Supplementary Information
|
|
|
- Reynard Boyd
- 5 years ago
- Views:
Transcription
1 Supplementary Information Supplementary Figure 1 Supplementary Figure 1 Scheme of isolating broadly neutralizing Abs against influenza viruses from human memory B cell repertoire. Representative fluorescence-labeled cell sorting (FACS) data showing frequency of H3-reactive memory B cells from total PBMCs.
2 Supplementary Figure 2 a b Supplementary Figure 2 Amino acid sequence alignments of 3I14 and related germline heavy chain (a) and light chain (b) variable region. The corresponding V, D and J sequences determined using the IMGT database are shown for comparison.
3 Supplementary Figure 3 H1-CA9 H3-UY7 H1-SI6 H3-VIC11 H1-PR8 H4-NL5 H5-VN4 H7-CA444 H5-IN5 H7-AH13 H9-HK99 H7-NL219 H3-PE9 H14-AS82
4 Supplementary Figure 3 3I14 scfvfc binding to recombinant HAs. Blue curves are the experimental trace obtained from biolayer interferometry experiments, and red curves are the best global fits to the data used to calculate the K d s presented in Fig 1c.
5 Supplementary Figure 4 A/Brisbane/1/7 (H3N2) A/Wisconsin/67/5 x A/Puerto Rico/8/1934 (H3N2) A/Aichi/2/1968 x A/Puerto Rico/8/1934 (H3N2) I14 scfvfc F1 scfvfc A/Sydney/5/1997 (H3N2) A/Aichi/2/1968 (H3N2) A/Nanchang/993/1995 (H3N2) I14 scfvfc F1 scfvfc A/Hong Kong/8/1968 (H3N2) A/Perth/16/9 (H3N2) A/California/4/9 (H1N1) I14 scfvfc F1 scfvfc A/Puerto Rico/8/1934 (H1N1) 1 A/Anhui/1/13 (H7N9) A/Netherlands/219/3 (H7N1) Pseudotyped Virus 1 3I14 scfvfc F1 scfvfc A/FPV/Rostock/1934 (H7N1) Pseudotyped Virus A/Vietnam/13/4 (H5N1) Pseudotyped Virus A/Hong Kong/156/97 (H5N1) Pseudotyped Virus I14 scfvfc F1 scfvfc Supplementary Figure 4 3I14 scfvfc Ab neutralized influenza viruses infection and HApseudotyped luciferase reporter viruses. MAb 3I14 (black) and Anti-group 1 mab F1 (red) neutralized different stains of infectious viruses and pseudotyped viruses. The data represent average neutralization titers from 2-3 independent experiments.
6 Figure 5 Supplementary Figure 5 3I14 scfvfc Ab binding to full-length or HA1 of recombinant H3- PE9. Blue curves are the experimental trace obtained from biolayer interferometry experiments, and red curves are the best global fits to the data. The curves show 3I14 binds H3-PE9 full-length but dose not bind H3-PE9 HA1 subunit.
7 Supplementary Figure 6 Supplementary Fig. 6 3I14 mediates a surrogate reporter-based ADCC assay 3I14 and other anti-stem bnabs, FI6v3, CR9114, 39.29, F1 and CR induced ADCC in H3- and H5- expressed 293T cells /well H3 or H5-expressed 293T cells were attached to the plates prior to assay, and the medium was then replaced with low IgG serum assay buffer. Each well were added different bnabs at concentration of 5, 1,.2 and.4 µg ml -1. After one-hour, Jurkat effector cells were added for 6.x1 4 /well to assay plates and incubated for 6 hours. The supernatants were measured luminescence using Bio-GloTM Luciferase Assay kits (Promega). Bars represent mean ± S.E.M. P value was calculated using two-way ANOVA, compared to F1 (upper panel) and CR (lower panel). * represents p value for each comparison <.1. Data represent a representative experiment from three independent experiments, and all tests were performed in triplicate.
8 Supplementary Figure 7 Supplementary Figure 7 Binding of the 3I14 IgG1 variants to recombinant H1, H3 and H5. Blue curves are the experimental trace obtained from biolayer interferometry experiments, and red curves are the best global fits to the data used to calculate the K d s presented in Table 2.
9 Supplementary Figure 8 a b Supplementary Figure 8 3I14 WT and VLD94N IgG1 variants binding to recombinant H5- VN4 (a) and H3-PE9 (b). Green or blue curves are the experimental trace obtained from biolayer interferometry experiments, and red curves are the best global fits to the data used to calculate the K d values. Affinity measurements (K d values) for the binding curves were reported in Table 3.
10 Supplementary Figure 9 Supplementary Figure 9 The interactions of G31 of 3I14 light chain with H3 in the H3/3I14 complex model. The helix A of HA2 domain of H3 is shown as ribbon in cyan; the light chain of 3I14 is shown as ribbon in orange; the main chain atoms of G31 are shown in stick and the side chain atoms of Q42 and D46 of H3 HA2 are shown in stick; the distance between G31 and H3 are illustrated by green dash lines and labeled in black. (The PyMOL Molecular Graphics System, Version.99 rc6 Schrödinger, LLC).
11 Supplementary Figure 1 Supplementary Figure 1 The representative FACS plots for all of the gating schemes. Fresh PBMCs were sequentially gated by lymphocytes gate, SSC-H/W gate and FSC-H/W gate. The single B cells were isolated by CD19/CD27 double positive gate and H3-FITC gate and were sorted into 384-well plate.
12 Supplementary Figure 11 Supplementary Figure 11 Trypsin Cleavage Inhibition Assay (full-size images)..4 µg recombinant H3-histidine (H3-BR7) was incubated in the presence of 2.5 µg 3I14 or Fm-6 IgG1, or in the absence of antibody in Tris-HCl buffer at ph 8. containing 2 µg ml -1 Trypsin at 37 C. Trypsin digestion was stopped at several time-points by boiling the sample in a C water bath. Samples were run on 1% reduced SDS-PAGE and blotted using a HisProbe-HRP Abs. Uncleaved HA (HA) is indicated on the right side.
13 Supplementary Table 1 Contact residues at H3/3I14 and H5/3I14 interfaces 3I14 HCDR3 LCDR1 LCDR2 LCDR3 Y13 Y14 F15* D16 F19* G31* N32 T33 N52 S53* D94 H3 I18 W21 D19 D19 L38 Q42 Q42 Q42 N49 K39 L38 L38 D46 I45 H5 D19 D19 D19 D19 K38 Q42 Q42 T49 N53 E39 I45 K38 K38 E39 D46 T41 I45 Contact residues defined by interatomic distances < 4 Å, except residue D94 in H3 and H5 complexes defined by distances < 5 Å and < 7 Å, respectively. The color scheme indicates contributions to the binding energy: very favorable (red); favorable (orange); neutral (blue) and unfavorable (black). *Residues indicate the somatic mutations of germline-encoded residues.
14 Supplementary Table 2 Sequence comparison of 3I14 epitope among 16 HA subtypes Group Strains K d * Relative K d ** Fusion peptide Helix A (nm) to CR Group 1 H1-CA V D G W A L K T Q I D T N H1-SI6.3 - V D G W A Q K T Q I N T N H1-PR8.8 - I D G W A Q K T Q I N T N H2-JP V D G W A K E T Q F D T N H5-VN V D G W A K E T Q I D T N H5-IN V D G W A K E T Q I D T N H6-NY V D G W A K E T Q I D T N H11-MEM I N G W A K E T Q I D T N H13-MD I N G W A K E T Q I D T N H16-SE6 - > nm I N G W A K A T Q I D T N H8-ON I D G W A Q K T Q I D T N H9-HK V A G W A R D T Q I D T N H12-AB76 - > 2.9 nm V A G W A R D T Q I D Q N Group 2 H3-PE V D G W A L K T Q I D N N H3-UY V D G W A L K T Q I D N N H3-VIC V D G W A L K T Q I D N N H4-NL I D G W A L K T Q I D N N H14-AS I D G W A L K T Q I D N N H7-NL I D G W A Y K T Q I D T N H7-AH I D G W A Y K T Q I D T N H1-SE2 - - V D G W A Y K T Q I D T N H15-WA79 - < 1 nm I D G W A Y K T Q I D T N *K d determined by Surface Plasmon Resonance (SPR) Biosensor (Fig 1c). **Relative K d determined by Flow Cytometry (Fig 1b) and reference 16. Residues carrying positively charged side chain are labeled orange; while negatively charged side chain residues are labeled blue.
15 Supplementary Table 3 The SHMs of IGHV1-69 and IGHV3-3-encoded bnabs in V regions Germline gene usage bnabs SHMs in V region VH VL IGHV3-3-encoded 3I FI MAb IGHV1-69-encoded CR CR F MAb
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Map of pct-vhvl-k1 native V H :V L display vector.
 Supplementary Figure 1 Map of pct-vhvl-k1 native V H :V L display vector. Natively-paired V H :V L sequences are cloned en masse into this vector for human antibody repertoire mining, and their corresponding
Supplementary Figure 1 Map of pct-vhvl-k1 native V H :V L display vector. Natively-paired V H :V L sequences are cloned en masse into this vector for human antibody repertoire mining, and their corresponding
Supplementary Figure 1.
 9 8 5 6F12 IgG2a Isotype Control 5 1 15 Supplementary Figure 1. Passive treatment with isotype control mab does not affect viral challenge. Wild type mice were treated with 4 mg/kg of mouse IgG2a 6F12
9 8 5 6F12 IgG2a Isotype Control 5 1 15 Supplementary Figure 1. Passive treatment with isotype control mab does not affect viral challenge. Wild type mice were treated with 4 mg/kg of mouse IgG2a 6F12
Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL*
 Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL* Site AA Sequence 3x10 6 3x10 6 20x10 6 20x10 6 100x10 6 100x10 6 Post-1 st Post-2 nd Post-1 st Post-2 nd Post-1 st Post-2
Supplementary Table 1: Antigenic regions/sites on Ebola-GP identified using GFPDL* Site AA Sequence 3x10 6 3x10 6 20x10 6 20x10 6 100x10 6 100x10 6 Post-1 st Post-2 nd Post-1 st Post-2 nd Post-1 st Post-2
Nature Biotechnology: doi: /nbt.4086
 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade
 Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Data Sheet PD-1 / NFAT - Reporter - Jurkat Recombinant Cell Line Catalog #: 60535
 Data Sheet PD-1 / NFAT - Reporter - Jurkat Recombinant Cell Line Catalog #: 60535 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
Data Sheet PD-1 / NFAT - Reporter - Jurkat Recombinant Cell Line Catalog #: 60535 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. dcas9-mq1 fusion protein induces de novo
Supplementary Figure 1.
 Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Figure 1. Assessment of quaternary structure of soluble RSV F proteins. Soluble variants of F proteins from A2 and B1 RSV strains were expressed in HEK293 cells. The cell culture supernatants
Supplementary Information. Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications
 Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
Chapter 17: Immunization & Immune Testing. 1. Immunization 2. Diagnostic Immunology
 Chapter 17: Immunization & Immune Testing 1. Immunization 2. Diagnostic Immunology 1. Immunization Chapter Reading pp. 505-511 What is Immunization? A method of inducing artificial immunity by exposing
Chapter 17: Immunization & Immune Testing 1. Immunization 2. Diagnostic Immunology 1. Immunization Chapter Reading pp. 505-511 What is Immunization? A method of inducing artificial immunity by exposing
1. Immunization. What is Immunization? 12/9/2016. Chapter 17: Immunization & Immune Testing. 1. Immunization 2. Diagnostic Immunology
 Chapter 17: Immunization & Immune Testing 1. Immunization 2. Diagnostic Immunology 1. Immunization Chapter Reading pp. 505-511 What is Immunization? A method of inducing artificial immunity by exposing
Chapter 17: Immunization & Immune Testing 1. Immunization 2. Diagnostic Immunology 1. Immunization Chapter Reading pp. 505-511 What is Immunization? A method of inducing artificial immunity by exposing
Supplementary Figure 1. Botrocetin induces binding of human VWF to human
 Supplementary Figure 1: Supplementary Figure 1. Botrocetin induces binding of human VWF to human platelets in the absence of elevated shear and induces platelet agglutination as detected by flow cytometry.
Supplementary Figure 1: Supplementary Figure 1. Botrocetin induces binding of human VWF to human platelets in the absence of elevated shear and induces platelet agglutination as detected by flow cytometry.
Supplemental Figure 1 Kam et al., 2017
 nti-zikv IgG response (OD 450 nm) % Infectivity (Relative to virus only) 3 2 1 2 0 1 0 0 8 0 6 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C-LL 1 4 0 2 0 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C -LL 0 0.1 1 1 0 1 0 0 [b],
nti-zikv IgG response (OD 450 nm) % Infectivity (Relative to virus only) 3 2 1 2 0 1 0 0 8 0 6 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C-LL 1 4 0 2 0 0 1-H1L1 2F-H1L3 SIgN-3C SIgN-3C -LL 0 0.1 1 1 0 1 0 0 [b],
Discovery and Humanization of Novel High Affinity Neutralizing Monoclonal Antibodies to Human IL-17A
 Discovery and Humanization of Novel High Affinity Neutralizing Monoclonal Antibodies to Human IL-17A Contacts: Marty Simonetti martysimonetti@gmail.com Kirby Alton kirby.alton@abeomecorp.com Rick Shimkets
Discovery and Humanization of Novel High Affinity Neutralizing Monoclonal Antibodies to Human IL-17A Contacts: Marty Simonetti martysimonetti@gmail.com Kirby Alton kirby.alton@abeomecorp.com Rick Shimkets
Supplementary information Bi-specific Aptamers mediating Tumour Cell Lysis
 Supplementary information Bi-specific Aptamers mediating Tumour Cell Lysis Figure S1 SELEX scheme of different CD16 DNA selection approaches. Nine rounds of filter SELEX were conducted in parallel, only
Supplementary information Bi-specific Aptamers mediating Tumour Cell Lysis Figure S1 SELEX scheme of different CD16 DNA selection approaches. Nine rounds of filter SELEX were conducted in parallel, only
Exceptional Human Antibody Discovery. Corporate Overview
 Exceptional Human Antibody Discovery Corporate Overview Co 1 1 Our Mission Trianni is a biotech company with the scientific mission of creating optimized and highly versatile platforms for generation of
Exceptional Human Antibody Discovery Corporate Overview Co 1 1 Our Mission Trianni is a biotech company with the scientific mission of creating optimized and highly versatile platforms for generation of
Box 7905, Engineering Building I, North Carolina State University, Raleigh, NC 27695, Phone: Fax:
 Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Inefficient ribosomal skipping enables simultaneous secretion and display of proteins in Saccharomyces cerevisiae Carlos A. Cruz-Teran 1, Karthik Tiruthani 1, Adam Mischler, and Balaji M. Rao * Department
Data Sheet. TCR activator / PD-L1 - CHO Recombinant Cell line Cat. #: 60536
 Data Sheet TCR activator / PD-L1 - CHO Recombinant Cell line Cat. #: 60536 Product Description Recombinant CHO-K1 cells constitutively expressing human PD-L1 (Programmed Cell Death 1 Ligand 1, CD274, B7
Data Sheet TCR activator / PD-L1 - CHO Recombinant Cell line Cat. #: 60536 Product Description Recombinant CHO-K1 cells constitutively expressing human PD-L1 (Programmed Cell Death 1 Ligand 1, CD274, B7
Supporting Information
 Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
Supplemental Figure 1
 Supplemental Figure 1 Supplemental Fig. 1. (A) The binding avidity between Fz4-FcH6 and MBP-Norrin dimers measured by biolayer interferometry. 5 µg/ml Fz4-Fc protein was loaded onto anti human Fc capture
Supplemental Figure 1 Supplemental Fig. 1. (A) The binding avidity between Fz4-FcH6 and MBP-Norrin dimers measured by biolayer interferometry. 5 µg/ml Fz4-Fc protein was loaded onto anti human Fc capture
Programmable nucleic acid nanoswitches for the rapid, single-step detection of antibodies in bodily fluids
 Supporting Information: Programmable nucleic acid nanoswitches for the rapid, single-step detection of antibodies in bodily fluids Alessandro Porchetta 1, #, Rudy Ippodrino 2, #, Bruna Marini 2, Arnaldo
Supporting Information: Programmable nucleic acid nanoswitches for the rapid, single-step detection of antibodies in bodily fluids Alessandro Porchetta 1, #, Rudy Ippodrino 2, #, Bruna Marini 2, Arnaldo
Supporting Information
 Supporting Information Table S1. Overview of samples used for sequencing, and the number of sequences obtained from each sample. Visit 1 is day 0, Visit 2 is day 7, Visit 3 is day 28, and Visit 4 is day
Supporting Information Table S1. Overview of samples used for sequencing, and the number of sequences obtained from each sample. Visit 1 is day 0, Visit 2 is day 7, Visit 3 is day 28, and Visit 4 is day
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot
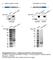 Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Figure 1. Antigens generated for mab development (a) K9M1P1-mIgG and hgh-k9m1p1 antigen (~37 kda) expression verified by western blot (vector: ~25 kda). (b) Silver staining was used to assess
Supplementary Online Material. Structural mimicry in transcription regulation of human RNA polymerase II by the. DNA helicase RECQL5
 Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Online Material Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5 Susanne A. Kassube, Martin Jinek, Jie Fang, Susan Tsutakawa and Eva Nogales
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin
 Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA
 Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Supplementary Figure S1. Binding of HSA mutants to hfcrn. (a) The levels of titrated amounts of HSA variants (5.0-0.002 μg/ml) directly coated in the wells at ph 6.0 were controlled using a horseradish
Supplementary methods
 Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Supplementary methods Cell culture, infection, transfection, and RNA interference HEK293 cells and its derivatives were grown in DMEM supplemented with 10% FBS. Various constructs were introduced into
Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure
 ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
ME-10-0005 Conformation of the Mineralocorticoid Receptor N- terminal Domain: Evidence for Induced and Stable Structure Katharina Fischer 1, Sharon M. Kelly 2, Kate Watt 1, Nicholas C. Price 2 and Iain
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Single cell imaging of Bruton's Tyrosine Kinase using an irreversible inhibitor
 SUPPLEMENTARY INFORMATION Single cell imaging of Bruton's Tyrosine Kinase using an irreversible inhibitor Anna Turetsky 1,a, Eunha Kim 1,a, Rainer H. Kohler 1, Miles A. Miller 1, Ralph Weissleder 1,2,
SUPPLEMENTARY INFORMATION Single cell imaging of Bruton's Tyrosine Kinase using an irreversible inhibitor Anna Turetsky 1,a, Eunha Kim 1,a, Rainer H. Kohler 1, Miles A. Miller 1, Ralph Weissleder 1,2,
GC GC - AAV2/8-4E10
 SUPPLEMENTARY INFORMATION doi:.38/nature660 a Luciferase Expression (Photons/Sec) c f 12 11 11 GC - AAV2/8-Luciferase 9 GC - AAV2/8-Luciferase 0 8 16 24 32 40 48 56 64 Weeks Post-AAV Injection Human IgG
SUPPLEMENTARY INFORMATION doi:.38/nature660 a Luciferase Expression (Photons/Sec) c f 12 11 11 GC - AAV2/8-Luciferase 9 GC - AAV2/8-Luciferase 0 8 16 24 32 40 48 56 64 Weeks Post-AAV Injection Human IgG
APA105Hu01 100µg Active Nerve Growth Factor (NGF) Organism Species: Homo sapiens (Human) Instruction manual
 APA105Hu01 100µg Active Nerve Growth Factor (NGF) Organism Species: Homo sapiens (Human) Instruction manual FOR RESEARCH USE ONLY NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES [ PROPERTIES ] 1th Edition
APA105Hu01 100µg Active Nerve Growth Factor (NGF) Organism Species: Homo sapiens (Human) Instruction manual FOR RESEARCH USE ONLY NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES [ PROPERTIES ] 1th Edition
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Maximizing Assembly and Yield of Unmodified Bispecific Antibodies
 Maximizing Assembly and Yield of Unmodified Bispecific Antibodies Pauline Malinge Head of Molecular Interaction Facility 1 Outline Introduction The κλ body format The κλ body platform Bispecific Antibody
Maximizing Assembly and Yield of Unmodified Bispecific Antibodies Pauline Malinge Head of Molecular Interaction Facility 1 Outline Introduction The κλ body format The κλ body platform Bispecific Antibody
Quantitative analysis of recombination in YFP and CFP gene of FRET biosensor induced by lentiviral or retroviral gene transfer.
 Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
Supplementary Information: Quantitative analysis of recombination in and gene of FRET biosensor induced by lentiviral or retroviral gene transfer. Akira T. Komatsubara, Michiyuki Matsuda,, and Kazuhiro
SUPPLEMENTARY MATERIAL. Viral variants that initiate and drive maturation of V1V2- directed HIV-1 broadly neutralizing antibodies
 SUPPLEMENTARY MATERIAL Viral variants that initiate and drive maturation of V1V2- directed HIV-1 broadly neutralizing antibodies Jinal N. Bhiman 1,2, Colin Anthony 3, Nicole A. Doria-Rose 4, Owen Karimanzira
SUPPLEMENTARY MATERIAL Viral variants that initiate and drive maturation of V1V2- directed HIV-1 broadly neutralizing antibodies Jinal N. Bhiman 1,2, Colin Anthony 3, Nicole A. Doria-Rose 4, Owen Karimanzira
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than. SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with
 Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Supplementary Fig. S1. SAMHD1c has a more potent dntpase activity than SAMHD1c. Purified recombinant SAMHD1c and SAMHD1c proteins (with concentration of 800nM) were incubated with 1mM dgtp for the indicated
Implementation of the Next Generation Effector Function Assays for Comparability Assessments
 Implementation of the Next Generation Effector Function Assays for Comparability Assessments March 25, 2014 Poonam Aggarwal Bioassays 2014: Scientific Approaches & Regulatory Strategies to be held March
Implementation of the Next Generation Effector Function Assays for Comparability Assessments March 25, 2014 Poonam Aggarwal Bioassays 2014: Scientific Approaches & Regulatory Strategies to be held March
Phage Antibody Selection With Reichert SPR System
 Phage Antibody Selection With Reichert SPR System Reichert Technologies Webinar April 4, 2016 Mark A. Sullivan, Ph.D. Department of Microbiology and Immunology University of Rochester Presentation Outline
Phage Antibody Selection With Reichert SPR System Reichert Technologies Webinar April 4, 2016 Mark A. Sullivan, Ph.D. Department of Microbiology and Immunology University of Rochester Presentation Outline
SUPPLEMENTARY INFORMATION. Supplementary Figures 1-8
 SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
SUPPLEMENTARY INFORMATION Supplementary Figures 1-8 Supplementary Figure 1. TFAM residues contacting the DNA minor groove (A) TFAM contacts on nonspecific DNA. Leu58, Ile81, Asn163, Pro178, and Leu182
Supplementary Figure. S1
 Supplementary Figure. S1 Supplementary Figure S1. Correlation of phagocytic ability measured with YG and YO beads. Fresh human monocytes (2 10 6 /ml) were labelled with APC conjugated anti CD14 mab alone
Supplementary Figure. S1 Supplementary Figure S1. Correlation of phagocytic ability measured with YG and YO beads. Fresh human monocytes (2 10 6 /ml) were labelled with APC conjugated anti CD14 mab alone
Supplemental Material for: Delineating antibody recognition against Zika
 Supplemental Material for: Delineating antibody recognition against Zika virus during natural infection Authors: Lei Yu 1,8, Ruoke Wang 2,8, Fei Gao 2, Min Li 3, Jianying Liu 4, Jian Wang 1,Wenxin Hong
Supplemental Material for: Delineating antibody recognition against Zika virus during natural infection Authors: Lei Yu 1,8, Ruoke Wang 2,8, Fei Gao 2, Min Li 3, Jianying Liu 4, Jian Wang 1,Wenxin Hong
Supplementary Figure 1: Sequence alignment of partial stem region of flaviviruses
 Supplementary Figure 1: Sequence alignment of partial stem region of flaviviruses E prtoeins. Polyprotein sequences of viruses were downloaded from GenBank and aligned by CLC Sequence Viewer software.
Supplementary Figure 1: Sequence alignment of partial stem region of flaviviruses E prtoeins. Polyprotein sequences of viruses were downloaded from GenBank and aligned by CLC Sequence Viewer software.
Data Sheet HVEM/NF-κB Reporter Jurkat Recombinant Cell Line Catalog # 79310
 Data Sheet HVEM/NF-κB Reporter Jurkat Recombinant Cell Line Catalog # 79310 Product Description Recombinant clonal stable Jurkat T cell line expressing firefly luciferase gene under the control of 4 copies
Data Sheet HVEM/NF-κB Reporter Jurkat Recombinant Cell Line Catalog # 79310 Product Description Recombinant clonal stable Jurkat T cell line expressing firefly luciferase gene under the control of 4 copies
Nature Structural & Molecular Biology: doi: /nsmb.3018
 Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Supplementary Figure 1 Validation of genetic complementation assay in Bmal1 / Per2 Luc fibroblasts. (a) Only Bmal1, not Bmal2, rescues circadian rhythms from cells. Cells expressing various Bmal constructs
Recombinant, Insect Cell-Derived RSV Nanoparticle Vaccine
 Recombinant, Insect Cell-Derived RSV Nanoparticle Vaccine Gregory Glenn Chief Medical Officer MVADS-Copenhagen 4 July 2012 1 Agenda for RSV Discussion Overview of Insect Cell Technology Respiratory Syncytial
Recombinant, Insect Cell-Derived RSV Nanoparticle Vaccine Gregory Glenn Chief Medical Officer MVADS-Copenhagen 4 July 2012 1 Agenda for RSV Discussion Overview of Insect Cell Technology Respiratory Syncytial
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection
 Brief Report Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection David J. DiLillo, 1 Peter Palese, 2 Patrick C. Wilson, 3 and Jeffrey V. Ravetch 1 1 The
Brief Report Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection David J. DiLillo, 1 Peter Palese, 2 Patrick C. Wilson, 3 and Jeffrey V. Ravetch 1 1 The
Data Sheet CD137/NF-κB Reporter - HEK293 Recombinant Cell Line Catalog # 79289
 Data Sheet CD137/NF-κB Reporter - HEK293 Recombinant Cell Line Catalog # 79289 Background Human CD137 (4-1BB; TNFRS9) is an inducible co-stimulatory molecule that activates T cells. CD137:CD137L-mediated
Data Sheet CD137/NF-κB Reporter - HEK293 Recombinant Cell Line Catalog # 79289 Background Human CD137 (4-1BB; TNFRS9) is an inducible co-stimulatory molecule that activates T cells. CD137:CD137L-mediated
Enhance Your Protein Interaction Research with A New Level of Bench-Top Productivity
 LAMDAGEN C O R P O R A T I O N Enhance your research with real-time monitoring and quantitation of biomolecular binding with highly sensitive detection in a label-free format 9 Enhance Your Protein Interaction
LAMDAGEN C O R P O R A T I O N Enhance your research with real-time monitoring and quantitation of biomolecular binding with highly sensitive detection in a label-free format 9 Enhance Your Protein Interaction
Phenotypic screening reveals TNFR2 as a promising target for cancer immunotherapy
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Phenotypic screening reveals TNFR2 as a promising target for cancer immunotherapy Supplementary Materials SUPPLEMENTARY MATERIALS
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Phenotypic screening reveals TNFR2 as a promising target for cancer immunotherapy Supplementary Materials SUPPLEMENTARY MATERIALS
Supplementary Information
 Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
Supplementary Information Supplementary Figure 1: Over-expression of CD300f in NIH3T3 cells enhances their capacity to phagocytize AC. (a) NIH3T3 cells were stably transduced by EV, CD300f WT or CD300f
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Data Sheet ICOS/NFAT Reporter-Jurkat Recombinant Cell Line Catalog #: 79668
 Data Sheet ICOS/NFAT Reporter-Jurkat Recombinant Cell Line Catalog #: 79668 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
Data Sheet ICOS/NFAT Reporter-Jurkat Recombinant Cell Line Catalog #: 79668 Product Description Recombinant Jurkat T cell expressing firefly luciferase gene under the control of NFAT response elements
Luteinizing Hormone ELISA kit
 55R-IB19104 Luteinizing Hormone ELISA kit Enzyme Immunoassay for the determination of LH in human serum 1. Principle of the test: The LH ELISA kit is a solid phase direct sandwich method. The samples and
55R-IB19104 Luteinizing Hormone ELISA kit Enzyme Immunoassay for the determination of LH in human serum 1. Principle of the test: The LH ELISA kit is a solid phase direct sandwich method. The samples and
High throughput screening: Huh-7 cells were seeded into 96-well plate (2000
 1 SUPPLEMENTARY INFORMATION METHODS 6 7 8 9 1 11 1 1 1 1 16 17 18 19 High throughput screening: Huh-7 cells were seeded into 96-well plate ( cells/well) and infected with MOI of DENV-. One hour post-infection
1 SUPPLEMENTARY INFORMATION METHODS 6 7 8 9 1 11 1 1 1 1 16 17 18 19 High throughput screening: Huh-7 cells were seeded into 96-well plate ( cells/well) and infected with MOI of DENV-. One hour post-infection
A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated
 A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated cancer therapeutic antibodies Supplemental Data File in this Data Supplement: Supplementary Figure 1-4
A novel therapeutic strategy to rescue the immune effector function of proteolyticallyinactivated cancer therapeutic antibodies Supplemental Data File in this Data Supplement: Supplementary Figure 1-4
T-cell response. Taken from NIAID: s.aspx
 T-cell receptor T-cell response 1. Macrophage or dendritic cell digest antigen bacteria, virus 2. Fragments of Ag bind to major histo-compatiblity (MHC) proteins in macrophage. 3. MHC I-Ag fragment expressed
T-cell receptor T-cell response 1. Macrophage or dendritic cell digest antigen bacteria, virus 2. Fragments of Ag bind to major histo-compatiblity (MHC) proteins in macrophage. 3. MHC I-Ag fragment expressed
HEK293T. Fig. 1 in the
 Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Supplementary Information Supplementary Figure 1 Zinc uptake assay of hzip4 and hzip4-δecd transiently expressed in HEK293T cells. The results of one representative e experiment are shown in Fig. 1 in
Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells
 SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
SUPPLEMENTARY FIGURES Recruitment of Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells Niklas Engels, Lars Morten König, Christina Heemann, Johannes
OmniAb. Naturally optimized human antibodies
 OmniAb Naturally optimized human antibodies Transgenic animals for hmab discovery Only company to offer three platforms Patented technology with freedom to operate V L V H C C H 1 hinge C H 2 C H 3 2 28
OmniAb Naturally optimized human antibodies Transgenic animals for hmab discovery Only company to offer three platforms Patented technology with freedom to operate V L V H C C H 1 hinge C H 2 C H 3 2 28
Pre-made Reporter Lentivirus for p53 Signal Pathway
 Pre-made Reporter for p53 Signal Pathway Cat# Product Name Amounts LVP977-P or: LVP977-P-PBS P53-GFP (Puro) LVP978-P or: LVP978-P-PBS P53-RFP (Puro) LVP979-P or: LVP979-P-PBS P53-Luc (Puro) LVP980-P or:
Pre-made Reporter for p53 Signal Pathway Cat# Product Name Amounts LVP977-P or: LVP977-P-PBS P53-GFP (Puro) LVP978-P or: LVP978-P-PBS P53-RFP (Puro) LVP979-P or: LVP979-P-PBS P53-Luc (Puro) LVP980-P or:
How to Screen a Billion Drug Candidates?
 How to Screen a Billion Drug Candidates? Single Cell Functional Assays: Ultra-sensitive, Ultra-fast, Ultrahigh-throughput Next-Generation Drug Discovery The advantages of de novo protein engineering for
How to Screen a Billion Drug Candidates? Single Cell Functional Assays: Ultra-sensitive, Ultra-fast, Ultrahigh-throughput Next-Generation Drug Discovery The advantages of de novo protein engineering for
ADCC Reporter Bioassay: A Novel, Bioluminescent Cell-Based Assay for Quantifying Fc Effector Function of Antibodies
 ADCC Reporter Bioassay: A Novel, Bioluminescent Cell-Based Assay for Quantifying Fc Effector Function of Antibodies Richard Somberg, Ph.D. October 2012 Outline Introduction to ADCC Problem with classic
ADCC Reporter Bioassay: A Novel, Bioluminescent Cell-Based Assay for Quantifying Fc Effector Function of Antibodies Richard Somberg, Ph.D. October 2012 Outline Introduction to ADCC Problem with classic
For quantitative detection of mouse IGF-1 in serum, body fluids, tissue lysates or cell culture supernatants.
 Mouse IGF-1 ELISA Kit (migf-1-elisa) Cat. No. EK0378 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Mouse IGF-1 ELISA Kit (migf-1-elisa) Cat. No. EK0378 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Application Note AN001
 Testing hybridoma supernatants with the Spots On Dots Antibody Screening Kit Application Note AN1 Table of Contents Overview... 2 Figure 1. Screening of hybridomas raised against peptide antigens... 3
Testing hybridoma supernatants with the Spots On Dots Antibody Screening Kit Application Note AN1 Table of Contents Overview... 2 Figure 1. Screening of hybridomas raised against peptide antigens... 3
Supplementary information
 Supplementary information Identification of E-cadherin signature sites functioning as cleavage sites for Helicobacter pylori HtrA Thomas P. Schmidt 1*, Anna M. Perna 2*, Tim Fugmann 3, Manja Böhm 4, Jan
Supplementary information Identification of E-cadherin signature sites functioning as cleavage sites for Helicobacter pylori HtrA Thomas P. Schmidt 1*, Anna M. Perna 2*, Tim Fugmann 3, Manja Böhm 4, Jan
ELISPOT and FLUOROSPOT kits
 ELISPOT and FLUOROSPOT kits Interleukins Interferons Granzymes and perforins TNF superfamily ligands and receptors Apoptosis markers And many more... ELISPOT and FLUOROSPOT: a cell-based assay to assess
ELISPOT and FLUOROSPOT kits Interleukins Interferons Granzymes and perforins TNF superfamily ligands and receptors Apoptosis markers And many more... ELISPOT and FLUOROSPOT: a cell-based assay to assess
Transcription Factor Runx3 Is Induced by Influenza A Virus and Double-Strand RNA and
 Transcription Factor Is Induced by Influenza A Virus and Double-Strand RNA and Mediates Airway Epithelial Cell Apoptosis Huachen Gan 1, Qin Hao 1, Steven Idell 1,2 & Hua Tang 1 1 Department of Cellular
Transcription Factor Is Induced by Influenza A Virus and Double-Strand RNA and Mediates Airway Epithelial Cell Apoptosis Huachen Gan 1, Qin Hao 1, Steven Idell 1,2 & Hua Tang 1 1 Department of Cellular
Who pairs with whom? High-throughput sequencing of the human paired heavy and light chain repertoire
 Who pairs with whom? High-throughput sequencing of the human paired heavy and light chain repertoire Technical Journal Club September 15 th Christina Müller Background - antibody repertoire is the sum
Who pairs with whom? High-throughput sequencing of the human paired heavy and light chain repertoire Technical Journal Club September 15 th Christina Müller Background - antibody repertoire is the sum
Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression
 Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression LVP336 LVP336-PBS LVP339 LVP339-PBS LVP297 LVP297-PBS LVP013 LVP013-PBS LVP338 LVP338-PBS LVP027 LVP027-PBS LVP337 LVP337-PBS
Pre-made Lentiviral Particles for Nuclear Permeant CRE Recombinase Expression LVP336 LVP336-PBS LVP339 LVP339-PBS LVP297 LVP297-PBS LVP013 LVP013-PBS LVP338 LVP338-PBS LVP027 LVP027-PBS LVP337 LVP337-PBS
Rat IGF-1 ELISA Kit (rigf-1-elisa)
 Rat IGF-1 ELISA Kit (rigf-1-elisa) Cat. No. EK0377 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
Rat IGF-1 ELISA Kit (rigf-1-elisa) Cat. No. EK0377 96 Tests in 8 x 12 divisible strips Background Insulin-like growth factor 1 (IGF-1), also known as somatomedin C, is a polypeptide protein hormone similar
p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1
 Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
Supplementary Information p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1 Bei Wang 1, Dandan Niu 1, Liyun Lai 1, & Ee Chee Ren 1,2 1 Singapore Immunology
15 June 2011 Supplementary material Bagriantsev et al.
 Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
Supplementary Figure S1 Characterization of K 2P 2.1 (TREK-1) GOF mutants A, Distribution of the positions of mutated nucleotides, represented by a red x, from a pool of 18 unselected K 2P 2.1 (KCNK2)
The Trianni Mouse: Best-In-Class Technology for Human Antibody Discovery
 The Trianni Mouse: Best-In-Class Technology for Human Antibody Discovery Corporate Overview Co David Meininger, PhD, MBA Chief Business Officer, Trianni 1 Our Mission Trianni is a biotech company with
The Trianni Mouse: Best-In-Class Technology for Human Antibody Discovery Corporate Overview Co David Meininger, PhD, MBA Chief Business Officer, Trianni 1 Our Mission Trianni is a biotech company with
Supplementary Figure 1. α-synuclein is truncated in PD and LBD brains. Nature Structural & Molecular Biology: doi: /nsmb.
 Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
Supplementary Figure 1 α-synuclein is truncated in PD and LBD brains. (a) Specificity of anti-n103 antibody. Anti-N103 antibody was coated on an ELISA plate and different concentrations of full-length
(a) Immunoblotting to show the migration position of Flag-tagged MAVS
 Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
Supplementary Figure 1 Characterization of six MAVS isoforms. (a) Immunoblotting to show the migration position of Flag-tagged MAVS isoforms. HEK293T Mavs -/- cells were transfected with constructs expressing
ACE2 (Human) ELISA Kit
 ACE2 (Human) ELISA Kit Catalog Number KA4223 96 assays Version: 10 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of the Assay...
ACE2 (Human) ELISA Kit Catalog Number KA4223 96 assays Version: 10 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of the Assay...
Western-GUARANTEED Antibody Service FAQ
 Western-GUARANTEED Antibody Service FAQ Content Q 1: When do I need a Western GUARANTEED Peptide Antibody Package?...2 Q 2: Can GenScript provide a Western blot guaranteed antibody?...2 Q 3: Does GenScript
Western-GUARANTEED Antibody Service FAQ Content Q 1: When do I need a Western GUARANTEED Peptide Antibody Package?...2 Q 2: Can GenScript provide a Western blot guaranteed antibody?...2 Q 3: Does GenScript
SUPPLEMENTAL MATERIAL. Supplemental Methods:
 SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
SUPPLEMENTAL MATERIAL Supplemental Methods: Immunoprecipitation- As we described but with some modifications [22]. As part of another ongoing project, lysate from human umbilical vein endothelial cells
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide,
 Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Supplementary Figure 1. Intracellular distribution of the EPE peptide. HeLa cells were serum-starved (16 h, 0.1%), and treated with EPE peptide, conjugated with either TAT or Myristic acid and biotin for
Biochemical Binding ADCC Assays Utilizing LANCE Toolbox Reagents for the Characterization of higgs and FcγR1A
 APPLICATION NOTE LANCE TR-FRET Author: Daniel Cardillo PerkinElmer, Inc. Hopkinton, MA Biochemical Binding ADCC Assays Utilizing LANCE Toolbox Reagents for the Characterization of higgs and FcγR1A Introduction
APPLICATION NOTE LANCE TR-FRET Author: Daniel Cardillo PerkinElmer, Inc. Hopkinton, MA Biochemical Binding ADCC Assays Utilizing LANCE Toolbox Reagents for the Characterization of higgs and FcγR1A Introduction
Biosensing Probe for Quality Control Monitoring of the Structural Integrity of Therapeutic Antibodies
 Supporting Information Biosensing Probe for Quality Control Monitoring of the Structural Integrity of Therapeutic Antibodies Hideki Watanabe, Seiki Yageta, Hiroshi Imamura, and Shinya Honda *,, Biomedical
Supporting Information Biosensing Probe for Quality Control Monitoring of the Structural Integrity of Therapeutic Antibodies Hideki Watanabe, Seiki Yageta, Hiroshi Imamura, and Shinya Honda *,, Biomedical
Alternate Approaches Addressing Variability in ADCC Assay. Prabhavathy Munagala, Ph.D. United States Pharmacopeia, India October 28, 2014
 Alternate Approaches Addressing Variability in ADCC Assay Prabhavathy Munagala, Ph.D. United States Pharmacopeia, India October 28, 2014 Disclaimer It is a scientific presentation and Conclusions presented
Alternate Approaches Addressing Variability in ADCC Assay Prabhavathy Munagala, Ph.D. United States Pharmacopeia, India October 28, 2014 Disclaimer It is a scientific presentation and Conclusions presented
Visualizing mechanical tension across membrane receptors with a fluorescent sensor
 Nature Methods Visualizing mechanical tension across membrane receptors with a fluorescent sensor Daniel R. Stabley, Carol Jurchenko, Stephen S. Marshall, Khalid S. Salaita Supplementary Figure 1 Fabrication
Nature Methods Visualizing mechanical tension across membrane receptors with a fluorescent sensor Daniel R. Stabley, Carol Jurchenko, Stephen S. Marshall, Khalid S. Salaita Supplementary Figure 1 Fabrication
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Decline in HI Titer After Vaccination
 Figure S Decline in HI Titer After Vaccination Geometric mean titers were determined for the second blood draw for each of the binned groups in both the and cohorts. The titers are expressed as a percentage
Figure S Decline in HI Titer After Vaccination Geometric mean titers were determined for the second blood draw for each of the binned groups in both the and cohorts. The titers are expressed as a percentage
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a)
 Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Supplementary Figure 1 - Characterization of rbag3 binding on macrophages cell surface.
 Supplementary Figure 1 - Characterization of rbag3 binding on macrophages cell surface. (a) Human PDAC cell lines were treated as indicated in Figure 1 panel F. Cells were analyzed for FITC-rBAG3 binding
Supplementary Figure 1 - Characterization of rbag3 binding on macrophages cell surface. (a) Human PDAC cell lines were treated as indicated in Figure 1 panel F. Cells were analyzed for FITC-rBAG3 binding
Human IL10RB ELISA Pair Set ( CRFB4 )
 Human IL10RB ELISA Pair Set ( CRFB4 ) Catalog Number : SEK10945 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in the
Human IL10RB ELISA Pair Set ( CRFB4 ) Catalog Number : SEK10945 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in the
Supplementary Material
 Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
Supplementary Material Supplementary Methods Cell synchronization. For synchronized cell growth, thymidine was added to 30% confluent U2OS cells to a final concentration of 2.5mM. Cells were incubated
(Hadlock, Journal of Virology, 2000), (Keck, Journal of Virology, 2008) CBH-5 Domain B 412, 416, 417, 418, 420, 421, 422, 423, 483, 484, 485, 488,
 Supplementary Table. mabs analyzed mab Antigenic Critical Binding Residues by Citations Domains Alanine Scanning 2 CBH-4B Domain A NA (Hadlock, Journal of Virology, 2), (Keck, Journal of Virology, 24)
Supplementary Table. mabs analyzed mab Antigenic Critical Binding Residues by Citations Domains Alanine Scanning 2 CBH-4B Domain A NA (Hadlock, Journal of Virology, 2), (Keck, Journal of Virology, 24)
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
To determine MRK-003 IC50 values, cell lines were plated in triplicate in 96-well plates at 3 x
 Supplementary methods: Cellular growth assay and cell cycle analysis To determine MRK-003 IC50 values, cell lines were plated in triplicate in 96-well plates at 3 x 10 3 cells/well (except for TALL1 and
Supplementary methods: Cellular growth assay and cell cycle analysis To determine MRK-003 IC50 values, cell lines were plated in triplicate in 96-well plates at 3 x 10 3 cells/well (except for TALL1 and
InhibiScreen Kinase Inhibitor Assay
 InhibiScreen Kinase Inhibitor Assay Flow Cytometric assessment of efficacy of PI3Kγ, PI3Kδ, BTK and SYK pathway inhibitors using the measurement of basophil activation in human EDTA and Heparin Whole Blood.
InhibiScreen Kinase Inhibitor Assay Flow Cytometric assessment of efficacy of PI3Kγ, PI3Kδ, BTK and SYK pathway inhibitors using the measurement of basophil activation in human EDTA and Heparin Whole Blood.
Supplementary Figure 1. Nature Structural & Molecular Biology: doi: /nsmb.3494
 Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Supplementary Figure 1 Pol structure-function analysis (a) Inactivating polymerase and helicase mutations do not alter the stability of Pol. Flag epitopes were introduced using CRISPR/Cas9 gene targeting
Porcine IL-12/IL-23 p40 ELISA kit
 Porcine IL-12/IL-23 p40 ELISA kit Catalog number: NB-E50014 (96 wells) The kit is designed to quantitatively detect the levels of Porcine IL-12/IL-23 p40 in cell culture supernatants. FOR RESEARCH USE
Porcine IL-12/IL-23 p40 ELISA kit Catalog number: NB-E50014 (96 wells) The kit is designed to quantitatively detect the levels of Porcine IL-12/IL-23 p40 in cell culture supernatants. FOR RESEARCH USE
Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546
 Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546 Description This cell line expresses a surface human GITR (glucocorticoid-induced TNFR family related gene; TNFRSF18;
Data Sheet GITR / NF-ĸB-Luciferase Reporter (Luc) - Jurkat Cell Line Catalog #60546 Description This cell line expresses a surface human GITR (glucocorticoid-induced TNFR family related gene; TNFRSF18;
