Hematopoietic stem cell transplantation without irradiation
|
|
|
- Jayson Dean
- 5 years ago
- Views:
Transcription
1 nature methods Hematopoietic stem cell transplantation without irradiation Claudia Waskow, Vikas Madan, Susanne Bartels, Céline Costa, Rosel Blasig & Hans-Reimer Rodewald Supplementary figures and text: Supplementary Figure 1 Reconstitution of non-irradiated Rag -/- -/- W/Wv γ c Kit mice by donor HSC for at least 16 weeks (long-term reconstitution) Supplementary Figure 2 HSC reconstitution of non-irradiated Rag -/- -/- W/Wv γ c Kit and irradiated C57BL/6 wild type mice Supplementary Figure 3 Cell-intrinsic defects in HSC maintenance in Bclaf1- deficient mice Supplementary Figure 4 Reduced absolute numbers of long-term HSC in Rag -/- -/- γ c Kit W/Wv mice Supplementary Table 1 Stem cell failure of Bclaf1 -/- HSC revealed in Rag -/- γ c -/- Supplementary Results Supplementary Discussion Supplementary Methods Kit W/Wv recipients
2 Supplementary Figure 1 legend Reconstitution of non-irradiated Rag2 -/- γ -/- c Kit W/Wv mice by donor HSCs for at least 16 weeks (long-term reconstitution). 1.5 x 10 5 purified Lin - cells from 129/Ola x C57BL/6 mice (see text) were injected into non-irradiated Rag2 -/- γ -/- c Kit W/Wv mice. 16 weeks after transfer, donor cells were quantified by flow cytometry using high Kit expression as donor marker. (a) The fraction of Kit + cells per Lin - cells was comparable in C57BL/6 (mean = 21.6 ± 6.4% [one standard deviation]), and in transplanted Rag2 -/- γ -/- c Kit W/Wv (19.8 ± 6.1%) (P = 0.32 comparing C57BL/6 versus transplanted Rag2 -/- γ -/- c Kit W/Wv; not significant). Kit + cells were undetectable in non-transplanted Rag2 -/- γ -/- c Kit W/Wv mice. (b). The fraction of Kit + Sca-1 + cells per Lin - cells was similar in C57BL/6 (2.6 ± 1.7%), and in transplanted Rag2 -/- γ -/- c Kit W/Wv (2.1 ± 1.5%) (P = 0.37 comparing C57BL/6 versus transplanted Rag2 -/- γ -/- c Kit W/Wv ; not significant). Kit + Sca-1 + cells were absent from non-transplanted Rag2 -/- γ -/- c Kit W/Wv mice.
3 Supplementary Figure 2 legend HSC reconstitution of non-irradiated Rag2 -/- γ -/- c Kit W/Wv and irradiated C57BL/6 wildtype mice. (a) Total bone marrow cells from B6.SJL (CD ) mice were injected into lethally irradiated wild-type C57BL/6 (CD ) mice (closed squares), or non-irradiated Rag2 -/- γ -/- c Kit W/Wv (CD ) mice (open squares), and analyzed after 7 weeks. The graph shows percentages of the donor cell contribution to Lin - bone marrow cells as a function of the injected number of total bone marrow cells as indicated in the figure. *10 4 congenic B6.SJL bone marrow cells did not protect lethally irradiated wild-type recipients precluding analysis of donor contribution. Each symbol represents an individual mouse, and the bar indicates the mean for each experimental group. (b) Red blood cell parameters in the peripheral blood of reconstituted Rag2 -/- γ -/- c Kit W/Wv mice were improved in a donor cell dose-dependent manner at 7 weeks after bone marrow transfer. Graphs show mean cellular volume (MCV) and red blood cell numbers (RBC) in wild-type (wt) mice, and in non-injected Rag2 -/- γ -/- c Kit W/Wv mice and in Rag2 -/- γ -/- c Kit W/Wv mice injected with the indicated number of total B6.SJL bone marrow cells.
4 Each symbol represents an individual mouse, and the bar indicates the mean for each experimental group.
5 Supplementary Figure 3 legend Cell-intrinsic defects in HSC maintenance in Bclaf1-deficient mice. (a) The genotype of Bclaf1-deficient mice was analyzed by Southern blot (Supplementary Methods). The probe detected a 10 kb fragment for the wild-type allele and a 5 kb fragment for the mutated allele. Bclaf1 -/-, Bclaf1 +/-, Bclaf1 +/+ genotypes are shown. (b) Kinetics of the increase in RBC numbers in Rag2 -/- γ -/- c Kit W/Wv mice transplanted with 10 4 Kit + Lin - bone marrow cells from wild-type (circles) or Bclaf1 -/- (squares) mice. As controls, RBC numbers were determined in wild-type mice (inversed triangles) and non-transplanted Rag2 -/- γ -/- c Kit W/Wv mice (triangles). The symbols represent the mean ± one standard deviation of two to five analyzed mice per time point and genotype. (c) Hematopoietic progenitor cells (10 4 Kit + Lin - ) of 10 days old wild-type or Bclaf1 -/- bone marrow cells were transplanted into non-irradiated Rag2 -/- γ -/- c Kit W/Wv mice. After 4.5 months, HSC reconstitution of primary recipients was analyzed (1 st transfer). At this time point, 1000 Kit + Sca-1 + Lin - cells were sorted from the primary recipients, and were transferred into secondary recipients. After 3 months, HSC reconstitution was analyzed in secondary recipients (2 nd transfer). Dot plots show expression of Kit and Sca-1 on Lin - bone marrow cells. Percentages of Kit + Sca-1 + Lin - cells are indicated next to the gate, and absolute numbers of Kit + Sca-1 + Lin - cells, given within each plot, refer to the mean ± one standard deviation for the number (n) of mice analyzed.
6
7 Supplementary Figure 4 legend Reduced absolute numbers of long-term HSCs in Rag2 -/- γ -/- c Kit W/Wv mice. (a) Total bone marrow cellularity per two femura is shown for wild-type (WB-Kit +/+ ) mice, Rag2 -/- γ -/- c mice, Kit W/Wv mice, and Rag2 -/- γ -/- c Kit W/Wv mice. Each symbol represents an individual mouse, and the bar represents the mean of each experimental group. (b) Bone marrow cells, gated as Kit +/lo Sca-1 + Lineage (CD3, CD4, CD8, B220, CD19, Ter119, CD11b, Gr-1) cells were further analyzed for expression of CD34 and Flk2. The same Kit +/lo gate was used for all genotypes to accommodate stem cells from all strains, regardless of their Kit expression level. Considering total bone marrow cellularity, and according to percentages obtained by flow cytometry, absolute numbers of Kit + Sca-1 + Lin - CD34 - Flk2 - long-term-hsc-containing cells were determined for wild-type (WB-Kit +/+ ) mice, Rag2 -/- γ -/- c mice, Kit W/Wv mice, and Rag2 -/- γ -/- c Kit W/Wv mice. Each symbol represents an individual mouse, and the bars represent the mean of each experimental group. Supplementary Table 1 Stem cell failure of Bclaf1 -/- HSCs revealed in Rag2 -/- γ c -/- Kit W/Wv recipients Organ Cell type Spleen T cells Spleen B cells Thymus Pre T cells Bone marrow HSCs wt KO wt KO wt KO wt KO 1 st transfer 3/3 4/4 3/3 4/4 3/3 4/4 3/3 4/4 2 nd transfer 3/3 2/5 3/3 3/5 ND ND 3/3 0/5 Transplantation of Kit + Lin - and Kit + Sca-1 + Lin - cells was performed as described in Supplementary Fig. 3. Donor-derived mature spleen T cells (CD45 + CD3 + ), B cells (CD45 + CD19 + ), thymic pre T cells (Lin - CD25 + ), and bone marrow HSCs (Kit + Sca- 1 + Lin - ) were analyzed in primary (1 st transfer) and secondary (2 nd transfer) recipients of wild-type (wt) or Bclaf1 -/- (KO) cells. Numbers show successful reconstitution of the
8 indicated cell lineage per total number of recipients. Supplementary Results Thymocyte numbers in mice analyzed in Fig. 1a-d Consistent with strong HSC engraftment in transplanted Rag2 -/- γ -/- c Kit W/Wv mice, thymocyte numbers were comparable in two non-reconstituted wild-type mice (6.7x10 7, 3.6x10 7 ), and in two reconstituted Rag2 -/- γ -/- c Kit W/Wv (1.29x10 8, 3.7x10 7 ). In contrast, thymocyte numbers were more than 10-fold lower in two reconstituted Rag2 -/- γ -/- c mice (1.5x10 8, 5x10 5 ), indicating transient, or only marginal thymopoiesis in the absence of robust HSC engraftment in Rag2 -/- γ -/- c mice. Reconstitution efficiency of Rag2 -/- γ -/- c Kit W/Wv mice Reconstitution efficiency was assessed by comparing lethally irradiated wild-type (C57BL/6) and non-irradiated Rag2 -/- γ -/- c Kit W/Wv mice. Donor cells (B6.SJL; H-2 b ) were syngeneic for C57BL/6 cells (H-2 b ), and shared the parental H-2 b with Rag2 -/- γ -/- c Kit W/Wv mice. For each combination, donor B6.SJL (CD ) and host (CD ) differed in the CD45 allele. The contribution of donor cells to the Lin - compartment of the host was donor cell dose-dependent, and comparable between lethally irradiated wildtype and Rag2 -/- γ -/- c Kit W/Wv mice (Supplementary Fig. 2a). Engraftment following very low numbers of HSCs (10 4 total bone marrow cells) was only measurable in Rag2 -/- γ -/- c Kit W/Wv mice, but not in irradiated wild-type mice, because this low cell number failed to provide sufficient radioprotection for lethally irradiated wild-type mice (Supplementary Fig. 2a).
9 HSC cell dose-dependent cure of the macrocytic anemia in Rag2 -/- γ -/- c Kit W/Wv mice Because Rag2 -/- γ -/- c Kit W/Wv mice, as Kit W/Wv mice 1, suffer from a characteristic macrocytic anemia, we analyzed the impact of titrated numbers of donor bone marrow cells on anemic parameters. Peripheral blood (Supplementary Fig. 2b) was analyzed using an automated hemocytometer according to the manufacturer s instructions (ADVIA120; Bayer, Leverkusen, Germany). Both mean cellular volume (MCV) and red blood cell (RBC) numbers were corrected in a dose-dependent manner, and as few as 10 4 total bone marrow cells improved the anemia (Supplementary Fig. 2b). Analysis of Bclaf1 -/- HSCs in Rag2 -/- γ -/- c Kit W/Wv mice To provide a second example for a stem cell mutant analysis, HSCs from knockout mice deficient for the the Bcl2-associated transcription factor 1 (Bclaf1) gene 2 were serially transplanted into non-irradiated Rag2 -/- γ -/- c Kit W/Wv mice (Supplementary Fig. 3). Bclaf1 - /- mice were generated by homologous recombination in ES cells, and the correct targeting was verified by Southern blot hybridization (Supplementary Fig. 3a; and Blasig and Horak, manuscript in preparation). Lethality of Bclaf1 -/- mice within the first 3 weeks after birth precluded analysis of HSCs in adult Bclaf1 -/- mice. However, a 2-fold reduction of the absolute number of HSCs in 10 days old mutants (Supplementary Fig. 3c) suggested that hematopoietic failure may be an underlying cause of the premature death, possibly pointing at a role for Bclaf1 in the generation or function of HSCs. Reconstitution of the HSC compartment in primary Rag2 -/- γ -/- c Kit W/Wv recipients was monitored by the kinetics of the cure of the anemia which was comparable from wild-type or Bclaf1 -/- bone marrow (Supplementary Fig. 3b). In addition, 4.5 months after transplantation, both wild-type and Bclaf1 -/- bone marrow had successfully given rise to donor-derived T and B cells in all transplanted mice (Supplementary Table 1). Similar to HSCs in young Bclaf1 -/- mice, numbers of HSCs in primary Rag2 -/- γ -/- c Kit W/Wv mice were reduced 2-fold compared to wild-type and Bclaf1-deficient HSCs (Supplementary Fig. 3c). More dramatic defects were uncovered in secondary Rag2 -/-
10 γ -/- c Kit W/Wv recipients that were transplanted with 1000 sorted wild-type or Bclaf1 -/- Kit + Sca-1 + Lin - cells retrieved from primary Rag2 -/- γ -/- c Kit W/Wv recipients 4.5 months after the first transplantation. Analysis of the bone marrow in secondary Rag2 -/- γ -/- c Kit W/Wv recipients 3 months after transplantation still revealed HSC engraftment in the case of wild-type HSCs (Supplementary Fig. 3c, left). In contrast, HSCs, and, in fact, any Kit + cells were absent from secondary Rag2 -/- γ -/- c Kit W/Wv bone marrow after serial transplantation of Bclaf1 -/- HSCs (Supplementary Fig. 3c, right). Consistent with stem cell exhaustion between the first and second transplantation, Bclaf1 -/- HSCs failed to reconstitute the HSC compartment in all secondary recipients (Supplementary Fig. 3c and Supplementary Table 1). In keeping with this lack of sustained hematopoiesis, T and B cells were reconstituted in only 2 or 3 out of 5 secondary recipients, respectively, receiving Bclaf1 -/- HSCs (Supplementary Table 1). In contrast, wild-type HSCs successfully generated T and B cells as well as HSCs in all secondary recipient mice. We conclude that Bclaf1 -/- HSCs failed to engraft in the HSC compartment of secondary recipients, suggesting a crucial function for Bclaf1 in HSC maintenance. These findings call for a detailed characterization of the underlying defect. Supplementary Discussion Considerations on parent into F1 transplantations in Kit W/Wv recipients We state in the manuscript that non-irradiated Kit W/Wv mice, which are F1 hybrids between WB-Kit W/+ (H-2 j ) and C57Bl/6-Kit Wv/+ (H-2 b ) parents, accept syngeneic F1 (H-2 j x b ), and, partially, parental HSCs. While Kit W/Wv mice fully reject WB bone marrow, they partially accept C57BL/6 bone marrow 3. Of note, both for CFU-S and for long-term engraftment, transplantation of C57BL/6 (parental) bone marrow into Kit W/Wv mice is known to be inferior to transplantation of WBB6 (= syngeneic F1) bone marrow into Kit W/Wv mice 3. This has been attributed to F1 hybrid resistance 3, and it is, along with the rejection of allogeneic HSCs by Kit W/Wv mice 1, probably the major reason for why Kit W/Wv mice have not been routinely used for adoptive transfer analyses of HSCs.
11 Size of the endogenous HSC compartments in wild-type and mutant mice Comparison of absolute numbers of the endogenous long-term HSC-containing Kit + Sca-1 + Lin - CD34 - Flk2 - population in wild-type mice, Rag2 -/- γ -/- c mice, Kit W/Wv mice and Rag2 -/- γ -/- c Kit W/Wv mice revealed a 4.5 fold reduction comparing Rag2 -/- γ -/- c Kit W/Wv mice and wild-type mice (Supplementary Fig. 4). Based on our engraftment studies, and on CFU-S formation, we conclude that these endogenous HSCs in Rag2 -/- γ -/- c Kit W/Wv mice cannot compete in their niches against Kit + HSCs even in the absence of any exogenous myeloablation. As a result of this deficiency, and the reduced stem cell numbers, bone marrow and spleen niches of non-irradiated Rag2 -/- γ -/- c Kit W/Wv mice are freely accessible for donor stem and progenitor cells. In addition, engraftment of these niches by allogeneic HSCs is absolutely dependent on lack of Rag2 and γ c expression in Rag2 -/- γ -/- c Kit W/Wv mice. Supplementary Methods Mice DBA/2 (H-2 d ), BALB/c (H-2 d ), C57BL/6 (B6) H-2 b ), C57BL/6.SJL (B6.SJL; H-2 b ) were from Jackson Laboratory, Bar Harbour, USA, and Rag2 -/- γ -/- c were from Taconic. To generate Rag2 -/- γ -/- c Kit W/Wv mice, we used WB-Kit W/+ and C57BL/6-Kit Wv/+ which we obtained in 1994 from Japan-SLC, Shizuoka, Japan. These strains are termed WB-S/+ for WB-Kit W/+ and C57BL/6-Wv/+ for C57BL/6-Kit Wv/+ by Japan-SLC, which obtained the mice in 1981 from the Institute of Cancer Research, Osaka University School of Medicine. The parental strains of these mice were WB/ReJ-Kit W /J and C57BL/6J-Kit W-v /J from the from Jackson Laboratory 4. WBB6F1-Kit W/Wv (Kit W/Wv ) mice, and Rag2 -/- γ -/- c Kit W/Wv mice were continuously bred in our animal facility as described before 5. By way of their generation, Rag2 -/- γ -/- c Kit W/Wv mice co-express the parental haplotypes H-2 j and H-2 b.
12 Flow Cytometry Bone marrow cells were flushed from 2 femura, counted and either directly used (total bone marrow) or labeled in 100 µl PBS with 5% FCS for 40 on ice and sorted for HSC containing Kit + Sca-1 + Lineage (CD3, CD4, CD8, B220, CD19, Ter119, CD11b, Gr-1) cells. All sorts and analysis were performed using a FACS Aria, or FACS Calibur or CantoII instruments (all BD). Antibodies used were: Gr-1 (RB6-8C5), Mac-1 (M1/70.15), CD3 (145-2C11) CD4 (H129.19) CD8 (53-6.7), CD19 (1D3), CD34 (RAM34), CD45.1 (A20-1.7), CD45R (RA3.6B2), CD117 (2B8), CD135 (A2F10.1), H-2K d (SF1-1.1), Ter119 (Ter119) Sca-1 (E ) (all BD-Pharmingen). Flow cytometry and cell sorting were done as described 6. HSC transplantation and CFU-S assay C57BL/6 recipient mice were lethally irradiated (950 cgy; split dose with 2h time gap between both doses) and 2 hours later injected with cells in 200 µl PBS with 5% FCS. For Fig. 1a-d, 2000 purified Kit + Sca-1 + Lin - from BALB/c mice (H-2 d ) were injected into non-irradiated wild-type C57BL/6 (H-2 b ) mice, Rag2 -/- γ -/- c (H-2 b ) mice, Kit W/Wv (H-2 j x b ) mice and Rag2 -/- γ -/- c Kit W/Wv (H-2 j x b ) mice. In this combination, donors and recipients were hence fully MHC-mismatched. The donor contribution to bone marrow and spleen was analyzed by flow cytometry using the indicated phenotypes. In the CFU-S assay, mice were irradiated (950 cgy), or left non-irradiated as indicated, and spleens were removed 11 days later, fixed in Bouin s solution and photographed for visible colonies. In Fig. 1e-j, C57BL/6 and DBA/2 donor cells were 2x10 5 total bone marrow cells, and BALB/c donor cells were 2000 sorted Kit + Lin - cells. HSC mutant mice Mll5-deficient mice have been described 7. Bclaf1-deficient mice were generated in the FMP, Berlin, Germany (Blasig and Horak, manuscript in preparation) by homologous recombination in ES cells, and the genotype of mutant mice was analyzed by Southern blot hybridization after digestion of genomic mouse tail DNA with PstI. Blots were hybridized simultaneously with two probes generated by PCR using ES cell DNA as
13 template. Probe 1: forward primer: 5 -CCATCACTCTTGGCTTA - 3, reverse primer: 5 GCA TGC ATG GAG GCA AAT CAC 3 ; Probe 2: forward primer: 5 ACA GTC TAG ATA TGA GAA ATC AGA 3, reverse primer: 5 CCA GCA CTC TTA TTC AGA AAA CTG 3. Both fragments were labelled with 32 P-dCTP using the Amersham TM Rediprime TM II Random Primer Labelling System (GE Healthcare, Buckinghanshire, UK). Supplementary References 1. Russell, E. S. Adv. Genet 20, (1979). 2. Kasof, G. M., Goyal, L. & White, E. Mol. Cell. Biol. 19, (1999). 3. Harrison, D. E. Immunogenetics 13, (1981). 4. Kitamura, Y., Go, S. & Hatanaka, K. Blood 52, (1978). 5. Waskow, C., Bartels, S., Schlenner, S. M., Costa, C. & Rodewald, H. R. Blood 109, (2007). 6. Rodewald, H. R., Ogawa, M., Haller, C., Waskow, C. & DiSanto, J. P. Immunity 6, (1997). 7. Madan, V. et al. Blood 113, (2009).
Supplemental Information Inventory
 Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Cell Stem Cell, Volume 6 Supplemental Information Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1 Grant A. Challen, Nathan C. Boles, Stuart M. Chambers, and Margaret A.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1: Phenotypically defined hematopoietic stem and progenitor cell populations show distinct mitochondrial activity and mass. (A) Isolation by FACS of commonly
Supplementary Figures Supplementary Figure 1: Phenotypically defined hematopoietic stem and progenitor cell populations show distinct mitochondrial activity and mass. (A) Isolation by FACS of commonly
Flow cytometry Stained cells were analyzed and sorted by SORP FACS Aria (BD Biosciences).
 Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
Mice C57BL/6-Ly5.1 or -Ly5.2 congenic mice were used for LSK transduction and competitive repopulation assays. Animal care was in accordance with the guidelines of Keio University for animal and recombinant
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
DOI: 1.138/ncb3342 EV O/E 13 4 B Relative expression 1..8.6.4.2 shctl Pcdh2_1 C Pcdh2_2 Number of shrns 12 8 4 293T mterc +/+ G3 mterc -/- D % of GFP + cells 1 8 6 4 2 Per2 p=.2 p>
We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and
 Mice We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and an Frt flanked neo cassette (Fig. S1A). Targeted BA1 (C57BL/6 129/SvEv) hybrid embryonic stem cells
Mice We designed the targeting vector in such a way that the first exon was flanked by two LoxP sites and an Frt flanked neo cassette (Fig. S1A). Targeted BA1 (C57BL/6 129/SvEv) hybrid embryonic stem cells
88.7 <GFP-A>: EGFP GFP. Null. % of Cells! <APC-A> B220!
 B. C. 1 Cell Number (X18) A. % of Cells % of Max 8 Con 6 4 88.7 1 13 14 : EGFP 4 6 4 1 WT 8 % of% ofcells Max 6 6 4 G. 1 13 B Hdac3-/- 14 15 1 13 14 15 Ter119 Ly6C Wild type WT 4
B. C. 1 Cell Number (X18) A. % of Cells % of Max 8 Con 6 4 88.7 1 13 14 : EGFP 4 6 4 1 WT 8 % of% ofcells Max 6 6 4 G. 1 13 B Hdac3-/- 14 15 1 13 14 15 Ter119 Ly6C Wild type WT 4
Dierks Supplementary Fig. S1
 Dierks Supplementary Fig. S1 ITK SYK PH TH K42R wt K42R (kinase deficient) R29C E42K Y323F R29C E42K Y323F (reduced phospholipid binding) (enhanced phospholipid binding) (reduced Cbl binding) E42K Y323F
Dierks Supplementary Fig. S1 ITK SYK PH TH K42R wt K42R (kinase deficient) R29C E42K Y323F R29C E42K Y323F (reduced phospholipid binding) (enhanced phospholipid binding) (reduced Cbl binding) E42K Y323F
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
Chen et al., page 1 MicroRNAs Modulate Hematopoietic Lineage Differentiation Chang-Zheng Chen, Ling Li, Harvey F. Lodish, David. Bartel Supplemental Online Material Methods Cell isolation Murine bone marrow
MLN8237 induces proliferation arrest, expression of differentiation markers and
 Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supplementary Figure Legends Supplementary Figure 1 827 induces proliferation arrest, expression of differentiation markers and polyploidization of a human erythroleukemia cell line with the activating
Supporting Information
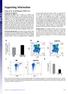 (xe number cells) LSK (% gated) Supporting Information Youm et al../pnas. SI Materials and Methods Quantification of sjtrecs. CD + T subsets were isolated from splenocytes using mouse CD + T cells positive
(xe number cells) LSK (% gated) Supporting Information Youm et al../pnas. SI Materials and Methods Quantification of sjtrecs. CD + T subsets were isolated from splenocytes using mouse CD + T cells positive
Revised: RG-RV2 by Fukuhara et al.
 Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
Supplemental Figure 1 The generation of Spns2 conditional knockout mice. (A) Schematic representation of the wild type Spns2 locus (Spns2 + ), the targeted allele, the floxed allele (Spns2 f ) and the
7-amino actinomycin D (7ADD) was added to all samples 10 minutes prior to analysis on the flow cytometer in order to gate 7AAD viable cells.
 Antibody staining for Ho uptake analyses For HSC staining, 10 7 BM cells from Ho perfused mice were stained with biotinylated lineage antibodies (CD3, CD5, B220, CD11b, Gr-1, CD41, Ter119), anti Sca-1-PECY7,
Antibody staining for Ho uptake analyses For HSC staining, 10 7 BM cells from Ho perfused mice were stained with biotinylated lineage antibodies (CD3, CD5, B220, CD11b, Gr-1, CD41, Ter119), anti Sca-1-PECY7,
Supplementary Figure 1. Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings.
 Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Supplementary Figure 1 Homozygous rag2 E450fs mutants are healthy and viable similar to wild-type and heterozygous siblings. (left) Representative bright-field images of wild type (wt), heterozygous (het)
Multiple layers of B cell memory with different effector functions. Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos,
 Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Multiple layers of B cell memory with different effector functions Ismail Dogan, Barbara Bertocci, Valérie Vilmont, Frédéric Delbos, Jérome Mégret, Sébastien Storck, Claude-Agnès Reynaud & Jean-Claude
Nature Immunology: doi: /ni.3694
 Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplementary Figure 1 Expression of Bhlhe41 and Bhlhe40 in B cell development and mature B cell subsets. (a) Scatter plot showing differential expression of genes between splenic B-1a cells and follicular
Supplemental Data Supplemental Figure 1.
 Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplemental Data Supplemental Figure 1. Silique arrangement in the wild-type, jhs, and complemented lines. Wild-type (WT) (A), the jhs1 mutant (B,C), and the jhs1 mutant complemented with JHS1 (Com) (D)
Supplementary Fig. 1: Characterization of Asxl2 -/- mouse model. (a) HSCs and their
 Supplementary Fig. 1: Characterization of Asxl2 -/- mouse model. (a) HSCs and their differentiated cell populations were sorted from the BM of WT mice using respective surface markers, and Asxl2 mrna expression
Supplementary Fig. 1: Characterization of Asxl2 -/- mouse model. (a) HSCs and their differentiated cell populations were sorted from the BM of WT mice using respective surface markers, and Asxl2 mrna expression
Supplemental Data. mir156-regulated SPL Transcription. Factors Define an Endogenous Flowering. Pathway in Arabidopsis thaliana
 Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Cell, Volume 138 Supplemental Data mir156-regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana Jia-Wei Wang, Benjamin Czech, and Detlef Weigel Table S1. Interaction
Supplementary Figure 1A A404 Cells +/- Retinoic Acid
 Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
Supplementary Figure 1A A44 Cells +/- Retinoic Acid 1 1 H3 Lys4 di-methylation SM-actin VEC cfos (-) RA (+) RA 14 1 1 8 6 4 H3 Lys79 di-methylation SM-actin VEC cfos (-) RA (+) RA Supplementary Figure
(A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: WT; lower
 Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
Supplementary Figures S1. (A-B) P2ry14 expression was assessed by (A) genotyping (upper arrow: ; lower arrow: KO) and (B) q-pcr analysis with Lin- cells, The white vertical line in panel A indicates that
2111: ALL Post-HCT. Add/ Remove/ Modify. Manual Section. Date. Description. Comprehensive Disease- Specific Manuals
 2111: ALL Post-HCT The Acute Lymphoblastic Leukemia Post-HCT Data Form is one of the Comprehensive Report Forms. This form captures ALL-specific post-hct data such as: planned treatments post-hct, the
2111: ALL Post-HCT The Acute Lymphoblastic Leukemia Post-HCT Data Form is one of the Comprehensive Report Forms. This form captures ALL-specific post-hct data such as: planned treatments post-hct, the
PCR analysis was performed to show the presence and the integrity of the var1csa and var-
 Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Supplementary information: Methods: Table S1: Primer Name Nucleotide sequence (5-3 ) DBL3-F tcc ccg cgg agt gaa aca tca tgt gac tg DBL3-R gac tag ttt ctt tca ata aat cac tcg c DBL5-F cgc cct agg tgc ttc
Hes6. PPARα. PPARγ HNF4 CD36
 SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
SUPPLEMENTARY INFORMATION Supplementary Table Positions and Sequences of ChIP primers -63 AGGTCACTGCCA -79 AGGTCTGCTGTG Hes6-0067 GGGCAaAGTTCA ACOT -395 GGGGCAgAGTTCA PPARα -309 GGCTCAaAGTTCAaGTTCA CPTa
Solutions to Problem Set 6
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Name: Question 1 Solutions to 7.012 Problem Set 6 The
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Name: Question 1 Solutions to 7.012 Problem Set 6 The
Gene Sequence Annealing Temperature. Actin 5 : 5 -CATCACTATTGGCAACGAGC-3 60 C 3 : 5 -ACGCAGCTCAGTAACAGTCC-3 PCR product: 410 bp
 Supporting Materials and Methods RT-PCR RT-PCR was performed as described in ref. 1 using the following oligonucleotides and PCR conditions: 2 min at 95 C, (20 s at 95 C, 20 s at the annealing temp, and
Supporting Materials and Methods RT-PCR RT-PCR was performed as described in ref. 1 using the following oligonucleotides and PCR conditions: 2 min at 95 C, (20 s at 95 C, 20 s at the annealing temp, and
Nature Biotechnology: doi: /nbt.4086
 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3 p815 p815-hcd2 Ag (-) anti-cd3
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Mid-term review. Recitation5 03/31/2014. Stem Cell Biology and Function W4193 1
 Mid-term review Recitation5 03/31/2014 Stem Cell Biology and Function W4193 1 What to expect in the midterm2 It will be all so far but mainly whatever was NOT covered by the previous mid-term- not a huge
Mid-term review Recitation5 03/31/2014 Stem Cell Biology and Function W4193 1 What to expect in the midterm2 It will be all so far but mainly whatever was NOT covered by the previous mid-term- not a huge
SUPPLEMENTAL METHODS. Chromatin immunoprecipitation (ChIP) assay. Cell culture. Non-ablated mouse transplantation. Hematology and flow cytometry
 SUPPLEMENTAL METHODS Cell culture EML C1 cells were cultured in IMDM 20% horse serum, 1% antibiotic-antimycotic (Gibco by Life Technologies) 15% SCF conditioned medium from BHK/ MKL cells or 50 100 ng/ml
SUPPLEMENTAL METHODS Cell culture EML C1 cells were cultured in IMDM 20% horse serum, 1% antibiotic-antimycotic (Gibco by Life Technologies) 15% SCF conditioned medium from BHK/ MKL cells or 50 100 ng/ml
hcd1tg/hj1tg/ ApoE-/- hcd1tg/hj1tg/ ApoE+/+
 ApoE+/+ ApoE-/- ApoE-/- H&E (1x) Supplementary Figure 1. No obvious pathology is observed in the colon of diseased ApoE-/me. Colon samples were fixed in 1% formalin and laid out in Swiss rolls for paraffin
ApoE+/+ ApoE-/- ApoE-/- H&E (1x) Supplementary Figure 1. No obvious pathology is observed in the colon of diseased ApoE-/me. Colon samples were fixed in 1% formalin and laid out in Swiss rolls for paraffin
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information Dissecting binding of a β-barrel outer membrane
Supplemental Information
 Supplemental Information DLA-matched bone marrow transplantation reverses the immunodeficiency of SCID dogs. Bone marrow transplantation studies were initiated with the goal of reversing the immunodeficiency
Supplemental Information DLA-matched bone marrow transplantation reverses the immunodeficiency of SCID dogs. Bone marrow transplantation studies were initiated with the goal of reversing the immunodeficiency
Tumour 2. Tumour 9. Tumour 25 Tumour 26. Tumour 27. t(6;16) doi: /nature06159 SUPPLEMENTARY INFORMATION Supplementary Figure 1
 doi: 1.18/nature6159 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Tumour Tumour 9 t(6;16) Tumour 5 Tumour 6 Tumour 7 www.nature.com/nature 1 doi: 1.18/nature6159 SUPPLEMENTARY INFORMATION Supplementary
doi: 1.18/nature6159 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Tumour Tumour 9 t(6;16) Tumour 5 Tumour 6 Tumour 7 www.nature.com/nature 1 doi: 1.18/nature6159 SUPPLEMENTARY INFORMATION Supplementary
SUPPLEMENTARY MATERIALS AND METHODS. E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1)
 SUPPLEMENTARY MATERIALS AND METHODS E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1) dinb::kan (lab stock) derivative was used as wild-type. MG1655 alka tag dinb (2) is
SUPPLEMENTARY MATERIALS AND METHODS E. coli strains, plasmids, and growth conditions. Escherichia coli strain P90C (1) dinb::kan (lab stock) derivative was used as wild-type. MG1655 alka tag dinb (2) is
Interferon- -producing immature myeloid cells confer protection. against severe invasive group A Streptococcus infections. Supplementary Information
 Supplementary Information Interferon- -producing immature myeloid cells confer protection against severe invasive group A Streptococcus infections Takayuki Matsumura, Manabu Ato, Tadayoshi Ikebe, Makoto
Supplementary Information Interferon- -producing immature myeloid cells confer protection against severe invasive group A Streptococcus infections Takayuki Matsumura, Manabu Ato, Tadayoshi Ikebe, Makoto
Supplemental Figure 1
 Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
Supplemental Figure 1 Supplemental figure 1. Generation of gene targeted mice expressing an anti-id BCR. (A,B) Generation of the VDJ aid H KI mouse. (A) Targeting Construct. Top: Targeting construct for
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of
 Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
Supplementary Figure 1. Localization of MST1 in RPE cells. Proliferating or ciliated HA- MST1 expressing RPE cells (see Fig. 5b for establishment of the cell line) were immunostained for HA, acetylated
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1 Histogram displaying the distribution of DNA barcode copy numbers for each virus library. 100,000 BB88 cells were infected by lentivirus that carried the corresponding
SUPPLEMENTARY FIGURES Supplementary Figure 1 Histogram displaying the distribution of DNA barcode copy numbers for each virus library. 100,000 BB88 cells were infected by lentivirus that carried the corresponding
show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras ex3op/ex3op mice
 Supplemental Data Supplemental Figure Legends Supplemental Figure 1. Hematologic profile of Kras / mice. (A) Scatter plots show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras / mice
Supplemental Data Supplemental Figure Legends Supplemental Figure 1. Hematologic profile of Kras / mice. (A) Scatter plots show PB hemoglobin concentration (Hgb) and platelet counts (Plt) in Kras / mice
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
DOI: 10.1038/ncb3240 Supplementary Figure 1 GBM cell lines display similar levels of p100 to p52 processing but respond differentially to TWEAK-induced TERT expression according to TERT promoter mutation
The GOODELL laboratory
 The GOODELL laboratory Author Nathan Boles Feb.5, 2009 Title Hematopoietic Progenitor Staining Introduction This protocol describes the analysis or isolation of the early hematopoietic progenitors. HBSS+
The GOODELL laboratory Author Nathan Boles Feb.5, 2009 Title Hematopoietic Progenitor Staining Introduction This protocol describes the analysis or isolation of the early hematopoietic progenitors. HBSS+
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Validation of the monoclonal antibody to mouse ACKR1 and expression of ACKR1 by BM hematopoietic cells. (a to d) Comparison of immunostaining of BM cells by anti-mouse ACKR1 antibodies:
Supplementary Figure 1 Validation of the monoclonal antibody to mouse ACKR1 and expression of ACKR1 by BM hematopoietic cells. (a to d) Comparison of immunostaining of BM cells by anti-mouse ACKR1 antibodies:
-Immune phenotype -Cancer xenografts -Immune system humanization -Services
 -Immune phenotype -Cancer xenografts -Immune system humanization -Services services@herabiolabs.com 859-414-0648 About Hera BioLabs Precision Toxicology & Efficacy: utilizing precisely gene-edited models
-Immune phenotype -Cancer xenografts -Immune system humanization -Services services@herabiolabs.com 859-414-0648 About Hera BioLabs Precision Toxicology & Efficacy: utilizing precisely gene-edited models
Rorα is essential for nuocyte development
 Rorα is essential for nuocyte development See Heng Wong,,, Jennifer A. Walker,, Helen E. Jolin,, Lesley F. Drynan, Emily Hams 3, Ana Camelo, Jillian L. Barlow, Daniel R. Neill,6, Veera Panova, Ute Koch,
Rorα is essential for nuocyte development See Heng Wong,,, Jennifer A. Walker,, Helen E. Jolin,, Lesley F. Drynan, Emily Hams 3, Ana Camelo, Jillian L. Barlow, Daniel R. Neill,6, Veera Panova, Ute Koch,
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11496 Cl. 8 Cl. E93 Rag1 -/- 3H9 + BM Rag1 -/- BM CD CD c-kit c-kit c-kit wt Spleen c-kit B22 B22 IgM IgM IgM Supplementary Figure 1. FACS analysis of single-cell-derived pre-b cell clones.
doi:1.138/nature11496 Cl. 8 Cl. E93 Rag1 -/- 3H9 + BM Rag1 -/- BM CD CD c-kit c-kit c-kit wt Spleen c-kit B22 B22 IgM IgM IgM Supplementary Figure 1. FACS analysis of single-cell-derived pre-b cell clones.
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 PPAR-γ is dispensable for the development of tissue macrophages in the heart, kidneys, lamina propria and white adipose tissue. Plots show the expression of F4/80 and CD11b (a) or
Supplementary Figure 1 PPAR-γ is dispensable for the development of tissue macrophages in the heart, kidneys, lamina propria and white adipose tissue. Plots show the expression of F4/80 and CD11b (a) or
Supplementary Figure 1
 Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Supplementary Figure 1 Ex2 promotor region Cre IRES cherry pa Ex4 Ex5 Ex1 untranslated Ex3 Ex5 untranslated EYFP pa Rosa26 STOP loxp loxp Cre recombinase EYFP pa Rosa26 loxp 1 kb Interleukin-9 fate reporter
Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis
 1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
1 2 3 4 5 6 7 8 9 10 11 12 Figure S1. Characterization of the irx9l-1 mutant. (A) Diagram of the Arabidopsis IRX9L gene drawn based on information from TAIR (the Arabidopsis Information Research). Exons
ΔPDD1 x ΔPDD1. ΔPDD1 x wild type. 70 kd Pdd1. Pdd3
 Supplemental Fig. S1 ΔPDD1 x wild type ΔPDD1 x ΔPDD1 70 kd Pdd1 50 kd 37 kd Pdd3 Supplemental Fig. S1. ΔPDD1 strains express no detectable Pdd1 protein. Western blot analysis of whole-protein extracts
Supplemental Fig. S1 ΔPDD1 x wild type ΔPDD1 x ΔPDD1 70 kd Pdd1 50 kd 37 kd Pdd3 Supplemental Fig. S1. ΔPDD1 strains express no detectable Pdd1 protein. Western blot analysis of whole-protein extracts
Percent survival. Supplementary fig. S3 A.
 Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary fig. S3 A. B. 100 Percent survival 80 60 40 20 Ml 0 0 100 C. Fig. S3 Comparison of leukaemia incidence rate in the triple targeted chimaeric mice and germline-transmission translocator mice
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM
 Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Supplementary Figure 1: Analysis of monocyte subsets and lineage relationships. (a) Gating strategy for definition of MDP and cmop populations in BM of Cx3cr1 GFP/+ mice related to Fig. 1a. MDP was defined
Mouse expression data were normalized using the robust multiarray algorithm (1) using
 Supplementary Information Bioinformatics statistical analysis of microarray data Mouse expression data were normalized using the robust multiarray algorithm (1) using a custom probe set definition that
Supplementary Information Bioinformatics statistical analysis of microarray data Mouse expression data were normalized using the robust multiarray algorithm (1) using a custom probe set definition that
Fundamental properties of Stem Cells
 Stem cells Learning Goals: Define what a stem cell is and describe its general properties, using hematopoietic stem cells as an example. Describe to a non-scientist the current progress of human stem cell
Stem cells Learning Goals: Define what a stem cell is and describe its general properties, using hematopoietic stem cells as an example. Describe to a non-scientist the current progress of human stem cell
19F MRI cellular tracer preserves the differentiation potential of hematopoietic stem cells. Brooke Helfer, PhD Celsense, Inc Pittsburgh PA
 19F MRI cellular tracer preserves the differentiation potential of hematopoietic stem cells Brooke Helfer, PhD Celsense, Inc Pittsburgh PA Financial disclosure I have the following relationships to disclose:
19F MRI cellular tracer preserves the differentiation potential of hematopoietic stem cells Brooke Helfer, PhD Celsense, Inc Pittsburgh PA Financial disclosure I have the following relationships to disclose:
Arabidopsis actin depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB
 Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Arabidopsis actin depolymerizing factor mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB Files in this Data Supplement: Supplemental Table S1 Supplemental Table
Supplementary Figure 1. Phenotype, morphology and distribution of embryonic GFP +
 Supplementary Figure 1 Supplementary Figure 1. Phenotype, morphology and distribution of embryonic GFP + haematopoietic cells in the intestine. GFP + cells from E15.5 intestines were obtained after collagenase
Supplementary Figure 1 Supplementary Figure 1. Phenotype, morphology and distribution of embryonic GFP + haematopoietic cells in the intestine. GFP + cells from E15.5 intestines were obtained after collagenase
Disease and selection in the human genome 3
 Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Disease and selection in the human genome 3 Ka/Ks revisited Please sit in row K or forward RBFD: human populations, adaptation and immunity Neandertal Museum, Mettman Germany Sequence genome Measure expression
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure 1 Supplementary Figure 1: Vector maps of TRMPV and TRMPVIR variants. Many derivatives of TRMPV have been generated and tested. Unless otherwise noted, experiments in this paper use
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. CD11b expression on different B cell subsets in anti-snrnp Ig Tg mice. (a) Highly purified follicular (FO) and marginal zone (MZ) B cells were sorted from
Supplementary Figure S1 Supplementary Figure S1. CD11b expression on different B cell subsets in anti-snrnp Ig Tg mice. (a) Highly purified follicular (FO) and marginal zone (MZ) B cells were sorted from
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11809 Supplementary Figure 1. Antibiotic treatment reduces intestinal bacterial load and allows access of non-pathogenic bacteria to the MLN, inducing intestinal
SUPPLEMENTARY INFORMATION doi:10.1038/nature11809 Supplementary Figure 1. Antibiotic treatment reduces intestinal bacterial load and allows access of non-pathogenic bacteria to the MLN, inducing intestinal
administration of tamoxifen. Bars show mean ± s.e.m (n=10-11). P-value was determined by
 Supplementary Figure 1. Chimerism of CD45.2 + GFP + cells at 1 month post transplantation No significant changes were detected in chimerism of CD45.2 + GFP + cells between recipient mice repopulated with
Supplementary Figure 1. Chimerism of CD45.2 + GFP + cells at 1 month post transplantation No significant changes were detected in chimerism of CD45.2 + GFP + cells between recipient mice repopulated with
(i) A trp1 mutant cell took up a plasmid containing the wild type TRP1 gene, which allowed that cell to multiply and form a colony
 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been isolated that cannot grow
Wu et al., Determination of genetic identity in therapeutic chimeric states. We used two approaches for identifying potentially suitable deletion loci
 SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
SUPPLEMENTARY METHODS AND DATA General strategy for identifying deletion loci We used two approaches for identifying potentially suitable deletion loci for PDP-FISH analysis. In the first approach, we
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
advances.sciencemag.org/cgi/content/full/4/9/eaat5401/dc1 Supplementary Materials for GLK-IKKβ signaling induces dimerization and translocation of the AhR-RORγt complex in IL-17A induction and autoimmune
GENOME 371, Problem Set 6
 GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
GENOME 371, Problem Set 6 1. S. pombe is a distant relative of baker s yeast (which you used in quiz section). Wild type S. pombe can grow on plates lacking tryptophan (-trp plates). A mutant has been
Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC
 Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Supplementary Appendixes Supplement 1: Sequences of Capture Probes. Capture probes were /5AmMC6/CTG TAG GTG CGG GTG GAC GTA GTC ACG TAG CTC CGG CTG GA-3 for vimentin, /5AmMC6/TCC CTC GCG CGT GGC TTC CGC
Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR
 Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR 1 The problem We wish to clone a yet unknown gene from a known
Lecture 10, 20/2/2002: The process of solution development - The CODEHOP strategy for automatic design of consensus-degenerate primers for PCR 1 The problem We wish to clone a yet unknown gene from a known
isolated from ctr and pictreated mice. Activation of effector CD4 +
 Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
Supplementary Figure 1 Bystander inflammation conditioned T reg cells have normal functional suppressive activity and ex vivo phenotype. WT Balb/c mice were treated with polyi:c (pic) or PBS (ctr) via
The transcrip-on factor NR4A1 (Nur77) controls bone marrow differen-a-on and survival of Ly6C monocytes
 Supplementary Informa0on The transcrip-on factor NR4A1 (Nur77) controls bone marrow differen-a-on and survival of Ly6C monocytes Richard N. Hanna 1, Leo M. Carlin 2, Harper G. Hubbeling 3, Dominika Nackiewicz
Supplementary Informa0on The transcrip-on factor NR4A1 (Nur77) controls bone marrow differen-a-on and survival of Ly6C monocytes Richard N. Hanna 1, Leo M. Carlin 2, Harper G. Hubbeling 3, Dominika Nackiewicz
a Lamtor1 (gene) b Lamtor1 (mrna) c WT Lamtor1 Lamtor1 flox Lamtor2 A.U. p = LysM-Cre Lamtor3 Lamtor4 Lamtor5 BMDMs: Φ WT Φ KO β-actin WT BMDMs
 a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
a Lamtor (gene) b Lamtor (mrna) c BMDMs: Φ WT Φ KO Lamtor flox 8 bp LysM-Cre 93 bp..5 p =.4 WT BMDMs: Φ WT Φ KO Lamtor (protein) BMDMs: Φ WT Φ KO Lamtor 8 kda Lamtor 4 kda Lamtor3 4 kda Lamtor4 kda Lamtor5.5
Multiplexing Genome-scale Engineering
 Multiplexing Genome-scale Engineering Harris Wang, Ph.D. Department of Systems Biology Department of Pathology & Cell Biology http://wanglab.c2b2.columbia.edu Rise of Genomics An Expanding Toolbox Esvelt
Multiplexing Genome-scale Engineering Harris Wang, Ph.D. Department of Systems Biology Department of Pathology & Cell Biology http://wanglab.c2b2.columbia.edu Rise of Genomics An Expanding Toolbox Esvelt
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full//458/eaas956/dc Supplementary Materials for Thrombopoietin receptor independent stimulation of hematopoietic stem cells by eltrombopag Yun-Ruei Kao,
www.sciencetranslationalmedicine.org/cgi/content/full//458/eaas956/dc Supplementary Materials for Thrombopoietin receptor independent stimulation of hematopoietic stem cells by eltrombopag Yun-Ruei Kao,
Dnmt3a is Essential for Hematopoietic Stem Cell Differentiation
 Supplementary Information Dnmt3a is Essential for Hematopoietic Stem Cell Differentiation Grant A. Challen 1,2,3, Deqiang Sun 9*, Mira Jeong 1,2,5*, Min Luo 5*, Jaroslav Jelinek 8*, Jonathan S. Berg 10,12*,
Supplementary Information Dnmt3a is Essential for Hematopoietic Stem Cell Differentiation Grant A. Challen 1,2,3, Deqiang Sun 9*, Mira Jeong 1,2,5*, Min Luo 5*, Jaroslav Jelinek 8*, Jonathan S. Berg 10,12*,
Supplementary Figure 1. Characterization of the POP2 transcriptional and post-transcriptional regulatory elements. (A) POP2 nucleotide sequence
 1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
1 5 6 7 8 9 10 11 1 1 1 Supplementary Figure 1. Characterization of the POP transcriptional and post-transcriptional regulatory elements. (A) POP nucleotide sequence depicting the consensus sequence for
Table S1. Antibodies and recombinant proteins used in this study
 Table S1. Antibodies and recombinant proteins used in this study Labeled Antibody Clone Cat. no. Streptavidin-PerCP BD Biosciences 554064 Biotin anti-mouse CD25 7D4 BD Biosciences 553070 PE anti-mouse
Table S1. Antibodies and recombinant proteins used in this study Labeled Antibody Clone Cat. no. Streptavidin-PerCP BD Biosciences 554064 Biotin anti-mouse CD25 7D4 BD Biosciences 553070 PE anti-mouse
Mark J. Kiel, Melih Acar, Glenn L. Radice, and Sean J. Morrison
 Cell Stem Cell, Volume 4 Supplemental Data Hematopoietic Stem Cells Do Not Depend on N-Cadherin to Regulate Their Maintenance Mark J. Kiel, Melih Acar, Glenn L. Radice, and Sean J. Morrison SUPPLEMENTAL
Cell Stem Cell, Volume 4 Supplemental Data Hematopoietic Stem Cells Do Not Depend on N-Cadherin to Regulate Their Maintenance Mark J. Kiel, Melih Acar, Glenn L. Radice, and Sean J. Morrison SUPPLEMENTAL
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang *
 In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
In vivo BrdU Incorporation Assay for Murine Hematopioetic Stem Cells Ningfei An, Yubin Kang * Division of Hematology-Oncology, Department of Medicine, Medical University of South Carolina, Charleston,
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms
 Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Genetics - Problem Drill 19: Dissection of Gene Function: Mutational Analysis of Model Organisms No. 1 of 10 1. The mouse gene knockout is based on. (A) Homologous recombination (B) Site-specific recombination
Supplementary Figure 1. Brwd1 transcripts in Brwd1 mut B cells. Nature Immunology: doi: /ni.3249
 Supplementary Figure 1 Brwd1 transcripts in Brwd1 mut B cells. (a) Analysis of Brwd1 transcripts in isolated small pre-b (Lin B220 + CD43 IgM FSC lo ) and splenic B (B220 + ) cells from WT and Brwd1 mut
Supplementary Figure 1 Brwd1 transcripts in Brwd1 mut B cells. (a) Analysis of Brwd1 transcripts in isolated small pre-b (Lin B220 + CD43 IgM FSC lo ) and splenic B (B220 + ) cells from WT and Brwd1 mut
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
% Viability. isw2 ino isw2 ino isw2 ino isw2 ino mM HU 4-NQO CPT
 a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
a Drug concentration b 1.3% MMS nhp1 nhp1 8 nhp1 mag1.5% MMS.3% MMS nhp1 nhp1 ino8 9 ino8 9 % Viability 4.5% MMS ino8 9 ino8 9 2.5.1.15 % MMS c d nhp1 nhp1 nhp1 nhp1 nhp1 nhp1 Control (YPD) γ IR (1 gy)
Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR
 Supplemental Dataset Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. DNA sequence Amino acid sequence WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR Allele 1 CCTGTC------------------GATAGC
Supplemental Dataset Supplemental Table 1. Mutant ADAMTS3 alleles detected in HEK293T clone 4C2. DNA sequence Amino acid sequence WT CCTGTCACTTTGGTTGATAGC MVLLSLWLIAAALVEVR Allele 1 CCTGTC------------------GATAGC
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature13420 Estimating SRC number by the Poisson distribution of random events Schematic flowchart explaining the calculation made to estimate the number of transplanted SCID repopulating cells
doi:10.1038/nature13420 Estimating SRC number by the Poisson distribution of random events Schematic flowchart explaining the calculation made to estimate the number of transplanted SCID repopulating cells
Table S1. Oligonucleotide primer sequences
 Table S1. Oligonucleotide primer sequences Primer Name DNA Sequence (5 -> 3 ) Description CD19c CD19d Cre7 Sfpi1lox1 Sfpi1lox2 Sfpi1lox3 5 -AACCAGTCAACACCCTTCC-3 5 -CCAGACTAGATACAGACCAG-3 5 -TCAGCTACACCAGAGACGG-3
Table S1. Oligonucleotide primer sequences Primer Name DNA Sequence (5 -> 3 ) Description CD19c CD19d Cre7 Sfpi1lox1 Sfpi1lox2 Sfpi1lox3 5 -AACCAGTCAACACCCTTCC-3 5 -CCAGACTAGATACAGACCAG-3 5 -TCAGCTACACCAGAGACGG-3
Analysis of gene function
 Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Genome 371, 22 February 2010, Lecture 12 Analysis of gene function Gene knockouts PHASE TWO: INTERPRETATION I THINK I FOUND A CORNER PIECE. 3 BILLION PIECES Analysis of a disease gene Gene knockout or
Nature Immunology: doi: /ni Supplementary Figure 1. Time course of OT-II T reg cell development in thymic slices.
 Supplementary Figure 1 Time course of OT-II T reg cell development in thymic slices. Time course of T reg cell development following addition of OT-II TCR transgenic Rag2 -/- mice (OT-II) thymocytes to
Supplementary Figure 1 Time course of OT-II T reg cell development in thymic slices. Time course of T reg cell development following addition of OT-II TCR transgenic Rag2 -/- mice (OT-II) thymocytes to
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin
 Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Supplementary Figure 1 Generation of migg1-yf mice. (A) Targeting strategy. Upper panel: schematic organization of the murine ɣ1 immunoglobulin locus. The EcoRI restriction site between exons M1 and M2
Nature Immunology: doi: /ni Supplementary Figure 1. Construction of lnckdm2b RFP reporter and lnckdm2b-knockout mice.
 Supplementary Figure 1 Construction of lnckdm2b RFP reporter and lnckdm2b-knockout mice. (a) ILC3s (Lin CD45 lo CD90 hi ) were isolated from small intestines, followed by immunofluorescence staining. LncKdm2b
Supplementary Figure 1 Construction of lnckdm2b RFP reporter and lnckdm2b-knockout mice. (a) ILC3s (Lin CD45 lo CD90 hi ) were isolated from small intestines, followed by immunofluorescence staining. LncKdm2b
Supplemental Figure 1
 Supplemental Figure 1 A P1 NOD - SCID(X*X*)(0/0) NOD - SCID(X*Y)(A*0201/A*0201) B F1 (X*X)(A*0201/0) (X*X*)(0/0) F2 (X*X*)(0/0) (X*Y)(0/0) (X*X*)(A*0201/0) (X*X*)(A*0201/0) F3 (X*X*)(0/0) (X*X)(A*0201/0)
Supplemental Figure 1 A P1 NOD - SCID(X*X*)(0/0) NOD - SCID(X*Y)(A*0201/A*0201) B F1 (X*X)(A*0201/0) (X*X*)(0/0) F2 (X*X*)(0/0) (X*Y)(0/0) (X*X*)(A*0201/0) (X*X*)(A*0201/0) F3 (X*X*)(0/0) (X*X)(A*0201/0)
Supplemental material
 Supplemental material Diversity of O-antigen repeat-unit structures can account for the substantial sequence variation of Wzx translocases Yaoqin Hong and Peter R. Reeves School of Molecular Bioscience,
Supplemental material Diversity of O-antigen repeat-unit structures can account for the substantial sequence variation of Wzx translocases Yaoqin Hong and Peter R. Reeves School of Molecular Bioscience,
GENOTYPING BY PCR PROTOCOL FORM MUTANT MOUSE REGIONAL RESOURCE CENTER North America, International
 Please provide the following information required for genetic analysis of your mutant mice. Please fill in form electronically by tabbing through the text fields. The first 2 pages are protected with gray
Please provide the following information required for genetic analysis of your mutant mice. Please fill in form electronically by tabbing through the text fields. The first 2 pages are protected with gray
Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the
 Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the portal vein for digesting adult livers, whereas it was
Supplementary Fig. 1. Isolation and in vitro expansion of EpCAM + cholangiocytes. For collagenase perfusion, enzyme solution was injected from the portal vein for digesting adult livers, whereas it was
strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular
![strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular strain devoid of the aox1 gene [1]. Thus, the identification of AOX1 in the intracellular](/thumbs/72/66361212.jpg) Additional file 2 Identification of AOX1 in P. pastoris GS115 with a Mut s phenotype Results and Discussion The HBsAg producing strain was originally identified as a Mut s (methanol utilization slow) strain
Additional file 2 Identification of AOX1 in P. pastoris GS115 with a Mut s phenotype Results and Discussion The HBsAg producing strain was originally identified as a Mut s (methanol utilization slow) strain
Supplemental Table S1. RT-PCR primers used in this study
 Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
Supplemental Table S1. RT-PCR primers used in this study -----------------------------------------------------------------------------------------------------------------------------------------------
embryos. Asterisk represents loss of or reduced expression. Brackets represent
 Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
Supplemental Figures Supplemental Figure 1. tfec expression is highly enriched in tail endothelial cells (A- B) ISH of tfec at 15 and 16hpf in WT embryos. (C- D) ISH of tfec at 36 and 38hpf in WT embryos.
II 0.95 DM2 (RPP1) DM3 (At3g61540) b
 Table S2. F 2 Segregation Ratios at 16 C, Related to Figure 2 Cross n c Phenotype Model e 2 Locus A Locus B Normal F 1 -like Enhanced d Uk-1/Uk-3 149 64 36 49 DM2 (RPP1) DM1 (SSI4) a Bla-1/Hh-0 F 3 111
Table S2. F 2 Segregation Ratios at 16 C, Related to Figure 2 Cross n c Phenotype Model e 2 Locus A Locus B Normal F 1 -like Enhanced d Uk-1/Uk-3 149 64 36 49 DM2 (RPP1) DM1 (SSI4) a Bla-1/Hh-0 F 3 111
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16108 DOI: 10.1038/NMICROBIOL.2016.108 The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia Carrie A. Cowardin,
SUPPLEMENTARY INFORMATION ARTICLE NUMBER: 16108 DOI: 10.1038/NMICROBIOL.2016.108 The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia Carrie A. Cowardin,
Application Note. Abstract. Reynolds S, 1 Edinger M, 1 Gray J, 1 Tanaka S, 2 Collins P 1 1
 Identification and Functionality of Adult Mouse Hematopoietic Stem Cell Side Populations after Enrichment on the BD FACSAria II Flow Cytometer Equipped with a 375-nm Near UV Laser Reynolds S, 1 Edinger
Identification and Functionality of Adult Mouse Hematopoietic Stem Cell Side Populations after Enrichment on the BD FACSAria II Flow Cytometer Equipped with a 375-nm Near UV Laser Reynolds S, 1 Edinger
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
www.sciencesignaling.org/cgi/content/full/10/494/eaan6284/dc1 Supplementary Materials for Activation of master virulence regulator PhoP in acidic ph requires the Salmonella-specific protein UgtL Jeongjoon
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
Supplementary Figures Supplementary Fig. 1 Characterization of GSCs. a. Immunostaining of primary GSC spheres from GSC lines. Nestin (neural progenitor marker, red), TLX (green). Merged images of nestin,
