Nanoflow LC Q-TOF MS for De Novo Peptide Sequencing in Microbial Proteomics
|
|
|
- Asher Clark
- 5 years ago
- Views:
Transcription
1 COUPLING MATTERS Nanoflow LC Q-TOF MS for De Novo Peptide Sequencing in Microbial Proteomics Bart Devreese and Jozef Van Beeumen, Laboratory of Protein Biochemistry and Protein Engineering, Ghent University, Belgium. The publication of several genome sequences has dramatically changed the scope of biochemical sciences. The availability of the libraries of genes that an organism can use to develop and to adapt to external stress has shifted biochemical research to the analysis of the final gene products; that is, proteins, their posttranslational modifications and their interactions with other biomolecules. The first step in this proteomics 1 approach is the identification of proteins involved in a particular biological process. This work is generally based on so-called differential expression analysis, which generates information about genes that are switched on or off in the presence of a particular external signal. Some of the methods (e.g., microarray techniques) for differential analysis focus on messenger ribonucleic acid (mrna) expression, and take advantage of the array of molecular biology tools available (e.g., polymerase chain reaction technology to amplify the transcripts of genes expressed at low levels). However, critics state that quantitative analysis of mrna levels does not always reflect the presence of a functional protein. Indeed, many proteins are post-translationally modified, or need to be translocated to specific cell organelles before they can exhibit their function. Therefore, it is widely accepted that analysis of gene expression at the protein level is necessary to study the functional role of individual genes in depth. Several techniques are available to perform differential analysis of protein expression. Novel approaches describe the use of protein arrays 2 or multidimensional separation techniques 3 of which some use differential isotopic labelling methods. 4 Despite these new developments, two-dimensional gel electrophoresis is still the most widespread method for the routine separation of proteins expressed by individual cells. Proteins are separated first on the basis of charge by isoelectrofocusing, using immobilized ph-gradient polyacrylamide strips now commercially available in various lengths and ph gradients. In the second dimension proteins are separated by mass using the well-known technique of sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein spots are routinely visualized using Coomassie blue or silver staining techniques, although the use of fluorescent dyes is emerging. In addition, several manufacturers have developed software tools for the quantitative analysis of these gel profiles, allowing the identification of up- or down-regulation of gene expression when comparing protein extracts from cells grown under different conditions. The final task of this proteomics approach is the identification of protein spots for which differential expression has been demonstrated. Mass spectrometry (MS), using either electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI), is the key technology for this purpose. Generally, individual protein spots are digested using trypsin and, after extraction, the peptide mixture is analysed via MS. Different approaches for protein identification from the MS data have been developed. The most widely used is peptide mass fingerprinting (PMF). 5 In this technique the search is simply based on an algorithm that looks in a database for a simulated tryptic digest of a protein that best matches with the experimental one: masses of the obtained tryptic peptides are compared with those from peptides theoretically expected from a cleavage after lysine and arginine residues of all the proteins present in the database. MALDItime of flight (TOF) MS is the most appropriate method to perform PMF because of the low demands on sample preparation, and particularly because of the low complexity of the spectra; that is, the one peptide one peak result in contrast to the multiple charging events in ESI. While PMF is certainly useful for highthroughput approaches, because it is easily automated, its scoring rate is often limited. It accounts poorly for post-translational modifications, handles poorly with mixtures, and certain groups of proteins are difficult to identify. For example, the scoring algorithms are mainly based on counting the number of hits (the more peptides matching, the higher the score). This way, one easily overlooks proteins with limited numbers of tryptic peptides, such as small proteins (<1 kda) and large hydrophobic (membrane) proteins. It has been shown that by including some sequence information, be it only a few residues, in the database search increases the reliability and the scoring rates by one order of magnitude. Although there are new developments coupling TOF-TOF and Q-TOF technology with MALDI, 6,7 most of the MS MS approaches in proteomics are still performed using ESI on ion trap or Q-TOF instruments. The disadvantage of ESI is its low tolerance to buffer salts, and sample clean-up is often a necessity. Some manual tools using small spinning columns or micropipette tips with chromatographic media have been developed, but none are useful in highthroughput environments. Therefore, on-line chromatographic separations prefacing mass spectrometric analyses are 2 LC GC Europe October 2
2 the best choice. We have implemented a nanoflow liquid chromatography (LC) approach to combine high throughput with the sensitivity necessary to allow protein identification from 2D-PAGE separations. The Nano LC System An Ultimate micro LC system (LC Packings A Dionex Company, Amsterdam, The Netherlands) is used (Figure 1). A Famos autosampler (LC Packings) is used for the injection of samples. The samples are first injected onto a micro precolumn, a procedure that has two advantages. First, it allows a significant reduction in injection times (nanoflow LC is performed at flowrates of 15 nl/min and so it would take at least 3 min to inject a 3 µl sample). The sample is injected at a flowrate of 1 µl/min using an external pump. The second advantage is the preliminary desalting: samples are cleaned up prior to injecting onto the separating column, thereby extending the lifetime of the nano LC column. After the injection/desalting step, the valve is switched to make a connection with the nano LC column. The peptides stored on the precolumn are eluted to the separating column and then separated using a standard reversed-phase (RP) gradient. The endcapping of the columns (PEPMAP, Dionex) allows the use of formic acid rather than trifluoroacetic acid (TFA) as the counter ion in the RP-LC separation, without loss of resolution. This has an important impact on the sensitivity in the mass spectrometer because TFA dramatically reduces signal intensities in ESI. In our system, the outlet of the column is connected to a homemade nano-esi interface for which fused-silica capillary needles are used (New Objective, Woburn, Massachusetts, USA). Mass spectral data are acquired on a Micromass Q-TOF instrument (Manchester, UK). The instrument is run in the data-dependent MS to MS MS switching mode, which means that any peptide eluting from the column with a mass peak above a certain threshold is automatically selected for MS MS analysis. Although there are new developments coupling TOF-TOF and Q-TOF technology with MALDI, most of the MS MS approaches in proteomics are still performed using ESI on ion trap or Q-TOF instruments. Figure 1: Schematic of the nanoflow LC system: flow scheme during sample loading on the micro precolumn and flow scheme during gradient elution of the peptides. ABI pump, 1 L/min Sample vial ABI pump, 1 L/min Sample vial Sample loop Sample loop Waste Ultimate pump.1 L/min Waste Ultimate pump.1 L/min precolumn precolumn Nano LC column Nano LC column Detector Home designed ES device with fused-silica capillaries Detector Home designed ES device with fused-silica capillaries Figure 2: Details of the 2D-PAGE of a total lysate of Shewanella hanedai grown at 12 C and 4 C. Arrows highlight the two analysed proteins. De Novo Sequence Analysis and Microbial Proteomics The Q-TOF MS MS data quality not only allows the use of MS MS data for supporting PMF in database searching for identifying proteins from organisms of which the genome has been sequenced, but also allows partial sequence analysis (de novo sequencing). The sequence data 3
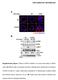
3 Figure 3: Total ion current chromatogram for the MS MS scan of the tryptic digest of spot 1 shown in Figure 2. The numbers on top of the peaks refer to the mass of the molecular ion selected for fragmentation Scan Figure 4: MS MS spectra of two peptides with molecular ions of m/z 664 and 624 automatically selected for fragmentation as seen in Figure Ala Mass Glu Pro His Glu Lxx Lxx Asp Asn Val Val Gln His Mass can then be used in similarity searches to identify proteins from model organisms of which no genome sequencing efforts have been undertaken. Today, according to the database maintained by the Institute of Genomic Research, the genomes of some 6 bacterial strains have been completely sequenced, and more than other sequencing efforts are underway. Nevertheless, many model organisms of biomedical or environmental importance remain of which no genome sequencing effort has been initiated. Our mass spectrometric approach for de novo sequence analysis allows the initiation of proteomic efforts without needing genomic information on the bacterium of interest. The increasing number of protein sequences in the databases, covering a wide range of bacterial subfamilies, provides the data necessary for efficient similarity searching approaches for protein identification. Example De Novo Sequencing of Cold-Induced Proteins One of our study targets is a psychrophilic bacterium, Shewanella hanedai. It is a marine Antarctic bacterium and a potential source of poly-unsaturated fatty acids, widely used as food supplements. 8 We have initiated a study of the cold adaptation of this organism, because the synthesis of these fatty acids seems to stem from a mechanism to increase membrane flexibility, which is an essential way to maintain motility and to ensure transmembrane transport of nutrients at near-zero degrees Celsius. The protein and DNA sequence data available on this organism are very limited, and we must rely on similarity searching for database searching. Figure 2 shows the 2D-PAGE gels of a total lysate of S. hanedai grown at different temperatures. Clearly, two protein spots are higher in expression when the organism is grown at lower temperatures. The spots were cut from the gel, digested, and prepared for nano LC MS MS analysis. For a detailed description of the sample preparation we refer to a forthcoming paper. 9 The mass spectrometer was used in automated MS to MS MS switching mode, in which it first generates a survey scan. Each time a peptide elutes from the column and gives an ESI signal above a certain threshold it is selected for collision-induced dissociation analysis. Figure 3 shows the total ion current chromatogram display for this MS MS analysis. After the peptide analysis is finished (either because the signal drops 4 LC GC Europe October 2
4 The Q-TOF MS MS data quality not only allows the use of MS MS data for supporting PMF in database searching for identifying proteins from organisms of which the genome has been sequenced, but also allows partial sequence analysis (de novo sequencing). below a threshold level, or because of a set time limit) the instrument switches back to a survey scan. Selection of an individual peak from the TIC chromatogram allows the analysis of the MS MS spectrum of the corresponding peptide, as shown in Figure 4. The quality of Q-TOF MS MS is generally sufficient to read at least half of the sequence of a 15 2 amino acid residue peptide, delivering a portion of the sequence that allows a significant database searching based on similarity. We have mainly used the Blast-search routine provided by the National Center for Biotechnology Information ( freely accessible for the academic world. We thus take advantage of the growing number of proteins available in this database, as provided by the genome project efforts on several micro-organisms. Figure 5 provides the identification of proteins based on the stretches of sequence determined from the MS MS spectra. The first protein, alkylhydroperoxidase C, belongs to a family of stress response proteins that is expressed by many micro-organisms during exposure to external stress. The second protein is similar to a member of the Fe-superoxide dismutase family. This clearly shows that S. hanedai deals with stress responses in a very similar way to mesophilic organisms, because similar proteins are expressed when providing cold shock to organisms such as E. coli. The difference is that the onset of the stress response occurs at a much lower temperature in the psychrohilic organism. Conclusions The use of nano LC, incorporating an on-line microconcentration/desalting step, has proven to be an excellent tool for sample introduction in ESI Q-TOF technology. It provides both the sample clean-up (important for prolonging the lifetime of the column as well as the stability of the electrospray) and the sensitivity necessary to analyse tryptic digests from single spots of 2D-PAGE gels, generally providing femtomole amounts of peptides. The Q-TOF MS MS spectra are of sufficient quality to allow (partial) de novo sequence analysis of the peptides, with sequence stretches of sufficient length for identification of peptides based on similarity searching. References 1. M.R. Wilkins et al., Bio/Technology, 14, 61 (1996). 2. A. Mirzabekow and A. Kolchinsky, Current Opin. Chem. Biol., 6, 7 (2). 3. G.J. Opiteck et al., Anal. Biochem., 258, 349 (1998). 4. S.P. Gigy et al., Nature Biotechnol., 17, 994 (1999). 5. D.J.C. Pappin et al., Curr. Biol., 3, 327 (1993). Figure 5: Identification of the two proteins highlighted in Figure 2 based on similarity searching. The sequences shown in red are those obtained from mass spectral analysis and were used in the Blast search. The peptides were found to possess the highest similarities to proteins from Vibrio cholerae, which is evolutionary related to Shewanella hanedai. Spot 1: Alkyl hydroperoxidase C MLRSKKMVLV GRKAPDFTAA AVLGNGEIVD NFNFAEFTKG KKAVVFFYPL DFTFVCPSEL IAFDNRYEDF LAKGVEVIGV SIDSQFSHNA WRNTAVENGG IGQVKYPLVA DVKHEICKAY DVEHPEAGVA FRGSFLIDEE GMVRHQVVND LPLGRNIDEM LRMVDALNFH QTHGEVCPAQ AY DVEHPEAGVA HQVVND LLPGR WEAGKAGMEA SPKGVAAFLS QYSADLK Spot 2: Fe-superoxidase dismutase MAFELPALPY AKDALEPHIS AETLDFHHGK HHNTYVVKLN GLI P GTEFEN KSLEEIIKTS LD GLVEGTELAE K TGGIFNNAAQ VWNHTFYWHC LSPNGGGEPT GAVAEAINAA FGSFADFKAK FTDSAINNFG SPDG DDA FGSFA TDSAVNNFG SSWTWLVKKA DGTLAITNTS NAATPLTEEG VTPLLTVDLW EHAYYIDYRN VRPDYMNGFW SAWTWLVK ALVNWDFVAQ NLAK 5
5 6. K.F. Medzhiradszky et al., Anal. Chem., 72, 552 (). 7. P. Verhaert et al., Proteomics, 1, 118 (1). 8. J.P. Bowman et al., Int. J. Sys. Bacteriol., 47, 14 (1997). 9. B. Devreese et al., J. Chromatogr. A, in press (2). Bart Devreese obtained his PhD degree in 1997 in the laboratory of Professor Van Beeumen, having used the novel soft ionization mass spectrometric techniques in the elucidation of the primary structures of redox proteins from sulfate reducing bacteria. In 1999 he obtained a Postdoctoral Fellowship from the Fund for Scientific Research-Flanders, Belgium and initiated proteomic analysis of psychrophilic and other extremophilic micro-organisms. Jozef Van Beeumen is head of the laboratory of Protein Biochemistry and Protein Engineering at Ghent University, Belgium. His scientific career is centered around protein structure analysis, for the major part on bacterial redox systems, antibiotic resistance and extremophilic organisms. He was the first to use the techniques of ESI-MS and MALDI-MS in the field in Belgium in the early 199s. His laboratory is also equipped with X-ray crystallographic equipment to study the 3-dimensional structure of proteins. He recently became president of the Department of Biochemistry, Physiology and Microbiology at Ghent University. 6 LC GC Europe October 2
Proteomics. Proteomics is the study of all proteins within organism. Challenges
 Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Proteomics Proteomics is the study of all proteins within organism. Challenges 1. The proteome is larger than the genome due to alternative splicing and protein modification. As we have said before we
Advances in analytical biochemistry and systems biology: Proteomics
 Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Advances in analytical biochemistry and systems biology: Proteomics Brett Boghigian Department of Chemical & Biological Engineering Tufts University July 29, 2005 Proteomics The basics History Current
Proteomics and some of its Mass Spectrometric Applications
 Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Proteomics and some of its Mass Spectrometric Applications What? Large scale screening of proteins, their expression, modifications and interactions by using high-throughput approaches 2 1 Why? The number
Computing with large data sets
 Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Computing with large data sets Richard Bonneau, spring 2009 Lecture 14 (week 8): genomics 1 Central dogma Gene expression DNA RNA Protein v22.0480: computing with data, Richard Bonneau Lecture 14 places
Proteomics and Cancer
 Proteomics and Cancer Japan Society for the Promotion of Science (JSPS) Science Dialogue Program at Niitsu Senior High School Niitsu, Niigata September 4th 2006 Vladimir Valera, M.D, PhD JSPS Postdoctoral
Proteomics and Cancer Japan Society for the Promotion of Science (JSPS) Science Dialogue Program at Niitsu Senior High School Niitsu, Niigata September 4th 2006 Vladimir Valera, M.D, PhD JSPS Postdoctoral
Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics
 Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics INTRODUCTION Peptide sequencing by nano LC-ESI-MS/MS is a widespread technique used for protein identification in proteomics.
Parallel LC with Capillary PS-DVB Monolithic Columns for High-Throughput Proteomics INTRODUCTION Peptide sequencing by nano LC-ESI-MS/MS is a widespread technique used for protein identification in proteomics.
Mass Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System
 GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
GUIDE TO LC MS - December 21 1 Spectrometry Analysis of Liquid Chromatography Fractions using Ettan LC MS System Henrik Wadensten, Inger Salomonsson, Staffan Lindqvist, Staffan Renlund, Amersham Biosciences,
A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery Using HPLC-Chip/ MS-Enabled ESI-TOF MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS A Highly Accurate Mass Profiling Approach to Protein Biomarker Discovery
Improving Productivity with Applied Biosystems GPS Explorer
 Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Product Bulletin TOF MS Improving Productivity with Applied Biosystems GPS Explorer Software Purpose GPS Explorer Software is the application layer software for the Applied Biosystems 4700 Proteomics Discovery
Nano LC at 20 nl/min Made Easy: A Splitless Pump Combined with Fingertight UHPLC Nano Column to Boost LC-MS Sensitivity in Proteomics
 Nano LC at 20 nl/min Made Easy: A Splitless Pump Combined with Fingertight UHPLC Nano Column to Boost LC-MS Sensitivity in Proteomics Rieux L 1., De Pra M 1., Köcher T 2., Mechtler K 2., Swart R 1 1 Thermo
Nano LC at 20 nl/min Made Easy: A Splitless Pump Combined with Fingertight UHPLC Nano Column to Boost LC-MS Sensitivity in Proteomics Rieux L 1., De Pra M 1., Köcher T 2., Mechtler K 2., Swart R 1 1 Thermo
SGN-6106 Computational Systems Biology I
 SGN-6106 Computational Systems Biology I A View of Modern Measurement Systems in Cell Biology Kaisa-Leena Taattola 21.2.2007 1 The cell a complex system (Source: Lehninger Principles of Biochemistry 4th
SGN-6106 Computational Systems Biology I A View of Modern Measurement Systems in Cell Biology Kaisa-Leena Taattola 21.2.2007 1 The cell a complex system (Source: Lehninger Principles of Biochemistry 4th
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis
 Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Laboratory Water Quality Affects Protein Separation by 2D Gel Electrophoresis 2D gel electrophoresis remains a dominant technique in proteomics. Obtaining high quality gels requires careful and tedious
Solutions Guide. MX Series II Modular Automation for Nano and Analytical Scale HPLC And Low Pressure Fluid Switching Applications
 Solutions Guide MX Series II Modular Automation for Nano and Analytical Scale HPLC And Low Pressure Fluid Switching Applications Page 1 of 12 Table of Contents Sample Injection... 3 Two- Selection... 4
Solutions Guide MX Series II Modular Automation for Nano and Analytical Scale HPLC And Low Pressure Fluid Switching Applications Page 1 of 12 Table of Contents Sample Injection... 3 Two- Selection... 4
LECTURE-3. Protein Chemistry to proteomics HANDOUT. Proteins are the most dynamic and versatile macromolecules in a living cell, which
 LECTURE-3 Protein Chemistry to proteomics HANDOUT PREAMBLE Proteins are the most dynamic and versatile macromolecules in a living cell, which regulates essential activities of the cell. The classical protein
LECTURE-3 Protein Chemistry to proteomics HANDOUT PREAMBLE Proteins are the most dynamic and versatile macromolecules in a living cell, which regulates essential activities of the cell. The classical protein
columns PepSwift and ProSwift Capillary Monolithic Reversed-Phase Columns
 columns PepSwift and ProSwift Capillary Monolithic Reversed-Phase Columns PepSwift and ProSwift monolithic columns are specially designed for high-resolution LC/MS analysis in protein identification, biomarker
columns PepSwift and ProSwift Capillary Monolithic Reversed-Phase Columns PepSwift and ProSwift monolithic columns are specially designed for high-resolution LC/MS analysis in protein identification, biomarker
The Role of Chemistry Consumables in Biopharmaceutical Product Development
 Technology & Services The Role of Chemistry Consumables in Biopharmaceutical Product Development a report by John C Gebler, Jeff R Mazzeo and William J Warren Director, Life Sciences Reasearch and Development,
Technology & Services The Role of Chemistry Consumables in Biopharmaceutical Product Development a report by John C Gebler, Jeff R Mazzeo and William J Warren Director, Life Sciences Reasearch and Development,
Hichrom Limited 1 The Markham Centre, Station Road, Theale, Reading, Berkshire, RG7 4PE, UK Tel: +44 (0) Fax: +44 (0)
 Hichrom Limited 1 The Markham Centre, Station Road, Theale, Reading, Berkshire, RG7 4PE, UK Tel: +44 (0)118 930 3660 Fax: +44 (0)118 932 3484 Email: www.hichrom.co.uk Table of Contents Sample Injection...
Hichrom Limited 1 The Markham Centre, Station Road, Theale, Reading, Berkshire, RG7 4PE, UK Tel: +44 (0)118 930 3660 Fax: +44 (0)118 932 3484 Email: www.hichrom.co.uk Table of Contents Sample Injection...
Strategies in proteomics
 Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
Strategies in proteomics Systems biology - understand cellpathways, network, and complex interacting (includes Genomics, Proteomics, Metabolomics..) Biological processes - characterize protein complexes,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
SUPPLEMENTARY INFORMATION Supplementary Figure 1 Effect of ROCK inhibition on lumen abnormality in MDCK cysts. (A) MDCK cells as indicated cultured in Matrigel were treated with and without Y27632 (10
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins
 Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Application Note # ET-20 BioPharma Compass: A fully Automated Solution for Characterization and QC of Intact and Digested Proteins BioPharma Compass TM is a fully automated solution for the rapid characterization
Proteomics And Cancer Biomarker Discovery. Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar. Overview. Cancer.
 Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Proteomics And Cancer Biomarker Discovery Dr. Zahid Khan Institute of chemical Sciences (ICS) University of Peshawar Overview Proteomics Cancer Aims Tools Data Base search Challenges Summary 1 Overview
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Tuesday May 29 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Proteomics and Vaccine Potency Testing
 Proteomics and Vaccine Potency Testing Louisa B. Tabatabai, PhD Respiratory Diseases of Livestock Research Unit National Animal Disease Center, ARS, USDA, Ames, Iowa What is proteomics Methodologies of
Proteomics and Vaccine Potency Testing Louisa B. Tabatabai, PhD Respiratory Diseases of Livestock Research Unit National Animal Disease Center, ARS, USDA, Ames, Iowa What is proteomics Methodologies of
Lecture 5: 8/31. CHAPTER 5 Techniques in Protein Biochemistry
 Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Lecture 5: 8/31 CHAPTER 5 Techniques in Protein Biochemistry Chapter 5 Outline The proteome is the entire set of proteins expressed and modified by a cell under a particular set of biochemical conditions.
Automated On-Line SDS Removal from Diminutive Protein Samples Prior to Capillary HPLC Application Note 221
 Automated On-Line SDS Removal from Diminutive Protein Samples Prior to Capillary HPLC Application Note 221 Joan Stevens, Ph.D.; Luke Roenneburg; Tim Hegeman (Gilson, Inc.) Introduction Removing detergent
Automated On-Line SDS Removal from Diminutive Protein Samples Prior to Capillary HPLC Application Note 221 Joan Stevens, Ph.D.; Luke Roenneburg; Tim Hegeman (Gilson, Inc.) Introduction Removing detergent
Tools and considerations to increase resolution of complex proteome samples by two-dimensional offline LC/MS. Application
 Tools and considerations to increase resolution of complex proteome samples by two-dimensional offline LC/MS Application Martin Vollmer Patric Hörth Edgar Nägele Abstract Off-line 2D-HPLC applying a continuous
Tools and considerations to increase resolution of complex proteome samples by two-dimensional offline LC/MS Application Martin Vollmer Patric Hörth Edgar Nägele Abstract Off-line 2D-HPLC applying a continuous
Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research
 PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
PO-CON138E Development and evaluation of Nano-ESI coupled to a triple quadrupole mass spectrometer for quantitative proteomics research ASMS 213 ThP 115 Shannon L. Cook 1, Hideki Yamamoto 2, Tairo Ogura
Medicinal Chemistry of Modern Antibiotics
 Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2012 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2012 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Medicinal Chemistry of Modern Antibiotics
 Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2008 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
Chemistry 259 Medicinal Chemistry of Modern Antibiotics Spring 2008 Lecture 5: Modern Target Discovery & MOA Thomas Hermann Department of Chemistry & Biochemistry University of California, San Diego Drug
A highly sensitive and robust 150 µm column to enable high-throughput proteomics
 APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
APPLICATION NOTE 21744 Robust LC Separation Optimized MS Acquisition Comprehensive Data Informatics A highly sensitive and robust 15 µm column to enable high-throughput proteomics Authors Xin Zhang, 1
PLRP-S Polymeric Reversed-Phase Column for LC/MS Separation of mabs and ADC
 PLRP-S Polymeric Reversed-Phase Column for LC/MS Separation of mabs and ADC Analysis of Intact and Fragmented mabs and ADC Application Note Biotherapeutics and Biologics Author Suresh Babu C.V. Agilent
PLRP-S Polymeric Reversed-Phase Column for LC/MS Separation of mabs and ADC Analysis of Intact and Fragmented mabs and ADC Application Note Biotherapeutics and Biologics Author Suresh Babu C.V. Agilent
ApplicationNOTE THE USE OF A NANOLOCKSPRAY ELECTROSPRAY INTERFACE FOR EXACT MASS PROTEOMICS STUDIES
 OVERVIEW This application note describes the implementation of a dual sprayer NanoLockSpray TM source on the Waters Micromass Q-Tof micro TM mass spectrometer This source consists of a dual sprayer arrangement;
OVERVIEW This application note describes the implementation of a dual sprayer NanoLockSpray TM source on the Waters Micromass Q-Tof micro TM mass spectrometer This source consists of a dual sprayer arrangement;
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B
 Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
Improving Retention Time Precision and Chromatography of Early Eluting Peptides with Acetonitrile/Water Blends as Solvent B Stephan Meding, Aran Paulus, and Remco Swart ¹Thermo Fisher Scientific, Germering,
2D difference gel electrophoresis (2D-DIGE) Spot picking and in-gel digestion
 2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the
2D difference gel electrophoresis (2D-DIGE) Proteins were labeled with spectrally resolvable fluorescent cyanine dyes prior to further separation according to their charge and size. For each one of the
Application of Agilent AdvanceBio Desalting-RP Cartridges for LC/MS Analysis of mabs A One- and Two-dimensional LC/MS Study
 Application of Agilent AdvanceBio Desalting-RP Cartridges for LC/MS Analysis of mabs A One- and Two-dimensional LC/MS Study Application note Biotherapeutics and Biologics Authors Suresh Babu C.V., Anne
Application of Agilent AdvanceBio Desalting-RP Cartridges for LC/MS Analysis of mabs A One- and Two-dimensional LC/MS Study Application note Biotherapeutics and Biologics Authors Suresh Babu C.V., Anne
Exam MOL3007 Functional Genomics
 Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Thursday December 20 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Faculty of Medicine Department of Cancer Research and Molecular Medicine Exam MOL3007 Functional Genomics Thursday December 20 th 9.00-13.00 ECTS credits: 7.5 Number of pages (included front-page): 5 Supporting
Proteomics Technology Note
 Targeted Proteomics for the Identification of Nucleic Acid Binding Proteins in E. coli: Finding more low abundant proteins using the QSTAR LC/MS/MS System, Multi- Dimensional Liquid Chromatography, Pro
Targeted Proteomics for the Identification of Nucleic Acid Binding Proteins in E. coli: Finding more low abundant proteins using the QSTAR LC/MS/MS System, Multi- Dimensional Liquid Chromatography, Pro
Kinetics Review. Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets)
 Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Quiz 1 Kinetics Review Tonight at 7 PM Phys 204 We will do two problems on the board (additional ones than in the problem sets) I will post the problems with solutions on Toolkit for those that can t make
Automation of MALDI-TOF Analysis for Proteomics
 Application Note #MT-50 BIFLEX III / REFLEX III Automation of MALDI-TOF Analysis for Proteomics D. Suckau, C. Köster, P.Hufnagel, K.-O. Kräuter and U. Rapp, Bruker Daltonik GmbH, Bremen, Germany Introduction
Application Note #MT-50 BIFLEX III / REFLEX III Automation of MALDI-TOF Analysis for Proteomics D. Suckau, C. Köster, P.Hufnagel, K.-O. Kräuter and U. Rapp, Bruker Daltonik GmbH, Bremen, Germany Introduction
Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS
 Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS Sensitive workflow for the detection of PTMs in biological samples from small sample volumes Klaus Faserl, 1 Bettina Sarg, 1
Detecting Challenging Post Translational Modifications (PTMs) using CESI-MS Sensitive workflow for the detection of PTMs in biological samples from small sample volumes Klaus Faserl, 1 Bettina Sarg, 1
Protein Microarrays?
 Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Protein Microarrays? Protein Chemistry/Proteomics Unit and the Neuroscience Program,Biomedicum Helsinki E-Mail: marc.baumann@helsinki.fi (http://research.med.helsinki.fi/corefacilities/proteinchem) What
Application Note TOF/MS
 Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Application Note TOF/MS New Level of Confidence for Protein Identification: Results Dependent Analysis and Peptide Mass Fingerprinting Using the 4700 Proteomics Discovery System Purpose The Applied Biosystems
Lecture 8: Affinity Chromatography-III
 Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Lecture 8: Affinity Chromatography-III Key words: Chromatography; Affinity chromatography; Protein Purification During this lecture, we shall be studying few more examples of affinity chromatography. The
Proteomics: A Challenge for Technology and Information Science. What is proteomics?
 Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Proteomics: A Challenge for Technology and Information Science CBCB Seminar, November 21, 2005 Tim Griffin Dept. Biochemistry, Molecular Biology and Biophysics tgriffin@umn.edu What is proteomics? Proteomics
Developing a natural partnership for rapid, sensitive identification of binding partners
 GE Healthcare Technology Note 24 Biacore systems Developing a natural partnership for rapid, sensitive identification of binding partners Biacore 3000 and mass spectrometry Summary The coupling of label-free
GE Healthcare Technology Note 24 Biacore systems Developing a natural partnership for rapid, sensitive identification of binding partners Biacore 3000 and mass spectrometry Summary The coupling of label-free
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS
 Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Využití cílené proteomiky pro kontrolu falšování potravin: identifikace peptidových markerů v mase pomocí LC- Q Exactive MS/MS Michal Godula Ph.D. Thermo Fisher Scientific The world leader in serving science
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore, India
 Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Quantitation of Oxidation Sites on a Monoclonal Antibody Using an Agilent 1260 Infinity HPLC-Chip/MS System Coupled to an Accurate-Mass 6520 Q-TOF LC/MS Application Note Authors Ravindra Gudihal Agilent
Faster, easier, flexible proteomics solutions
 Agilent HPLC-Chip LC/MS Faster, easier, flexible proteomics solutions Our measure is your success. products applications soft ware services Phospho- Anaysis Intact Glycan & Glycoprotein Agilent s HPLC-Chip
Agilent HPLC-Chip LC/MS Faster, easier, flexible proteomics solutions Our measure is your success. products applications soft ware services Phospho- Anaysis Intact Glycan & Glycoprotein Agilent s HPLC-Chip
Key questions of proteomics. Bioinformatics 2. Proteomics. Foundation of proteomics. What proteins are there? Protein digestion
 s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture X roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
s s Key questions of proteomics What proteins are there? Bioinformatics 2 Lecture X roteomics How much is there of each of the proteins? - Absolute quantitation - Stoichiometry What (modification/splice)
Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS
 Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest and Lei Xiong SCIEX, Redwood City,
Improving Sensitivity in Bioanalysis using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest and Lei Xiong SCIEX, Redwood City,
Zakopane, April, 2011
 Clinical Proteomics: A Technology to shape the future? Zakopane, April, 2011 Protein Chemistry/Proteomics/Peptide Synthesis and Array Unit Institute of Biomedicine/Anatomy Biomedicum Helsinki, 00014 University
Clinical Proteomics: A Technology to shape the future? Zakopane, April, 2011 Protein Chemistry/Proteomics/Peptide Synthesis and Array Unit Institute of Biomedicine/Anatomy Biomedicum Helsinki, 00014 University
Algorithm for Matching Additional Spectra
 Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
Improved Methods for Comprehensive Sample Analysis Using Protein Prospector Peter R. Baker 1, Katalin F. Medzihradszky 1 and Alma L. Burlingame 1 1 Mass Spectrometry Facility, Dept. of Pharmaceutical Chemistry,
Quantitative mass spec based proteomics
 Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
Quantitative mass spec based proteomics Tuula Nyman Institute of Biotechnology tuula.nyman@helsinki.fi THE PROTEOME The complete protein complement expressed by a genome or by a cell or a tissue type (M.
Two-Dimensional LC Protein Separation on Monolithic Columns in a Fully Automated Workflow
 Technical Note Two-Dimensional LC Protein Separation on Monolithic Columns in a Fully Automated Workflow Introduction The complex nature of proteomics samples requires high-resolution separation methods
Technical Note Two-Dimensional LC Protein Separation on Monolithic Columns in a Fully Automated Workflow Introduction The complex nature of proteomics samples requires high-resolution separation methods
timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics
 timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics The timstof Pro powered by PASEF enables sequencing speed of > 2 Hz, high sensitivity
timstof Pro with PASEF and Evosep One: Maximizing throughput, robustness and analytical depth for shotgun proteomics The timstof Pro powered by PASEF enables sequencing speed of > 2 Hz, high sensitivity
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH)
 User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
User Rules & Regulations Core Facility for Mass Spectrometry and Proteomics (CFMP) at the Centre for Molecular Biology of Heidelberg University (ZMBH) Accepted on 30. January 2017 1 Definition and aims
Proteomics 6/4/2009 WESTERN BLOT ANALYSIS
 SDS-PAGE (PolyAcrylamide Gel Electrophoresis) Proteomics WESTERN BLOT ANALYSIS Presented by: Nuvee Prapasarakul Veterinary Microbiology Chulalongkorn University Proteomics has been said to be the next
SDS-PAGE (PolyAcrylamide Gel Electrophoresis) Proteomics WESTERN BLOT ANALYSIS Presented by: Nuvee Prapasarakul Veterinary Microbiology Chulalongkorn University Proteomics has been said to be the next
Benefits of 2D-LC/MS/MS in Pharmaceutical Bioanalytics
 Application Note Small Molecule Pharmaceuticals Benefits of 2D-LC/MS/MS in Pharmaceutical Bioanalytics Avoiding Matrix Effects Increasing Detection Sensitivity Authors Jonas Dinser Daiichi-Sankyo Europe
Application Note Small Molecule Pharmaceuticals Benefits of 2D-LC/MS/MS in Pharmaceutical Bioanalytics Avoiding Matrix Effects Increasing Detection Sensitivity Authors Jonas Dinser Daiichi-Sankyo Europe
NanoLC-Ultra system. data sheet. Introducing the NanoLC-Ultra family of high pressure HPLCs for proteomics research
 data sheet NanoLC-Ultra system The new NanoLC-Ultra is Eksigent s third generation system, delivering superior gradient precision at pressures up to 10,000 psi. Introducing the NanoLC-Ultra family of high
data sheet NanoLC-Ultra system The new NanoLC-Ultra is Eksigent s third generation system, delivering superior gradient precision at pressures up to 10,000 psi. Introducing the NanoLC-Ultra family of high
Antibody Analysis by ESI-TOF LC/MS
 Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS Antibody Analysis by ESI-TOF LC/MS Authors Lorenzo Chen, Merck & Company,
Application Note PROTEOMICS METABOLOMICS GENOMICS INFORMATICS GLYILEVALCYSGLUGLNALASERLEUASPARG CYSVALLYSPROLYSPHETYRTHRLEUHISLYS Antibody Analysis by ESI-TOF LC/MS Authors Lorenzo Chen, Merck & Company,
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis
 Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Enabling Systems Biology Driven Proteome Wide Quantitation of Mycobacterium Tuberculosis SWATH Acquisition on the TripleTOF 5600+ System Samuel L. Bader, Robert L. Moritz Institute of Systems Biology,
Multidimensional LC MS for Proteomics Present and Future
 Multidimensional LC MS for Proteomics Present and Future Martin Vollmer, Patric Hörth, Cornelia Vad and Edgar Nägele, Agilent Technologies R&D and Marketing GmbH & Co. KG, Waldbronn, Germany. Today, two-dimensional
Multidimensional LC MS for Proteomics Present and Future Martin Vollmer, Patric Hörth, Cornelia Vad and Edgar Nägele, Agilent Technologies R&D and Marketing GmbH & Co. KG, Waldbronn, Germany. Today, two-dimensional
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications
![MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1. Lecture 25: Mass Spectrometry Applications](/thumbs/74/71368870.jpg) MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
MBios 478: Mass Spectrometry Applications [Dr. Wyrick] Slide #1 Lecture 25: Mass Spectrometry Applications Measuring Protein Abundance o ICAT o DIGE Identifying Post-Translational Modifications Protein-protein
Automated platform of µlc-ms/ms using SAX trap column for
 Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
Analytical and Bioanalytical Chemistry Electronic Supplementary Material Automated platform of µlc-ms/ms using SAX trap column for highly efficient phosphopeptide analysis Xionghua Sun, Xiaogang Jiang
CaptiveSpray nanobooster
 CaptiveSpray nanobooster The Revolution in Proteomics Ionization Innovation with Integrity LC-MS Source The Key to Performance and Reliability for nano flow MS Bruker CaptiveSpray principle: Stable and
CaptiveSpray nanobooster The Revolution in Proteomics Ionization Innovation with Integrity LC-MS Source The Key to Performance and Reliability for nano flow MS Bruker CaptiveSpray principle: Stable and
Fast and Efficient Peptide Mapping of a Monoclonal Antibody (mab): UHPLC Performance with Superficially Porous Particles
 Fast and Efficient Peptide Mapping of a Monoclonal Antibody (mab): UHPLC Performance with Superficially Porous Particles Application Note Biotherapeutics and Biosimilars Authors James Martosella, Alex
Fast and Efficient Peptide Mapping of a Monoclonal Antibody (mab): UHPLC Performance with Superficially Porous Particles Application Note Biotherapeutics and Biosimilars Authors James Martosella, Alex
Separation of Native Monoclonal Antibodies and Identification of Charge Variants:
 Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Separation of Native Monoclonal Antibodies and Identification of Charge Variants: Teamwork of the Agilent 31 OFFGEL Fractionator, Agilent 21 Bioanalyzer and Agilent LC/MS Systems Application Note Biosimilar
Analysis of Monoclonal Antibody (mab) Using Agilent 1290 Infinity LC System Coupled to Agilent 6530 Accurate-Mass Quadrupole Time-of-Flight (Q-TOF)
 Analysis of Monoclonal Antibody (mab) Using Agilent 9 Infinity LC System Coupled to Agilent Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) Application Note Authors Ravindra Gudihal and Suresh Babu CV
Analysis of Monoclonal Antibody (mab) Using Agilent 9 Infinity LC System Coupled to Agilent Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) Application Note Authors Ravindra Gudihal and Suresh Babu CV
Thermo Scientific EASY-nLC 1000 System. Effortless, split-free nanoflow UHPLC Top performance in LC/MS
 Thermo Scientific EASY-nLC 1000 System Effortless, split-free nanoflow UHPLC Top performance in LC/MS Effortless split-free, nanoflow UHPLC The Thermo Scientific EASY-nLC 1000 is a fully integrated, split-free,
Thermo Scientific EASY-nLC 1000 System Effortless, split-free nanoflow UHPLC Top performance in LC/MS Effortless split-free, nanoflow UHPLC The Thermo Scientific EASY-nLC 1000 is a fully integrated, split-free,
Proteomics. Manickam Sugumaran. Department of Biology University of Massachusetts Boston, MA 02125
 Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
Proteomics Manickam Sugumaran Department of Biology University of Massachusetts Boston, MA 02125 Genomic studies produced more than 75,000 potential gene sequence targets. (The number may be even higher
MagSi Beads. Magnetic Silica Beads for Life Science and Biotechnology study
 MagSi Beads Magnetic Silica Beads for Life Science and Biotechnology study MagnaMedics Diagnostics B.V. / Rev. 9.2 / 2012 Wide range of products for numerous applications MagnaMedics separation solutions
MagSi Beads Magnetic Silica Beads for Life Science and Biotechnology study MagnaMedics Diagnostics B.V. / Rev. 9.2 / 2012 Wide range of products for numerous applications MagnaMedics separation solutions
Application Note. Authors. Abstract. Ravindra Gudihal Agilent Technologies India Pvt. Ltd. Bangalore India
 Characterization of bacteriophage derived anti-staphylococcal protein (P28) from production to purification using Agilent HPLC-Chip Q-TOF LC/MS system Application Note Authors Ravindra Gudihal Agilent
Characterization of bacteriophage derived anti-staphylococcal protein (P28) from production to purification using Agilent HPLC-Chip Q-TOF LC/MS system Application Note Authors Ravindra Gudihal Agilent
Thermo Scientific EASY-nLC 1200 System. Leading in simplicity. and performance
 Thermo Scientific EASY-nLC 1200 System Leading in simplicity and performance Peak performance made EASY Effortless ultra high performance for everybody A straightforward LC-MS solution Optimized and integrated
Thermo Scientific EASY-nLC 1200 System Leading in simplicity and performance Peak performance made EASY Effortless ultra high performance for everybody A straightforward LC-MS solution Optimized and integrated
BIOLOGY 429 PROTEOMICS LABORATORY Fall 2004
 Dr. Karin Sauer Dr. Anna Tan-Wilson BIOLOGY 429 PROTEOMICS LABORATORY Fall 2004 COURSE BULLETIN DESCRIPTION: Proteomics techniques for the analysis of global changes in protein patterns. Methods include
Dr. Karin Sauer Dr. Anna Tan-Wilson BIOLOGY 429 PROTEOMICS LABORATORY Fall 2004 COURSE BULLETIN DESCRIPTION: Proteomics techniques for the analysis of global changes in protein patterns. Methods include
Basic protein and peptide science for proteomics. Henrik Johansson
 Basic protein and peptide science for proteomics Henrik Johansson Proteins are the main actors in the cell Membranes Transport and storage Chemical factories DNA Building proteins Structure Proteins mediate
Basic protein and peptide science for proteomics Henrik Johansson Proteins are the main actors in the cell Membranes Transport and storage Chemical factories DNA Building proteins Structure Proteins mediate
Strategies for Quantitative Proteomics. Atelier "Protéomique Quantitative" La Grande Motte, France - June 26, 2007
 Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
Strategies for Quantitative Proteomics Atelier "Protéomique Quantitative", France - June 26, 2007 Bruno Domon, Ph.D. Institut of Molecular Systems Biology ETH Zurich Zürich, Switzerland OUTLINE Introduction
1 Ten Years of the Proteome
 1 Ten Years of the Proteome MARC R. WILKINS AND RON D. APPEL Abstract The concept of the proteome is now over 10 years old. As with all anniversaries, it is a good time to look back and reflect on what
1 Ten Years of the Proteome MARC R. WILKINS AND RON D. APPEL Abstract The concept of the proteome is now over 10 years old. As with all anniversaries, it is a good time to look back and reflect on what
Monoclonal Antibody Analysis on a Reversed-Phase C4 Polymer Monolith Column
 Monoclonal Antibody Analysis on a Reversed-Phase C4 Polymer Monolith Column Shane Bechler 1, Ken Cook 2, and Kelly Flook 1 1 Thermo Fisher Scientific, Sunnyvale, CA, USA; 2 Thermo Fisher Scientific, Runcorn,
Monoclonal Antibody Analysis on a Reversed-Phase C4 Polymer Monolith Column Shane Bechler 1, Ken Cook 2, and Kelly Flook 1 1 Thermo Fisher Scientific, Sunnyvale, CA, USA; 2 Thermo Fisher Scientific, Runcorn,
De novo sequencing in the identification of mass data. Wang Quanhui Liu Siqi Beijing Institute of Genomics, CAS
 De novo sequencing in the identification of mass data Wang Quanhui Liu Siqi Beijing Institute of Genomics, CAS The difficulties in mass data analysis Although the techniques of genomic sequencing are being
De novo sequencing in the identification of mass data Wang Quanhui Liu Siqi Beijing Institute of Genomics, CAS The difficulties in mass data analysis Although the techniques of genomic sequencing are being
Proteins. Patrick Boyce Biopharmaceutical Marketing Manager Waters Corporation 1
 Routine Characterization of mabs and Other Proteins Patrick Boyce Biopharmaceutical Marketing Manager Europe and India 2011 Waters Corporation 1 Agenda Why? What scientific challenges? Technology Example
Routine Characterization of mabs and Other Proteins Patrick Boyce Biopharmaceutical Marketing Manager Europe and India 2011 Waters Corporation 1 Agenda Why? What scientific challenges? Technology Example
Clone Selection Using the Agilent 1290 Infinity Online 2D-LC/MS Solution
 Clone Selection Using the Agilent 9 Infinity Online D-LC/MS Solution Application Note Biopharmaceuticals Authors Abstract Ravindra Gudihal and This Application Note describes the Agilent 9 Infinity online
Clone Selection Using the Agilent 9 Infinity Online D-LC/MS Solution Application Note Biopharmaceuticals Authors Abstract Ravindra Gudihal and This Application Note describes the Agilent 9 Infinity online
Spectrum Mill MS Proteomics Workbench. Comprehensive tools for MS proteomics
 Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
Spectrum Mill MS Proteomics Workbench Comprehensive tools for MS proteomics Meeting the challenge of proteomics data analysis Mass spectrometry is a core technology for proteomics research, but large-scale
QSTAR XL Hybrid LC/MS/MS System. Extended Performance Plus Exceptional Flexibility. QSTAR XL. Hybrid LC/MS/MS System
 QSTAR XL Hybrid LC/MS/MS System Extended Performance Plus Exceptional Flexibility. QSTAR XL Hybrid LC/MS/MS System Extended performance and flexibility for your most challenging analyses. Applied Biosystems/MDS
QSTAR XL Hybrid LC/MS/MS System Extended Performance Plus Exceptional Flexibility. QSTAR XL Hybrid LC/MS/MS System Extended performance and flexibility for your most challenging analyses. Applied Biosystems/MDS
Jet Stream Proteomics for Sensitive and Robust Standard Flow LC/MS
 Jet Stream Proteomics for Sensitive and Robust Standard Flow LC/MS Technical Overview Authors Yanan Yang, Vadiraj hat, and Christine Miller Agilent Technologies, Inc. Santa Clara, California Introduction
Jet Stream Proteomics for Sensitive and Robust Standard Flow LC/MS Technical Overview Authors Yanan Yang, Vadiraj hat, and Christine Miller Agilent Technologies, Inc. Santa Clara, California Introduction
Rapid Peptide Mapping via Automated Integration of On-line Digestion, Separation and Mass Spectrometry for the Analysis of Therapeutic Proteins
 Rapid Peptide Mapping via Automated Integration of On-line Digestion, Separation and Mass Spectrometry for the Analysis of Therapeutic Proteins Esther Lewis, 1 Kevin Meyer, 2 Zhiqi Hao, 1 Nick Herold,
Rapid Peptide Mapping via Automated Integration of On-line Digestion, Separation and Mass Spectrometry for the Analysis of Therapeutic Proteins Esther Lewis, 1 Kevin Meyer, 2 Zhiqi Hao, 1 Nick Herold,
Towards unbiased biomarker discovery
 Towards unbiased biomarker discovery High-throughput molecular profiling technologies are routinely applied for biomarker discovery to make the drug discovery process more efficient and enable personalised
Towards unbiased biomarker discovery High-throughput molecular profiling technologies are routinely applied for biomarker discovery to make the drug discovery process more efficient and enable personalised
Characterizing Antibody-Drug Conjugates Using Micro Pillar Array Columns Combined with Mass Spectrometry (µpac -MS)
 Characterizing Antibody-Drug Conjugates Using Micro Pillar Array Columns Combined with Mass Spectrometry (µpac -MS) Abstract MARKET Pharma, Biotech KEYWORDS The use of micro pillar array columns (µpac
Characterizing Antibody-Drug Conjugates Using Micro Pillar Array Columns Combined with Mass Spectrometry (µpac -MS) Abstract MARKET Pharma, Biotech KEYWORDS The use of micro pillar array columns (µpac
PROTEOMICS. Timothy Palzkill Baylor College of Medicine. KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow
 PROTEOMICS PROTEOMICS by Timothy Palzkill Baylor College of Medicine KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow ebook ISBN: 0-306-46895-6 Print ISBN: 0-792-37565-3 2002
PROTEOMICS PROTEOMICS by Timothy Palzkill Baylor College of Medicine KLUWER ACADEMIC PUBLISHERS New York / Boston / Dordrecht / London / Moscow ebook ISBN: 0-306-46895-6 Print ISBN: 0-792-37565-3 2002
What are proteomics? And what can they tell us about seed maturation and germination?
 Danseed Symposium Kobæk Strand, Skelskør 14 March, 2017 What are proteomics? And what can they tell us about seed maturation and germination? Ian Max Møller Department of Molecular Biology and Genetics
Danseed Symposium Kobæk Strand, Skelskør 14 March, 2017 What are proteomics? And what can they tell us about seed maturation and germination? Ian Max Møller Department of Molecular Biology and Genetics
Improving Sensitivity for an Immunocapture LC-MS Assay of Infliximab in Rat Plasma Using Trap-and-Elute MicroLC-MS
 Improving Sensitivity for an Immunocapture LC-MS Assay of in Rat Plasma Using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest
Improving Sensitivity for an Immunocapture LC-MS Assay of in Rat Plasma Using Trap-and-Elute MicroLC-MS Using the SCIEX M3 MicroLC system for Increased Sensitivity in Antibody Quantitation Remco van Soest
Characterize Fab and Fc Fragments by Cation-Exchange Chromatography
 Characterize Fab and Fc Fragments by Cation-Exchange Chromatography Application Note Biologics and Biosimilars Authors Isabel Vandenheede, Emmie Dumont, Pat Sandra, and Koen Sandra Research Institute for
Characterize Fab and Fc Fragments by Cation-Exchange Chromatography Application Note Biologics and Biosimilars Authors Isabel Vandenheede, Emmie Dumont, Pat Sandra, and Koen Sandra Research Institute for
Purification: Step 1. Lecture 11 Protein and Peptide Chemistry. Cells: Break them open! Crude Extract
 Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1 Lecture 11 Protein and Peptide Chemistry Cells: Break them open! Crude Extract Total contents of cell Margaret A. Daugherty Fall 2003 Big Problem: Crude extract is not the natural
Purification: Step 1. Protein and Peptide Chemistry. Lecture 11. Big Problem: Crude extract is not the natural environment. Cells: Break them open!
 Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
Lecture 11 Protein and Peptide Chemistry Margaret A. Daugherty Fall 2003 Purification: Step 1 Cells: Break them open! Crude Extract Total contents of cell Big Problem: Crude extract is not the natural
Protein analysis. Dr. Mamoun Ahram Summer semester, Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5
 Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Protein analysis Dr. Mamoun Ahram Summer semester, 2015-2016 Resources This lecture Campbell and Farrell s Biochemistry, Chapters 5 Bases of protein separation Proteins can be purified on the basis Solubility
Identification of human serum proteins detectable after Albumin removal with Vivapure Anti-HSA Kit
 Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Andreas Kocourek, Pieter Eyckerman, Robert Zeidler, Pascal Bolon and Birgit Thome-Kromer Introduction The analysis of the complete proteome is a major interest of many researchers. Of particular importance
Increasing Throughput and Efficiency with Exactive LC/MS and Triple Quadrupole LC/MS/MS
 Increasing Throughput and Efficiency with Exactive LC/MS and Triple Quadrupole LC/MS/MS Nicholas Duczak Thermo Scientific Annual Mass Spectrometry Users Meeting Somerset, NJ October 12th, 2011 Lead Finding
Increasing Throughput and Efficiency with Exactive LC/MS and Triple Quadrupole LC/MS/MS Nicholas Duczak Thermo Scientific Annual Mass Spectrometry Users Meeting Somerset, NJ October 12th, 2011 Lead Finding
A Combination of Liquid Chromatography and Mass Spectrometry for Peptidomic Research
 522-529 LE Oct04 14/10/04 2:54 pm Page 2 OUPLING MATTERS A ombination of Liquid hromatography and Mass Spectrometry for Peptidomic Research Elke lynen and Liliane Schoofs, Laboratory for Developmental
522-529 LE Oct04 14/10/04 2:54 pm Page 2 OUPLING MATTERS A ombination of Liquid hromatography and Mass Spectrometry for Peptidomic Research Elke lynen and Liliane Schoofs, Laboratory for Developmental
Overview. Tools for Protein Sample Preparation, 2-D Electrophoresis, and Imaging and Analysis
 Expression Proteomics // Tools for Protein Separation and Analysis www.expressionproteomics.com 1 2 3 4 Overview Tools for Protein Sample Preparation, 2-D Electrophoresis, and Imaging and Analysis overview
Expression Proteomics // Tools for Protein Separation and Analysis www.expressionproteomics.com 1 2 3 4 Overview Tools for Protein Sample Preparation, 2-D Electrophoresis, and Imaging and Analysis overview
Mass Spectrometry. Kyle Chau and Andrew Gioe
 Mass Spectrometry Kyle Chau and Andrew Gioe Computation of Molecular Mass - Mass Spectrum is a plot of intensity as a function of masscharge ratio, m/z. - Mass-charge ratio can be determined through accelerating
Mass Spectrometry Kyle Chau and Andrew Gioe Computation of Molecular Mass - Mass Spectrum is a plot of intensity as a function of masscharge ratio, m/z. - Mass-charge ratio can be determined through accelerating
Expand Your Research with Metabolomics and Proteomics. Christine Miller Omics Market Manager ASMS 2017
 Expand Your Research with Metabolomics and Proteomics Christine Miller Omics Market Manager ASMS 2017 New Additions to Agilent Omics Workflows Acquisition Ion Mobility Q-TOF IM All Ions MS/MS Find Features
Expand Your Research with Metabolomics and Proteomics Christine Miller Omics Market Manager ASMS 2017 New Additions to Agilent Omics Workflows Acquisition Ion Mobility Q-TOF IM All Ions MS/MS Find Features
