Reporting Checklist for Nature Neuroscience
|
|
|
- Andrew Franklin
- 5 years ago
- Views:
Transcription
1 Corresponding Author: Manuscript Number: Manuscript Type: Feltri NNA54268B Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 5 # Supplementary Tables: 1 # Supplementary Videos: 0 This checklist is used to ensure good reporting standards and to improve the reproducibility of published results. For more information, please read Reporting Life Sciences Research. Please note that in the event of publication, it is mandatory that authors include all relevant methodological and statistical information in the manuscript. Statistics reporting, by figure example example Please specify the following information for each panel reporting quantitative data, and where each item is reported (section, e.g. Results, & paragraph number). Each figure should ideally contain an exact sample size (n) for each experimental group/condition, where n is an exact number and not a range, a clear definition of how n is defined (for example x cells from x slices from x animals from x litters, collected over x days), a description of the statistical test used, the results of the tests, any descriptive statistics and clearly defined error bars if applicable. For any experiments using custom statistics, please indicate the test used and stats obtained for each experiment. Each figure should include a statement of how many times the experiment shown was replicated in the lab; the details of sample collection should be sufficiently clear so that the replicability of the experiment is obvious to the reader. For experiments reported in the text but not in the figures, please use the paragraph number instead of the figure number. Note: Mean and standard deviation are not appropriate on small samples, and plotting independent data points is usually more informative. When technical replicates are reported, error and significance measures reflect the experimental variability and not the variability of the biological process; it is misleading not to state this clearly. FIGURE NUMBER 1a results, TEST USED WHICH TEST? oneway ANOVA unpaired t test Results EXACT VALUE 9, 9, 10, 15 n DEFINED? mice from at least 3 litters/group 15 slices from 10 mice Methods para 8 Results DESCRIPTIVE STATS (AVERAGE, VARIANCE) REPORTED? Results P VALUE EXACT VALUE p = p = Results DEGREES OF FREEDOM & F/t/z/R/ETC VALUE VALUE F(3, 36) = 2.97 t(28) = Results 2e Fisher exact test 777, 531 cells p < 1
2 FIGURE NUMBER 3n 3o 4b 4d 5a 5b TEST USED WHICH TEST? unpaired t test, bonferroni corrected unpaired t test, bonferroni corrected One way unpaired t test Two ways One way EXACT VALUE n DEFINED? 3, 3, 2 mice 3, 3, 3, 3, 3, 2 mice 6, 4, 3 mice 3, 3 mice 3 independent experiments 3, 3, 3 mice DESCRIPTIVE STATS (AVERAGE, VARIANCE) REPORTED? mean / SD P VALUE EXACT VALUE p(tazckoyapc Het) < p(tazckoyapc KO) < p(tazcko) = 0.013; p(yapcko) = ; p(tazckoyapc Het) = 0.011; p(tazchetyap cko) = p(tazcko) = p(double mutant) = p(tunel) = 0.21 p(ph3) = ANOVA p < p(dag1) < ; < p(itga6) < ; < p(dag1) = p(itga6) = ; = DEGREES OF FREEDOM & F/t/z/R/ETC VALUE VALUE t=15.08 df=4, t=26.55 df=3 t=6.261 df=4; t=13.10 df=4; t=6.572 df=4; t=7.545 df=4 F (2, 10) = t=1.461 df=4 t=4.341 df=4 F (2, 12) = F (2, 6) = F (2, 6) =
3 5c 5d S1b S1b S2b Two ways Two ways One way unpaired t test Two ways 3 5, 5, 4; 4, 4, 4; 3, 3, 3 6, 3, 4, 4, 5, 3 independent experiments mice mice 3,3 mice 3, 3, 3 independent experiments mean / SD mean / SD ANOVA p < Sox10: p(itga6) < ; = p(dag1) = ; = Tead1: p(itga6) < p(dag1) < p(dag1) = p(itga6) = ; < p(itgb4) = 0.011; = TUNEL ANOVA p = p(tazcko;yap chet) = Nuclei/area ANOVA p < p(tazcko) = ; p(tazcko;yap chet) = ; p(tazcko;yap cko) = p = 0.05 ANOVA p < 2uM: p(erbb2) = ; p(cdc42) = ; p(egr2) = ; p(sox10) = uM: p(erbb2) < ; p(cdc42) < ; p(egr2) < ; p(sox10) < F (2, 30) = F (2, 32) = F (5, 15) = F (5, 18) = t=3.694 df=4 F (2, 24) =
4 S2c S3b One way Two ways 3, 3, 3 mice 3, 3, 3 Representative figures independent experiments 1. Are any representative images shown (including Western blots and immunohistochemistry/staining) in the paper? If so, what figure(s)? 2. For each representative image, is there a clear statement of how many times this experiment was successfully repeated and a discussion of any limitations in repeatability? If so, where is this reported (section, paragraph #)? Statistics and general methods 1. Is there a justification of the sample size? If so, how was it justified? Even if no sample size calculation was performed, authors should report why the sample size is adequate to measure their effect size. 2. Are statistical tests justified as appropriate for every figure? mean / SD Figure 1a, c, Figure 2ae Figure 3ad, fm, Figure 4a, c, Figure 5di, Figure 7ac, Figure S1a, Figure S2d, p(erbb2) = p(cdc42) = ANOVA p < Sox10: p(20kb) < ; p(7.6kb) = ; p(30bp) < Tead1: p(20kb) < F (2, 6) = F (2, 6) = F (2, 42) = Each representative figure is one of the figures that used for statistical analysis, the number of repeating is the n described in individual figure s. We chose the sample size based on literatures in the field. Details are indicated in the Material and method, section "Statistical analyses". The statistics were used based on the properties of the data points, and described in individual figure s. a. If there is a section summarizing the statistical methods in the methods, is the statistical test for each experiment clearly defined? Yes, each statistical test is defined in each figure s. 4
5 b. Do the data meet the assumptions of the specific statistical test you chose (e.g. normality for a parametric test)? Where is this described (section, paragraph #)? c. Is there any estimate of variance within each group of data? Is the variance similar between groups that are being statistically compared? Where is this described (section, paragraph #)? d. Are tests specified as one or twosided? Two sided e. Are there adjustments for multiple comparisons? Yes,. 3. Are criteria for excluding data points reported? Was this criterion established prior to data collection? Where is this described (section, paragraph #)? 4. Define the method of randomization used to assign subjects (or samples) to the experimental groups and to collect and process data. If no randomization was used, state so. Where does this appear (section, paragraph #)? 5. Is a statement of the extent to which investigator knew the group allocation during the experiment and in assessing outcome included? If no blinding was done, state so. 6. For experiments in live vertebrates, is a statement of compliance with ethical guidelines/regulations included? 7. Is the species of the animals used reported? 8. Is the strain of the animals (including background strains of KO/ transgenic animals used) reported? 9. Is the sex of the animals/subjects used reported? Based on previous literature, we assumed the data points have a normal distribution. Yes. Either standard deviation or standard error of mean were plotted in each graph to analyze our data. No data points were excluded "Animal models and morphology", "Immunochemistry" and "TUNEL and proliferation assays". "Animal models and morphology". "Animal models and morphology". "Animal models and morphology". "Animal models and morphology". 10. Is the age of the animals/subjects reported? Yes, in each figure s. 5
6 11. For animals housed in a vivarium, is the light/dark cycle reported? 12. For animals housed in a vivarium, is the housing group (i.e. number of animals per cage) reported? 13. For behavioral experiments, is the time of day reported (e.g. light or dark cycle)? 14. Is the previous history of the animals/subjects (e.g. prior drug administration, surgery, behavioral testing) reported? a. If multiple behavioral tests were conducted in the same group of animals, is this reported? 15. If any animals/subjects were excluded from analysis, is this reported? Reagents a. How were the criteria for exclusion defined? Where is this described (section, paragraph #)? b. Specify reasons for any discrepancy between the number of animals at the beginning and end of the study. Where is this described (section, paragraph #)? 1. Have antibodies been validated for use in the system under study (assay and species)? a. Is antibody catalog number given? Where does this appear (section, paragraph #)? b. Where were the validation data reported (citation, supplementary information, Antibodypedia)? Yes "Western blot" and "Immunohistochemistry". Reported from the companies that provide the antibodies Where does this appear (section, paragraph #)? 6
7 2. Cell line identity a. Are any cell lines used in this paper listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample? b. If yes, include in the Methods section a scientific justification of their useindicate here in which section and paragraph the justification can be found. c. For each cell line, include in the Methods section a statement that specifies: the source of the cell lines have the cell lines been authenticated? If so, by which method? have the cell lines been tested for mycoplasma contamination? Data deposition Data deposition in a public repository is mandatory for: a. Protein, DNA and RNA sequences b. Macromolecular structures c. Crystallographic data for small molecules d. Microarray data "Cell culture" and "Verteprofin treatment" "Cell culture" "Cell culture" and "Verteprofin treatment" Deposition is strongly recommended for many other datasets for which structured public repositories exist; more details on our data policy are available here. We encourage the provision of other source data in supplementary information or in unstructured repositories such as Figshare and Dryad. We encourage publication of Data Descriptors (see Scientific Data) to maximize data reuse. 1. Are accession codes for deposit dates provided? Computer code/software Yes, details will be indicated in the Material and method, section "RNA seq analysis" NCBI GEO :GSE pvalue and FDR of gene enrichment (Supplementary dataset S1) and GO category enrichment ( 6b) were calculated using ROAST Any custom algorithm/software that is central to the methods must be supplied by the authors in a usable and readable form for readers at the time of publication. However, referees may ask for this information at any time during the review process. 1. Identify all custom software or scripts that were required to conduct the study and where in the procedures each was used. 7
8 2. If computer code was used to generate results that are central to the paper's conclusions, include a statement in the Methods section under "Code availability" to indicate whether and how the code can be accessed. Include version information as necessary and any restrictions on availability. Human subjects 1. Which IRB approved the protocol? Where is this stated (section, paragraph #)? 2. Is demographic information on all subjects provided? 3. Is the number of human subjects, their age and sex clearly defined? 4. Are the inclusion and exclusion criteria (if any) clearly specified? 5. How well were the groups matched? Where is this information described (section, paragraph #)? 6. Is a statement included confirming that informed consent was obtained from all subjects? 7. For publication of patient photos, is a statement included confirming that consent to publish was obtained? fmri studies For papers reporting functional imaging (fmri) results please ensure that these minimal reporting guidelines are met and that all this information is clearly provided in the methods: 1. Were any subjects scanned but then rejected for the analysis after the data was collected? a. If yes, is the number rejected and reasons for rejection described? 8
9 2. Is the number of blocks, trials or experimental units per session and/ or subjects specified? 3. Is the length of each trial and interval between trials specified? 4. Is a blocked, eventrelated, or mixed design being used? If applicable, please specify the block length or how the eventrelated or mixed design was optimized. 5. Is the task design clearly described? 6. How was behavioral performance measured? 7. Is an ANOVA or factorial design being used? 8. For data acquisition, is a whole brain scan used? If not, state area of acquisition. a. How was this region determined? 9. Is the field strength (in Tesla) of the MRI system stated? a. Is the pulse sequence type (gradient/spin echo, EPI/spiral) stated? b. Are the fieldofview, matrix size, slice thickness, and TE/TR/ flip angle clearly stated? 10. Are the software and specific parameters (model/functions, smoothing kernel size if applicable, etc.) used for data processing and preprocessing clearly stated? 11. Is the coordinate space for the anatomical/functional imaging data clearly defined as subject/native space or standardized stereotaxic space, e.g., original Talairach, MNI305, ICBM152, etc? Where (section, paragraph #)? 12. If there was data normalization/standardization to a specific space template, are the type of transformation (linear vs. nonlinear) used and image types being transformed clearly described? Where (section, paragraph #)? 13. How were anatomical locations determined, e.g., via an automated labeling algorithm (AAL), standardized coordinate database (Talairach daemon), probabilistic atlases, etc.? 9
10 14. Were any additional regressors (behavioral covariates, motion etc) used? 15. Is the contrast construction clearly defined? 16. Is a mixed/random effects or fixed inference used? a. If fixed effects inference used, is this justified? 17. Were repeated measures used (multiple measurements per subject)? a. If so, are the method to account for within subject correlation and the assumptions made about variance clearly stated? 18. If the threshold used for inference and visualization in figures varies, is this clearly stated? 19. Are statistical inferences corrected for multiple comparisons? a. If not, is this labeled as uncorrected? 20. Are the results based on an ROI (region of interest) analysis? a. If so, is the rationale clearly described? b. How were the ROI s defined (functional vs anatomical localization)? 21. Is there correction for multiple comparisons within each voxel? 22. For clusterwise significance, is the clusterdefining threshold and the corrected significance level defined? Additional comments Additional Comments 10
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Leonard Petrucelli NNA52530C Article Reporting Checklist for Nature Neuroscience # Main s: 8 # Supplementary s: 10 # Supplementary Tables: 3 #
Corresponding Author: Manuscript Number: Manuscript Type: Leonard Petrucelli NNA52530C Article Reporting Checklist for Nature Neuroscience # Main s: 8 # Supplementary s: 10 # Supplementary Tables: 3 #
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: McColl NNRS50586AZ Resource Reporting Checklist for Nature Neuroscience # Main Figures: 8 # lementary Figures: 7 # lementary Tables: 13 # lementary
Corresponding Author: Manuscript Number: Manuscript Type: McColl NNRS50586AZ Resource Reporting Checklist for Nature Neuroscience # Main Figures: 8 # lementary Figures: 7 # lementary Tables: 13 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Michael Cole NNA51 Article Reporting Checklist for Nature Neuroscience # Main Figures: 5 # lementary Figures: 4 # lementary Tables: 0 # lementary
Corresponding Author: Manuscript Number: Manuscript Type: Michael Cole NNA51 Article Reporting Checklist for Nature Neuroscience # Main Figures: 5 # lementary Figures: 4 # lementary Tables: 0 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: xiangdong fu, yuanchao xue NNA52804A Article Reporting Checklist Nature Neuroscience # Main Figures: 5 # Supplementary Figures: 7 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: xiangdong fu, yuanchao xue NNA52804A Article Reporting Checklist Nature Neuroscience # Main Figures: 5 # Supplementary Figures: 7 # Supplementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Yang Dan NNA56598 Article Reporting Checklist for Nature Neuroscience # Main Figures: 8 # Supplementary Figures: 12 # Supplementary Tables: 0 #
Corresponding Author: Manuscript Number: Manuscript Type: Yang Dan NNA56598 Article Reporting Checklist for Nature Neuroscience # Main Figures: 8 # Supplementary Figures: 12 # Supplementary Tables: 0 #
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Liu NNA52890T Article Reporting Checklist for Nature Neuroscience # Main Figures: 4 # Supplementary Figures: 3 # Supplementary Tables: 1 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Liu NNA52890T Article Reporting Checklist for Nature Neuroscience # Main Figures: 4 # Supplementary Figures: 3 # Supplementary Tables: 1 # Supplementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Chay T. Kuo NNA47008 Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 11 # Supplementary Tables:
Corresponding Author: Manuscript Number: Manuscript Type: Chay T. Kuo NNA47008 Article Reporting Checklist for Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 11 # Supplementary Tables:
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Robert Froemke NNA7412T Article Reporting Checklist for Nature Neuroscience # Main s: 7 # Supplementary s: 14 # Supplementary Tables: 0 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Robert Froemke NNA7412T Article Reporting Checklist for Nature Neuroscience # Main s: 7 # Supplementary s: 14 # Supplementary Tables: 0 # Supplementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Lennart Mucke NNRS49607AZ Resource Reporting Checklist for Nature Neuroscience # Main s: # lementary s: 4 # lementary Tables: 9 # lementary Videos:
Corresponding Author: Manuscript Number: Manuscript Type: Lennart Mucke NNRS49607AZ Resource Reporting Checklist for Nature Neuroscience # Main s: # lementary s: 4 # lementary Tables: 9 # lementary Videos:
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: David Dupret NNBC5329A Brief Communication Reporting Checklist for Nature Neuroscience # s: 3 # s: 11 # Tables: 0 # Videos: 0 This checklist is
Corresponding Author: Manuscript Number: Manuscript Type: David Dupret NNBC5329A Brief Communication Reporting Checklist for Nature Neuroscience # s: 3 # s: 11 # Tables: 0 # Videos: 0 This checklist is
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Rachel Wilson NNA48246B Article Reporting Checklist for Nature Neuroscience # Main Figures: 8 # Supplementary Figures: 7 # Supplementary Tables:
Corresponding Author: Manuscript Number: Manuscript Type: Rachel Wilson NNA48246B Article Reporting Checklist for Nature Neuroscience # Main Figures: 8 # Supplementary Figures: 7 # Supplementary Tables:
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: RuRong Ji NNA59063A Article Reporting Checklist for Nature Neuroscience # Main ures: 8 # Supplementary ures: 15 # Supplementary Tables: 0 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: RuRong Ji NNA59063A Article Reporting Checklist for Nature Neuroscience # Main ures: 8 # Supplementary ures: 15 # Supplementary Tables: 0 # Supplementary
Reporting Checklist for Nature Neuroscience
 Corponding Author: Manuscript Number: Manuscript Type: Gregory C. DeAngelis NNA49009A Article Reporting Checklist for Nature Neuroscience # Main Figu: 7 # lementary Figu: # lementary Tables: 0 # lementary
Corponding Author: Manuscript Number: Manuscript Type: Gregory C. DeAngelis NNA49009A Article Reporting Checklist for Nature Neuroscience # Main Figu: 7 # lementary Figu: # lementary Tables: 0 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Hillel Adesnik NNA58169T Article Reporting Checklist for Nature Neuroscience # Main ures: 5 # lementary ures: 6 # lementary Tables: 1 # lementary
Corresponding Author: Manuscript Number: Manuscript Type: Hillel Adesnik NNA58169T Article Reporting Checklist for Nature Neuroscience # Main ures: 5 # lementary ures: 6 # lementary Tables: 1 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Marcelo Coba Manuscript Number: NNRS3314D Manuscript Type: Resource Reporting Checklist for Nature Neuroscience # Main Figures: 7 # lementary Figures: 4 # lementary Tables: 11 # lementary
Corresponding Author: Marcelo Coba Manuscript Number: NNRS3314D Manuscript Type: Resource Reporting Checklist for Nature Neuroscience # Main Figures: 7 # lementary Figures: 4 # lementary Tables: 11 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Ofer Yizhar, PhD NNA54616C Article Reporting Checklist for Nature Neuroscience # Main s: 5 # lementary s: 10 # lementary Tables: 0 # lementary
Corresponding Author: Manuscript Number: Manuscript Type: Ofer Yizhar, PhD NNA54616C Article Reporting Checklist for Nature Neuroscience # Main s: 5 # lementary s: 10 # lementary Tables: 0 # lementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Atsushi Miyawaki NNT51493A Technical Report Reporting Checklist Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 12 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Atsushi Miyawaki NNT51493A Technical Report Reporting Checklist Nature Neuroscience # Main Figures: 7 # Supplementary Figures: 12 # Supplementary
Reporting Checklist for Nature Neuroscience
 Corresponding Author: Manuscript Number: Manuscript Type: Benjamin R. Rost NNT51693 Technical Report Reporting Checklt for Nature Neuroscience # Main Figures: 6 # Supplementary Figures: 9 # Supplementary
Corresponding Author: Manuscript Number: Manuscript Type: Benjamin R. Rost NNT51693 Technical Report Reporting Checklt for Nature Neuroscience # Main Figures: 6 # Supplementary Figures: 9 # Supplementary
Reporting Checklist for Nature Neuroscience
 Correspondg Author: Manuscript Number: Manuscript Type: Spencer Smith NNA48000A Article Reportg Checklist for Nature Neuroscience # Ma Figures: 6 # Supplementary Figures: 8 # Supplementary Tables: 1 #
Correspondg Author: Manuscript Number: Manuscript Type: Spencer Smith NNA48000A Article Reportg Checklist for Nature Neuroscience # Ma Figures: 6 # Supplementary Figures: 8 # Supplementary Tables: 1 #
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Adams Waldorf, Rajagopal, Gale Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Life Sciences Reporting Summary Adams Waldorf, Rajagopal, Gale Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Our web collection on statistics for biologists may be useful.
 Reporting Summary Corresponding author(s): FELIPE CAVA Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency and transparency in
Reporting Summary Corresponding author(s): FELIPE CAVA Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency and transparency in
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Rich, Jeremy N. Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Rich, Jeremy N. Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Jeffrey W. Elias, PhD UCD SOM Office of Research Grants Facilitation.
 NIH IS CHANGING/ENHANCING THE REVIEW CRITERIA AND FOCUSING ON MISSING OPPORTUNITIES TO EXAMINE SEX DIFFERENCES JOURNALS ARE CHANGING/ENHANCING THE REVIEW CRITERIA Jeffrey W. Elias, PhD UCD SOM Office of
NIH IS CHANGING/ENHANCING THE REVIEW CRITERIA AND FOCUSING ON MISSING OPPORTUNITIES TO EXAMINE SEX DIFFERENCES JOURNALS ARE CHANGING/ENHANCING THE REVIEW CRITERIA Jeffrey W. Elias, PhD UCD SOM Office of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb3575 In the format provided by the authors and unedited. Supplementary Figure 1 Validation of key reagents and assays a, top, IHC with antibody recognizing specifically
Efforts to Improve Data Transparency at the JBC
 Efforts to Improve Data Transparency at the JBC Roger J. Colbran Associate Editor Credit: Amanda Fosang, Associate Editor 2 Why Improve Transparency? Irreproducibility of pre-clinical studies Negative
Efforts to Improve Data Transparency at the JBC Roger J. Colbran Associate Editor Credit: Amanda Fosang, Associate Editor 2 Why Improve Transparency? Irreproducibility of pre-clinical studies Negative
Our web collection on statistics for biologists may be useful.
 Reporting Summary Corresponding author(s): Qi-jing Li, Lilin Ye and Bo Zhu Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Summary Corresponding author(s): Qi-jing Li, Lilin Ye and Bo Zhu Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Summary. Statistical parameters. Software and code. nature research reporting summary April Corresponding author(s):
 Reporting Summary Corresponding author(s): Alexander Flügel 2018-07-09758C Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Reporting Summary Corresponding author(s): Alexander Flügel 2018-07-09758C Nature Research wishes to improve the reproducibility of the work that we publish. This form provides structure for consistency
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Stein Aerts Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Stein Aerts Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines
 Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
Revision Checklist for Science Signaling Research Manuscripts: Data Requirements and Style Guidelines Further information can be found at: http://stke.sciencemag.org/sites/default/files/researcharticlerevmsinstructions_0.pdf.
PROMOTING TRANSPARENT RESEARCH METHODS, PROTOCOLS AND DATA TO REDUCE IRREPRODUCIBILITY
 PROMOTING TRANSPARENT RESEARCH METHODS, PROTOCOLS AND DATA TO REDUCE IRREPRODUCIBILITY December 2015 STM Innovations Seminar Andrew L Hufton, PhD Managing Editor, Scientific Data Nature Publishing Group
PROMOTING TRANSPARENT RESEARCH METHODS, PROTOCOLS AND DATA TO REDUCE IRREPRODUCIBILITY December 2015 STM Innovations Seminar Andrew L Hufton, PhD Managing Editor, Scientific Data Nature Publishing Group
Reporting Checklist for Nature Neuroscience
 Coesponding Autho: Manuscipt Numbe: Manuscipt Type: Yuki Oka NNA5991A Aticle Repoting Checklist fo Natue Neuoscience # Main Figues: 7 # Supplementay Figues: # Supplementay Tables: 0 # Supplementay Videos:
Coesponding Autho: Manuscipt Numbe: Manuscipt Type: Yuki Oka NNA5991A Aticle Repoting Checklist fo Natue Neuoscience # Main Figues: 7 # Supplementay Figues: # Supplementay Tables: 0 # Supplementay Videos:
BIOEQUIVALENCE TRIAL INFORMATION
 PRESENTATION OF BIOEQUIVALENCE TRIAL INFORMATION BIOEQUIVALENCE TRIAL INFORMATION GENERAL INSTRUCTIONS: Please review all the instructions thoroughly and carefully prior to completing the Bioequivalence
PRESENTATION OF BIOEQUIVALENCE TRIAL INFORMATION BIOEQUIVALENCE TRIAL INFORMATION GENERAL INSTRUCTIONS: Please review all the instructions thoroughly and carefully prior to completing the Bioequivalence
Cerebrospinal fluid tau, neurogranin and neurofilament light in Alzheimerís disease
 Cerebrospinal fluid tau, neurogranin and neurofilament light in Alzheimerís disease Niklas Mattsson, Philip Insel, Sebastian Palmqvist, Erik Portelius, Henrik Zetterberg, Michael Weiner, Kaj Blennow, Oskar
Cerebrospinal fluid tau, neurogranin and neurofilament light in Alzheimerís disease Niklas Mattsson, Philip Insel, Sebastian Palmqvist, Erik Portelius, Henrik Zetterberg, Michael Weiner, Kaj Blennow, Oskar
BIOEQUIVALENCE TRIAL INFORMATION FORM (Medicines and Allied Substances Act [No. 3] of 2013 Part V Section 39)
![BIOEQUIVALENCE TRIAL INFORMATION FORM (Medicines and Allied Substances Act [No. 3] of 2013 Part V Section 39) BIOEQUIVALENCE TRIAL INFORMATION FORM (Medicines and Allied Substances Act [No. 3] of 2013 Part V Section 39)](/thumbs/90/102231627.jpg) ZAMRA BTIF BIOEQUIVALENCE TRIAL INFORMATION FORM (Medicines and Allied Substances Act [No. 3] of 2013 Part V Section 39) The Guidelines on Bioequivalence Studies to be consulted in completing this form.
ZAMRA BTIF BIOEQUIVALENCE TRIAL INFORMATION FORM (Medicines and Allied Substances Act [No. 3] of 2013 Part V Section 39) The Guidelines on Bioequivalence Studies to be consulted in completing this form.
Alternative TSSs are co-regulated in single cells in the mouse brain
 Alternative TSSs are co-regulated in single cells in the mouse brain Kasper Karlsson, Peter Lönnerberg, Sten Linnarsson Corresponding author: Sten Linnarsson, Karolinska Institutet Review timeline: Submission
Alternative TSSs are co-regulated in single cells in the mouse brain Kasper Karlsson, Peter Lönnerberg, Sten Linnarsson Corresponding author: Sten Linnarsson, Karolinska Institutet Review timeline: Submission
Medicines Control Authority Of Zimbabwe
 Medicines Control Authority Of Zimbabwe BIOEQUIVALENCE APPLICATION FORM Form: EVF03 This application form is designed to facilitate information exchange between the Applicant and the MCAZ for bioequivalence
Medicines Control Authority Of Zimbabwe BIOEQUIVALENCE APPLICATION FORM Form: EVF03 This application form is designed to facilitate information exchange between the Applicant and the MCAZ for bioequivalence
Real-time RT PCR. for identification of differentially expressed genes. (with Schizophrenia application) Rolf Sundberg, Stockholm Univ.
 Real-time RT PCR for identification of differentially expressed genes. (with Schizophrenia application) Rolf Sundberg, Stockholm Univ. Göteborg, May 2006 Real-time PCR for mrna quantitation Review paper
Real-time RT PCR for identification of differentially expressed genes. (with Schizophrenia application) Rolf Sundberg, Stockholm Univ. Göteborg, May 2006 Real-time PCR for mrna quantitation Review paper
iphemap: An atlas of Phenotype to genotype relationships of human ipsc models of neurological diseases
 iphemap: An atlas of Phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth, Jacob E. Vaughn, Josh C. Orack, Chelsea Skinner, Jamil Khouri, Sofia B. Lizarraga,
iphemap: An atlas of Phenotype to genotype relationships of human ipsc models of neurological diseases Ethan W. Hollingsworth, Jacob E. Vaughn, Josh C. Orack, Chelsea Skinner, Jamil Khouri, Sofia B. Lizarraga,
Microarray Gene Expression Analysis at CNIO
 Microarray Gene Expression Analysis at CNIO Orlando Domínguez Genomics Unit Biotechnology Program, CNIO 8 May 2013 Workflow, from samples to Gene Expression data Experimental design user/gu/ubio Samples
Microarray Gene Expression Analysis at CNIO Orlando Domínguez Genomics Unit Biotechnology Program, CNIO 8 May 2013 Workflow, from samples to Gene Expression data Experimental design user/gu/ubio Samples
Methods in Clinical Cancer Research Workshop Format for Protocol Concept Synopsis Sheet (blank) SYNOPSIS
 B Methods in Clinical Cancer Research Workshop Format for Protocol Concept Synopsis Sheet (blank) elow is the format to follow for the concept sheet for protocols. It is based on the International Committee
B Methods in Clinical Cancer Research Workshop Format for Protocol Concept Synopsis Sheet (blank) elow is the format to follow for the concept sheet for protocols. It is based on the International Committee
A normal genetic variation modulates synaptic MMP-9 protein levels and the severity of schizophrenia symptoms
 A normal genetic variation modulates synaptic MMP-9 protein levels and the severity of schizophrenia symptoms Katarzyna Lepeta, Katarzyna J. Purzycka, Katarzyna Pachulska-Wieczorek, Marina Mitjans, Martin
A normal genetic variation modulates synaptic MMP-9 protein levels and the severity of schizophrenia symptoms Katarzyna Lepeta, Katarzyna J. Purzycka, Katarzyna Pachulska-Wieczorek, Marina Mitjans, Martin
Lab 1: A review of linear models
 Lab 1: A review of linear models The purpose of this lab is to help you review basic statistical methods in linear models and understanding the implementation of these methods in R. In general, we need
Lab 1: A review of linear models The purpose of this lab is to help you review basic statistical methods in linear models and understanding the implementation of these methods in R. In general, we need
David M. Rocke Division of Biostatistics and Department of Biomedical Engineering University of California, Davis
 David M. Rocke Division of Biostatistics and Department of Biomedical Engineering University of California, Davis Outline RNA-Seq for differential expression analysis Statistical methods for RNA-Seq: Structure
David M. Rocke Division of Biostatistics and Department of Biomedical Engineering University of California, Davis Outline RNA-Seq for differential expression analysis Statistical methods for RNA-Seq: Structure
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): George Q. Daley Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): George Q. Daley Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Precision in Quantitative Imaging: Trial Development and Quality Assurance
 Precision in Quantitative Imaging: Trial Development and Quality Assurance Susanna I Lee MD, PhD Thanks to: Mitchell Schnall, Mark Rosen. Dan Sullivan, Patrick Bossuyt Imaging Chain: Patient Data Raw data
Precision in Quantitative Imaging: Trial Development and Quality Assurance Susanna I Lee MD, PhD Thanks to: Mitchell Schnall, Mark Rosen. Dan Sullivan, Patrick Bossuyt Imaging Chain: Patient Data Raw data
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Christer Betsholtz Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we
Life Sciences Reporting Summary Corresponding author(s): Christer Betsholtz Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we
Edinburgh Research Explorer
 Edinburgh Research Explorer Protocol for a retrospective, controlled cohort study of the impact of a change in Nature journals' editorial policy for life sciences research on the completeness of reporting
Edinburgh Research Explorer Protocol for a retrospective, controlled cohort study of the impact of a change in Nature journals' editorial policy for life sciences research on the completeness of reporting
Archives of Scientific Psychology Reporting Questionnaire for Manuscripts Describing Primary Data Collections
 (Based on APA Journal Article Reporting Standards JARS Questionnaire) 1 Archives of Scientific Psychology Reporting Questionnaire for Manuscripts Describing Primary Data Collections JARS: ALL: These questions
(Based on APA Journal Article Reporting Standards JARS Questionnaire) 1 Archives of Scientific Psychology Reporting Questionnaire for Manuscripts Describing Primary Data Collections JARS: ALL: These questions
Important Information for MCP Authors
 Guidelines to Authors for Publication of Manuscripts Describing Development and Application of Targeted Mass Spectrometry Measurements of Peptides and Proteins and Submission Checklist The following Guidelines
Guidelines to Authors for Publication of Manuscripts Describing Development and Application of Targeted Mass Spectrometry Measurements of Peptides and Proteins and Submission Checklist The following Guidelines
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Integrative Genomics 1a. Introduction
 2016 Course Outline Integrative Genomics 1a. Introduction ggibson.gt@gmail.com http://www.cig.gatech.edu 1a. Experimental Design and Hypothesis Testing (GG) 1b. Normalization (GG) 2a. RNASeq (MI) 2b. Clustering
2016 Course Outline Integrative Genomics 1a. Introduction ggibson.gt@gmail.com http://www.cig.gatech.edu 1a. Experimental Design and Hypothesis Testing (GG) 1b. Normalization (GG) 2a. RNASeq (MI) 2b. Clustering
Exploration and Analysis of DNA Microarray Data
 Exploration and Analysis of DNA Microarray Data Dhammika Amaratunga Senior Research Fellow in Nonclinical Biostatistics Johnson & Johnson Pharmaceutical Research & Development Javier Cabrera Associate
Exploration and Analysis of DNA Microarray Data Dhammika Amaratunga Senior Research Fellow in Nonclinical Biostatistics Johnson & Johnson Pharmaceutical Research & Development Javier Cabrera Associate
Aims and challenges of clinical trial data sharing
 Aims and challenges of clinical trial data sharing Facilitating data access to non-industry funded research UCL and Yale meeting: 9 October 2015 London Dr Trish Groves Head of research,
Aims and challenges of clinical trial data sharing Facilitating data access to non-industry funded research UCL and Yale meeting: 9 October 2015 London Dr Trish Groves Head of research,
Gene Expression Data Analysis
 Gene Expression Data Analysis Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu BMIF 310, Fall 2009 Gene expression technologies (summary) Hybridization-based
Gene Expression Data Analysis Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu BMIF 310, Fall 2009 Gene expression technologies (summary) Hybridization-based
Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate
 Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
Supplementary Figure Legends Supplementary Fig. 1 related to Fig. 1 Clinical relevance of lncrna candidate BC041951 in gastric cancer. (A) The flow chart for selected candidate lncrnas in 660 up-regulated
New NIH 2-page Rigor and Transparency Document
 New NIH 2-page Rigor and Transparency Document Grants affected in 2016 include: NOT-OD-16-081 NOT-OD-16-058 NOT-OD-16-034 NOT-OD-16-011 NOT-OD-16-031 NOT-OD-16-012 NOT-OD-16-005 NOT-OD-16-004 NOT-OD-15-103
New NIH 2-page Rigor and Transparency Document Grants affected in 2016 include: NOT-OD-16-081 NOT-OD-16-058 NOT-OD-16-034 NOT-OD-16-011 NOT-OD-16-031 NOT-OD-16-012 NOT-OD-16-005 NOT-OD-16-004 NOT-OD-15-103
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding Author: Date: Wanjun Chen & Keji Zhao Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life science
Life Sciences Reporting Summary Corresponding Author: Date: Wanjun Chen & Keji Zhao Nature Research wishes to improve the reproducibility of the work we publish. This form is published with all life science
Designing a Complex-Omics Experiments. Xiangqin Cui. Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham
 Designing a Complex-Omics Experiments Xiangqin Cui Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham 1/7/2015 Some slides are from previous lectures of Grier
Designing a Complex-Omics Experiments Xiangqin Cui Section on Statistical Genetics Department of Biostatistics University of Alabama at Birmingham 1/7/2015 Some slides are from previous lectures of Grier
BIOSTATISTICS AND MEDICAL INFORMATICS (B M I)
 Biostatistics and Medical Informatics (B M I) 1 BIOSTATISTICS AND MEDICAL INFORMATICS (B M I) B M I/POP HLTH 451 INTRODUCTION TO SAS PROGRAMMING FOR 2 credits. Use of the SAS programming language for the
Biostatistics and Medical Informatics (B M I) 1 BIOSTATISTICS AND MEDICAL INFORMATICS (B M I) B M I/POP HLTH 451 INTRODUCTION TO SAS PROGRAMMING FOR 2 credits. Use of the SAS programming language for the
GeneQuery: A phenotype search tool based on gene co-expression clustering. Alexander Predeus 21-oct-2015
 GeneQuery: A phenotype search tool based on gene co-expression clustering Alexander Predeus 21-oct-2015 About myself Graduated from Moscow state University (1998-2003) PhD: Michigan State University (2003-2009):
GeneQuery: A phenotype search tool based on gene co-expression clustering Alexander Predeus 21-oct-2015 About myself Graduated from Moscow state University (1998-2003) PhD: Michigan State University (2003-2009):
Gene expression: Microarray data analysis. Copyright notice. Outline: microarray data analysis. Schedule
 Gene expression: Microarray data analysis Copyright notice Many of the images in this powerpoint presentation are from Bioinformatics and Functional Genomics by Jonathan Pevsner (ISBN -47-4-8). Copyright
Gene expression: Microarray data analysis Copyright notice Many of the images in this powerpoint presentation are from Bioinformatics and Functional Genomics by Jonathan Pevsner (ISBN -47-4-8). Copyright
QPCR ASSAYS FOR MIRNA EXPRESSION PROFILING
 TECH NOTE 4320 Forest Park Ave Suite 303 Saint Louis, MO 63108 +1 (314) 833-9764 mirna qpcr ASSAYS - powered by NAWGEN Our mirna qpcr Assays were developed by mirna experts at Nawgen to improve upon previously
TECH NOTE 4320 Forest Park Ave Suite 303 Saint Louis, MO 63108 +1 (314) 833-9764 mirna qpcr ASSAYS - powered by NAWGEN Our mirna qpcr Assays were developed by mirna experts at Nawgen to improve upon previously
JGK TRAINING PROGRAMME MODULE 2: METHOD VALIDATION AND MEASUREMENT OF UNCERTAINTY IN VETERINARY LABORATORIES PRACTICAL COURSE
 Registered as JKG Lab AfriQA (Pty) Ltd Reg. No. 2012/154570/07 P.O Box 1581 Potchefstroom 2520 3 Sylvia Street `` Potchefstroom 2531 Tel: +27 18 294 4528 Fax: +27 86 650 1716 Cell: +27 82 562 1858 Email
Registered as JKG Lab AfriQA (Pty) Ltd Reg. No. 2012/154570/07 P.O Box 1581 Potchefstroom 2520 3 Sylvia Street `` Potchefstroom 2531 Tel: +27 18 294 4528 Fax: +27 86 650 1716 Cell: +27 82 562 1858 Email
CHAPTER 17 Brain Image Analysis and Atlas Construction
 CHAPTER 17 Brain Image Analysis and Atlas Construction Paul M. Thompson Michael S. Mega Katherine L. Narr Elizabeth R. Sowell Rebecca E. Blanton Arthur W. Toga Contents 17.1 Challenges in brain image analysis
CHAPTER 17 Brain Image Analysis and Atlas Construction Paul M. Thompson Michael S. Mega Katherine L. Narr Elizabeth R. Sowell Rebecca E. Blanton Arthur W. Toga Contents 17.1 Challenges in brain image analysis
Web-based tools for Bioinformatics; A (free) introduction to (freely available) NCBI, MUSC and World-wide.
 Page 1 of 24 Web-based tools for Bioinformatics; A (free) introduction to (freely available) NCBI, MUSC and World-wide. When and Where---Wednesdays at 1pm-2pmRoom 438 Library Admin Building Beginning September
Page 1 of 24 Web-based tools for Bioinformatics; A (free) introduction to (freely available) NCBI, MUSC and World-wide. When and Where---Wednesdays at 1pm-2pmRoom 438 Library Admin Building Beginning September
Animal Research Ethics Committee. Guidelines for Submitting Protocols for ethical approval of research or teaching involving live animals
 Animal Research Ethics Committee Guidelines for Submitting Protocols for ethical approval of research or teaching involving live animals AREC Document No: 3 Revised Guidelines for Submitting Protocols
Animal Research Ethics Committee Guidelines for Submitting Protocols for ethical approval of research or teaching involving live animals AREC Document No: 3 Revised Guidelines for Submitting Protocols
Figure S1. TT and MZ-CRC-1 cell viability and proliferation after Palbociclib treatment. Cell viability assays (A) and cell counting assays (B) were
 Figure S1. TT and MZ-CRC-1 cell viability and proliferation after Palbociclib treatment. Cell viability assays (A) and cell counting assays (B) were performed with TT and MZ-CRC-1 cells treated with Palbociclib
Figure S1. TT and MZ-CRC-1 cell viability and proliferation after Palbociclib treatment. Cell viability assays (A) and cell counting assays (B) were performed with TT and MZ-CRC-1 cells treated with Palbociclib
Practical What & Where of Rigor and Reproducibility in the NIH Application. SIU School of Medicine Office of Grants & Contracts with Sophia Ran, PhD
 Practical What & Where of Rigor and Reproducibility in the NIH Application SIU School of Medicine Office of Grants & Contracts with Sophia Ran, PhD Recent Training Sessions NIH Regional Seminar Rigor and
Practical What & Where of Rigor and Reproducibility in the NIH Application SIU School of Medicine Office of Grants & Contracts with Sophia Ran, PhD Recent Training Sessions NIH Regional Seminar Rigor and
Views of a Clinical Study Report
 Out-of-(CSR)-Body Experiences Tips on Assembling Appendices, Datasets, and CRFs Susan C Sisk, PhD, RAC 1 Views of a Clinical Study Report OR Photos courtesy of Leigh Vaughan and RAPS, 2008 2 Topics Process
Out-of-(CSR)-Body Experiences Tips on Assembling Appendices, Datasets, and CRFs Susan C Sisk, PhD, RAC 1 Views of a Clinical Study Report OR Photos courtesy of Leigh Vaughan and RAPS, 2008 2 Topics Process
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Vinod Balachandran Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we
Life Sciences Reporting Summary Corresponding author(s): Vinod Balachandran Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we
High-density SNP Genotyping Analysis of Broiler Breeding Lines
 Animal Industry Report AS 653 ASL R2219 2007 High-density SNP Genotyping Analysis of Broiler Breeding Lines Abebe T. Hassen Jack C.M. Dekkers Susan J. Lamont Rohan L. Fernando Santiago Avendano Aviagen
Animal Industry Report AS 653 ASL R2219 2007 High-density SNP Genotyping Analysis of Broiler Breeding Lines Abebe T. Hassen Jack C.M. Dekkers Susan J. Lamont Rohan L. Fernando Santiago Avendano Aviagen
ENCODE RBP Antibody Characterization Guidelines
 ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
ENCODE RBP Antibody Characterization Guidelines Approved on November 18, 2016 Background An integral part of the ENCODE Project is to characterize the antibodies used in the experiments. This document
Supplementary Data 1.
 Supplementary Data 1. Evaluation of the effects of number of F2 progeny to be bulked (n) and average sequencing coverage (depth) of the genome (G) on the levels of false positive SNPs (SNP index = 1).
Supplementary Data 1. Evaluation of the effects of number of F2 progeny to be bulked (n) and average sequencing coverage (depth) of the genome (G) on the levels of false positive SNPs (SNP index = 1).
Analysis of Microarray Data
 Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review
Analysis of Microarray Data Lecture 3: Visualization and Functional Analysis George Bell, Ph.D. Senior Bioinformatics Scientist Bioinformatics and Research Computing Whitehead Institute Outline Review
Identification of biological themes in microarray data from a mouse heart development time series using GeneSifter
 Identification of biological themes in microarray data from a mouse heart development time series using GeneSifter VizX Labs, LLC Seattle, WA 98119 Abstract Oligonucleotide microarrays were used to study
Identification of biological themes in microarray data from a mouse heart development time series using GeneSifter VizX Labs, LLC Seattle, WA 98119 Abstract Oligonucleotide microarrays were used to study
Experimental Design and Statistical Methods. Workshop GENERAL OVERVIEW. Jesús Piedrafita Arilla.
 Experimental Design and Statistical Methods Workshop GENERAL OVERVIEW Jesús Piedrafita Arilla jesus.piedrafita@uab.cat Departament de Ciència Animal i dels Aliments TWO TYPES OF STUDIES EXPERIMENT: it
Experimental Design and Statistical Methods Workshop GENERAL OVERVIEW Jesús Piedrafita Arilla jesus.piedrafita@uab.cat Departament de Ciència Animal i dels Aliments TWO TYPES OF STUDIES EXPERIMENT: it
Supplementary Material Cohort Description
 Supplementary Material Cohort Description The following longitudinal twin cohorts were included: BrainSCALE: Brain Structure and Cognition: an Adolescent Longitudinal Twin Study into genetic etiology University
Supplementary Material Cohort Description The following longitudinal twin cohorts were included: BrainSCALE: Brain Structure and Cognition: an Adolescent Longitudinal Twin Study into genetic etiology University
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Goel Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Life Sciences Reporting Summary Corresponding author(s): Goel Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish. This
Supplementary Figure 1. Serial deletion mutants of BLITz.
 Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Supplementary Figure 1 Serial deletion mutants of BLITz. (a) Design of TEVseq insertion into J -helix. C-terminal end of J -helix was serially deleted and replaced by TEVseq. TEV cleavage site is labeled
Crowe Critical Appraisal Tool (CCAT) User Guide
 Crowe Critical Appraisal Tool (CCAT) User Guide Version 1.4 (19 November 2013) Use with the CCAT Form version 1.4 only Michael Crowe, PhD michael.crowe@my.jcu.edu.au This work is licensed under the Creative
Crowe Critical Appraisal Tool (CCAT) User Guide Version 1.4 (19 November 2013) Use with the CCAT Form version 1.4 only Michael Crowe, PhD michael.crowe@my.jcu.edu.au This work is licensed under the Creative
Some Principles for the Design and Analysis of Experiments using Gene Expression Arrays and Other High-Throughput Assay Methods
 Some Principles for the Design and Analysis of Experiments using Gene Expression Arrays and Other High-Throughput Assay Methods BST 226 Statistical Methods for Bioinformatics January 8, 2014 1 The -Omics
Some Principles for the Design and Analysis of Experiments using Gene Expression Arrays and Other High-Throughput Assay Methods BST 226 Statistical Methods for Bioinformatics January 8, 2014 1 The -Omics
Whole Transcriptome Analysis of Illumina RNA- Seq Data. Ryan Peters Field Application Specialist
 Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Whole Transcriptome Analysis of Illumina RNA- Seq Data Ryan Peters Field Application Specialist Partek GS in your NGS Pipeline Your Start-to-Finish Solution for Analysis of Next Generation Sequencing Data
Life Sciences Reporting Summary
 Life Sciences Reporting Summary Corresponding author(s): Craig M. Powell Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Life Sciences Reporting Summary Corresponding author(s): Craig M. Powell Initial submission Revised version Final submission Nature Research wishes to improve the reproducibility of the work that we publish.
Psy 420 Midterm 2 Part 2 (Version A) In lab (50 points total)
 Psy 40 Midterm Part (Version A) In lab (50 points total) A researcher wants to know if memory is improved by repetition (Duh!). So he shows a group of five participants a list of 0 words at for different
Psy 40 Midterm Part (Version A) In lab (50 points total) A researcher wants to know if memory is improved by repetition (Duh!). So he shows a group of five participants a list of 0 words at for different
PROPENSITY SCORE MATCHING A PRACTICAL TUTORIAL
 PROPENSITY SCORE MATCHING A PRACTICAL TUTORIAL Cody Chiuzan, PhD Biostatistics, Epidemiology and Research Design (BERD) Lecture March 19, 2018 1 Outline Experimental vs Non-Experimental Study WHEN and
PROPENSITY SCORE MATCHING A PRACTICAL TUTORIAL Cody Chiuzan, PhD Biostatistics, Epidemiology and Research Design (BERD) Lecture March 19, 2018 1 Outline Experimental vs Non-Experimental Study WHEN and
Jiali Li, Sukhvir P Mahal, Cheryl A Demczyk and Charles Weissmann
 Manuscript EMBOR-2011-35144 Mutability of prions Jiali Li, Sukhvir P Mahal, Cheryl A Demczyk and Charles Weissmann Corresponding author: Charles Weissmann, Scripps Florida Review timeline: Submission date:
Manuscript EMBOR-2011-35144 Mutability of prions Jiali Li, Sukhvir P Mahal, Cheryl A Demczyk and Charles Weissmann Corresponding author: Charles Weissmann, Scripps Florida Review timeline: Submission date:
RNA-Seq Analysis. Simon Andrews, Laura v
 RNA-Seq Analysis Simon Andrews, Laura Biggins simon.andrews@babraham.ac.uk @simon_andrews v2018-10 RNA-Seq Libraries rrna depleted mrna Fragment u u u u NNNN Random prime + RT 2 nd strand synthesis (+
RNA-Seq Analysis Simon Andrews, Laura Biggins simon.andrews@babraham.ac.uk @simon_andrews v2018-10 RNA-Seq Libraries rrna depleted mrna Fragment u u u u NNNN Random prime + RT 2 nd strand synthesis (+
Supplementary Figure 1. Utilization of publicly available antibodies in different applications.
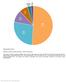 Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
Supplementary Figure 1 Utilization of publicly available antibodies in different applications. The fraction of publicly available antibodies toward human protein targets is shown with data for the following
BINF 6010 ITSC 8010 Spring 2010 Biotechnology & Genomics Lab Experimental Design- Technical.
 BINF 6010 ITSC 8010 Spring 2010 Biotechnology & Genomics Lab Experimental Design- Technical http://webpages.uncc.edu/~jweller2 Topics Experimental Design - sources of technical variation Library construction
BINF 6010 ITSC 8010 Spring 2010 Biotechnology & Genomics Lab Experimental Design- Technical http://webpages.uncc.edu/~jweller2 Topics Experimental Design - sources of technical variation Library construction
Discussion Solution Mollusks and Litter Decomposition
 Discussion Solution Mollusks and Litter Decomposition. Is the rate of litter decomposition affected by the presence of mollusks? 2. Does the effect of mollusks on litter decomposition differ among the
Discussion Solution Mollusks and Litter Decomposition. Is the rate of litter decomposition affected by the presence of mollusks? 2. Does the effect of mollusks on litter decomposition differ among the
Bios 6648: Design & conduct of clinical research
 Bios 6648: Design & conduct of clinical research Section 3 - Essential principle (randomization) 3.4 Trial monitoring: Interim decision and group sequential designs Bios 6648- pg 1 (a) Recruitment and
Bios 6648: Design & conduct of clinical research Section 3 - Essential principle (randomization) 3.4 Trial monitoring: Interim decision and group sequential designs Bios 6648- pg 1 (a) Recruitment and
Random matrix analysis for gene co-expression experiments in cancer cells
 Random matrix analysis for gene co-expression experiments in cancer cells OIST-iTHES-CTSR 2016 July 9 th, 2016 Ayumi KIKKAWA (MTPU, OIST) Introduction : What is co-expression of genes? There are 20~30k
Random matrix analysis for gene co-expression experiments in cancer cells OIST-iTHES-CTSR 2016 July 9 th, 2016 Ayumi KIKKAWA (MTPU, OIST) Introduction : What is co-expression of genes? There are 20~30k
Estoril Education Day
 Estoril Education Day -Experimental design in Proteomics October 23rd, 2010 Peter James Note Taking All the Powerpoint slides from the Talks are available for download from: http://www.immun.lth.se/education/
Estoril Education Day -Experimental design in Proteomics October 23rd, 2010 Peter James Note Taking All the Powerpoint slides from the Talks are available for download from: http://www.immun.lth.se/education/
Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers
 Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers Daniela Tosoni, Sarah Pambianco, Blanche Ekalle Soppo, Silvia Zecchini, Giovanni Bertalot1,
Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers Daniela Tosoni, Sarah Pambianco, Blanche Ekalle Soppo, Silvia Zecchini, Giovanni Bertalot1,
MRI Protocols in Experimental Stroke. Xenios Milidonis Centre for Clinical Brain Sciences The University of Edinburgh
 MRI Protocols in Experimental Stroke Xenios Milidonis Centre for Clinical Brain Sciences The University of Edinburgh MultiPART Meeting, Barcelona, 2 October 204 Objective In MultiPART magnetic resonance
MRI Protocols in Experimental Stroke Xenios Milidonis Centre for Clinical Brain Sciences The University of Edinburgh MultiPART Meeting, Barcelona, 2 October 204 Objective In MultiPART magnetic resonance
Calculation of Spot Reliability Evaluation Scores (SRED) for DNA Microarray Data
 Protocol Calculation of Spot Reliability Evaluation Scores (SRED) for DNA Microarray Data Kazuro Shimokawa, Rimantas Kodzius, Yonehiro Matsumura, and Yoshihide Hayashizaki This protocol was adapted from
Protocol Calculation of Spot Reliability Evaluation Scores (SRED) for DNA Microarray Data Kazuro Shimokawa, Rimantas Kodzius, Yonehiro Matsumura, and Yoshihide Hayashizaki This protocol was adapted from
Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation
 Manuscript EMBO-2016-43146 Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation Susana Abreu, Franziska Kriegenburg, Rubén Gómez-Sánchez, Muriel
Manuscript EMBO-2016-43146 Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation Susana Abreu, Franziska Kriegenburg, Rubén Gómez-Sánchez, Muriel
Design of Experiments (DOE) Instructor: Thomas Oesterle
 1 Design of Experiments (DOE) Instructor: Thomas Oesterle 2 Instructor Thomas Oesterle thomas.oest@gmail.com 3 Agenda Introduction Planning the Experiment Selecting a Design Matrix Analyzing the Data Modeling
1 Design of Experiments (DOE) Instructor: Thomas Oesterle 2 Instructor Thomas Oesterle thomas.oest@gmail.com 3 Agenda Introduction Planning the Experiment Selecting a Design Matrix Analyzing the Data Modeling
Accelerating Gene Set Enrichment Analysis on CUDA-Enabled GPUs. Bertil Schmidt Christian Hundt
 Accelerating Gene Set Enrichment Analysis on CUDA-Enabled GPUs Bertil Schmidt Christian Hundt Contents Gene Set Enrichment Analysis (GSEA) Background Algorithmic details cudagsea Performance evaluation
Accelerating Gene Set Enrichment Analysis on CUDA-Enabled GPUs Bertil Schmidt Christian Hundt Contents Gene Set Enrichment Analysis (GSEA) Background Algorithmic details cudagsea Performance evaluation
AccuSEQ v2.1 Real-Time PCR Detection and Analysis Software
 DATA SHEET AccuSEQ Real-Time PCR Detection and Analysis Software AccuSEQ v2.1 Real-Time PCR Detection and Analysis Software Introduction Automated presence or absence calls for Applied Biosystems MycoSEQ
DATA SHEET AccuSEQ Real-Time PCR Detection and Analysis Software AccuSEQ v2.1 Real-Time PCR Detection and Analysis Software Introduction Automated presence or absence calls for Applied Biosystems MycoSEQ
