8 patients (PMID: ). Following our initial analysis in the MSKCC cohort (n=58), we extended our findings to one of the largest cohorts of human pancreatic cancer with available next generation sequencing, and that is unselected by survival (n=166). MSKCC PDAC Cohort: All tumor samples were surgically resected primary pancreatic ductal adenocarcinomas. Patients treated with neoadjuvant therapy were excluded. All tumors were subjected to pathologic re-review and histologic confirmation by two expert PDAC pathologists prior to analyses. Long term survivors were defined as patients with overall survival > 3 years from surgery, short term survivors as patients with survival >3 months and < 1 year from surgery to exclude perioperative mortalities. (Reported on page # 8, paragraph # 1, line # ) All experimental findings reported in this manuscript were reliably reproducible. MSKCC PDAC Cohort: Long term survivors were defined as patients with overall survival > 3 years from surgery, short term survivors as patients with survival >3 months and < 1 year from surgery to exclude perioperative mortalities. (Reported on page # 8 paragraph # 1, line # Tissue Microarray Multiplexed Immunohistochemistry and Immunofluorescence: Automated digital quantification of tissue microarrays was performed blinded to the investigator with respect to the short and long term cohorts. Whole Exome Sequencing and Neoantigen Prediction: Computational mutation calling and neoantigen prediction was performed blinded to the investigator with respect to the short and long term cohorts. Transcriptome analysis: Bulk tumor transcriptome analysis was performed blinded to the investigator with respect to the short and long term cohorts. T Cell Receptor Sequencing: TCR Sequencing was performed blinded to the investigator with respect to the short and long term cohorts. Neoantigen fitness modeling: Neoantigen fitness modeling was performed blinded 1
2 to the investigator with respect to the short and long term cohorts. Note: all studies involving animals and/or human research participants must disclose whether blinding and randomization were used. 6. Statistical parameters n/a For all figures and tables that use statistical methods, confirm that the following items are present in relevant figure legends (or in the Methods section if additional space is needed). Confirmed Software The exact sample size (n) for each experimental group/condition, given as a discrete number and unit of measurement (animals, litters, cultures, etc.) A description of how samples were collected, noting whether measurements were taken from distinct samples or whether the same sample was measured repeatedly A statement indicating how many times each experiment was replicated The statistical test(s) used and whether they are one- or two-sided (note: only common tests should be described solely by name; more complex techniques should be described in the Methods section) A description of any assumptions or corrections, such as an adjustment for multiple comparisons The test results (e.g. P values) given as exact values whenever possible and with confidence intervals noted A clear description of statistics including central tendency (e.g. median, mean) and variation (e.g. standard deviation, interquartile range) Clearly defined error bars Policy information about availability of computer code 7. Software Describe the software used to analyze the data in this study. See the web collection on statistics for biologists for further resources and guidance. For statistical measurements in Figure 3D, the glm function in R version 3.4 was used for model fitting these models. All data analysis was performed using Prism 7.0, GraphPad Software unless otherwise indicated. Cox regression was performed using STATA Mutations were called using Mutect ( download/mutect), Mutect Rescue ( and HaplotypeCaller ( software.broadinstitute.org/gatk/documentation/tooldocs/current/ org_broadinstitute_gatk_tools_walkers_haplotypecaller_haplotypecaller.php_. Neoantigens were called using NASeek (reference # 9), and pvac_seq ( github.com/griffithlab/pvac-seq). MHC amplitude calculations were performed using NetMHC 3.4 ( TCR recognition probability was calculated using blastp Biopairwise 2 ( github.com/biopython/biopython.github.io/). Tumor clonal structure was calculated using PhyloWGS. ( morrislab/phylowgs) For manuscripts utilizing custom algorithms or software that are central to the paper but not yet described in the published literature, software must be made available to editors and reviewers upon request. We strongly encourage code deposition in a community repository (e.g. GitHub). Nature Methods guidance for providing algorithms and software for publication provides further information on this topic. 2
3 Materials and reagents Policy information about availability of materials 8. Materials availability Indicate whether there are restrictions on availability of unique materials or if these materials are only available for distribution by a for-profit company. 9. Antibodies Describe the antibodies used and how they were validated for use in the system under study (i.e. assay and species). There are no restrictions on availability of unique materials and there are no materials available for distribution by a for-profit company. Immunohistochemistry: Human specific antibodies to MUC16 (clone OCT125, dilution 1:130), WT1 (clone CAN-R9 (IHC)-56-2, dilution 1:30), and Annexin A2 (ab54771, 5 ug/ml) were purchased from Abcam (MA, USA). Antibodies to MUC1 (clone M695, dilution 1:100), and Mesothelin (clone 5B2, dilution 1:50) were purchased from Vector laboratories (CA, USA). Multiplexed Immunohistochemistry: Antibodies to Granzyme B (clone GrB-7, Dako), CD3 (clone 2GV6, Ventana), CD8, (clone C8/144b, Dako), FoxP3 (clone 236A/E7, Abcam), CD20 (clone L26, Dako), CD68 (clone KP1, Dako), DC-LAMP (clone 1010E1.01, Novus Biologicals), MHC class-i (HLA-ABC, clone EMR8-5, Abcam) and CK19 (clone EP1580Y, Abcam) were used. Immunofluorescence: Antibodies to CD4 (Ventana, cat# , 0.5ug/ml), FoxP3 (Abcam, cat#ab20034, 5 ug/ml), mouse IgG (Vector Labs, cat#mkb-22258), CK19 (Abcam, cat#ab52625, 1ug/ml), and DAPI (Sigma Aldrich, cat#d9542, 5 ug/ml) were used. Flow Cytometry: Human-specific antibodies used in all flow cytometric phenotyping included CD45 (clone HI30, BioLegend), CD3 (clone OKT3, BioLegend ), CD4 (clone SK3, BD Biosciences), CD8 (clone SK1, BioLegend), CD56 (clone B159, BD Biosciences), CD69 (clone FN50, BD Biosciences), CD19 (clone SJ25C1, BD Biosciences), PD1 (clone MIH4, BD Biosciences), CD45RA (clone HI100, BD Biosciences), CD45 RO (clone UCHL1, BD Biosciences), CD56 (clone B159, BD Biosciences) and CD107a (clone H4A3, BD Biosciences). 10. Eukaryotic cell lines a. State the source of each eukaryotic cell line used. HEK293T cells were obtained from the ATCC Human Primary Cell collection ( b. Describe the method of cell line authentication used. Cell line authentication was performed using Short Tandem Repeat (STR) analysis. c. Report whether the cell lines were tested for mycoplasma contamination. d. If any of the cell lines used are listed in the database of commonly misidentified cell lines maintained by ICLAC, provide a scientific rationale for their use. Animals and human research participants HEK293T cells were tested for mycoplasma contamination prior to use. Not applicable Policy information about studies involving animals; when reporting animal research, follow the ARRIVE guidelines 11. Description of research animals Provide details on animals and/or animal-derived materials used in the study. Not applicable. 3
4 Policy information about studies involving human research participants 12. Description of human research participants Describe the covariate-relevant population characteristics of the human research participants. All clinicopathologic and population characteristics of all human research participants are provided in detail in Extended Data Tables 1-6. Briefly, all patients had surgically resected primary pancreatic ductal adenocarcinomas. Patients treated with neoadjuvant therapy were excluded. All tumors were subjected to pathologic re-review and histologic confirmation by two expert PDAC pathologists prior to analyses. Long term survivors were defined as patients with overall survival > 3 years from surgery, short term survivors as patients with survival >3m and < 1 year from surgery to exclude perioperative mortalities. Patients were not excluded based on age or gender. For patients included in the Rapid Autopsy cohort, primary and metastatic tumor samples were collected posthumously from four patients as part of the Gastrointestinal Cancer Rapid Medical Donation program. Informed consent was obtained from all subjects. 4
5 Flow Cytometry Reporting Summary Form fields will expand as needed. Please do not leave fields blank. Data presentation For all flow cytometry data, confirm that: 1. The axis labels state the marker and fluorochrome used (e.g. CD4-FITC). Corresponding author(s): Vinod Balachandran Initial submission Revised version Final submission 2. The axis scales are clearly visible. Include numbers along axes only for bottom left plot of group (a 'group' is an analysis of identical markers). 3. All plots are contour plots with outliers or pseudocolor plots. 4. A numerical value for number of cells or percentage (with statistics) is provided. Methodological details 5. Describe the sample preparation. Fresh blood and tumor samples from patients undergoing elective surgery at Memorial Hospital were collected. Peripheral-blood mononuclear cells were isolated by density centrifugation of 25 ml of whole blood over 25 ml of Ficoll-Paque Plus (GE Healthcare). Tumors were mechanically dissociated by mincing with scalpels, scissors, and forceps. Minced tumors were then enzymatically dissociated in a 2.5 ml solution containing collagenase IV (Sigma SKU# C5138, 5mg/ml), DNase I (Roche SKU# , 1ul/ml), and a protease inhibitor cocktail mini tablet (Roche SKU# ) in HBSS, for 30 minutes in a shaker incubator at 37C. Enzymatic dissociation was quenched with 5 ml Fetal Bovine Serum (USA Scientific Ref# # , now discontinued). The cell suspension was then passed through a 100-micron strainer (Falcon cell strainer Ref# ) using the flat portion of a plunger from a 3-ml syringe, and filtered again through a 40-micron strainer (Falcon cell strainer Ref# ) prior to flow cytometric analysis. Tumor draining lymph nodes were mechanically dissociated using the flat portion of a plunger from a 3-ml syringe through a 100-micron strainer (Falcon cell strainer Ref# ), and filtered again through a 40-micron strainer (Falcon cell strainer Ref# ) prior to flow cytometric analysis. 6. Identify the instrument used for data collection. Flow cytometry was performed on an LSRFortessa (BD Biosciences; Catalog # ; Serial # H ). 7. Describe the software used to collect and analyze the flow cytometry data. 8. Describe the abundance of the relevant cell populations within post-sort fractions. Data were analyzed using FlowJo Software (Tree Star). No cell sorting was performed in this report. 9. Describe the gating strategy used. Relevant gating strategies used are indicated in Figures 3c, 4i, Extended Data Figure 2b, and Extended Data Figure 10. nature research flow cytometry reporting summary June 2017 Tick this box to confirm that a figure exemplifying the gating strategy is provided in the Supplementary Information. 1
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Adams Waldorf, Rajagopal, Gale Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Life Sciences Reporting Summary Adams Waldorf, Rajagopal, Gale Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Rich, Jeremy N. Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Rich, Jeremy N. Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Goel Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Life Sciences Reporting Summary Corresponding author(s): Goel Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding Author: Date: Wanjun Chen & Keji Zhao Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life science
Life Sciences Reporting Summary Corresponding Author: Date: Wanjun Chen & Keji Zhao Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life science
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Christer Betsholtz Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we
Life Sciences Reporting Summary Corresponding author(s): Christer Betsholtz Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Stein Aerts Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Stein Aerts Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Our web collection on statistics for biologists may be useful.
 Reporting Summary Corresponding author(s): FELIPE CAVA Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency and transparency in
Reporting Summary Corresponding author(s): FELIPE CAVA Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency and transparency in
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding Author: Johanna A. Joyce Date: Jun 13, 2017 Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life
Life Sciences Reporting Summary Corresponding Author: Johanna A. Joyce Date: Jun 13, 2017 Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): George Q. Daley Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): George Q. Daley Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Eduard Batlle Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Eduard Batlle Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Reporting Summary. Statistical parameters. Software and code. nature research reporting summary April Corresponding author(s):
 Reporting Summary Corresponding author(s): Alexander Flügel 2018-07-09758C Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Summary Corresponding author(s): Alexander Flügel 2018-07-09758C Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: RuRong Ji NNA59063A Article Reporting Checklist for Nature Neuroscience # Main ures: 8 # Supplementary ures: 15 # Supplementary Tables: 0 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: RuRong Ji NNA59063A Article Reporting Checklist for Nature Neuroscience # Main ures: 8 # Supplementary ures: 15 # Supplementary Tables: 0 # Supplementary
Supporting Information
 Supporting Information of Laser Immunotherapy in Combination with Perdurable PD-1 Blocking for Treatment of Metastatic Tumor Lihua Luo, Chunqi Zhu, Hang Yin, Mengshi Jiang, Junlei Zhang, Bing Qin, Zhenyu
Supporting Information of Laser Immunotherapy in Combination with Perdurable PD-1 Blocking for Treatment of Metastatic Tumor Lihua Luo, Chunqi Zhu, Hang Yin, Mengshi Jiang, Junlei Zhang, Bing Qin, Zhenyu
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Antonio Iavarone Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Antonio Iavarone Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Our web collection on statistics for biologists may be useful.
 Reporting Summary Corresponding author(s): Qi-jing Li, Lilin Ye and Bo Zhu Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Summary Corresponding author(s): Qi-jing Li, Lilin Ye and Bo Zhu Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Marcelo Coba Manuscript Number: NNRS3314D Manuscript Type: Resource Reporting Checklist for Nature Neuroscience # Main Figures: 7 # lementary Figures: 4 # lementary Tables: 11 # lementary
Corresponding Author: Marcelo Coba Manuscript Number: NNRS3314D Manuscript Type: Resource Reporting Checklist for Nature Neuroscience # Main Figures: 7 # lementary Figures: 4 # lementary Tables: 11 # lementary
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding Author: Date: Jun 30, 2017 Henrique Veiga-Fernandes Nature Research wishes to improve the reproducibility of the work we publish. This form is published with
Life Sciences Reporting Summary Corresponding Author: Date: Jun 30, 2017 Henrique Veiga-Fernandes Nature Research wishes to improve the reproducibility of the work we publish. This form is published with
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Leonard Petrucelli NNA52530C Article Reporting Checklist for Nature Neuroscience # Main s: 8 # Supplementary s: 10 # Supplementary Tables: 3 #
Corresponding Author: Manuscript Number: Manuscript Type: Leonard Petrucelli NNA52530C Article Reporting Checklist for Nature Neuroscience # Main s: 8 # Supplementary s: 10 # Supplementary Tables: 3 #
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Justin Ichida NMED-A84701B Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work
Life Sciences Reporting Summary Corresponding author(s): Justin Ichida NMED-A84701B Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Michael Cole NNA51 Article Reporting Checklist for Nature Neuroscience # Main Figures: 5 # lementary Figures: 4 # lementary Tables: 0 # lementary
Corresponding Author: Manuscript Number: Manuscript Type: Michael Cole NNA51 Article Reporting Checklist for Nature Neuroscience # Main Figures: 5 # lementary Figures: 4 # lementary Tables: 0 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: McColl NNRS50586AZ Resource Reporting Checklist for Nature Neuroscience # Main Figures: 8 # lementary Figures: 7 # lementary Tables: 13 # lementary
Corresponding Author: Manuscript Number: Manuscript Type: McColl NNRS50586AZ Resource Reporting Checklist for Nature Neuroscience # Main Figures: 8 # lementary Figures: 7 # lementary Tables: 13 # lementary
Application Protocol: CD45 CK Immunostaining for patient blood
 REPRODUCTION AND USE This document is protected by copyright and it cannot be used or shared without permission from Vortex Biosciences, Inc. Such permission is given on condition that Vortex Biosciences
REPRODUCTION AND USE This document is protected by copyright and it cannot be used or shared without permission from Vortex Biosciences, Inc. Such permission is given on condition that Vortex Biosciences
Initial genotyping of all new litters was performed by Transnetyx (Memphis,
 SUPPLEMENTAL INFORMATION MURILLO ET AL. SUPPLEMENTAL EXPERIMENTAL PROCEDURES Mouse Genotyping Initial genotyping of all new litters was performed by Transnetyx (Memphis, USA). Assessment of LoxPpα recombination
SUPPLEMENTAL INFORMATION MURILLO ET AL. SUPPLEMENTAL EXPERIMENTAL PROCEDURES Mouse Genotyping Initial genotyping of all new litters was performed by Transnetyx (Memphis, USA). Assessment of LoxPpα recombination
a KYSE270-CON KYSE270-Id1
 a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
a KYSE27-CON KYSE27- shcon shcon sh b Human Mouse CD31 Relative MVD 3.5 3 2.5 2 1.5 1.5 *** *** c KYSE15 KYSE27 sirna (nm) 5 1 Id2 Id2 sirna 5 1 sirna (nm) 5 1 Id2 sirna 5 1 Id2 [h] (pg per ml) d 3 2 1
TF-1a lymphoblastic leukemia cell line: marking with GFP, phenotyping and sorting
 Supplemental Material Supplemental Methods TF-1a lymphoblastic leukemia cell line: marking with GFP, phenotyping and sorting In order to determine if the multi-parameter FACS approach would be successful
Supplemental Material Supplemental Methods TF-1a lymphoblastic leukemia cell line: marking with GFP, phenotyping and sorting In order to determine if the multi-parameter FACS approach would be successful
Antibody Titrations. Institut für HIV Forschung SOP #06-03 (Nov BS) Background. Reagents
 Institut für HIV Forschung SOP #06-03 (Nov 2017 - BS) Antibody Titrations Reagents Reagent Vendor Catalogue # Stock Conc. FACS Tubes BD Falcon 352054 - Anti-CD28/CD49d Antibody BD Fastimmune 347690 - GolgiPlug
Institut für HIV Forschung SOP #06-03 (Nov 2017 - BS) Antibody Titrations Reagents Reagent Vendor Catalogue # Stock Conc. FACS Tubes BD Falcon 352054 - Anti-CD28/CD49d Antibody BD Fastimmune 347690 - GolgiPlug
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Feltri NNA54268B Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 5 # Supplementary Tables: 1 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Feltri NNA54268B Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 5 # Supplementary Tables: 1 # Supplementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Chay T. Kuo NNA47008 Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 11 # Supplementary Tables:
Corresponding Author: Manuscript Number: Manuscript Type: Chay T. Kuo NNA47008 Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 11 # Supplementary Tables:
The acquired blood sample was placed on a rotator in room temperature until the
 SUPPLEMENTARY METHODS CTCs Isolation and Enrichment with the ISET assay The acquired blood sample was placed on a rotator in room temperature until the isolation process (t
SUPPLEMENTARY METHODS CTCs Isolation and Enrichment with the ISET assay The acquired blood sample was placed on a rotator in room temperature until the isolation process (t
Nature Biotechnology: doi: /nbt.4086
 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
The RT-qPCR analysis of selected type-i IFNs related genes, IRF7 and Oas3. The
 SUPPLEMENTARY MATERIALS AND METHODS Real time quantitative PCR The RT-qPCR analysis of selected type-i IFNs related genes, IRF7 and Oas3. The RT-qPCR was performed on the Applied Biosystems StepOne TM
SUPPLEMENTARY MATERIALS AND METHODS Real time quantitative PCR The RT-qPCR analysis of selected type-i IFNs related genes, IRF7 and Oas3. The RT-qPCR was performed on the Applied Biosystems StepOne TM
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Yang Dan NNA56598 Article Reporting Checklist for Nature Neuroscience # Main Figures: 8 # Supplementary Figures: 12 # Supplementary Tables: 0 #
Corresponding Author: Manuscript Number: Manuscript Type: Yang Dan NNA56598 Article Reporting Checklist for Nature Neuroscience # Main Figures: 8 # Supplementary Figures: 12 # Supplementary Tables: 0 #
A human immunodeficiency caused by mutations in the PIK3R1 gene. Whole exome sequencing. Whole exome sequencing libraries were prepared from 3
 A human immunodeficiency caused by mutations in the PIK3R1 gene. Supplementary Methods Whole exome sequencing. Whole exome sequencing libraries were prepared from 3 µg of genomic DNA extracted from total
A human immunodeficiency caused by mutations in the PIK3R1 gene. Supplementary Methods Whole exome sequencing. Whole exome sequencing libraries were prepared from 3 µg of genomic DNA extracted from total
PITT ISL METHODS 1) Flow-based CTL assay 2) Gut Mucosal Processing
 PITT ISL METHODS 1) Flow-based CTL assay a. Purified CD8 + T cells are co-cultured with fresh, autologous, infected CD4 + T cells at various effector/target ratios for 18 hours at 37 o C. b. Baseline percentage
PITT ISL METHODS 1) Flow-based CTL assay a. Purified CD8 + T cells are co-cultured with fresh, autologous, infected CD4 + T cells at various effector/target ratios for 18 hours at 37 o C. b. Baseline percentage
Autocrine Complement Inhibits IL10-Dependent T-Cell Mediated. Antitumor Immunity to Promote Tumor Progression
 Supplemental Information Autocrine Complement Inhibits IL10-Dependent T-Cell Mediated Antitumor Immunity to Promote Tumor Progression Yu Wang, Sheng-Nan Sun, Qing Liu, Yang-Yang Yu, Jian Guo, Kun Wang,
Supplemental Information Autocrine Complement Inhibits IL10-Dependent T-Cell Mediated Antitumor Immunity to Promote Tumor Progression Yu Wang, Sheng-Nan Sun, Qing Liu, Yang-Yang Yu, Jian Guo, Kun Wang,
Murine in vivo CD8 + T Cell Killing Assay Myoungjoo V. Kim 1*, Weiming Ouyang 2, Will Liao 3, Michael Q. Zhang 4 and Ming O. Li 5
 Murine in vivo CD8 + T Cell Killing Assay Myoungjoo V. Kim 1*, Weiming Ouyang 2, Will Liao 3, Michael Q. Zhang 4 and Ming O. Li 5 1 Department of Immunobiology, Yale University School of Medicine, New
Murine in vivo CD8 + T Cell Killing Assay Myoungjoo V. Kim 1*, Weiming Ouyang 2, Will Liao 3, Michael Q. Zhang 4 and Ming O. Li 5 1 Department of Immunobiology, Yale University School of Medicine, New
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding Author: Ana Domingos Date: 7.09.2017 Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life science
Life Sciences Reporting Summary Corresponding Author: Ana Domingos Date: 7.09.2017 Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life science
Materials and Reagents
 Isolation of CD34 + Cells from Human Fetal Liver and Cord Blood Institute of Molecular and Cell Biology, 138673, Singapore Qingfeng Chen 1* and Jianzhu Chen 2* 1 Humanized Mouse Unit, Institute of Molecular
Isolation of CD34 + Cells from Human Fetal Liver and Cord Blood Institute of Molecular and Cell Biology, 138673, Singapore Qingfeng Chen 1* and Jianzhu Chen 2* 1 Humanized Mouse Unit, Institute of Molecular
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Liu NNA52890T Article Reporting Checklist for Nature Neuroscience # Main Figures: 4 # Supplementary Figures: 3 # Supplementary Tables: 1 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Liu NNA52890T Article Reporting Checklist for Nature Neuroscience # Main Figures: 4 # Supplementary Figures: 3 # Supplementary Tables: 1 # Supplementary
Supplementary Figure 1 A green: cytokeratin 8
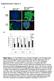 Supplementary Figure 1 A green: cytokeratin 8 green: α-sma red: α-sma blue: DAPI blue: DAPI Panc-1 Panc-1 Panc-1+hPSC Panc-1+hPSC monoculture coculture B Suppl. Figure 1: A, Immunofluorescence staining
Supplementary Figure 1 A green: cytokeratin 8 green: α-sma red: α-sma blue: DAPI blue: DAPI Panc-1 Panc-1 Panc-1+hPSC Panc-1+hPSC monoculture coculture B Suppl. Figure 1: A, Immunofluorescence staining
Application Note. Assay Portability on the BD FACSVerse System. Summary. Maria Jaimes, Yibing Wang, Catherine McIntyre, and Dev Mittar
 September Assay Portability on the BD FACSVerse System Maria Jaimes, Yibing Wang, Catherine McIntyre, and Dev Mittar Contents Summary Introduction 3 Objective 4 Methods 6 Results Discussion Conclusions
September Assay Portability on the BD FACSVerse System Maria Jaimes, Yibing Wang, Catherine McIntyre, and Dev Mittar Contents Summary Introduction 3 Objective 4 Methods 6 Results Discussion Conclusions
Quick-Spin the tetramer tubes before opening, due to the change in altitude and the low volume.
 Quick-Spin the tetramer tubes before opening, due to the change in altitude and the low volume. Staining ex vivo samples with IA g7 Ins10-23 RE#3 Tetramers, from Kappler/Marrack Laboratory Howard Hughes
Quick-Spin the tetramer tubes before opening, due to the change in altitude and the low volume. Staining ex vivo samples with IA g7 Ins10-23 RE#3 Tetramers, from Kappler/Marrack Laboratory Howard Hughes
Data Sheet CD137/NF-κB Reporter - HEK293 Recombinant Cell Line Catalog # 79289
 Data Sheet CD137/NF-κB Reporter - HEK293 Recombinant Cell Line Catalog # 79289 Background Human CD137 (4-1BB; TNFRS9) is an inducible co-stimulatory molecule that activates T cells. CD137:CD137L-mediated
Data Sheet CD137/NF-κB Reporter - HEK293 Recombinant Cell Line Catalog # 79289 Background Human CD137 (4-1BB; TNFRS9) is an inducible co-stimulatory molecule that activates T cells. CD137:CD137L-mediated
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature24473 1. Supplementary Information Computational identification of neoantigens Neoantigens from the three datasets were inferred using a consistent pipeline
SUPPLEMENTARY INFORMATION doi:10.1038/nature24473 1. Supplementary Information Computational identification of neoantigens Neoantigens from the three datasets were inferred using a consistent pipeline
Product datasheet. ARG30119 Pro-B Cell Marker Antibody panel (CD19, CD34, CD38, CD40, CD45)(FACS)
 Product datasheet info@arigobio.com Package: 1 kit ARG30119 Pro-B Cell Marker Antibody panel (CD19, CD34, CD38, CD40, CD45)(FACS) Component Cat. No. Component Name ARG62820 anti-cd34 antibody [4H11(APG)]
Product datasheet info@arigobio.com Package: 1 kit ARG30119 Pro-B Cell Marker Antibody panel (CD19, CD34, CD38, CD40, CD45)(FACS) Component Cat. No. Component Name ARG62820 anti-cd34 antibody [4H11(APG)]
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: xiangdong fu, yuanchao xue NNA52804A Article Reporting Checklist Nature Neuroscience # Main Figures: 5 # Supplementary Figures: 7 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: xiangdong fu, yuanchao xue NNA52804A Article Reporting Checklist Nature Neuroscience # Main Figures: 5 # Supplementary Figures: 7 # Supplementary
CFSE Cell Division Assay Kit
 CFSE Cell Division Assay Kit Item No. 10009853 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
CFSE Cell Division Assay Kit Item No. 10009853 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Isaac M Chiu Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Isaac M Chiu Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Figure legend. The expression of BAFF and its receptors in macrophages was detected at lesion areas of atherosclerosis and arthritis.
 Data supplement #1 Figure legend. The expression of BAFF and its receptors in macrophages was detected at lesion areas of atherosclerosis and arthritis. A. Consecutive sections of an atherosclerotic plaque
Data supplement #1 Figure legend. The expression of BAFF and its receptors in macrophages was detected at lesion areas of atherosclerosis and arthritis. A. Consecutive sections of an atherosclerotic plaque
Lifespan MOLECULAR PATHOLOGY CORE LABORATORY SERVICE & FEE SCHEDULE (Effective 09/01/17) ArcturusXT Laser Capture Microdissection (LCM)
 & FEE SCHEDULE (Effective 09/01/17) ArcturusXT Laser Capture Microdissection (LCM) : : Initial training includes LCM operation and $145.00 flat fee* No charge (first 4 hours; cap handling plus 3 hr free
& FEE SCHEDULE (Effective 09/01/17) ArcturusXT Laser Capture Microdissection (LCM) : : Initial training includes LCM operation and $145.00 flat fee* No charge (first 4 hours; cap handling plus 3 hr free
Supplementary Figure 1
 S U P P L E M E N TA R Y I N F O R M AT I O N In the format provided by the authors and unedited. DOI: 10.1038/ncb3632 Supplementary Figure 1 a PEG-4MAL macromer Functionalized PEG-4MAL Adhesive ligand
S U P P L E M E N TA R Y I N F O R M AT I O N In the format provided by the authors and unedited. DOI: 10.1038/ncb3632 Supplementary Figure 1 a PEG-4MAL macromer Functionalized PEG-4MAL Adhesive ligand
BD IMag. Streptavidin Particles Plus - DM. Technical Data Sheet. Product Information
 Technical Data Sheet Streptavidin Particles Plus - DM Product Information Material Number: Size: Storage Buffer: 557812 5 ml Aqueous buffered solution containing BSA and 0.09% sodium azide. Description
Technical Data Sheet Streptavidin Particles Plus - DM Product Information Material Number: Size: Storage Buffer: 557812 5 ml Aqueous buffered solution containing BSA and 0.09% sodium azide. Description
Our web collection on statistics for biologists may be useful.
 Reporting Summary Corresponding author(s): Charles E. Murry Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency and transparency
Reporting Summary Corresponding author(s): Charles E. Murry Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency and transparency
Immunofluorescent staining and flow cytometric analysis of cells
 UCAN-U 0003 Version 1 November 2010 Page 1 of 7 CMCI (Center for molecular and Cellular Intervention) University Medical Center Utrecht Written By Name Function Date Signature Mark Klein Lab manager Conformation
UCAN-U 0003 Version 1 November 2010 Page 1 of 7 CMCI (Center for molecular and Cellular Intervention) University Medical Center Utrecht Written By Name Function Date Signature Mark Klein Lab manager Conformation
Real-time PCR. Total RNA was isolated from purified splenic or LP macrophages using
 Supplementary Methods Real-time PCR. Total RNA was isolated from purified splenic or LP macrophages using the Qiagen RNeasy Mini Kit, according to the manufacturer s protocol with on-column DNase digestion
Supplementary Methods Real-time PCR. Total RNA was isolated from purified splenic or LP macrophages using the Qiagen RNeasy Mini Kit, according to the manufacturer s protocol with on-column DNase digestion
Reagent Titration for the ICS Assay
 Purpose This standard operating procedure (SOP) describes how to titrate new lots of fluorochromeconjugated antibody reagents and the cell viability marker for use in the intracellular cytokine staining
Purpose This standard operating procedure (SOP) describes how to titrate new lots of fluorochromeconjugated antibody reagents and the cell viability marker for use in the intracellular cytokine staining
efluor Organic Dyes 450/50 BP Fluorescence Intensity Wavelength (nm)
 efluor Organic Dyes efluor Organic Dyes Catalog No. Description Clone Application Anti-Mouse efluor 450 Products 48-0042 Anti-mouse CD4 RM4-5 Flow Cytometry 48-0081 Anti-mouse CD8 53-6.7 Flow Cytometry
efluor Organic Dyes efluor Organic Dyes Catalog No. Description Clone Application Anti-Mouse efluor 450 Products 48-0042 Anti-mouse CD4 RM4-5 Flow Cytometry 48-0081 Anti-mouse CD8 53-6.7 Flow Cytometry
Single Cell Sample Submission Guideline
 Document NO.: SOP-SMM-016 Version: A1 Effective Date: 2016-11-23 Document NO:SOP-SMM-016 Version: A1 Page 1 of 8 Content CHAPTERⅠREQUIREMENTS FOR SINGLE CELL OR FEW CELLS ( CELLS ISOLATED BY CUSTOMERS)...
Document NO.: SOP-SMM-016 Version: A1 Effective Date: 2016-11-23 Document NO:SOP-SMM-016 Version: A1 Page 1 of 8 Content CHAPTERⅠREQUIREMENTS FOR SINGLE CELL OR FEW CELLS ( CELLS ISOLATED BY CUSTOMERS)...
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang *
 In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
Supplementary File 3: DNA and RNA isolation
 Supplementary File 3: DNA and RNA isolation Q-CROC-02 Biopsy protocol For the purposes of this protocol, four needle core biopsies (NCBs) of lymph node tissue are isolated from each patient using a 16G
Supplementary File 3: DNA and RNA isolation Q-CROC-02 Biopsy protocol For the purposes of this protocol, four needle core biopsies (NCBs) of lymph node tissue are isolated from each patient using a 16G
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Lennart Mucke NNRS49607AZ Resource Reporting Checklist for Nature Neuroscience # Main s: # lementary s: 4 # lementary Tables: 9 # lementary Videos:
Corresponding Author: Manuscript Number: Manuscript Type: Lennart Mucke NNRS49607AZ Resource Reporting Checklist for Nature Neuroscience # Main s: # lementary s: 4 # lementary Tables: 9 # lementary Videos:
Isolation of ILC2 from Mouse Liver Tamar Mchedlidze and Stefan Wirtz *
 Isolation of ILC2 from Mouse Liver Tamar Mchedlidze and Stefan Wirtz * Medical Department 1, FAU Erlangen-Nuremberg, Erlangen, Germany *For correspondence: stefan.wirtz@uk-erlangen.de [Abstract] Group
Isolation of ILC2 from Mouse Liver Tamar Mchedlidze and Stefan Wirtz * Medical Department 1, FAU Erlangen-Nuremberg, Erlangen, Germany *For correspondence: stefan.wirtz@uk-erlangen.de [Abstract] Group
Proteasome activity is required for the initiation of precancerous pancreatic
 Proteasome activity is required for the initiation of precancerous pancreatic lesions Takaki Furuyama 1,2, Shinji Tanaka 1,2*, Shu Shimada 1, Yoshimitsu Akiyama 1, Satoshi Matsumura 2, Yusuke Mitsunori
Proteasome activity is required for the initiation of precancerous pancreatic lesions Takaki Furuyama 1,2, Shinji Tanaka 1,2*, Shu Shimada 1, Yoshimitsu Akiyama 1, Satoshi Matsumura 2, Yusuke Mitsunori
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
Supplementary Figures Supplementary Figure 1. Description of the observed lymphatic metastases in two different SIX1-induced MCF7 metastasis models (Nude and NOD/SCID). Supplementary Figure 2. MCF7-SIX1
IMMUNOPATHOLOGY. This SOP will be applied to npod paraffin samples stained by immunohistochemistry.
 1 PURPOSE IMMUNOPATHOLOGY The purpose of this Standard Operating Procedure (SOP) is to outline procedures for immunopathology preparation and analysis of npod samples. 2 SCOPE This SOP will be applied
1 PURPOSE IMMUNOPATHOLOGY The purpose of this Standard Operating Procedure (SOP) is to outline procedures for immunopathology preparation and analysis of npod samples. 2 SCOPE This SOP will be applied
Protein and transcriptome quantitation using BD AbSeq Antibody-Oligonucleotide
 Protein and transcriptome quantitation using BD AbSeq Antibody-Oligonucleotide technology and the 10X Genomics Chromium Single Cell Gene Expression Solution Jocelyn G. Olvera, Brigid S. Boland, John T.
Protein and transcriptome quantitation using BD AbSeq Antibody-Oligonucleotide technology and the 10X Genomics Chromium Single Cell Gene Expression Solution Jocelyn G. Olvera, Brigid S. Boland, John T.
Supporting Information
 Supporting Information Cheng et al. 10.1073/pnas.1207354109 SI Materials and Methods Generation of Stim1 Stim2 Double-KO Mice and Assessment of Saliva Secretion. T-cell specific deletion of Stim1 and Stim2
Supporting Information Cheng et al. 10.1073/pnas.1207354109 SI Materials and Methods Generation of Stim1 Stim2 Double-KO Mice and Assessment of Saliva Secretion. T-cell specific deletion of Stim1 and Stim2
The measurement of telomere length was performed by the same method as in the previous study (11).
 1 SUPPLEMENTAL DATA 2 3 METHODS 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 Telomere length measurement by quantitative real-time PCR (q-pcr) The measurement of telomere length was performed
1 SUPPLEMENTAL DATA 2 3 METHODS 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 Telomere length measurement by quantitative real-time PCR (q-pcr) The measurement of telomere length was performed
UNIVERSITY OF ROCHESTER MEDICAL CENTER
 Date: 04 October 2005 Authors: Sally Quataert and Jennifer Hossler Approval: Sally A. Quataert 1. Purpose: To enumerate the total number of or IgM secreting plasma B cells in human peripheral blood mononuclear
Date: 04 October 2005 Authors: Sally Quataert and Jennifer Hossler Approval: Sally A. Quataert 1. Purpose: To enumerate the total number of or IgM secreting plasma B cells in human peripheral blood mononuclear
Competitive Bone-marrow Transplantations Maria Maryanovich and Atan Gross *
 Competitive Bone-marrow Transplantations Maria Maryanovich and Atan Gross * Biological Regulation, Weizmann Institute of Science, Rehovot, Israel *For correspondence: atan.gross@weizmann.ac.il [Abstract]
Competitive Bone-marrow Transplantations Maria Maryanovich and Atan Gross * Biological Regulation, Weizmann Institute of Science, Rehovot, Israel *For correspondence: atan.gross@weizmann.ac.il [Abstract]
Corporate Tutorial IBA T-CATCH. Cell isolation in pipette tips. ISCT, Paris Lothar Germeroth
 Corporate Tutorial IBA T-CATCH Cell isolation in pipette tips ISCT, Paris 2014 Lothar Germeroth Workshop IBA Introduction of the Streptamer technology Staining with Streptamers off rate assay T-CATCH Cell
Corporate Tutorial IBA T-CATCH Cell isolation in pipette tips ISCT, Paris 2014 Lothar Germeroth Workshop IBA Introduction of the Streptamer technology Staining with Streptamers off rate assay T-CATCH Cell
CFSE Cell Division Assay Kit
 CFSE Cell Division Assay Kit Catalog Number KA1302 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
CFSE Cell Division Assay Kit Catalog Number KA1302 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin
 Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin Carla Tripisciano 1, René Weiss 1, Tanja Eichhorn 1,
Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin Carla Tripisciano 1, René Weiss 1, Tanja Eichhorn 1,
Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.
 LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
LEGENDS TO SUPPLEMENTARY FIGURES Figure S1. Phenotypic characterization of AND-1_WASKO cell lines. AND- 1_WASKO_C1.1 (WASKO_C1.1) and AND-1_WASKO_C1.2 (WASKO_C1.2) were stained with the antibodies oct3/4
Supplementary Figure 1. Effect of FRC-specific ablation of Myd88 on PP and mln organization.
 Supplementary Figure 1 Effect of FRC-specific ablation of Myd88 on PP and mln organization. (a) PP numbers in 8 10 week old Cre-negative littermate (Ctrl) and Myd88-cKO mice (n = 11 mice; each dot represents
Supplementary Figure 1 Effect of FRC-specific ablation of Myd88 on PP and mln organization. (a) PP numbers in 8 10 week old Cre-negative littermate (Ctrl) and Myd88-cKO mice (n = 11 mice; each dot represents
Gene Sequence Annealing Temperature. Actin 5 : 5 -CATCACTATTGGCAACGAGC-3 60 C 3 : 5 -ACGCAGCTCAGTAACAGTCC-3 PCR product: 410 bp
 Supporting Materials and Methods RT-PCR RT-PCR was performed as described in ref. 1 using the following oligonucleotides and PCR conditions: 2 min at 95 C, (20 s at 95 C, 20 s at the annealing temp, and
Supporting Materials and Methods RT-PCR RT-PCR was performed as described in ref. 1 using the following oligonucleotides and PCR conditions: 2 min at 95 C, (20 s at 95 C, 20 s at the annealing temp, and
3D Mammary Colony-Forming Cell Assay Giusy Tornillo 1* and Sara Cabodi 2
 3D Mammary Colony-Forming Cell Assay Giusy Tornillo 1* and Sara Cabodi 2 1 Cardiff School of Biosciences, European Cancer Stem Cell Research Institute, Cardiff University, Cardiff, UK; 2 Department of
3D Mammary Colony-Forming Cell Assay Giusy Tornillo 1* and Sara Cabodi 2 1 Cardiff School of Biosciences, European Cancer Stem Cell Research Institute, Cardiff University, Cardiff, UK; 2 Department of
Centre for Innovative Cancer Research, Ottawa Hospital Research Institute, Ottawa, Canada;
 Ex vivo Natural Killer Cell Cytotoxicity Assay Lee-Hwa Tai 1, Christiano Tanese de Souza 1, Andrew P. Makrigiannis 2 and Rebecca Ann C. Auer 3* 1 Centre for Innovative Cancer Research, Ottawa Hospital
Ex vivo Natural Killer Cell Cytotoxicity Assay Lee-Hwa Tai 1, Christiano Tanese de Souza 1, Andrew P. Makrigiannis 2 and Rebecca Ann C. Auer 3* 1 Centre for Innovative Cancer Research, Ottawa Hospital
IsoFluxTM. System. The next generation of CTC Analysis is here
 IsoFluxTM System The next generation of CTC Analysis is here Product Overview IsoFlux System The next generation of circulating tumor cell analysis is here The IsoFlux System enriches intact rare cells
IsoFluxTM System The next generation of CTC Analysis is here Product Overview IsoFlux System The next generation of circulating tumor cell analysis is here The IsoFlux System enriches intact rare cells
detailed description. Cell proliferation and soft agar colony numbers were shown as relative values normalized to
 Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma Supplementary Material Supplemental Figure S1. Schematic overview of mouse kinome sirna screen. See Materials and Methods for detailed
Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma Supplementary Material Supplemental Figure S1. Schematic overview of mouse kinome sirna screen. See Materials and Methods for detailed
Supplemental material and methods
 Supplemental material and methods Antibodies: Primary antibodies used for these stainings were 174/2 1 (migg1 against PV-1), PAL-E 2 (migg2a; Abcam, Cambridge, UK), anti-nrp-1 (monoclonal migg2a or polyclonal
Supplemental material and methods Antibodies: Primary antibodies used for these stainings were 174/2 1 (migg1 against PV-1), PAL-E 2 (migg2a; Abcam, Cambridge, UK), anti-nrp-1 (monoclonal migg2a or polyclonal
Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546
 Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546 Description This cell line expresses a surface human GITR (glucocorticoid-induced TNFR family related gene; TNFRSF18;
Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546 Description This cell line expresses a surface human GITR (glucocorticoid-induced TNFR family related gene; TNFRSF18;
Jeffrey W. Elias, PhD UCD SOM Office of Research Grants Facilitation.
 NIH IS CHANGING/ENHANCING THE REVIEW CRITERIA AND FOCUSING ON MISSING OPPORTUNITIES TO EXAMINE SEX DIFFERENCES JOURNALS ARE CHANGING/ENHANCING THE REVIEW CRITERIA Jeffrey W. Elias, PhD UCD SOM Office of
NIH IS CHANGING/ENHANCING THE REVIEW CRITERIA AND FOCUSING ON MISSING OPPORTUNITIES TO EXAMINE SEX DIFFERENCES JOURNALS ARE CHANGING/ENHANCING THE REVIEW CRITERIA Jeffrey W. Elias, PhD UCD SOM Office of
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Robert Froemke NNA7412T Article Reporting Checklist for Nature Neuroscience # Main s: 7 # Supplementary s: 14 # Supplementary Tables: 0 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Robert Froemke NNA7412T Article Reporting Checklist for Nature Neuroscience # Main s: 7 # Supplementary s: 14 # Supplementary Tables: 0 # Supplementary
Adult Neuron Isolation Protocol (R. Kaletsky 9/2014)
 Notes before starting: Adult Neuron Isolation Protocol (R. Kaletsky 9/2014) Begin protocol ~45min before sort time to minimize time between cell harvesting and sorting Prepare pronase solution immediately
Notes before starting: Adult Neuron Isolation Protocol (R. Kaletsky 9/2014) Begin protocol ~45min before sort time to minimize time between cell harvesting and sorting Prepare pronase solution immediately
Nature Methods: doi: /nmeth.4237 SUPPLEMENTARY PROTOCOL: DETAILED ARTIFICIAL THYMIC ORGANOID (ATO) METHOD REAGENTS
 SUPPLEMENTARY PROTOCOL: DETAILED ARTIFICIAL THYMIC ORGANOID (ATO) METHOD REAGENTS CD34+ HSPC isolation Fresh human umbilical cord blood (CB), bone marrow (BM), mobilized peripheral blood (MPB), peripheral
SUPPLEMENTARY PROTOCOL: DETAILED ARTIFICIAL THYMIC ORGANOID (ATO) METHOD REAGENTS CD34+ HSPC isolation Fresh human umbilical cord blood (CB), bone marrow (BM), mobilized peripheral blood (MPB), peripheral
Nature Medicine: doi: /nm.4464
 Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Supplementary Fig. 1. Amino acid transporters and substrates used for selectivity screening. (A) Common transporters and amino acid substrates shown. Amino acids designated by one-letter codes. Transporters
Title: Micro-immunofluorescence Staining for Multichromatic FACS Analysis with Human Single Cell Suspensions
 Date: 27-December-2006 Authors: Sally A. Quataert and Matthew Cochran Approved: Title: Micro-immunofluorescence Staining for Multichromatic FACS Analysis with Human Single Cell Suspensions Purpose and
Date: 27-December-2006 Authors: Sally A. Quataert and Matthew Cochran Approved: Title: Micro-immunofluorescence Staining for Multichromatic FACS Analysis with Human Single Cell Suspensions Purpose and
Data Sheet PD-1 / NFAT - Reporter - Jurkat Recombinant Cell Line Catalog #: 60535
 Data Sheet PD-1 / NFAT - Reporter - Jurkat Recombinant Cell Line Catalog #: 60535 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
Data Sheet PD-1 / NFAT - Reporter - Jurkat Recombinant Cell Line Catalog #: 60535 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
ab pdc Subset Phenotyping Kit
 ab177772 pdc Subset Phenotyping Kit Instructions for Use A set of antibodies designed to identify pdcs and subsets of pdcs by multi-color flow cytometry. This product is for research use only and is not
ab177772 pdc Subset Phenotyping Kit Instructions for Use A set of antibodies designed to identify pdcs and subsets of pdcs by multi-color flow cytometry. This product is for research use only and is not
Measurement of peritoneal macrophage apoptosis by Celigo plate imaging cytometer
 SUPPLEMENTAL METHODS Measurement of peritoneal macrophage apoptosis by Celigo plate imaging cytometer For Celigo experiments, 0.1 ml containing 5 x 10 4 cells was seeded into 96 well plates for 30 min
SUPPLEMENTAL METHODS Measurement of peritoneal macrophage apoptosis by Celigo plate imaging cytometer For Celigo experiments, 0.1 ml containing 5 x 10 4 cells was seeded into 96 well plates for 30 min
Supplementary Figure 1
 Supplementary Figure 1 CITE-seq library preparation. (a) Illustration of the DNA-barcoded antibodies used in CITE-seq. (b) Antibody-oligonucleotide complexes appear as a high-molecular-weight smear when
Supplementary Figure 1 CITE-seq library preparation. (a) Illustration of the DNA-barcoded antibodies used in CITE-seq. (b) Antibody-oligonucleotide complexes appear as a high-molecular-weight smear when
ImmunoTag Human HIV ( antibodies plus p24 antigen ELISA Kit
 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name ImmunoTag Human HIV (1+ 0 + 2 antibodies plus p24 antigen ELISA Kit A Complete ELISA kit
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name ImmunoTag Human HIV (1+ 0 + 2 antibodies plus p24 antigen ELISA Kit A Complete ELISA kit
Isolation of Cells from Human Intestinal Tissue Heli Uronen-Hansson, Emma Persson, Petra Nilsson and William Agace *
 Isolation of Cells from Human Intestinal Tissue Heli Uronen-Hansson, Emma Persson, Petra Nilsson and William Agace * Immunology Section, Lund University, Lund, Sweden *For correspondence: william.agace@med.lu.se
Isolation of Cells from Human Intestinal Tissue Heli Uronen-Hansson, Emma Persson, Petra Nilsson and William Agace * Immunology Section, Lund University, Lund, Sweden *For correspondence: william.agace@med.lu.se
Efforts to Improve Data Transparency at the JBC
 Efforts to Improve Data Transparency at the JBC Roger J. Colbran Associate Editor Credit: Amanda Fosang, Associate Editor 2 Why Improve Transparency? Irreproducibility of pre-clinical studies Negative
Efforts to Improve Data Transparency at the JBC Roger J. Colbran Associate Editor Credit: Amanda Fosang, Associate Editor 2 Why Improve Transparency? Irreproducibility of pre-clinical studies Negative
Supporting Information
 Supporting Information Table S1. Overview of samples used for sequencing, and the number of sequences obtained from each sample. Visit 1 is day 0, Visit 2 is day 7, Visit 3 is day 28, and Visit 4 is day
Supporting Information Table S1. Overview of samples used for sequencing, and the number of sequences obtained from each sample. Visit 1 is day 0, Visit 2 is day 7, Visit 3 is day 28, and Visit 4 is day
HIC STANDARD OPERATING PROCEDURE: Immunofluorescence Staining for Multichromatic FACS Analysis with Human Single Cell Suspensions
 Date: 15-December-2005 Authors: Sally A. Quataert and Matthew Cochran Approved: Sally A. Quataert Purpose and Scope: This procedure describes a method for multichromatic staining of human single cell suspensions
Date: 15-December-2005 Authors: Sally A. Quataert and Matthew Cochran Approved: Sally A. Quataert Purpose and Scope: This procedure describes a method for multichromatic staining of human single cell suspensions
Data Sheet ICOS/NFAT Reporter-Jurkat Recombinant Cell Line Catalog #: 79668
 Data Sheet ICOS/NFAT Reporter-Jurkat Recombinant Cell Line Catalog #: 79668 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
Data Sheet ICOS/NFAT Reporter-Jurkat Recombinant Cell Line Catalog #: 79668 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
