the same host. (a) Experimental design. In order to compare the reconstituting ability of p18 -/-
|
|
|
- Gregory Watts
- 5 years ago
- Views:
Transcription
1 1
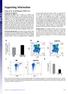
2 Supplementary Figure 1 Direct comparison between p18 -/- and p27 -/- hematopoietic cells in the same host. (a) Experimental design. In order to compare the reconstituting ability of p18 -/- and p27 -/- HSCs, we mixed BMNCs from p18 and p27 knockout mice at a ratio of 1 to 1 ( each), and then injected the cells into lethally irradiated B6.2 hosts. (b) cbmt results. The y-axis shows the ratio of p18 -/- versus p27 -/- cells. p18 -/- HSCs outcompeted p27 -/- HSCs by more than 4 times at 1 year post-bmt (n=10) (p>0.05). (c) Data from one representative recipient of cbmt, as shown in figure S1a. The values in the corner show the ratio of p18 -/- cells versus p27 -/- cells. As shown in the upper panel, p18 -/- dominated in blood, B cell, and myeloid lineages, while p27 -/- dominated in the T cell lineage. The lower panel shows the contribution of both p18 -/- and p27 -/- to different stages of hematopoiesis. 2
3 3
4 4
5 Supplementary Figure 2 Proliferative rates of HSCs and HPCs. (a) Age-matched p18 -/- and p18 +/+ mice drank water containing 1 mg/ml BrdU for 13 weeks. BrdU incorporation (%) in each population of p18 -/- and p18 +/+ mice at different time points (left). Linear regression for the LKS population was analyzed based on the equation: Ln (% of Brdu negative cells) = a + b (Time). The average turnover time is determined when the negative fraction declines to 37% (right). (b) BrdU incorporation in vivo after BMT. BMNCs ( ) from p18 -/- and p18 +/+ were transplanted into lethally irradiated mice, respectively. The mice received 100 µg/g of BrdU via I.P. injection and were sacrificed 16 hours afterward. Donor BMNCs were assayed for BrdU incorporation in different hematopoietic cell subsets using flow cytometry at different time 5
6 points, as indicated. No statistically significant difference between p18 +/+ and p18 -/- was observed at any time point (n=3-5/each). (c-f) Proliferative kinetics of single HSCs in vitro. Single CD34 - KLS cells were sorted into Terasaki plates with serum free medium containing 50 ng/ml SCF, 25 ng/ml FL, and 10 ng/ml TPO. A total of 147 cells from p18 +/+ mice and 168 cells from p18 -/- mice were monitored. Cells in each well were counted daily for 3 days. (c) Frequencies of divided cells ( 2 cells/well); (d) Frequencies of undivided cells (0 o ) and cells that divided once (1 o ) or more (2 o ) at day 3. (e) Distribution of total cell numbers per well at day 3. (f) Overall expansion of the original single cells at day 3. (g) the total number of homed cells (CD45.2) in the BM 17 hours after lin - Sca1 + c-kit + (LSK) cells were injected into the recipients.(h) Flow cytometry analysis of the average percentages of G0, G1, and S/G2/M fractions in CD LKS cells. 6
7 7
8 Supplementary Figure 3 (a) 3D structural model of the p18 protein and docking poses of all tested compounds at the p18 surface binding cavity. (b) Overview of the 3D structural model of 8
9 the p18 protein and docking poses of P18IN011. Purple dashed lines highlight the hydrogen binding. Yellow and blue dashed lines indicate the hydrophobic interaction with F37 and the charge interaction with R39, respectively. (c) The limit dilution analysis of groups treated with P18IN03A or P18IN005. (d) CDK6 activity assay in vitro. Recombinant Rb fragments were used as substrates for the activated complexes of CDK6/ CyclinD1 32. P-labeled GST-Rb was quantitatively analyzed by a scintillation counter and normalized with the mean of the CDK6-Rb group. Supplementary Figure 4 Frequency of CD34 - LKS cells measured by limit dilution assay (LDA). 9
10 Supplementary Figure 5 General chemistry syntheses route of P18IN011 and its analogs. 10
11 Supplementary Table 1 Function of HSCs (CD34-LKS) in the Secondary Recipients (6 months post-cbmt) Genotype Input Output Progeny/HSC (HSCs/mouse) (BMNCs/mouse) p18 -/- 2, ,000 p27 -/ ,600 WT ,000 11
12 Supplementary Note p18 inhibitors discovered. To overcome the intrinsic limitation and low throughput of the current bioassays for screening small molecules and testing HSC self-renewal, we have developed alternative technologies, which have enabled us to discover the first p18 inhibitor using our established target-focused computational chemical genomics screening and in vitro and ex vivo validation approaches. First, we have modeled the 3D protein structure of p18 and its binding surface cavity using our reported protocol 1, 2 based on X-ray crystallographic data 3, 4, 5, 6. The X-ray data show that p18 protein, which consists of five ankyrin repeats, binds next to the ATP binding site of the CDK6 catalytic cleft (residues ), and interacts with both the N-lobe (residues 9-104, rich in β-sheet) and C-lobe (residues, , predominantly α-helix) of the CDK6 kinase. According to structural biology data 7, p18 binding to CDK6 may lead to the dissociation of cyclin D, which binds at the site opposite the CDK6 catalytic cleft (PASTAIRE helix residues: 55-66). In addition, the data show that when the CDK6 is bound to p18, its N- and C-lobes are rotated 13Å away from each other, resulting in the misalignment of ATP binding residues. As a result, the N-lobe PASTAIRE helix domain of CDK6, which contains an invariant active site residue (Glu61), is displaced by 64.5 Å away from the active site, and is rotated by 16 Å. In addition, p18 displaces the N-lobe relative to the C lobe of CDK6, causing the hydrophobic residues (Ile19, 12
13 Val27, Ala41, Leu152) that sandwich the adenine ring of ATP to move by up to 4.5 Å. p18 inhibition of CDK6 also distorts the edge of the CDK6 active site via Phe82, affecting H-bonding interactions with the edge of the ATP ring. Taken together, these results cause us to speculate that binding of p18 to CDK6 disrupts the ATP binding site, as well as the cyclin D binding site in CDK6, thus blocking the activation effects of the CDK6/cyclin complex on the HSC cell cycle. Such important phenomena led us to the hypotheses that identifying a p18 protein blocker can impede p18 binding to CDK6, distort the CDK6 cell cycle, and stimulate the activation effect of the kinase complex on HSC cell self-renewal. Having defined the binding cavity at p18 above, then we conducted 3D docking virtual screening using our reported 3D virtual screening approach 1, 8 and compound library 9. The top 15% of docking score ranked compounds were subjected to manual inspection according to the conserved hydrogen bond with Arg39 or Asp76 of p18. Among them, the lead compounds P18IN003 and P18IN011 have a total score (equivalent logk d ) of 6.80 and 7.38, respectively, which are ranked in the top 5% of the in silico screened compounds. Subsequently, 12 compounds were obtained via MTA or commercially, and biologically tested in vitro for their ability to promote HSC self-renewal, among which 2 compounds (P18IN003 and 13
14 P18IN011) showed medium to great increased activity. The compound P18IN011 (MW , clogp 2.3) was identified as one of the virtual hits by the Surflex-dock database search. The docking pose shows that the hit compound P18IN011 has extensive H-bonding interactions with the key residues, including its amide NH group with D76 (1.90 Å), C=O with Q43 (3.16 Å) and D67 (3.20 Å), sulfonate O=S=O with and T69 (2.71 Å) and its charge interaction with R39 (charge interaction, 3.12 Å) of the p18 protein (Fig. 3c). In addition, hydrophobic interactions were also detected for its isoxazobenzine ring with F37 and V44 of p18 and also the acetamidobenzene ring surrounded by hydrophobic residues F71, A72 and V73 of p18 in a range of 3.3 ~ 3.7 Å. It is known that the residues F37, R39, Q43, V44, D67 and D76 of p18 are highly conserved residues located at the p18/cdk6 interface and they that play important roles in the H-bonding or hydrophobic interactions with CDK6. The identified additional binding residues T69, F71, A72 and V73, are exclusively for p18 based on homology modeling in comparison with other CKI proteins. Thus, by blocking these key residues, a compound is predicted to disrupt the interaction of p18 with CDK6, which has been further confirmed by the biological experiments. Finally, we carried out the CAFC assay with limiting dilution using our reported protocol 10, 11 to functionally validate the effects of the virtual p18 hits above. The confirmatory experiments with 14
15 dose response curves show that compounds P18IN011 and P18IN03 can significantly increase the frequency of HSCs (ED 50, 3.07 and 5.70 µm), suggesting a potential enhancement of HSC self-renewal. References: 1. Chen JZ, Wang J, Xie XQ. GPCR structure-based virtual screening approach for CB2 antagonist search. J Chem Inf Model 47, (2007). 2. Xie* XQ, Chen JZ, Billings EM. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins: Structure,Function,and Genetics 53, (2003). 3. Brotherton DH, et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19ink4d. Nature (London) 395, (1998). 4. Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes & Development 14, (2000). 5. Padmanabhan B, Adachi N, Kataoka K, Horikoshi M. Crystal Structure of the Homolog of the Oncoprotein Gankyrin, an Interactor of Rb and CDK4/6. J Biol Chem 279, (2004). 6. Venkataramani RN, MacLachlan TK, Chai X, El-Deiry WS, Marmorstein R. Structure-based Design of p18ink4c Proteins with Increased Thermodynamic Stability and Cell Cycle Inhibitory Activity. J Biol Chem 277, (2002). 7. Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev FIELD Full Journal Title:Genes & Development 14, (2000). 15
16 8. Chen J-Z, Han X-W, Liu Q, Makriyannis A, Wang J, Xie* X-Q. 3D-QSAR Studies of Arylpyrazole Antagonists of Cannabinoid Receptor Subtypes CB1 and CB2. A Combined NMR and CoMFA Approach. Journal of medicinal chemistry 49, (2006). 9. Xie* XQ, Chen JZ. Data mining a small molecule drug screening representative subset from NIH PubChem. J Chem Inf Model 48, (2008). 10. Yu H, Yuan Y, Shen H, Cheng T. Hematopoietic stem cell exhaustion impacted by p18 INK4C and p21 Cip1/Waf1 in opposite manners. Blood 107, (2006). 11. Yuan Y, Shen H, Franklin DS, Scadden DT, Cheng T. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18ink4c. Nat Cell Biol 6, (2004). 16
Supplemental Information Inventory
 Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1: Phenotypically defined hematopoietic stem and progenitor cell populations show distinct mitochondrial activity and mass. (A) Isolation by FACS of commonly
Supplementary Figures Supplementary Figure 1: Phenotypically defined hematopoietic stem and progenitor cell populations show distinct mitochondrial activity and mass. (A) Isolation by FACS of commonly
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Protocol S1: Supporting Information
 Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Protocol S1: Supporting Information Basis for the specificity of the kinase domain of Abl for peptide substrates The crystal structures reported in this work were obtained using two different ATP analog-peptide
Algorithms in Bioinformatics ONE Transcription Translation
 Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Algorithms in Bioinformatics ONE Transcription Translation Sami Khuri Department of Computer Science San José State University sami.khuri@sjsu.edu Biology Review DNA RNA Proteins Central Dogma Transcription
Clamping down on pathogenic bacteria how to shut down a key DNA polymerase complex
 Clamping down on pathogenic bacteria how to shut down a key DNA polymerase complex Bacterial DNA-replication machinery Pathogenic bacteria that are resistant to the current armoury of antibiotics are an
Clamping down on pathogenic bacteria how to shut down a key DNA polymerase complex Bacterial DNA-replication machinery Pathogenic bacteria that are resistant to the current armoury of antibiotics are an
Drug Discovery Targeting the GHKL Superfamily - HSP90, DNA Gyrase, Histidine Kinases
 Drug Discovery Targeting the GHKL Superfamily - HSP90, DNA Gyrase, Histidine Kinases (Jason) Xinshan Kang, Boris Klebansky, Richard Fine BioPredict, Inc., 660 KinderKamack Rd., radell, NJ 07649 232 nd
Drug Discovery Targeting the GHKL Superfamily - HSP90, DNA Gyrase, Histidine Kinases (Jason) Xinshan Kang, Boris Klebansky, Richard Fine BioPredict, Inc., 660 KinderKamack Rd., radell, NJ 07649 232 nd
STRUCTURAL BIOLOGY. α/β structures Closed barrels Open twisted sheets Horseshoe folds
 STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
STRUCTURAL BIOLOGY α/β structures Closed barrels Open twisted sheets Horseshoe folds The α/β domains Most frequent domain structures are α/β domains: A central parallel or mixed β sheet Surrounded by α
1) The penicillin family of antibiotics, discovered by Alexander Fleming in 1928, has the following general structure: O O
 ame: TF ame: LS1a Fall 06 Problem Set #3 Due Friday 10/13 at noon in your TF s drop box on the 2 nd floor of the Science Center All questions including the (*extra*) ones should be turned in 1) The penicillin
ame: TF ame: LS1a Fall 06 Problem Set #3 Due Friday 10/13 at noon in your TF s drop box on the 2 nd floor of the Science Center All questions including the (*extra*) ones should be turned in 1) The penicillin
Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity
 Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity Stanley G. Kimani 1 *, Sushil Kumar 1 *, Nitu Bansal 2 *, Kamalendra Singh 3, Vladyslav Kholodovych
Supplementary information for Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity Stanley G. Kimani 1 *, Sushil Kumar 1 *, Nitu Bansal 2 *, Kamalendra Singh 3, Vladyslav Kholodovych
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
Druggability & DruGUI. Ahmet Bakan Department of Computational and Systems Biology
 Druggability & DruGUI Ahmet Bakan ahb12@pitt.edu Department of Computational and Systems Biology Target Druggability Can a given biological target, such as a protein, bind with high affinity to a drug?
Druggability & DruGUI Ahmet Bakan ahb12@pitt.edu Department of Computational and Systems Biology Target Druggability Can a given biological target, such as a protein, bind with high affinity to a drug?
7-amino actinomycin D (7ADD) was added to all samples 10 minutes prior to analysis on the flow cytometer in order to gate 7AAD viable cells.
 Antibody staining for Ho uptake analyses For HSC staining, 10 7 BM cells from Ho perfused mice were stained with biotinylated lineage antibodies (CD3, CD5, B220, CD11b, Gr-1, CD41, Ter119), anti Sca-1-PECY7,
Antibody staining for Ho uptake analyses For HSC staining, 10 7 BM cells from Ho perfused mice were stained with biotinylated lineage antibodies (CD3, CD5, B220, CD11b, Gr-1, CD41, Ter119), anti Sca-1-PECY7,
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang *
 In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
88.7 <GFP-A>: EGFP GFP. Null. % of Cells! <APC-A> B220!
 B. C. 1 Cell Number (X18) A. % of Cells % of Max 8 Con 6 4 88.7 1 13 14 : EGFP 4 6 4 1 WT 8 % of% ofcells Max 6 6 4 G. 1 13 B Hdac3-/- 14 15 1 13 14 15 Ter119 Ly6C Wild type WT 4
B. C. 1 Cell Number (X18) A. % of Cells % of Max 8 Con 6 4 88.7 1 13 14 : EGFP 4 6 4 1 WT 8 % of% ofcells Max 6 6 4 G. 1 13 B Hdac3-/- 14 15 1 13 14 15 Ter119 Ly6C Wild type WT 4
Nature Biotechnology: doi: /nbt.4086
 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Solutions to 7.02 Quiz II 10/27/05
 Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
Solutions to 7.02 Quiz II 10/27/05 Class Average = 83 Standard Deviation = 9 Range Grade % 87-100 A 43 74-86 B 39 55-73 C 17 > 54 D 1 Question 1 (56 points) While studying deep sea bacteria, you discover
About ATCC. Established partner to global researchers and scientists
 Discovering ATCC Primary Immunology Cells - Model Systems to Study the Immune and Cardiovascular Systems James Clinton, Ph.D. Scientist, ATCC July 14, 2016 About ATCC Founded in 1925, ATCC is a non-profit
Discovering ATCC Primary Immunology Cells - Model Systems to Study the Immune and Cardiovascular Systems James Clinton, Ph.D. Scientist, ATCC July 14, 2016 About ATCC Founded in 1925, ATCC is a non-profit
PRIME-XV Hematopoietic Cell Basal XSFM
 PRIME-XV Hematopoietic Cell Basal XSFM Xeno-free, serum-free basal medium for human hematopoietic progenitor cell culture Optimized to support vigorous expansion of hematopoietic progenitor cells while
PRIME-XV Hematopoietic Cell Basal XSFM Xeno-free, serum-free basal medium for human hematopoietic progenitor cell culture Optimized to support vigorous expansion of hematopoietic progenitor cells while
In silico measurements of twist and bend. moduli for beta solenoid protein self-
 In silico measurements of twist and bend moduli for beta solenoid protein self- assembly units Leonard P. Heinz, Krishnakumar M. Ravikumar, and Daniel L. Cox Department of Physics and Institute for Complex
In silico measurements of twist and bend moduli for beta solenoid protein self- assembly units Leonard P. Heinz, Krishnakumar M. Ravikumar, and Daniel L. Cox Department of Physics and Institute for Complex
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Supplementary Table 1. Crystallographic statistics CRM1-SNUPN complex Space group P6 4 22 a=b=250.4, c=190.4 Data collection statistics: CRM1-selenomethionine SNUPN MAD data Peak Inflection Remote Native
Application of label-free technologies in a core facility research. Ewa Folta-Stogniew Yale University
 Application of label-free technologies in a core facility research Ewa Folta-Stogniew Yale University Biophysics Resource of Keck Laboratory: Yale School of Medicine Mission: quantitative characterization
Application of label-free technologies in a core facility research Ewa Folta-Stogniew Yale University Biophysics Resource of Keck Laboratory: Yale School of Medicine Mission: quantitative characterization
Products & Services. Customers. Example Customers & Collaborators
 Products & Services Eidogen-Sertanty is dedicated to delivering discovery informatics technologies that bridge the target-tolead knowledge gap. With a unique set of ligand- and structure-based drug discovery
Products & Services Eidogen-Sertanty is dedicated to delivering discovery informatics technologies that bridge the target-tolead knowledge gap. With a unique set of ligand- and structure-based drug discovery
NANYANG TECHNOLOGICAL UNIVERSITY SEMESTER I EXAMINATION CBC922 Medicinal Chemistry. NOVEMBER TIME ALLOWED: 120 min
 AYAG TECLGICAL UIVERSITY SEMESTER I EXAMIATI 2006-2007 CBC922 Medicinal Chemistry VEMBER 2006 - TIME ALLWED 120 min ISTRUCTIS T CADIDATES 1. This examination paper contains TW (2) parts and comprises SIX
AYAG TECLGICAL UIVERSITY SEMESTER I EXAMIATI 2006-2007 CBC922 Medicinal Chemistry VEMBER 2006 - TIME ALLWED 120 min ISTRUCTIS T CADIDATES 1. This examination paper contains TW (2) parts and comprises SIX
Scanning the protein surface to uncover new small molecule. binding sites using fragments. Gregg Siegal ZoBio
 Scanning the protein surface to uncover new small molecule binding sites using fragments Gregg Siegal ZoBio Increasing Interest in Alternative Small Molecule Binding Sites 1. Enhanced specificity 2. Adress
Scanning the protein surface to uncover new small molecule binding sites using fragments Gregg Siegal ZoBio Increasing Interest in Alternative Small Molecule Binding Sites 1. Enhanced specificity 2. Adress
Fig. 1. A schematic illustration of pipeline from gene to drug : integration of virtual and real experiments.
 1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
1 Integration of Structure-Based Drug Design and SPR Biosensor Technology in Discovery of New Lead Compounds Alexis S. Ivanov Institute of Biomedical Chemistry RAMS, 10, Pogodinskaya str., Moscow, 119121,
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Optimization of an Adapta Kinase Assay for Cdk9/cyclin T1
 Optimization of an Adapta Kinase Assay for Cdk9/cyclin T1 Overview This protocol describes how to perform an Adapta assay with the kinase Cdk9/cyclin T1. To maximize the ability of the assay to detect
Optimization of an Adapta Kinase Assay for Cdk9/cyclin T1 Overview This protocol describes how to perform an Adapta assay with the kinase Cdk9/cyclin T1. To maximize the ability of the assay to detect
Miniaturized Whole Cell-based GPCR camp Assay Using a Novel Detection System
 Miniaturized Whole Cell-based GPCR camp Assay Using a Novel Detection System Geetha Shankar, Ph.D. Associate Director New Lead Discovery Exelixis, Inc South San Francisco, CA Overview GPCR HTS assays ACT:One
Miniaturized Whole Cell-based GPCR camp Assay Using a Novel Detection System Geetha Shankar, Ph.D. Associate Director New Lead Discovery Exelixis, Inc South San Francisco, CA Overview GPCR HTS assays ACT:One
COMPUTER AIDED DRUG DESIGN: AN OVERVIEW
 Available online on 19.09.2018 at http://jddtonline.info Journal of Drug Delivery and Therapeutics Open Access to Pharmaceutical and Medical Research 2011-18, publisher and licensee JDDT, This is an Open
Available online on 19.09.2018 at http://jddtonline.info Journal of Drug Delivery and Therapeutics Open Access to Pharmaceutical and Medical Research 2011-18, publisher and licensee JDDT, This is an Open
Introduction to Protein Purification
 Introduction to Protein Purification 1 Day 1) Introduction to Protein Purification. Input for Purification Protocol Development - Guidelines for Protein Purification Day 2) Sample Preparation before Chromatography
Introduction to Protein Purification 1 Day 1) Introduction to Protein Purification. Input for Purification Protocol Development - Guidelines for Protein Purification Day 2) Sample Preparation before Chromatography
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Prediction of Protein-Protein Binding Sites and Epitope Mapping. Copyright 2017 Chemical Computing Group ULC All Rights Reserved.
 Prediction of Protein-Protein Binding Sites and Epitope Mapping Epitope Mapping Antibodies interact with antigens at epitopes Epitope is collection residues on antigen Continuous (sequence) or non-continuous
Prediction of Protein-Protein Binding Sites and Epitope Mapping Epitope Mapping Antibodies interact with antigens at epitopes Epitope is collection residues on antigen Continuous (sequence) or non-continuous
Ligand docking. CS/CME/Biophys/BMI 279 Oct. 22 and 27, 2015 Ron Dror
 Ligand docking CS/CME/Biophys/BMI 279 Oct. 22 and 27, 2015 Ron Dror 1 Outline Goals of ligand docking Defining binding affinity (strength) Computing binding affinity: Simplifying the problem Ligand docking
Ligand docking CS/CME/Biophys/BMI 279 Oct. 22 and 27, 2015 Ron Dror 1 Outline Goals of ligand docking Defining binding affinity (strength) Computing binding affinity: Simplifying the problem Ligand docking
Structure Based Virtual Screening for Discovery of Novel Human Neutrophil Elastase Inhibitors
 Structure Based Virtual Screening for Discovery of Novel Human Neutrophil Elastase Inhibitors Susana D. Lucas, a Lídia M. Gonçalves, a Teresa A. F. Cardote, a Henrique F. Correia, a Rui Moreira, a Rita
Structure Based Virtual Screening for Discovery of Novel Human Neutrophil Elastase Inhibitors Susana D. Lucas, a Lídia M. Gonçalves, a Teresa A. F. Cardote, a Henrique F. Correia, a Rui Moreira, a Rita
From mechanism to medicne
 From mechanism to medicne a look at proteins and drug design Chem 342 δ δ δ+ M 2009 δ+ δ+ δ M Drug Design - an Iterative Approach @ DSU Structural Analysis of Receptor Structural Analysis of Ligand-Receptor
From mechanism to medicne a look at proteins and drug design Chem 342 δ δ δ+ M 2009 δ+ δ+ δ M Drug Design - an Iterative Approach @ DSU Structural Analysis of Receptor Structural Analysis of Ligand-Receptor
Introduction to Assay Development
 Introduction to Assay Development A poorly designed assay can derail a drug discovery program before it gets off the ground. While traditional bench-top assays are suitable for basic research and target
Introduction to Assay Development A poorly designed assay can derail a drug discovery program before it gets off the ground. While traditional bench-top assays are suitable for basic research and target
Molecular design principles underlying β-strand swapping. in the adhesive dimerization of cadherins
 Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
Supplementary information for: Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins Jeremie Vendome 1,2,3,5, Shoshana Posy 1,2,3,5,6, Xiangshu Jin, 1,3 Fabiana
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid
 Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
Supplementary Figures Suppl. Figure 1: RCC1 sequence and sequence alignments. (a) Amino acid sequence of Drosophila RCC1. Same colors are for Figure 1 with sequence of β-wedge that interacts with Ran in
GSI Bovine TGF-β3 ELISA Kit-2 Plates DataSheet
 Transforming growth factor, beta 3 (TGF-β3) belongs to a large family of cytokines called the Transforming growth factor beta super family (1, 2), which includes the TGF-β family, bone morphogenetic proteins
Transforming growth factor, beta 3 (TGF-β3) belongs to a large family of cytokines called the Transforming growth factor beta super family (1, 2), which includes the TGF-β family, bone morphogenetic proteins
How Targets Are Chosen. Chris Wayman 12 th April 2012
 How Targets Are Chosen Chris Wayman 12 th April 2012 A few questions How many ideas does it take to make a medicine? 10 20 20-50 50-100 A few questions How long does it take to bring a product from bench
How Targets Are Chosen Chris Wayman 12 th April 2012 A few questions How many ideas does it take to make a medicine? 10 20 20-50 50-100 A few questions How long does it take to bring a product from bench
show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras ex3op/ex3op mice
 Supplemental Data Supplemental Figure Legends Supplemental Figure 1. Hematologic profile of Kras / mice. (A) Scatter plots show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras / mice
Supplemental Data Supplemental Figure Legends Supplemental Figure 1. Hematologic profile of Kras / mice. (A) Scatter plots show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras / mice
Supplementary Information. Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications
 Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
Supplementary Information Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications Zachary B Hill 1,4, Alexander J Martinko 1,2,4, Duy P Nguyen 1 & James A Wells *1,3 1 Department
Z -LYTE fluorescent kinase assay technology. Z -LYTE Kinase Assay Platform
 Z -LYTE fluorescent kinase assay technology Z -LYTE Kinase Assay Platform Z -LYTE Kinase Assay Platform Non-radioactive assay format for screening a diverse collection of over 185 kinase targets Assay
Z -LYTE fluorescent kinase assay technology Z -LYTE Kinase Assay Platform Z -LYTE Kinase Assay Platform Non-radioactive assay format for screening a diverse collection of over 185 kinase targets Assay
Analysis of gene function
 Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
MOLECULAR DOCKING STUDIES FOR IDENTIFICATION OF POTENT PHOSPHOINOSITIDE 3-KINASES (PI3KS) INHIBITORS
 CHAPTER 7 MLECULAR DCKIG STUDIES FR IDETIFICATI F PTET PHSPHISITIDE 3-KIASES (PI3KS) IHIBITRS 7.1 Introduction Phosphoinositide 3 kinases (PI3Ks) constitute a class of enzymes that catalyse phosphorylation
CHAPTER 7 MLECULAR DCKIG STUDIES FR IDETIFICATI F PTET PHSPHISITIDE 3-KIASES (PI3KS) IHIBITRS 7.1 Introduction Phosphoinositide 3 kinases (PI3Ks) constitute a class of enzymes that catalyse phosphorylation
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade
 Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade Supplementary Materials Supplementary methods Table S1-S Figure S1-S 1 1 1 1 1 1 1 1 1 0 1 0 Supplementary Methods Competitive
Application Note AN001
 Testing hybridoma supernatants with the Spots On Dots Antibody Screening Kit Application Note AN1 Table of Contents Overview... 2 Figure 1. Screening of hybridomas raised against peptide antigens... 3
Testing hybridoma supernatants with the Spots On Dots Antibody Screening Kit Application Note AN1 Table of Contents Overview... 2 Figure 1. Screening of hybridomas raised against peptide antigens... 3
Supporting Information
 (xe number cells) LSK (% gated) Supporting Information Youm et al../pnas. SI Materials and Methods Quantification of sjtrecs. CD + T subsets were isolated from splenocytes using mouse CD + T cells positive
(xe number cells) LSK (% gated) Supporting Information Youm et al../pnas. SI Materials and Methods Quantification of sjtrecs. CD + T subsets were isolated from splenocytes using mouse CD + T cells positive
Multi-Criteria Drug Discovery: using CoMFA models to drive target specificity. Lei Wang, Ph.D.
 Multi-Criteria Drug Discovery: using CoMFA models to drive target specificity Lei Wang, Ph.D. The Problem Compounds exist that have some desirable properties to become a drug but The known compounds are
Multi-Criteria Drug Discovery: using CoMFA models to drive target specificity Lei Wang, Ph.D. The Problem Compounds exist that have some desirable properties to become a drug but The known compounds are
Hematopoietic stem cell transplantation without irradiation
 nature methods Hematopoietic stem cell transplantation without irradiation Claudia Waskow, Vikas Madan, Susanne Bartels, Céline Costa, Rosel Blasig & Hans-Reimer Rodewald Supplementary figures and text:
nature methods Hematopoietic stem cell transplantation without irradiation Claudia Waskow, Vikas Madan, Susanne Bartels, Céline Costa, Rosel Blasig & Hans-Reimer Rodewald Supplementary figures and text:
DKK3 (Human) ELISA Kit
 DKK3 (Human) ELISA Kit Catalog Number KA1706 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information... 4
DKK3 (Human) ELISA Kit Catalog Number KA1706 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information... 4
Supplementary Figure 1. GST pull-down analysis of the interaction of GST-cIAP1 (A, B), GSTcIAP1
 Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Legends Supplementary Figure 1. GST pull-down analysis of the interaction of GST- (A, B), GST mutants (B) or GST- (C) with indicated proteins. A, B, Cell lysate from untransfected HeLa cells were loaded
Measurement of Hematopoietic Stem Cell Potency Prior to Transplantation
 WHITE PAPER Measurement of Hematopoietic Stem Cell Potency Prior to Transplantation February, 2009 This White Paper is a forward-looking statement. It represents the present state of the art and future
WHITE PAPER Measurement of Hematopoietic Stem Cell Potency Prior to Transplantation February, 2009 This White Paper is a forward-looking statement. It represents the present state of the art and future
Hgf (Mouse) ELISA Kit
 Hgf (Mouse) ELISA Kit Catalog Number KA1785 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay... 3 General
Hgf (Mouse) ELISA Kit Catalog Number KA1785 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Principle of the Assay... 3 General
Mid-term review. Recitation5 03/31/2014. Stem Cell Biology and Function W4193 1
 Mid-term review Recitation5 03/31/2014 Stem Cell Biology and Function W4193 1 What to expect in the midterm2 It will be all so far but mainly whatever was NOT covered by the previous mid-term- not a huge
Mid-term review Recitation5 03/31/2014 Stem Cell Biology and Function W4193 1 What to expect in the midterm2 It will be all so far but mainly whatever was NOT covered by the previous mid-term- not a huge
Cxcl13 (Mouse) ELISA Kit
 Cxcl13 (Mouse) ELISA Kit Catalog Number KA1778 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Cxcl13 (Mouse) ELISA Kit Catalog Number KA1778 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Combing molecular dynamics and machine learning for advanced pose and free-energy prediction
 Combing molecular dynamics and machine learning for advanced pose and free-energy prediction Oleksandr Yakovenko (Alex), PhD Chief Technical Officer IFOWON.CO (a spinoff from BC Cancer Agency) Vancouver,
Combing molecular dynamics and machine learning for advanced pose and free-energy prediction Oleksandr Yakovenko (Alex), PhD Chief Technical Officer IFOWON.CO (a spinoff from BC Cancer Agency) Vancouver,
Virtual Screening Manager for Grids
 Large-scale distributed in silico drug discovery using VSM-G Leo GHEMTIO : Phd Student (lghemtio@loria.fr) Bernard MAIGRET : Advisor (maigret@loria.fr) INRIA LORRAINE, http://www.loria.fr Virtual Screening
Large-scale distributed in silico drug discovery using VSM-G Leo GHEMTIO : Phd Student (lghemtio@loria.fr) Bernard MAIGRET : Advisor (maigret@loria.fr) INRIA LORRAINE, http://www.loria.fr Virtual Screening
RayBio Human TIMP-2 ELISA Kit
 RayBio Human TIMP-2 ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Human TIMP-2 ELISA Kit Protocol (Cat#: ELH-TIMP2-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service
RayBio Human TIMP-2 ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Human TIMP-2 ELISA Kit Protocol (Cat#: ELH-TIMP2-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service
Synergy NEO HTS Multi-Mode Microplate Reader and Life Technologies Reagents
 A p p l i c a t i o n G u i d e Synergy NEO HTS Multi-Mode Microplate Reader and Life Technologies Reagents High Performance Multi-Detection Capabilities that Span the Small Molecule Drug Discovery Process
A p p l i c a t i o n G u i d e Synergy NEO HTS Multi-Mode Microplate Reader and Life Technologies Reagents High Performance Multi-Detection Capabilities that Span the Small Molecule Drug Discovery Process
Ccl20 (Mouse) ELISA Kit
 Ccl20 (Mouse) ELISA Kit Catalog Number KA1805 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Ccl20 (Mouse) ELISA Kit Catalog Number KA1805 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Molecular Structures
 Molecular Structures 1 Molecular structures 2 Why is it important? Answers to scientific questions such as: What does the structure of protein X look like? Can we predict the binding of molecule X to Y?
Molecular Structures 1 Molecular structures 2 Why is it important? Answers to scientific questions such as: What does the structure of protein X look like? Can we predict the binding of molecule X to Y?
Flt3l (Mouse) ELISA Kit
 Flt3l (Mouse) ELISA Kit Catalog Number KA1784 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information...
Flt3l (Mouse) ELISA Kit Catalog Number KA1784 96 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information...
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information
 Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate toward tumor blood vessels Supplementary Information 1. Supplementary Figure S1-S10: Pages 2-11 2. Supplementary References:
Supporting Information
 Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
Supporting Information Chan et al. 10.1073/pnas.0903849106 SI Text Protein Purification. PCSK9 proteins were expressed either transiently in 2936E cells (1), or stably in HepG2 cells. Conditioned culture
SUPPLEMENTARY INFORMATION. Design and Characterization of Bivalent BET Inhibitors
 SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
SUPPLEMENTARY INFORMATION Design and Characterization of Bivalent BET Inhibitors Minoru Tanaka 1,2,#, Justin M. Roberts 1,#, Hyuk-Soo Seo 3, Amanda Souza 1, Joshiawa Paulk 1, Thomas G. Scott 1, Stephen
SensoLyte 440 West Nile Virus Protease Assay Kit *Fluorimetric*
 SensoLyte 440 West Nile Virus Protease Assay Kit *Fluorimetric* Revision Number: 1.1 Last updated: October 2014 Catalog # Kit Size AS-72079 500 Assays (96-well) Optimized Performance: This kit is optimized
SensoLyte 440 West Nile Virus Protease Assay Kit *Fluorimetric* Revision Number: 1.1 Last updated: October 2014 Catalog # Kit Size AS-72079 500 Assays (96-well) Optimized Performance: This kit is optimized
Computational Drug Design
 Computational Drug Design Univ.-Prof. Dr. Gerhard F. Ecker Pharmacoinformatics Research Group Department of Medicinal Chemistry, University of Vienna Althanstrasse 14, A-1090 Wien, Austria e-mail: gerhard.f.ecker@univie.ac.at;
Computational Drug Design Univ.-Prof. Dr. Gerhard F. Ecker Pharmacoinformatics Research Group Department of Medicinal Chemistry, University of Vienna Althanstrasse 14, A-1090 Wien, Austria e-mail: gerhard.f.ecker@univie.ac.at;
RayBio Human Leptin ELISA Kit
 RayBio Human Leptin ELISA Kit User Manual (Revised May 1, 2013) RayBio Human Leptin ELISA Kit Protocol (Cat#: ELH-Leptin-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service
RayBio Human Leptin ELISA Kit User Manual (Revised May 1, 2013) RayBio Human Leptin ELISA Kit Protocol (Cat#: ELH-Leptin-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
Ubiquitin (76 aa) UB genes encode linear fusions of UB either to itself (poly-ub genes) or to other proteins these fusions are cleaved by
 Ubiquitin (76 aa) UB genes encode linear fusions of UB either to itself (poly-ub genes) or to other proteins these fusions are cleaved by deubiquitylases (DUBs) yielding mature Ub deubiquitylase FLAG 3
Ubiquitin (76 aa) UB genes encode linear fusions of UB either to itself (poly-ub genes) or to other proteins these fusions are cleaved by deubiquitylases (DUBs) yielding mature Ub deubiquitylase FLAG 3
Solving Structure Based Design Problems using Discovery Studio 1.7 Building a Flexible Docking Protocol
 Solving Structure Based Design Problems using Discovery Studio 1.7 Building a Flexible Docking Protocol C. M. (Venkat) Venkatachalam Fellow, Life Sciences Dipesh Risal Marketing, Life Sciences Overview
Solving Structure Based Design Problems using Discovery Studio 1.7 Building a Flexible Docking Protocol C. M. (Venkat) Venkatachalam Fellow, Life Sciences Dipesh Risal Marketing, Life Sciences Overview
Polypharmacology. Giulio Rastelli Molecular Modelling and Drug Design Lab
 Polypharmacology Giulio Rastelli Molecular Modelling and Drug Design Lab www.mmddlab.unimore.it Dipartimento di Scienze della Vita Università di Modena e Reggio Emilia giulio.rastelli@unimore.it Magic
Polypharmacology Giulio Rastelli Molecular Modelling and Drug Design Lab www.mmddlab.unimore.it Dipartimento di Scienze della Vita Università di Modena e Reggio Emilia giulio.rastelli@unimore.it Magic
Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on
 Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
Supplemental Material Detailed Methods Cell culture and drug treatment. Lineage - Sca-1+ CD31+ EPCs were cultured on 5µg/mL human fibronectin coated plates in DMEM supplemented with 10% FBS and penicillin/streptomycin
1. Introduction. 2. Materials and Method
 American Journal of Pharmacological Sciences, 2016, Vol. 4, No. 1, 1-6 Available online at http://pubs.sciepub.com/ajps/4/1/1 Science and Education Publishing DOI:10.12691/ajps-4-1-1 In-silico Designing
American Journal of Pharmacological Sciences, 2016, Vol. 4, No. 1, 1-6 Available online at http://pubs.sciepub.com/ajps/4/1/1 Science and Education Publishing DOI:10.12691/ajps-4-1-1 In-silico Designing
466 Asn (N) to Ala (A) Generate beta dimer Interface
 Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
Table S1: Amino acid changes to the HexA α-subunit to convert the dimer interface from α to β and to introduce the putative GM2A binding surface from β- onto the α- subunit Residue position (α-numbering)
SUPPLEMENTARY INFORMATION. Small molecule activation of the TRAIL receptor DR5 in human cancer cells
 SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
SUPPLEMENTARY INFORMATION Small molecule activation of the TRAIL receptor DR5 in human cancer cells Gelin Wang 1*, Xiaoming Wang 2, Hong Yu 1, Shuguang Wei 1, Noelle Williams 1, Daniel L. Holmes 1, Randal
Molecular Structures
 Molecular Structures 1 Molecular structures 2 Why is it important? Answers to scientific questions such as: What does the structure of protein X look like? Can we predict the binding of molecule X to Y?
Molecular Structures 1 Molecular structures 2 Why is it important? Answers to scientific questions such as: What does the structure of protein X look like? Can we predict the binding of molecule X to Y?
Z -LYTE KINASE ASSAY KIT TYR 3 PEPTIDE PROTOCOL
 Z -LYTE KINASE ASSAY KIT TYR 3 PEPTIDE PROTOCOL Cat. no. PV3192 O-062193-r1 US 0405 TABLE OF CONTENTS 1.0 INTRODUCTION... 2 2.0 ASSAY THEORY... 2 3.0 Z9-LYTE KINASE ASSAY KIT TYR 3 PEPTIDE MATERIALS PROVIDED...
Z -LYTE KINASE ASSAY KIT TYR 3 PEPTIDE PROTOCOL Cat. no. PV3192 O-062193-r1 US 0405 TABLE OF CONTENTS 1.0 INTRODUCTION... 2 2.0 ASSAY THEORY... 2 3.0 Z9-LYTE KINASE ASSAY KIT TYR 3 PEPTIDE MATERIALS PROVIDED...
Supplementary Information for. Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction
 Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Information for Structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction Takamasa Teramoto, Yukari Fujikawa, Yoshirou Kawaguchi, Katsuhisa
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a)
 Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
Supplementary Figure 1. Electron microscopy of gb-698glyco/1g2 Fab complex. a) Representative images of 2D class averages of gb-698glyc bound to 1G2 Fab. Top views of the complex were underrepresented
RayBio Human IGF-BP-1
 RayBio Human IGF-BP-1 ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Human IGF-BP-1 ELISA Kit Protocol (Cat#: ELH-IGFBP1-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And
RayBio Human IGF-BP-1 ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Human IGF-BP-1 ELISA Kit Protocol (Cat#: ELH-IGFBP1-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And
19F MRI cellular tracer preserves the differentiation potential of hematopoietic stem cells. Brooke Helfer, PhD Celsense, Inc Pittsburgh PA
 19F MRI cellular tracer preserves the differentiation potential of hematopoietic stem cells Brooke Helfer, PhD Celsense, Inc Pittsburgh PA Financial disclosure I have the following relationships to disclose:
19F MRI cellular tracer preserves the differentiation potential of hematopoietic stem cells Brooke Helfer, PhD Celsense, Inc Pittsburgh PA Financial disclosure I have the following relationships to disclose:
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
RayBio Human bfgf ELISA Kit
 RayBio Human bfgf ELISA Kit User Manual (for Cell Lysate and Tissue Lysate) (Revised Mar 1, 2012) RayBio Human bfgf ELISA Kit Protocol (Cat#: ELH-bFGF-001C) RayBiotech, Inc. We Provide You With Excellent
RayBio Human bfgf ELISA Kit User Manual (for Cell Lysate and Tissue Lysate) (Revised Mar 1, 2012) RayBio Human bfgf ELISA Kit Protocol (Cat#: ELH-bFGF-001C) RayBiotech, Inc. We Provide You With Excellent
RayBio Human IL-3 ELISA Kit
 RayBio Human IL-3 ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Human IL-3 ELISA Kit Protocol (Cat#: ELH-IL3-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service Tel:(Toll
RayBio Human IL-3 ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Human IL-3 ELISA Kit Protocol (Cat#: ELH-IL3-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service Tel:(Toll
Recent years have witnessed an expansion in the disciplines encompassing drug
 Course overview Recent years have witnessed an expansion in the disciplines encompassing drug discovery outside the pharmaceutical industry. This is most notable with a significant number of Universities
Course overview Recent years have witnessed an expansion in the disciplines encompassing drug discovery outside the pharmaceutical industry. This is most notable with a significant number of Universities
Contribution and Mobilization of Mesenchymal Stem Cells in a mouse model of carbon tetrachloride-induced liver fibrosis
 Contribution and Mobilization of Mesenchymal Stem Cells in a mouse model of carbon tetrachloride-induced liver fibrosis Yan Liu 1,*, Zhipeng Han 1,*, Yingying Jing 1,*, Xue Yang 1, Shanshan Zhang 1, Chen
Contribution and Mobilization of Mesenchymal Stem Cells in a mouse model of carbon tetrachloride-induced liver fibrosis Yan Liu 1,*, Zhipeng Han 1,*, Yingying Jing 1,*, Xue Yang 1, Shanshan Zhang 1, Chen
 Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Problem: The GC base pairs are more stable than AT base pairs. Why? 5. Triple-stranded DNA was first observed in 1957. Scientists later discovered that the formation of triplestranded DNA involves a type
Supplementary Figure 1
 Supplementary Figure 1 2 Supplementary Figure 1: Sequence alignment of HsHSD17B8 and HsCBR4 of with KAR orthologs. The secondary structure elements as calculated by DSSP and residue numbers are displayed
Supplementary Figure 1 2 Supplementary Figure 1: Sequence alignment of HsHSD17B8 and HsCBR4 of with KAR orthologs. The secondary structure elements as calculated by DSSP and residue numbers are displayed
HUMAN IL-1 ELISA KIT. HUMAN IL-1β ELISA KIT PAGE 1 PURCHASE INFORMATION: ELISA NAME HUMAN IL-1 ELISA
 HUMAN IL-1β ELISA KIT PAGE 1 HUMAN IL-1 ELISA KIT PURCHASE INFORMATION: For the quantitative determination of human IL-1 concentrations in cell culture supernates, serum, and plasma ELISA NAME Catalog
HUMAN IL-1β ELISA KIT PAGE 1 HUMAN IL-1 ELISA KIT PURCHASE INFORMATION: For the quantitative determination of human IL-1 concentrations in cell culture supernates, serum, and plasma ELISA NAME Catalog
CHAPTER 4. Milestones of the drug discovery
 CHAPTER 4 Milestones of the drug discovery 4.Milestones of the drug discovery: Highlights the importance of the below critical milestones of the drug discovery and correlated to the current research. 1.
CHAPTER 4 Milestones of the drug discovery 4.Milestones of the drug discovery: Highlights the importance of the below critical milestones of the drug discovery and correlated to the current research. 1.
absorption - the movement of a drug from its site of administration to the bloodstream
 Glossary absorption - the movement of a drug from its site of administration to the bloodstream active pharmaceutical ingredient (API) - the chemical in an administered drug that is responsible for its
Glossary absorption - the movement of a drug from its site of administration to the bloodstream active pharmaceutical ingredient (API) - the chemical in an administered drug that is responsible for its
PF4 (Human) ELISA Kit
 PF4 (Human) ELISA Kit Catalog Number KA1761 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information... 4
PF4 (Human) ELISA Kit Catalog Number KA1761 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information... 4
Supplementary Material. for. A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis
 Supplementary Material for A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis Stephen Boulton 1, Madoka Akimoto 2, Rajeevan Selvaratnam 2, Amir Bashiri 2 and Giuseppe
Supplementary Material for A Tool Set to Map Allosteric Networks through the NMR Chemical Shift Covariance Analysis Stephen Boulton 1, Madoka Akimoto 2, Rajeevan Selvaratnam 2, Amir Bashiri 2 and Giuseppe
Ccl19 (Mouse) ELISA Kit
 Ccl19 (Mouse) ELISA Kit Catalog Number KA1806 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information...
Ccl19 (Mouse) ELISA Kit Catalog Number KA1806 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Principle of the Assay... 3 General Information...
RayBio Mouse TPO ELISA Kit
 RayBio Mouse TPO ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Mouse TPO ELISA Kit Protocol (Cat#: ELM-TPO-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service Tel:(Toll
RayBio Mouse TPO ELISA Kit User Manual (Revised Mar 1, 2012) RayBio Mouse TPO ELISA Kit Protocol (Cat#: ELM-TPO-001) RayBiotech, Inc. We Provide You With Excellent Protein Array System And Service Tel:(Toll
