Combinatorial gene regulation by Bmp and Wnt in zebrafish posterior mesoderm formation
|
|
|
- Ross Alan McGee
- 6 years ago
- Views:
Transcription
1 Corrigendum Combinatorial gene regulation by Bmp and Wnt in zebrafish posterior mesoderm formation Daniel P. Szeto and David Kimelman Development 131, The labelling in Fig. 6 of this article is incorrect. The label for A-F should read mut1-2.0-gfp/xex-bfp and the label for G-L should read mut1,2-2.0-gfp/xex-bfp The authors apologise to the readers for this mistake.
2 Research article 3751 Combinatorial gene regulation by Bmp and Wnt in zebrafish posterior mesoderm formation Daniel P. Szeto and David Kimelman* Department of Biochemistry, Box , University of Washington, Seattle, WA , USA *Author for correspondence ( Accepted 21 April 2004 Development 131, Published by The Company of Biologists 2004 doi: /dev Summary Combinatorial signaling is an important mechanism that allows the embryo to utilize overlapping signaling pathways to specify different territories. In zebrafish, the Wnt and Bmp pathways interact to regulate the formation of the posterior body. In order to understand how this works mechanistically, we have identified tbx6 as a posterior mesodermal gene activated by both of these signaling pathways. We isolated a genomic fragment from the tbx6 gene that recapitulates the endogenous tbx6 expression, and used this to ask how the Bmp and Wnt signaling pathways combine to regulate gene expression. We find that the tbx6 promoter utilizes distinct domains to integrate the signaling inputs from each pathway, including multiple Tcf/LEF sites and a novel Bmp-response element. Surprisingly, we found that overexpression of either signaling pathway can activate the tbx6 promoter and the endogenous gene, whereas inputs from both pathways are required for the normal pattern of expression. These results demonstrate that both Bmp and Wnt are present at submaximal levels, which allows the pathways to function combinatorially. We present a model in which overlapping Wnt and Bmp signals in the ventrolateral region activate the expression of tbx6 and other posterior mesodermal genes, leading to the formation of posterior structures. Supplemental data available online Key words: Bmp, Wnt, Posterior mesoderm, T-box genes, Zebrafish Introduction The posterior body of the zebrafish embryo is formed by a network of intercellular signaling molecules that pattern the formation of the ventrolateral mesoderm, which ultimately gives rise to the posterior (trunk and tail) mesoderm. These signaling factors include Bone morphogenetic proteins (Bmps) (reviewed by Hammerschmidt, 2002), Wnt8 (Kelly et al., 1995; Lekven et al., 2001), Fibroblast growth factors (Fgfs) (Amaya et al., 1993; Griffin et al., 1995; Rodaway et al., 1999; Draper et al., 2003; Griffin and Kimelman, 2003), and the Nodalrelated proteins (Schier and Shen, 2000; Schier and Talbot, 2001). Whereas the Nodals appear to be critical for mesoderm induction and the Fgfs are important for maintaining mesodermal gene expression (reviewed by Kimelman and Schier, 2002), the exact roles of Bmp and Wnt signaling in forming the ventral/posterior mesoderm have been uncertain. Three bmps are expressed in the mesoderm of the gastrulastage embryo and their expression continues at the most posterior end of the embryo throughout somitogenesis (Martinez-Barbera et al., 1997; Nikaido et al., 1997). As in Xenopus, a large body of evidence demonstrates that the Bmps are key regulators of the posterior mesoderm patterning in zebrafish (reviewed by Hammerschmidt, 2002). For example, bmp2b/swirl is expressed in a gradient along the dorsoventral axis (Barth et al., 1999; Wagner and Mullins, 2002) and zebrafish mutants lacking bmp2b/swirl do not form tails (Mullins et al., 1996; Kishimoto et al., 1997). Like the bmps, zebrafish wnt8 is also expressed in the ventrolateral mesoderm during gastrulation and at the posterior end of the embryo during somitogenesis (Kelly et al., 1995). In mutants lacking wnt8, the formation of the ventrolateral and posterior mesoderm is severely disrupted, demonstrating a requirement for Wnt signaling in the development of these mesodermal territories (Lekven et al., 2001). Two studies have suggested that the Wnt and Bmp pathways cooperate to form the ventral and posterior mesoderm in Xenopus (Hoppler and Moon, 1998; Marom et al., 1999). The importance of such a cooperative interaction between these pathways has been further highlighted in a recent zebrafish study, demonstrating that combined overexpression of Wnt8 and Bmp is capable of inducing ectopic tail formation (Agathon et al., 2003). How these pathways integrate to regulate mesodermal gene expression is unknown. In order to understand mechanistically how these two pathways interact, we sought a posterior mesodermal gene whose expression required input from both the Bmp and Wnt pathways. The T-box transcription factor tbx6 (Hug et al., 1997) was found to meet this criterion, and we therefore identified a minimal region of the tbx6 gene that reproduces the endogenous gene expression pattern. Analysis of the tbx6 promoter shows that Bmp and Wnt8 regulate tbx6 expression through different domains within the promoter, demonstrating that these pathways intersect by regulating transcription of specific target genes. Elimination of regions that respond to Bmp or Wnt signaling revealed that the promoter is able to respond to overexpression of a single pathway, whereas normally the promoter integrates responses to both pathways to provide normal tbx6 expression. Analysis of the expression
3 3752 Development 131 (15) Research article of the endogenous gene under conditions in which either Bmp or Wnt signaling is blocked together with analysis of the tbx6 promoter has allowed us to put forth a model in which submaximal levels of Bmp and Wnt signals cooperate to regulate the formation of the posterior mesoderm. Materials and methods Embryos and morpholino oligonucleotides injections Zebrafish embryos were obtained by natural spawning of adult AB strain zebrafish. Embryos were raised and maintained at 28.5 C in system water and staged as described (Westerfield, 1995). Wnt8- specific morpholino antisense oligonucleotides were a generous gift from Randy Moon. At the one-cell stage, each embryo was injected with approximately 1 nl volume of morpholino oligonucleotides (5 ng) using a Picospritzer II (Parker Hannifin). Embryos were collected at the appropriate stages and fixed in 4% paraformaldehyde, ph 7.0, in phosphate-buffered saline (PBS), overnight at 4 C. Fixed embryos were dechorionated, washed three times with PBS and stored in methanol at 20 C. Plasmids and constructs Various gfp reporter constructs were made by cloning the tbx6 promoter fragments into the BamHI restriction site of the gfp reporter plasmid vector. The dual fluorescence system was created by inserting the pxex-bfp DNA (gift of Stephen C. Ekker) into the existing gfp reporter constructs. The luciferase reporter constructs were made by cloning the tbx6 promoter fragments into the multiple cloning sites of the PGL3 promoter vector (Promega). Site-directed mutagenesis The two proximal Tcf site mutations in the tbx6 promoter were generated using the QuikChange procedure (Stratagene). Mut1-2.0 was constructed by using primers: 5 -gtgcacatacacacctctggcccccctcgaggtgatggaagaaaggtagaagcctag-3 and 5 -ctaggcttctaccttcttccatcacctcgaggggggccagaggtgtgtatgtgcac-3. Mut2-2.0 was constructed by using primers: 5 -gtgggctggctggagacaaaagacaggttaacgaggagactgatgttttgacagaag-3 and 5 -cttctgtcaaaacatcagtctcctcgttaacctgtcttttgtctccagccagcccac-3. Mut1,2-2.0 construct was generated by combining the two mutated Tcf sites. DNA and RNA injections DNA was isolated with a Qiagen midi kit and resuspended in RNasefree water. DNA at a concentration of 0.1 mg/ml was used for injection. RNAs were synthesized from SalI linearized SP64T-Xbmp4 (gift of Jim Smith), Asp718 linearized CS2-zwnt8 (gift of Randy Moon) and NotI linearized CS2-TVGR (gift of Paul Wilson) templates using the mmessage Machine Kit (Ambion) and dissolved in RNase-free sterile water. RNA (at concentrations indicated in the text) was injected in the presence or absence of 0.1 µg of reporter DNA into one-cell zebrafish embryos. The expression of the reporter gene was analyzed at the appropriate stages using bright field or fluorescence microscopy. In situ hybridization Whole-mount in situ hybridization was performed using digoxigeninlabeled antisense RNA probes and visualized using anti-digoxigenin Fab fragments conjugated with alkaline phosphatase (Roche Molecular Biochemicals) as described (Griffin et al., 1998). Riboprobes were made from DNA templates, which were linearized and transcribed with either SP6 or T7 RNA polymerases. Embryos were processed and hybridized as described (Griffin et al., 1998), except that 5 µg/ml of Proteinase K in PBS/0.1% Tween-20 was used for 5 to 10 minutes depending on the age of the collected embryos. Xenopus and zebrafish transgenesis PAC DNA was isolated with the Qiagen midi kit. To generate Xenopus transgenic embryos, we used 200 pg of PAC DNA for each injection as described (Amaya and Kroll, 1999) with the exception that no restriction enzyme was used. To generate stable transgenic zebrafish lines, reporter plasmid DNA was digested with BssHII to completion, and then separated on a 1% agarose preparative gel overnight. The gel slice containing the DNA insert was isolated and DNA was electroeluted using a Schleicher and Schuell ELUTRAP. DNA was extracted once with equal volume of phenol and once with an equal volume of chloroform. After extraction, DNA was ethanol precipitated and then resuspended in distilled water. For injection, 80 mg/ml of purified DNA insert was used to inject one-cell stage zebrafish embryos. Injected embryos were examined at the 15- to 18- somite stage for the presence of GFP fluorescence. Embryos showing specific GFP fluorescence in the tail region were collected and raised to sexual maturity as founders. Different combinations of founder crosses were set up for the identification of F1 embryos showing nonmosaic GFP fluorescence in the tail region at the 15- to 18-somite stage. Luciferase assays Injected embryos were collected at the shield stage and then separated into three pools of 10 embryos each for assay in triplicate. Experiments were repeated at least three times. Excess zebrafish embryo medium was removed, embryos were homogenized in 100 µl of 1 Cell Culture Lysis Reagent (Promega), and cleared by 10 minutes microcentrifugation at room temperature. Fifty microliters of the resulting supernatant was used for luciferase activity assays that were performed according to the Promega protocol with a Berthold luminometer. Results Regulation of tbx6 expression by Wnt and Bmp signals In our initial attempts to find a gene regulated by both the Wnt and Bmp pathways, we identified the T-box transcription factor tbx6 as a candidate. Tbx6, along with two other T-box genes, spadetail (spt) and no tail (ntl), is expressed in the posterior mesoderm from the gastrula stage through the end of somitogenesis (Schulte-Merker et al., 1994; Hug et al., 1997; Griffin et al., 1998). Wnt8 was previously shown to be required for the expression of tbx6 in the ventrolateral mesoderm (Lekven et al., 2001), which we confirmed using a pair of wnt8- specific morpholino oligonucleotides (MOs) (Fig. 1A,B) (n=55). Whereas the lateral expression was completely absent, some residual tbx6 expression was observed in the ventral region (Fig. 1B). In contrast, ntl expression was unaffected by the inhibition of Wnt8 function (Fig. 1E,F) and spt expression was only partially affected (Fig. 1C,D), indicating that tbx6 is the most dependent of the three mesodermal genes on Wnt function at the gastrula stage. To determine if tbx6 is regulated by Bmp signaling, we examined tbx6 expression in swirl mutants, which have a defective bmp2b gene (Kishimoto et al., 1997; Nikaido et al., 1997). In approximately one quarter of the embryos collected from the crosses of swirl heterozygotes, we observed a strong reduction in the expression of tbx6 (Fig. 1G,H) (n=75), whereas the expression of spt and ntl was normal in all the embryos examined (Fig. 1I-L). Interestingly, unlike the result observed with the wnt8mos (Fig. 1B), swirl embryos showed residual tbx6 expression in both the ventral and lateral regions (Fig. 1H). Similar results were obtained with embryos injected with RNA encoding the Bmp inhibitor Noggin (Fig. 2A,B)
4 Gene regulation in zebrafish 3753 Fig. 1. Wnt8 and Bmp2b are required for tbx6 expression. Animal pole views of shield stage embryos with dorsal to the top. The in situ probe is shown at the top of the Figure (ntl, no tail; spt, spadetail) and the genotype of the embryo is shown on the left side. Wnt8 morphants are wild-type embryos injected with a pair of wnt8- specific morpholino oligonucleotides. Note that residual tbx6 expression is only observed ventrally in the wnt8 morphant embryos whereas residual tbx6 expression is present laterally and ventrally in swirl / embryos. (85%, n=65). These results demonstrate that normal tbx6 expression requires input from both the Wnt and Bmp pathways, which is consistent with the observation that both Bmp and Wnt signals are required for the formation of the posterior body in zebrafish (Lekven et al., 2001; Agathon et al., 2003). To further examine the regulation of tbx6 by Bmp and Wnt signals, we overexpressed these factors in the early zebrafish embryo. We observed that overexpression of either signaling molecule caused ectopic tbx6 expression throughout the embryo (Fig. 2C,F) (80%, n=43). These results suggest that tbx6 can be separately activated by each of these signaling pathways alone, although it was possible that each of the overexpressed signals also activated the other signaling pathway. To eliminate the effect of the other pathway, we coinjected wnt8 with the Bmp inhibitor noggin, and coinjected Xenopus bmp4 (Xbmp4) RNA (Xbmp4 causes the same effects as the zebrafish Bmps) (Kishimoto et al., 1997) with the wnt8mos. Whereas 50 pg of noggin alone caused a strong reduction in tbx6 expression, as was observed in swr mutant embryos (Fig. 2B, compare with Fig. 1H), noggin only caused a partial reduction in the ectopic tbx6 induced by overexpression of wnt8 (Fig. 2D) (70%, n=35). This result suggests that only a part of the wnt8-mediated activation of tbx6 depends on the ability of Wnt8 to upregulate endogenous bmp2b expression (Agathon et al., 2003). Conversely, whereas the wnt8mos strongly downregulated tbx6 expression (Fig. 2G), the wnt8mos did not inhibit marginal tbx6 Fig. 2. Regulation of tbx6 by Wnt and Bmp. (A-H) Lateral views with the animal pole to the top. (A,B) Expression of endogenous tbx6 in an uninjected embryo (A) and in an embryo injected with 50 pg of noggin RNA (B). The injected embryo shows dramatic reduction of tbx6 expression. (C) An embryo injected with 75 pg of wnt8 RNA shows ectopic expression of tbx6. (D) An embryo coinjected with 75 pg of wnt8 RNA and 50 pg of noggin RNA showing ectopic expression of tbx6 in the animal pole. (E-H) Expression of endogenous tbx6 in an uninjected embryo (E) and in an embryo injected with 150 pg of Xbmp4 RNA (F). The injected embryo shows ectopic expression of tbx6. (G) An embryo injected with wnt8mos shows decreased expression of tbx6. (H) A zebrafish embryo coinjected with wnt8mos and 150 pg of Xbmp4 RNA showing strong marginal and ectopic dorsal domains of tbx6 expression. expression in Xbmp4-injected embryos, although it reduced the animal pole expression of tbx6 (Fig. 2H) (87%, n=45). Because bmp overexpression does not alter zygotic wnt8 expression (Agathon et al., 2003), our results suggest that maternal wnt8 (Kelly et al., 1995) is required for the high-level ectopic animal pole expression of tbx6 in Xbmp4-injected embryos, but it is not required for XBmp4 to activate tbx6 expression at the margin. In summary, our results show that both Wnt and Bmp can induce tbx6 expression when the other pathway is blocked, although the two factors alone can activate tbx6 in specific patterns. Identification and characterization of the tbx6 promoter In order to understand how these two pathways regulate tbx6 expression, we screened a zebrafish PAC genomic library and obtained three tbx6 positive clones. Using the frog transgenesis approach (Amaya and Kroll, 1999), we tested one of these tbx6 positive PAC clones (PAC-tbx6) for its ability to target expression of zebrafish tbx6 to the posterior mesoderm of developing Xenopus embryos. The advantage to using Xenopus is that we could examine the expression of the zebrafish tbx6 gene using in situ hybridization because the fish and frog tbx6 genes are sufficiently divergent (Uchiyama et al., 2001), without the need to make a reporter gene fusion in the PAC DNA. Approximately 20% of the transgenic embryos (7/36) showed zebrafish tbx6 transcripts in the posterior paraxial mesoderm (see Fig. S1A,B at supplemental), whereas the remainder of the embryos failed to express zebrafish tbx6 from the injected PAC (see Fig. S1C
5 3754 Development 131 (15) Research article Fig. 3. tbx6-gfp reporter constructs are correctly expressed in zebrafish embryos. (A) Schematic diagrams of gfp reporter constructs, ptbx6-1.7-gfp and ptbx6-2.0-gfp, which contain 1.7 and 2.0 kb of upstream sequence, respectively. The 1.7 kb region is subdivided into a proximal 900 bp domain and a distal 800 bp domain. The sequences of three putative Tcf binding sites (Tcf-1, Tcf-2 and Tcf-3) are indicated with the arrows showing the orientation of the sites. (B-E) Lateral views with animal pole to the top of shield stage (B,C) and 15-somite (D,E) embryos injected at the one-cell stage with ptbx6-1.7-gfp. Bright field (B,D) and GFP fluorescence (C,E) pictures showing specific GFP fluorescence at the margin at shield stage (C) and in the posterior mesoderm at the 15-somite stage (G). Note that because these embryos are not transgenic, the GFP is expressed mosaically in the embryos from the injected DNA. (F-O) Transgenic embryos containing ptbx6-1.7-gfp. (F,G) Lateral views with anterior to the top. Bright field (F) and fluorescence (G) pictures of a ptbx gfp transgenic embryo at the 15- to 18- somite stage. (H-O) In situ staining for gfp (H-K) and tbx6 (L-O). Embryos shown are at 70% epiboly (H,L; dorsal view with animal pole to the top), 95% epiboly (I,M; posterior view with dorsal to the top), 5-somite stage (J,N; posterior view with dorsal to the top) and 18-somite stage (K,O; lateral view with anterior to the top). The small inserts in K and O show a close-up posterior view of expression in the tip of the tail. Note the tbx6 in situ probe was from the 3 end of the gene and therefore does not bind to the transcripts from the transgene. E, exon sequence. at This expression pattern matches the endogenous Xenopus tbx6 (Uchiyama et al., 2001) as well as that of the ntl ortholog Xbra (Smith et al., 1991) (see Fig. S1D at These results demonstrate that PAC-tbx6 has all of the regulatory elements necessary for expression within the posterior mesoderm, and that these regulatory sites are conserved between fish and frogs. Having determined that PAC-tbx6 targets expression to the posterior mesoderm in developing Xenopus embryos, we subcloned smaller fragments and characterized them in zebrafish. We initially focused on two constructs fused in frame to GFP, ptbx6-1.7-gfp and ptbx6-2.0-gfp, which contain 1.7 and 2.0 kb of DNA upstream from the start of translation, respectively, and the first intron (Fig. 3A). These constructs were injected into one-cell stage zebrafish embryos and then analyzed for GFP fluorescence at different stages of development. At 60-75% epiboly, we observed GFP fluorescence at the margin where endogenous tbx6 is normally expressed (Fig. 3B,C, Table 1). At the 15- to 18-somite stage, GFP fluorescence was detected in the tail mesoderm, again matching the endogenous pattern of expression (Fig. 3D,E, Table 1). Although both reporter constructs were expressed in the same pattern, we noticed that ptbx6-2.0-gfp produced consistently higher fluorescence levels than ptbx6-1.7-gfp, suggesting the presence of a general enhancer between 1.7 and 2.0 kb. The fluorescence results were confirmed by in situ hybridization with a gfp probe on zebrafish embryos injected with either of the two reporter constructs (see Fig. S2 at Because expression from injected DNA is mosaic (nonuniform), we wanted to confirm that the tbx6 promoter constructs faithfully matched the endogenous gene expression by producing transgenic embryos. We therefore created stable transgenic zebrafish lines [Tg(tbx6:gfp)] expressing ptbx gfp. As shown in Fig. 3, the expression of transcripts from ptbx6-1.7-gfp was the same as the endogenous tbx6 gene expression pattern (Fig. 3H-O). The GFP fluorescence matched the transcript pattern, although it extended more anteriorly (Fig. 3F,G). This was expected because the posterior mesodermal cells originate at the most posterior end and are incorporated into somites as the posterior end extends (Kanki and Ho, 1997). Because GFP is a stable protein, it perdures for a while in more anterior regions as the posterior end extends caudally, producing a posterior to anterior gradient of fluorescence (Fig. 3G). We also tested ptbx6-1.7-gfp in transgenic Xenopus embryos and found that it produced the same pattern of expression as PAC-tbx6 (data not shown). These data demonstrate that ptbx6-1.7-gfp and ptbx6-2.0-gfp have all of the necessary regulatory elements to recapitulate
6 Gene regulation in zebrafish 3755 Table 1. Comparison of the targeting ability and promoter activity of different reporter constructs Reporter constructs GFP expression Relative promoter Responsiveness (GFP and Luciferase) Margin Tail Activity Bmp* Wnt ptbx % (n=90) 91% (n=90) 100% Yes Yes ptbx % (n=100) 88% (n=100) 34% Yes Yes mut % (n=50) 90% (n=50) n/d Yes Yes mut % (n=50) 85% (n=50) n/d Yes Yes mut1, % (n=50) 22% (n=70) 11% Yes Yes ptbx % (n=60) 0% (n=60) 6% No Yes n/d, not determined. *Tested with overexpression of XBmp4. Tested with overexpression of TVGR only. Tested with overexpression of wnt8 and TVGR. Only a few cells showed marginal GFP fluorescence. expression of the tbx6 gene in both transient expression assays and in transgenic embryos. Regulation of the tbx6 promoter by Wnt and Bmp During the development of the posterior mesoderm, the endogenous tbx6 promoter must be able to integrate inputs from both the Wnt and Bmp pathways, as shown above. We therefore tested the ability of ptbx6-1.7-gfp to respond to these signaling pathways by injecting the promoter with wnt8 or Xbmp4 and compared the response of the endogenous gene with the same levels of exogenous Wnt and Bmp signaling inputs. Similar to the response of the endogenous tbx6 gene in the presence of Wnt8 overexpression, we observed ectopic GFP fluorescence in embryos coinjected with wnt8 RNA and ptbx6-1.7-gfp (Fig. 4A-D), showing that the tbx6 promoter is capable of responding to Wnt8. Ectopic expression of Xbmp4 RNA also induced widespread expression of the ptbx6-1.7-gfp as in the case of the endogenous tbx6 gene (Fig. 4E-H). These results show that the promoter is activated by either Wnt or Bmp signals, similar to that of the endogenous tbx6 gene. Construction of a dual fluorescence system for promoter analysis in zebrafish We wanted to analyze the tbx6 promoter in detail by deleting and mutating specific regions to determine which parts of the promoter are necessary for normal expression. Because of the variable and mosaic patterns of expression when DNA is injected into zebrafish embryos, it is difficult to accurately quantitate results because some embryos express the injected DNA in only a few cells and/or in regions outside the area of interest (in our case the ventral and lateral mesoderm). In order to improve this situation, we added an additional feature to our reporter constructs. We placed a blue fluorescence protein (bfp) reporter gene under the control of a ubiquitous Xenopus EF1α (pxex) promoter, which has been characterized and shown to produce ubiquitous BFP fluorescence in developing zebrafish embryos (Finley et al., 2001). Our dual fluorescence (blue and green) system allows simultaneous detection of the regions of the embryo that inherited the injected DNA by examining the location of BFP fluorescence, in addition to revealing the specific expression of GFP under the control of the tbx6 promoter. For example, a construct containing ptbx6-2.0-gfp and pxex-bfp (Fig. 5A) showed scattered BFP fluorescence throughout the embryo at 70% epiboly with specific GFP expression at the margin (Fig. 5C,D). At later stages, the BFP was ubiquitous but the GFP was restricted to the most posterior end of the embryo (Fig. 5F,G). We used this system in our subsequent studies such that we only scored the presence or absence of GFP fluorescence in embryos in which there was substantial BFP fluorescence in the ventral-lateral mesoderm during the gastrula stages or in the posterior mesoderm during somitogenesis. We found that this was a much more reliable method for analyzing gene expression from exogenous promoters in transient zebrafish assays. Mutational analysis of the tbx6 promoter To understand how the tbx6 promoter is regulated by Wnt and Bmp signals, we undertook a systematic analysis of the tbx6 promoter. The canonical Wnt signaling pathway typically works through regulation of the Tcf transcription factor (reviewed by van Noort and Clevers, 2002). Consistent with this, ectopic expression of a constitutively activated Tcf (TVGR) (Darken and Wilson, 2001) caused widespread activation of ptbx6-1.7-gfp (Fig. 4I,J), as did ectopic wnt8 expression (Fig. 4C,D). Examination of the tbx6 promoter constructs revealed three consensus Tcf binding sites within the 900 bp proximal region of the promoter (Fig. 3A). The presence of multiple Tcf sites in the tbx6 promoter suggested that the expression of tbx6 is directly regulated by Wnt8 signaling during posterior mesoderm formation. Using our dual fluorescence system, we asked whether the activation of ptbx6-2.0-gfp in the posterior mesoderm required these Tcf DNA binding sites. To do this, we mutated the two proximal Tcf sites (Tcf-1 and Tcf-2) (Fig. 3A), generating mut1-2.0-gfp/xex-bfp containing a mutation in the first Tcf site, mut2-2.0-gfp/xex-bfp containing a mutation in the second Tcf site, and mut1,2-2.0-gfp/xex-bfp containing both mutations. We found that either single mutation alone did not dramatically affect the expression of the tbx6 promoter in the developing posterior mesoderm as indicated by the presence of strong GFP fluorescence (Fig. 6A-F, Table 1). In contrast, zebrafish embryos injected with the double Tcf site mutation construct showed almost no detectable GFP fluorescence in the majority of the embryos and the remainder had only occasional BFPpositive mesodermal cell expressing GFP (Fig. 6G-L, Table 1). The double Tcf site mutant promoter was still functional, however, because overexpression of the constitutively active Tcf was able to activate transcription from this promoter, probably through the remaining Tcf site (Table 1). In addition, this promoter was activated when coinjected with Xbmp4,
7 3756 Development 131 (15) Research article together, these data show that the normal expression of tbx6 in the developing mesoderm is regulated directly by Wnt activation through the Tcf sites in the proximal region of the promoter. Fig. 4. Regulation of ptbx gfp and endogenous tbx6 by Wnt and Bmp. (A-J) Lateral views with the animal pole to the top. (A-D) Overexpression of Wnt8. (A,B) Expression of endogenous tbx6 in an uninjected embryo (A) and in an embryo injected with 75 pg of wnt8 RNA (B). The injected embryo shows ectopic expression of tbx6. (C,D) Bright field (C) and fluorescence (D) pictures of a zebrafish embryo coinjected with ptbx6-1.7-gfp and 75 pg of wnt8 RNA showing ectopic GFP fluorescence in the animal pole. (E-H) Expression of endogenous tbx6 in an uninjected embryo (E) and in an embryo injected with 150 pg of Xbmp4 RNA (F). The injected embryo shows ectopic expression of tbx6. (G,H) Bright field (G) and fluorescence (H) pictures of a zebrafish embryo coinjected with ptbx6-1.7-gfp and 150 pg of Xbmp4 RNA at the shield stage showing ectopic GFP fluorescence in the animal pole. (I,J) Bright field (I) and fluorescence (J) pictures of a zebrafish embryo coinjected with ptbx6-1.7-gfp and 25 pg of TVGR RNA showing ectopic GFP fluorescence in the animal pole. TVGR is a constitutively active form of the Tcf transcription factor (Darken and Wilson, 2001). The in situ hybridizations are at shield stage whereas the bright field and fluorescence images are at 50% epiboly. as was the wild-type promoter (Table 1). These results demonstrate that either of the two most proximal Tcf sites are required for normal tbx6 expression, showing that Wnt signaling directly activates the tbx6 gene. Using a quantitative assay, we further confirmed the contribution of these Tcf binding sites to the overall promoter activity of ptbx We replaced the gfp gene with the luciferase gene to quantify the promoter activity of our reporter constructs. These luc reporter genes, ptbx6-2.0-luc, ptbx luc and mut1,2-2.0-luc, were injected into one-cell stage embryos and the luciferase activity of each reporter construct was determined in injected embryos at the 60% epiboly stage. Consistent with our early finding that ptbx6-2.0-gfp showed stronger GFP fluorescence than ptbx6-1.7-gfp, we found that the promoter activity of ptbx6-1.7-luc is 34% of ptbx6-2.0-luc. More importantly, we determined that the promoter activity of mut1,2-2.0-luc is 11% of ptbx6-2.0-luc (Table 1). Taken A 900 bp tbx6 promoter fragment is insufficient for normal expression Because all three Tcf DNA binding sites are located in the 900 bp proximal promoter region, we wanted to know if this region of the promoter is sufficient to correctly target expression to the developing posterior mesoderm. To answer that, we constructed the reporter gene, ptbx6-0.9-gfp/pxex-bfp, by placing the sequence of the 900 bp proximal region of the tbx6 promoter in front of the gfp gene, together with pxex-bfp. We observed no detectable GFP fluorescence in any of the embryos injected with ptbx6-0.9-gfp/pxex-bfp, even though these embryos had robust BFP expression (Table 1). In support of this, when the ptbx6-0.9 was fused to luciferase, we found that it was only 6% as active as ptbx6-2.0 (Table 1). To determine if the Tcf sites were still functional in this construct, we examined the responsiveness of ptbx6-0.9-gfp to the constitutively active Tcf and to ectopic Wnt8. In both cases we observed ectopic GFP fluorescence in the embryos, showing that ptbx6-0.9 is functional and capable of responding to Tcf activation (Table 1). Thus the 900 bp proximal region of the promoter is capable of responding to Tcf activation, but it is not sufficient to drive the expression of gfp in the developing posterior mesoderm, possibly because it lacks the ability to respond to Bmp signaling. Identification of a Bmp responsive domain within tbx6 promoter To determine whether ptbx6-0.9-gfp can respond to Bmp, we coinjected either ptbx6-1.7-gfp or ptbx6-0.9-gfp with Xbmp4 RNA into one-cell stage embryos and examined GFP fluorescence. At 50% epiboly, we observed no GFP fluorescence in embryos coinjected with ptbx6-0.9-gfp and Xbmp4 RNA (Table 1). In contrast, we observed ectopic GFP fluorescence in embryos coinjected with ptbx6-1.7-gfp and Xbmp4 RNA (Fig. 4G,H, Table 1). These results show that ptbx6-0.9-gfp has a Wnt response element but lacks a promoter element necessary for the Bmp response. Two regions were deleted in ptbx6-1.7 to produce ptbx6-0.9, the distal 800 bp region (dr800) and the first intron. To ask whether either of these regions can mediate Bmp responsiveness, we placed them in front of the SV40 minimal promoter driving the luciferase gene, generating dr800-sv40- luc and intron-sv40-luc. These reporter constructs were coinjected with and without Xbmp4 RNA into one-cell stage zebrafish embryos and luciferase activity was assayed at the shield stage. Addition of dr800 to SV40-luc enhanced the promoter activity approximately 3-fold, probably because of the endogenous Bmp present in the embryo (Fig. 7A). Coinjection of Xbmp4 RNA and dr800-sv40-luc resulted in a further 3-fold increase in activity. In contrast, with intron- SV40-luc the promoter activity was the same with or without the addition of Xbmp4 RNA (Fig. 7A). These data demonstrate that the Bmp response element is located in the 800 bp distal region of the tbx6 promoter. To better define the Bmp-responsive element of the tbx6 promoter, we generated a series of deletion constructs of the
8 Gene regulation in zebrafish 3757 Fig. 5. A dual fluorescence reporter for examining promoter expression in zebrafish embryos. (A) Schematic diagram of the dual fluorescence reporter construct, ptbx6-2.0-gfp/pxex-bfp. The black box shows the tbx6 first intron. (B-G) Pictures of embryos injected with the ptbx6-2.0-gfp/pxex-bfp promoter in bright field (B,E), showing GFP fluorescence (C,F) and BFP fluorescence (D,G). (B-D) Lateral views with animal pole to the top at 70% epiboly. (E-G) Ventral views with anterior to the top at the 16-somite stage. E, exon sequence. response (Fig. 7C). These data demonstrate that the core Bmp response element must be located within the 65 bp distal region of the tbx6 promoter and suggest that multiple sites within this region are necessary to obtain the Bmp response. Collectively, our data show that the tbx6 promoter has two distinct regulatory regions that confer the ability to respond to Wnt and Bmp signals. Moreover, we find that both regulatory elements must function cooperatively to confer full promoter activity in the developing posterior mesoderm. dr800 region. We initially found that the 3 -half of the dr800 region retains the ability to respond to Bmp activation, whereas the 5 -half did not (Fig. 7B). Further analysis of this 3 -half of the dr800 region revealed that a 65 bp domain is capable of conferring Bmp responsiveness, and that further reduction of this 65 bp region into smaller domains eliminated the Bmp Discussion We show here that tbx6 is regulated by combinatorial signaling from the Wnt and Bmp pathways. This fits well with a recent study demonstrating that the combination of Wnt and Bmp induces tail formation in zebrafish (Agathon et al., 2003), and suggests that tbx6 is one of a series of posterior mesodermal genes regulated by these two pathways. Indeed, eve1, which has a similar expression pattern to tbx6 (Joly et al., 1993), was also found to be jointly regulated by these two pathways (Agathon et al., 2003), although the mechanism of its regulation is at present unknown. Although the role of eve1 in posterior mesoderm formation is unclear, a recent study indicates that tbx6 functions as part of a network of T-box transcription factors to regulate domains within the posterior mesoderm (Goering et al., 2003). Although the normal expression of tbx6 is dependent on Wnt and Bmp signaling, we find that inhibition of either one of these signaling pathways does not lead to a complete loss of tbx6 expression in the ventrolateral mesoderm. Interestingly, wnt8 morphants show a graded reduction of tbx6 expression with the strongest effect in the lateral domains of the ventrolateral mesoderm. In embryos injected with wnt8mos, bmp expression becomes ventrally restricted even at the shield stage (see Fig. S3A-D at supplemental), and tbx6 expression is restricted to this region. In contrast, loss of Bmp function causes a strong uniform reduction in tbx6 expression, but does not significantly alter the expression of wnt8 (see Fig. S3E-H at dev.biologists.org/supplemental). These findings show that the Wnt and Bmp signaling pathways have Fig. 6. Tcf sites are required for normal tbx6 expression. Embryos were injected with the tbx6 promoter containing a mutation in Tcf site 1 (mut1-2.0-gfp/xex-bfp; A-F) or a promoter with mutations in Tcf sites 1 and 2 (mut1,2-2.0-gfp/xex-bfp; G-L). Embryos were viewed in bright field, for GFP fluorescence or for BFP fluorescence as indicated on the right side of the panels. Embryos were at 70-90% epiboly (A-C,G-I) or at the 5-somite stage (D-F,J-L).
9 3758 Development 131 (15) Research article A dr800-sv40-luc + bmp dr800-sv40-luc intron-sv40-luc + bmp intron-sv40-luc SV40-luc + bmp SV40-luc Luciferase Units ( 10 ) B Fig. 7. Regulation of the tbx6 promoter. (A) Luciferase activities of the indicated constructs measured at 50% epiboly, with and without the coinjection of Xbmp4 (bmp). dr800 is the distal 800 bp and intron is the first intron of the tbx6 promoter. (B) Luciferase activities at 50% epiboly of the indicated constructs coinjected with Xbmp4, showing a 65 bp region of the tbx6 promoter is fully capable of responding to Bmp activation. (C) Sequence and schematic diagram of the 65 bp region. Division of this region into domains of 25 bp [(1) to (5)] causes a complete loss of Bmp responsiveness. C DNA Constructs + bmp dr800 (800-bp) (5') 600-bp 660-bp (3') 230-bp 450-bp 110-bp 65-bp Vector only Luciferase Units ( 10 ) GATAGGGATGATAACCAATGTTAGCATTAAAGCCTACAACTAAAAAAGACATAAGATGAATGAAT 65 bp 25 bp (1) 25 bp (2) DNA Constructs 25 bp (3) 25 bp (4) 25 bp (5) Bmp Responsiveness ++ distinct impacts on the overall expression of tbx6 in the posterior mesoderm. Although the contribution of Bmp and Wnt8 signals has a qualitative and quantitative difference in their ability to activate tbx6 expression, their cooperation is essential to give the normal expression pattern during posterior mesoderm formation. This raises the question, why is combinatorial signaling used to regulate tbx6 and other mesodermal genes? The expression patterns of the bmp genes and wnt8 are very dynamic during early embryogenesis, yet tbx6 expression is maintained in the ventrolateral mesoderm throughout somitogenesis. Combinatorial signaling not only restricts the domain in which tbx6 is expressed to regions where both signals are present, but it permits tbx6 to be expressed in regions where Bmp signaling is very low (the lateral regions) and where Wnt8 signaling is very low (the ventral regions from 90% epiboly onwards). Thus as the levels of the signals vary, tbx6 expression is maintained. Interestingly, a family of three Drosophila genes, Dorsocross1-3, encode T-box genes most closely related to tbx6 (Reim et al., 2003). Although the expression patterns of these genes is complex, the expression of all three genes in the dorsal ectoderm and mesoderm requires both Wnt and Bmp signals (Reim et al., 2003). Whether this represents conservation of regulation or convergent evolution awaits further analysis, but it is an intriguing parallel. Analysis of the tbx6 promoter during posterior mesoderm development Two models can be used to account for the required input of both Wnt and Bmp signals for the normal tbx6 expression in the posterior mesoderm. One is that these pathways may signal independently of each other, but function together in a cooperative fashion to activate the expression of tbx6. Alternatively, these pathways may function in a linear pathway such that one signal is upstream of the other as was suggested from Xenopus studies (Hoppler and Moon, 1998; Marom et al., 1999) and a recent zebrafish study (Agathon et al., 2003). To delineate the interplay between Wnt8 and Bmp signaling pathways in regulating the expression of tbx6 during posterior mesoderm development, we isolated and characterized the promoter of tbx6. We found that a genomic fragment including sequences from 1.7 kb upstream of the start of translation through the second exon recapitulated the normal tbx6 expression pattern in both zebrafish and Xenopus embryos, demonstrating that the regulatory elements are conserved between these species. The normal domain of tbx6 expression was observed when the intron was removed from the promoter; however, we also observed some ectopic expression outside of the mesodermal domain, indicating that the intron contains a repressor of non-mesodermal expression (D.P.S. and D.K., unpublished). Analysis of the tbx6 promoter revealed independent
10 Gene regulation in zebrafish 3759 Fig. 8. Model for the combinatorial regulation of tbx6 by Bmp and Wnt. (A) In the wild-type embryo, tbx6 promoter utilizes two distinct domains to integrate the signaling inputs of both Bmp and Wnt (gray box, Bmp response domain; white box with three oval circles, Wnt response domain; oval circles, Tcf binding sites; black gradient, Bmp activity gradient; blue region, wnt8 expression domain; blue filled oval circle, occupied Tcf site; pink color, tbx6 gene and transcripts). wnt8 expressed at the margin works combinatorially with bmps expressed ventrally to activate the tbx6 promoter throughout the ventrolateral region. (B) In a bmp mutant or noggininjected embryo, endogenous Wnt8 only weakly activates expression throughout the margin. (C) In a A B wild type Bmp mutant C wnt8 mutant wnt8 mutant or in an embryo injected with wnt8mos, endogenous Bmp signaling activates tbx6 only in the most ventral region where Bmp activity is the highest. When Wnt8 signaling is inhibited, Bmp signaling is confined to a more ventral domain than in wild-type embryos. tbx6 tbx6 tbx6 elements that respond cooperatively to Wnt and Bmp signals. In the proximal 900 bp, we discovered three Tcf sites and demonstrated that mutation of both of the two most proximal sites prevents normal tbx6 expression. Because Tcf is activated by Wnt signaling, these results demonstrate that tbx6 is a direct target of Wnt signaling. We located the Bmp response element to a distinct region of the tbx6 promoter, located between 1.7 kb and 0.9 kb (dr800) from the start of translation. In the presence of high levels of Bmp signal, this region alone can activate transcription when attached to a SV40 minimal promoter, but the activity is much weaker than that of the full 1.7 kb promoter. Within this dr800 region, we identified a 65 bp domain, which is capable of responding to Bmp activation. Smaller fragments did not respond to Bmp signaling, suggesting that the Bmp response element is composed of more than one transcription factor binding site. How this element is activated by Bmp signaling is not yet known. A transcription factor required for the function of Bmp-type Smads called OAZ was shown to be required for the activation of a key Bmp target gene in Xenopus (Hata et al., 2000), but no OAZ site was found in the tbx6 promoter. Moreover, during somitogenesis OAZ is expressed anteriorly (Hata et al., 2000), whereas tbx6 is expressed at the most posterior end of the embryo (Hug et al., 1997), suggesting that a novel transcription factor regulates the Bmp response of tbx6. Regulation of tbx6 by combinatorial Wnt and Bmp signaling Because the normal regulation of tbx6 requires input from both the Wnt and Bmp pathways, we were surprised to find that ectopic expression of either signal at the level used to produce tails (Agathon et al., 2003) resulted in strong activation of the endogenous tbx6 gene and of the tbx6 promoter. Using inhibitors of Bmp and Wnt8 signaling, we found that overexpression of each signaling pathway alone is able to activate tbx6 expression when the other pathway is blocked, demonstrating that Bmp and Wnt8 have the potential to activate tbx6 without the contribution of the other signaling pathway. Because the endogenous Bmp and Wnt8 can not activate tbx6 when the other pathway is inhibited, our results suggest that Bmp and Wnt signals are present in the embryo at relatively low levels such that they can combinatorially activate tbx6 only when both are present. This mechanism ensures that tbx6 is only expressed in the regions where these two factors overlap. How this works in regulating the promoter remains to be determined. At endogenous levels of signaling, it could be that the Wnt and Bmp response sites are only active transiently when a single signal is present, and only when both signals are present does a stable transcription complex form. At higher levels of signaling through one factor, the sites are occupied continuously and this promotes transcription of the tbx6 gene. From all of this data we have put together a model to explain how tbx6 is regulated in the normal embryo and in mutants. In the normal embryo, wnt8 is expressed at the margin in the gastrula-stage embryo and bmps are expressed in a ventral to dorsal gradient (Fig. 8A). Together they combine to activate tbx6 at the margin, with both signals working at submaximal levels. In bmp mutants (and noggin-injected embryos), Wnt8 causes a low level of tbx6 expression in the lateral and ventral regions (Fig. 8B). In wnt8 mutants (or in embryos injected with wnt8mos), Bmps activate tbx6 in the ventral region where they are most abundant (Fig. 8C). Tbx6: a platform for multiple levels of regulation Our study has focused only on the regulation of tbx6 by the Wnt and Bmp pathways. Earlier analyses have shown that tbx6 is also strongly regulated by spt (Griffin et al., 1998) and weakly by ntl (Hug et al., 1997; Griffin et al., 1998). Although Agathon et al. (Agathon et al., 2003) demonstrated that tails can be induced by overexpression of Wnt and Bmp at levels similar to those used here, they also found that co-expression of a member of the Nodal pathway lowered the amount of Wnt and Bmp necessary to form tails. Because Nodal signaling regulates spt (Griffin and Kimelman, 2003), one explanation for the synergism between the Nodals and the Wnt and Bmp pathways is that Spadetail contributes directly or indirectly to tbx6 expression. Thus tbx6 will continue to serve as a useful paradigm for understanding how multiple regulatory inputs lead to the formation of the posterior mesoderm.
11 3760 Development 131 (15) Research article We thank Chris Bjornson and Wilson Clements for reading this manuscript; Mike Wu, John Gerhart and Enrique Amaya for helping us with the Xenopus transgenic method; and Randy Moon, Stephen Ekker, Jim Smith, Dave Raible and Paul Wilson for their generous gifts of plasmids and reagents. This work was supported by a NRSA fellowship (F32 HD08725) to D.P.S. and a grant from the NIH (1 P01 GM65469) to D.K. References Agathon, A., Thisse, C. and Thisse, B. (2003). The molecular nature of the zebrafish tail organizer. Nature 424, Amaya, E. and Kroll, K. L. (1999). A method for generating transgenic frog embryos. Methods Mol. Biol. 97, Amaya, E., Stein, P. A., Musci, T. J. and Kirschner, M. W. (1993). FGF signalling in the early specification of mesoderm in Xenopus. Development 118, Barth, K. A., Kishimoto, Y., Rohr, K. B., Seydler, C., Schulte-Merker, S. and Wilson, S. W. (1999). Bmp activity establishes a gradient of positional information throughout the entire neural plate. Development 126, Darken, R. S. and Wilson, P. A. (2001). Axis induction by wnt signaling: target promoter responsiveness regulates competence. Dev. Biol. 234, Draper, B. W., Stock, D. W. and Kimmel, C. B. (2003). Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development 130, Finley, K. R., Davidson, A. E. and Ekker, S. C. (2001). Three-color imaging using fluorescent proteins in living zebrafish embryos. BioTechniques 31, Goering, L. M., Hoshijima, K., Hug, B., Bisgrove, B., Kispert, A. and Grunwald, D. J. (2003). An interacting network of T-box genes directs gene expression and fate in the zebrafish mesoderm. Proc. Natl. Acad. Sci. USA 100, Griffin, K., Patient, R. and Holder, N. (1995). Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development 121, Griffin, K. J., Amacher, S. L., Kimmel, C. B. and Kimelman, D. (1998). Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 125, Griffin, K. J. and Kimelman, D. (2003). Interplay between FGF, one-eyed pinhead, and T-box transcription factors during zebrafish posterior development. Dev. Biol. 264, Hammerschmidt, M. and Mullins, M. C. (2002). Dorsoventral patterning in the zebrafish: bone morphogenetic proteins and beyond. In Pattern Formation in Zebrafish, Vol. 40 (ed. L. Solnica-Krezel), pp Berlin: Springer-Verlag. Hata, A., Seoane, J., Lagna, G., Montalvo, E., Hemmati-Brivanlou, A. and Massague, J. (2000). OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100, Hoppler, S. and Moon, R. T. (1998). BMP-2/-4 and Wnt-8 cooperatively pattern the Xenopus mesoderm. Mech. Dev. 71, Hug, B., Walter, V. and Grunwald, D. J. (1997). tbx6, a Brachyury-related gene expressed by ventral mesendodermal precursors in the zebrafish embryo. Dev. Biol. 183, Joly, J. S., Joly, C., Schulte-Merker, S., Boulekbache, H. and Condamine, H. (1993). The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development 119, Kanki, J. P. and Ho, R. K. (1997). The development of the posterior body in zebrafish. Development 124, Kelly, G. M., Greenstein, P., Erezyilmaz, D. F. and Moon, R. T. (1995). Zebrafish wnt8 and wnt8b share a common activity but are involved in distinct developmental pathways. Development 121, Kimelman, D. and Schier, A. F. (2002). Mesoderm induction and patterning. In Pattern Formation in Zebrafish. Vol. 40 (ed. L. Solinca-Krezel), pp Berlin: Springer-Verlag. Kishimoto, Y., Lee, K. H., Zon, L., Hammerschmidt, M. and Schulte- Merker, S. (1997). The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development 124, Lekven, A. C., Thorpe, C. J., Waxman, J. S. and Moon, R. T. (2001). Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell 1, Marom, K., Fainsod, A. and Steinbeisser, H. (1999). Patterning of the mesoderm involves several threshold responses to BMP-4 and Xwnt-8. Mech. Dev. 87, Martinez-Barbera, J. P., Toresson, H., Da Rocha, S. and Krauss, S. (1997). Cloning and expression of three members of the zebrafish Bmp family: Bmp2a, Bmp2b and Bmp4. Gene 198, Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., Brand, M., van Eeden, F. J., Furutani-Seiki, M., Granato, M., Haffter, P., Heisenberg, C. P. et al. (1996). Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development 123, Nikaido, M., Tada, M., Saji, T. and Ueno, N. (1997). Conservation of BMP signaling in zebrafish mesoderm patterning. Mech. Dev. 61, Reim, I., Lee, H. H. and Frasch, M. (2003). The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development 130, Rodaway, A., Takeda, H., Koshida, S., Broadbent, J., Price, B., Smith, J. C., Patient, R. and Holder, N. (1999). Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGF-beta family signals and discrimination of mesoderm and endoderm by FGF. Development 126, Schier, A. F. and Shen, M. M. (2000). Nodal signalling in vertebrate development. Nature 403, Schier, A. F. and Talbot, W. S. (2001). Nodal signaling and the zebrafish organizer. Int. J. Dev. Biol. 45, Schulte-Merker, S., van Eeden, F. J., Halpern, M. E., Kimmel, C. B. and Nusslein-Volhard, C. (1994). no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 120, Smith, J. C., Price, B. M., Green, J. B., Weigel, D. and Herrmann, B. G. (1991). Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67, Uchiyama, H., Kobayashi, T., Yamashita, A., Ohno, S. and Yabe, S. (2001). Cloning and characterization of the T-box gene Tbx6 in Xenopus laevis. Dev. Growth Differ. 43, van Noort, M. and Clevers, H. (2002). TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev. Biol. 244, 1-8. Wagner, D. S. and Mullins, M. C. (2002). Modulation of BMP activity in dorsal-ventral pattern formation by the chordin and ogon antagonists. Dev. Biol. 245, Westerfield, M. (1995). The Zebrafish Book. Eugene: University of Oregon Press.
The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains
 The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains Daniel P. Szeto 1 and David Kimelman 2 Department of Biochemistry,
The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains Daniel P. Szeto 1 and David Kimelman 2 Department of Biochemistry,
Mesoderm Formation. Fate map of early gastrula. Only two types of mesoderm are induced. Mesoderm induction by the vegetal hemisphere
 Fate map of early gastrula Mesoderm Formation Animal hemisphere forms ectoderm (lacks ) Sperm Entry Point dbl dbl brachyury Vegetal hemisphere forms endoderm (requires ) dbl goosecoid Marginal zone forms
Fate map of early gastrula Mesoderm Formation Animal hemisphere forms ectoderm (lacks ) Sperm Entry Point dbl dbl brachyury Vegetal hemisphere forms endoderm (requires ) dbl goosecoid Marginal zone forms
The role of the yolk syncytial layer in germ layer patterning in zebrafish
 Development 127, 4681-4689 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV3237 4681 The role of the yolk syncytial layer in germ layer patterning in zebrafish Shaw-Ree Chen and
Development 127, 4681-4689 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV3237 4681 The role of the yolk syncytial layer in germ layer patterning in zebrafish Shaw-Ree Chen and
Lecture 3 MOLECULAR REGULATION OF DEVELOPMENT
 Lecture 3 E. M. De Robertis, M.D., Ph.D. August 16, 2016 MOLECULAR REGULATION OF DEVELOPMENT GROWTH FACTOR SIGNALING, HOX GENES, AND THE BODY PLAN Two questions: 1) How is dorsal-ventral (D-V) cell differentiation
Lecture 3 E. M. De Robertis, M.D., Ph.D. August 16, 2016 MOLECULAR REGULATION OF DEVELOPMENT GROWTH FACTOR SIGNALING, HOX GENES, AND THE BODY PLAN Two questions: 1) How is dorsal-ventral (D-V) cell differentiation
Xenopus gastrulation. Dorsal-Ventral Patterning - The Spemann Organizer. Two mesoderm inducing signals
 Dorsal- atterning - The Spemann Organizer Two mesoderm inducing signals late-blastula early-gastrula nimal Blood Mesothelium Not Vegetal NC Endoderm Hans Spemann (1869-1941) Hilde Mangold (1898-1924) Dr
Dorsal- atterning - The Spemann Organizer Two mesoderm inducing signals late-blastula early-gastrula nimal Blood Mesothelium Not Vegetal NC Endoderm Hans Spemann (1869-1941) Hilde Mangold (1898-1924) Dr
+ + Development and Evolution Dorsoventral axis. Developmental Readout. Foundations. Stem cells. Organ formation.
 Development and Evolution 7.72 9.11.06 Dorsoventral axis Human issues Organ formation Stem cells Developmental Readout Axes Growth control Axon guidance 3D structure Analysis Model + + organisms Foundations
Development and Evolution 7.72 9.11.06 Dorsoventral axis Human issues Organ formation Stem cells Developmental Readout Axes Growth control Axon guidance 3D structure Analysis Model + + organisms Foundations
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/318/5851/794/dc1 Supporting Online Material for A Gene Regulatory Network Subcircuit Drives a Dynamic Pattern of Gene Expression Joel Smith, Christina Theodoris, Eric
www.sciencemag.org/cgi/content/full/318/5851/794/dc1 Supporting Online Material for A Gene Regulatory Network Subcircuit Drives a Dynamic Pattern of Gene Expression Joel Smith, Christina Theodoris, Eric
Pregnenolone Stabilizes Microtubules and Promotes Zebrafish Embryonic Cell Movement
 Pregnenolone Stabilizes Microtubules and Promotes Zebrafish Embryonic Cell Movement Hwei-Jan Hsu 1,2, Ming-Ren Liang 3, Chao-Tsen Chen 3, and Bon-Chu Chung 1 * Abstract Embryonic cell movement is essential
Pregnenolone Stabilizes Microtubules and Promotes Zebrafish Embryonic Cell Movement Hwei-Jan Hsu 1,2, Ming-Ren Liang 3, Chao-Tsen Chen 3, and Bon-Chu Chung 1 * Abstract Embryonic cell movement is essential
Biphasic wnt8a Expression Is Achieved Through Interactions of Multiple Regulatory Inputs
 a DEVELOPMENTAL DYNAMICS 241:1062 1075, 2012 RESEARCH ARTICLE Biphasic wnt8a Expression Is Achieved Through Interactions of Multiple Regulatory Inputs Anand Narayanan and Arne C. Lekven* Background: Vertebrate
a DEVELOPMENTAL DYNAMICS 241:1062 1075, 2012 RESEARCH ARTICLE Biphasic wnt8a Expression Is Achieved Through Interactions of Multiple Regulatory Inputs Anand Narayanan and Arne C. Lekven* Background: Vertebrate
Enzyme that uses RNA as a template to synthesize a complementary DNA
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Comparison of two or more protein or DNA sequence to ascertain similarities in sequences. If two genes have
Neural Induction. Chapter One
 Neural Induction Chapter One Fertilization Development of the Nervous System Cleavage (Blastula, Gastrula) Neuronal Induction- Neuroblast Formation Cell Migration Mesodermal Induction Lateral Inhibition
Neural Induction Chapter One Fertilization Development of the Nervous System Cleavage (Blastula, Gastrula) Neuronal Induction- Neuroblast Formation Cell Migration Mesodermal Induction Lateral Inhibition
Supporting Information
 Supporting Information Lu et al. 10.1073/pnas.1106801108 Fig. S1. Analysis of spatial localization of mrnas coding for 23 zebrafish Wnt genes by whole mount in situ hybridization to identify those present
Supporting Information Lu et al. 10.1073/pnas.1106801108 Fig. S1. Analysis of spatial localization of mrnas coding for 23 zebrafish Wnt genes by whole mount in situ hybridization to identify those present
HOW DOES Wnt SIGNALING POSTERIORIZE THE NEURAL PLATE?
 HOW DOES Wnt SIGNALING POSTERIORIZE THE NEURAL PLATE? An Undergraduate Research Scholars Thesis By JEFFREY JOON TAEK LEE Submitted to Honors and Undergraduate Research Texas A&M University In partial fulfillment
HOW DOES Wnt SIGNALING POSTERIORIZE THE NEURAL PLATE? An Undergraduate Research Scholars Thesis By JEFFREY JOON TAEK LEE Submitted to Honors and Undergraduate Research Texas A&M University In partial fulfillment
Readings. Lecture IV. Mechanisms of Neural. Neural Development. September 10, Bio 3411 Lecture IV. Mechanisms of Neural Development
 Readings Lecture IV. Mechanisms of Neural NEUROSCIENCE: References : 5 th ed, pp 477-506 (sorta) 4 th ed, pp 545-575 (sorta) Fainsod, A., Steinbeisser, H., & De Robertis, E. M. (1994). EMBO J, 13(21),
Readings Lecture IV. Mechanisms of Neural NEUROSCIENCE: References : 5 th ed, pp 477-506 (sorta) 4 th ed, pp 545-575 (sorta) Fainsod, A., Steinbeisser, H., & De Robertis, E. M. (1994). EMBO J, 13(21),
Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in. Zebrafish Embryos 1
 Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in Zebrafish Embryos 1 1. In vitro synthesis of capped Cas9 mrna The full length of humanized Cas9 cdnas with double NLS were cloned into pxt7
Protocol for Genome Editing via the RNA-guided Cas9 Nuclease in Zebrafish Embryos 1 1. In vitro synthesis of capped Cas9 mrna The full length of humanized Cas9 cdnas with double NLS were cloned into pxt7
Fertilization. Animal hemisphere. Sperm entry point
 Fertilization Animal hemisphere Sperm entry point Establishes the dorsal/ventral axis Ventral side sperm entry Dorsal side gray crescent Organized by sperm centriole Cleavage Unequal radial holoblastic
Fertilization Animal hemisphere Sperm entry point Establishes the dorsal/ventral axis Ventral side sperm entry Dorsal side gray crescent Organized by sperm centriole Cleavage Unequal radial holoblastic
Roche Molecular Biochemicals Technical Note No. LC 12/2000
 Roche Molecular Biochemicals Technical Note No. LC 12/2000 LightCycler Absolute Quantification with External Standards and an Internal Control 1. General Introduction Purpose of this Note Overview of Method
Roche Molecular Biochemicals Technical Note No. LC 12/2000 LightCycler Absolute Quantification with External Standards and an Internal Control 1. General Introduction Purpose of this Note Overview of Method
MCB 102 University of California, Berkeley August 11 13, Problem Set 8
 MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
MCB 102 University of California, Berkeley August 11 13, 2009 Isabelle Philipp Handout Problem Set 8 The answer key will be posted by Tuesday August 11. Try to solve the problem sets always first without
Contents... vii. List of Figures... xii. List of Tables... xiv. Abbreviatons... xv. Summary... xvii. 1. Introduction In vitro evolution...
 vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
vii Contents Contents... vii List of Figures... xii List of Tables... xiv Abbreviatons... xv Summary... xvii 1. Introduction...1 1.1 In vitro evolution... 1 1.2 Phage Display Technology... 3 1.3 Cell surface
Lecture 20: Drosophila embryogenesis
 Lecture 20: Drosophila embryogenesis Mitotic recombination/clonal analyses Embrygenesis Four classes of genes: Maternal genes Gap genes Pair-rule genes Segment polarity genes Homeotic genes Read 140-141;
Lecture 20: Drosophila embryogenesis Mitotic recombination/clonal analyses Embrygenesis Four classes of genes: Maternal genes Gap genes Pair-rule genes Segment polarity genes Homeotic genes Read 140-141;
Lecture 20: Drosophila melanogaster
 Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Lecture 20: Drosophila melanogaster Model organisms Polytene chromosome Life cycle P elements and transformation Embryogenesis Read textbook: 732-744; Fig. 20.4; 20.10; 20.15-26 www.mhhe.com/hartwell3
Chapter 14 Regulation of Transcription
 Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Chapter 14 Regulation of Transcription Cis-acting sequences Distance-independent cis-acting elements Dissecting regulatory elements Transcription factors Overview transcriptional regulation Transcription
Supplementary Methods
 Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Supplementary Methods Reverse transcribed Quantitative PCR. Total RNA was isolated from bone marrow derived macrophages using RNeasy Mini Kit (Qiagen), DNase-treated (Promega RQ1), and reverse transcribed
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM Part I: Definitions. Homology: Reverse transcriptase. Allostery: cdna library
 Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Biology 105: Introduction to Genetics PRACTICE FINAL EXAM 2006 Part I: Definitions Homology: Reverse transcriptase Allostery: cdna library Transformation Part II Short Answer 1. Describe the reasons for
Schematic representation of the endogenous PALB2 locus and gene-disruption constructs
 Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
Supplementary Figures Supplementary Figure 1. Generation of PALB2 -/- and BRCA2 -/- /PALB2 -/- DT40 cells. (A) Schematic representation of the endogenous PALB2 locus and gene-disruption constructs carrying
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
A Survey of Genetic Methods
 IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
IBS 8102 Cell, Molecular, and Developmental Biology A Survey of Genetic Methods January 24, 2008 DNA RNA Hybridization ** * radioactive probe reverse transcriptase polymerase chain reaction RT PCR DNA
Figure S1 Correlation in size of analogous introns in mouse and teleost Piccolo genes. Mouse intron size was plotted against teleost intron size for t
 Figure S1 Correlation in size of analogous introns in mouse and teleost Piccolo genes. Mouse intron size was plotted against teleost intron size for the pcloa genes of zebrafish, green spotted puffer (listed
Figure S1 Correlation in size of analogous introns in mouse and teleost Piccolo genes. Mouse intron size was plotted against teleost intron size for the pcloa genes of zebrafish, green spotted puffer (listed
UTR Reporter Vectors and Viruses
 UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
UTR Reporter Vectors and Viruses 3 UTR, 5 UTR, Promoter Reporter (Version 1) Applied Biological Materials Inc. #1-3671 Viking Way Richmond, BC V6V 2J5 Canada Notice to Purchaser All abm products are for
pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz
 pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz For research use only Version # 03B04-MT PRODUCT INFORMATION Content: - 20 µg of pbroad3-lacz provided as lyophilized
pbroad3-lacz An optimized vector for mouse and rat transgenesis Catalog # pbroad3-lacz For research use only Version # 03B04-MT PRODUCT INFORMATION Content: - 20 µg of pbroad3-lacz provided as lyophilized
In Situ Hybridization
 In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
In Situ Hybridization Modified from C. Henry, M. Halpern and Thisse labs April 17, 2013 Table of Contents Reagents... 2 AP Buffer... 2 Developing Solution... 2 Hybridization buffer... 2 PBT... 2 PI Buffer
Cold Fusion Cloning Kit. Cat. #s MC100A-1, MC101A-1. User Manual
 Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
Fusion Cloning technology Cold Fusion Cloning Kit Store the master mixture and positive controls at -20 C Store the competent cells at -80 C. (ver. 120909) A limited-use label license covers this product.
7.013 Practice Quiz
 MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel 7.013 Practice Quiz 2 2004 1 Question 1 A. The primer
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel 7.013 Practice Quiz 2 2004 1 Question 1 A. The primer
How To Choose a GeneCopoeia Luciferase System. Ed Davis, Ph.D.
 TECHNICAL NOTE How To Choose a GeneCopoeia Luciferase System Ed Davis, Ph.D. Introduction Luciferase reporter systems are invaluable tools for several applications, including regulation of gene expression
TECHNICAL NOTE How To Choose a GeneCopoeia Luciferase System Ed Davis, Ph.D. Introduction Luciferase reporter systems are invaluable tools for several applications, including regulation of gene expression
Problem Set 8. Answer Key
 MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
MCB 102 University of California, Berkeley August 11, 2009 Isabelle Philipp Online Document Problem Set 8 Answer Key 1. The Genetic Code (a) Are all amino acids encoded by the same number of codons? no
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
Supplementary Figure 1 Schematic and results of screening the combinatorial antibody library for Sox2 replacement activity. A single batch of MEFs were plated and transduced with doxycycline inducible
7.22 Example Problems for Exam 1 The exam will be of this format. It will consist of 2-3 sets scenarios.
 Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Massachusetts Institute of Technology Department of Biology 7.22, Fall 2005 - Developmental Biology Instructors: Professor Hazel Sive, Professor Martha Constantine-Paton 1 of 10 7.22 Fall 2005 sample exam
Supplementary Methods pcfd5 cloning protocol
 Supplementary Methods cloning protocol vermilion trna grna trna grna U6:3 Terminator AmpR attb is a vector for expressing one or multiple trna-flanked Cas9 grnas under the control of the strong, ubiquitous
Supplementary Methods cloning protocol vermilion trna grna trna grna U6:3 Terminator AmpR attb is a vector for expressing one or multiple trna-flanked Cas9 grnas under the control of the strong, ubiquitous
sides of the aleurone (Al) but it is excluded from the basal endosperm transfer layer
 Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
Supplemental Data. Gómez et al. (2009). The maize transcription factor MRP-1 (Myb-Related-Protein-1) is a key regulator of the differentiation of transfer cells. Supplemental Figure 1. Expression analyses
BMP signaling restricts hemato-vascular development from lateral mesoderm during somitogenesis
 RESEARCH ARTICLE 2177 Development 133, 2177-2187 (2006) doi:10.1242/dev.02386 BMP signaling restricts hemato-vascular development from lateral mesoderm during somitogenesis Sunny Gupta 1, Hao Zhu 2, Leonard
RESEARCH ARTICLE 2177 Development 133, 2177-2187 (2006) doi:10.1242/dev.02386 BMP signaling restricts hemato-vascular development from lateral mesoderm during somitogenesis Sunny Gupta 1, Hao Zhu 2, Leonard
Construction of plant complementation vector and generation of transgenic plants
 MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
MATERIAL S AND METHODS Plant materials and growth conditions Arabidopsis ecotype Columbia (Col0) was used for this study. SALK_072009, SALK_076309, and SALK_027645 were obtained from the Arabidopsis Biological
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Name AP Biology Mrs. Laux Take home test #11 on Chapters 14, 15, and 17 DUE: MONDAY, DECEMBER 21, 2009
 MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
MULTIPLE CHOICE QUESTIONS 1. Inducible genes are usually actively transcribed when: A. the molecule degraded by the enzyme(s) is present in the cell. B. repressor molecules bind to the promoter. C. lactose
Supplemental Information. Autoregulatory Feedback Controls. Sequential Action of cis-regulatory Modules. at the brinker Locus
 Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
Developmental Cell, Volume 26 Supplemental Information Autoregulatory Feedback Controls Sequential Action of cis-regulatory Modules at the brinker Locus Leslie Dunipace, Abbie Saunders, Hilary L. Ashe,
High Pure PCR Template Preparation Kit for preparation of 100 nucleic acid samples Cat. No
 for preparation of 100 nucleic acid samples Cat. No. 1 796 88 Principle Cells are lysed during a short incubation with Proteinase K in the presence of a chaotropic salt (guanidine HCl), which immediately
for preparation of 100 nucleic acid samples Cat. No. 1 796 88 Principle Cells are lysed during a short incubation with Proteinase K in the presence of a chaotropic salt (guanidine HCl), which immediately
To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR
 Plasmids To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR fragments downstream of firefly luciferase (luc) in pgl3 control (Promega). pgl3- CDK6 was made by amplifying a 2,886
Plasmids To generate the luciferase fusion to the human 3 UTRs, we sub-cloned the 3 UTR fragments downstream of firefly luciferase (luc) in pgl3 control (Promega). pgl3- CDK6 was made by amplifying a 2,886
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Design. Construction. Characterization
 Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
Design Construction Characterization DNA mrna (messenger) A C C transcription translation C A C protein His A T G C T A C G Plasmids replicon copy number incompatibility selection marker origin of replication
and rag1 as Markers for the Development of the Zebrafish Immune System
 bug journal vol. 2, 1999 early development of the immune system. For these purposes, the lab previously cloned the recombination activating genes, rag1 and rag2 (Willett et al., 1997 a). These genes are
bug journal vol. 2, 1999 early development of the immune system. For these purposes, the lab previously cloned the recombination activating genes, rag1 and rag2 (Willett et al., 1997 a). These genes are
Supplemental Information
 Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplemental Information Itemized List Materials and Methods, Related to Supplemental Figures S5A-C and S6. Supplemental Figure S1, Related to Figures 1 and 2. Supplemental Figure S2, Related to Figure
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and
 Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Supplementary Fig. S1. Building a training set of cardiac enhancers. (A-E) Empirical validation of candidate enhancers containing matches to Twi and Tin TFBS motifs and located in the flanking or intronic
Alternative Cleavage and Polyadenylation of RNA
 Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
Developmental Cell 18 Supplemental Information The Spen Family Protein FPA Controls Alternative Cleavage and Polyadenylation of RNA Csaba Hornyik, Lionel C. Terzi, and Gordon G. Simpson Figure S1, related
SCREENING AND PRESERVATION OF DNA LIBRARIES
 MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
MODULE 4 LECTURE 5 SCREENING AND PRESERVATION OF DNA LIBRARIES 4-5.1. Introduction Library screening is the process of identification of the clones carrying the gene of interest. Screening relies on a
SUPPLEMENTAL MATERIALS. Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary
 SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
SUPPLEMENTAL MATERIALS METHODS Chromatin immunoprecipitation assays. Single-cell suspensions of pituitary and liver cells were prepared from the indicated transgenic mice, as described (Ho et al, 2002).
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene. Andrew ElBardissi, The Pennsylvania State University
 Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Understanding the Cellular Mechanism of the Excess Microsporocytes I (EMSI) Gene Andrew ElBardissi, The Pennsylvania State University Abstract: Hong Ma, The Pennsylvania State University The Excess Microsporocytes
Expression Pattern of Xtshz1 and Xtshz3 During Xenopus Early Eye Formation
 SUNY College of Environmental Science and Forestry Digital Commons @ ESF Honors Theses 5-2012 Expression Pattern of Xtshz1 and Xtshz3 During Xenopus Early Eye Formation Katherine L. McKissick Follow this
SUNY College of Environmental Science and Forestry Digital Commons @ ESF Honors Theses 5-2012 Expression Pattern of Xtshz1 and Xtshz3 During Xenopus Early Eye Formation Katherine L. McKissick Follow this
TRANSGENIC ANIMALS. -transient transfection of cells -stable transfection of cells. - Two methods to produce transgenic animals:
 TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
TRANSGENIC ANIMALS -transient transfection of cells -stable transfection of cells - Two methods to produce transgenic animals: 1- DNA microinjection - random insertion 2- embryonic stem cell-mediated gene
Supplementary data. sienigma. F-Enigma F-EnigmaSM. a-p53
 Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
Supplementary data Supplemental Figure 1 A sienigma #2 sienigma sicontrol a-enigma - + ++ - - - - - - + ++ - - - - - - ++ B sienigma F-Enigma F-EnigmaSM a-flag HLK3 cells - - - + ++ + ++ - + - + + - -
To determine MRK-003 IC50 values, cell lines were plated in triplicate in 96-well plates at 3 x
 Supplementary methods: Cellular growth assay and cell cycle analysis To determine MRK-003 IC50 values, cell lines were plated in triplicate in 96-well plates at 3 x 10 3 cells/well (except for TALL1 and
Supplementary methods: Cellular growth assay and cell cycle analysis To determine MRK-003 IC50 values, cell lines were plated in triplicate in 96-well plates at 3 x 10 3 cells/well (except for TALL1 and
Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb
 Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
Cell Reports Supplemental Information Somatic Primary pirna Biogenesis Driven by cis-acting RNA Elements and Trans-Acting Yb Hirotsugu Ishizu, Yuka W. Iwasaki, Shigeki Hirakata, Haruka Ozaki, Wataru Iwasaki,
SUPPLEMENTARY INFORMATION
 Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers Wataru Sakai, Elizabeth M. Swisher, Beth Y. Karlan, Mukesh K. Agarwal, Jake Higgins, Cynthia Friedman, Emily Villegas,
supplementary information
 DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
DOI: 1.138/ncb1839 a b Control 1 2 3 Control 1 2 3 Fbw7 Smad3 1 2 3 4 1 2 3 4 c d IGF-1 IGF-1Rβ IGF-1Rβ-P Control / 1 2 3 4 Real-time RT-PCR Relative quantity (IGF-1/ mrna) 2 1 IGF-1 1 2 3 4 Control /
NAME TA SEC Problem Set 4 FRIDAY October 15, Answers to this problem set must be inserted into the box outside
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel NAME TA SEC 7.012 Problem Set 4 FRIDAY October 15,
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Figure 1 Ihh interacts preferentially with its upstream neighboring gene Nhej1. Genes are indicated by gray lines, and Ihh and Nhej1 are highlighted in blue. 4C seq performed in E14.5 limbs
Supplementary Information. Isl2b regulates anterior second heart field development in zebrafish
 Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
Supplementary Information Isl2b regulates anterior second heart field development in zebrafish Hagen R. Witzel 1, Sirisha Cheedipudi 1, Rui Gao 1, Didier Y.R. Stainier 2 and Gergana D. Dobreva 1,3* 1 Origin
ENDEXT Technology. Instruction manual for protein synthesis. with wheat germ cell-free system
 ENDEXT Technology Instruction manual for protein synthesis with wheat germ cell-free system 1 Protocol Overview Plasmid DNA construction (see Section 3.1) Preparation of plasmid DNA for transcription (see
ENDEXT Technology Instruction manual for protein synthesis with wheat germ cell-free system 1 Protocol Overview Plasmid DNA construction (see Section 3.1) Preparation of plasmid DNA for transcription (see
GENETICS - CLUTCH CH.15 GENOMES AND GENOMICS.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
!! www.clutchprep.com CONCEPT: OVERVIEW OF GENOMICS Genomics is the study of genomes in their entirety Bioinformatics is the analysis of the information content of genomes - Genes, regulatory sequences,
A Repressor Complex Governs the Integration of
 Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Developmental Cell 15 Supplemental Data A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis Dan Li, Chang Liu, Lisha Shen, Yang Wu, Hongyan Chen, Masumi Robertson, Chris A.
Efficient Method for Isolation of High Quality Concentrated Cellular RNA with Extremely Low Levels of Genomic DNA Contamination Application
 Efficient Method for Isolation of High Quality Concentrated Cellular RNA with Extremely Low Levels of Genomic DNA Contamination Application Gene Expression Authors Ilgar Abbaszade, Claudia Robbins, John
Efficient Method for Isolation of High Quality Concentrated Cellular RNA with Extremely Low Levels of Genomic DNA Contamination Application Gene Expression Authors Ilgar Abbaszade, Claudia Robbins, John
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.
 Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Fig. S1. Effect of p120-catenin overexpression on the interaction of SCUBE2 with E-cadherin. The expression plasmid encoding FLAG.SCUBE2, E-cadherin.Myc, or HA.p120-catenin was transfected in a combination
Nature Biotechnology: doi: /nbt.4166
 Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
Supplementary Figure 1 Validation of correct targeting at targeted locus. (a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained
ENDEXT TM Technology. Wheat Germ Premium Expression Kit. Ver 1.7. CellFree Sciences Co., Ltd.
 ENDEXT TM Technology Wheat Germ Premium Expression Kit Ver 1.7 CellFree Sciences Co., Ltd. 1. Purpose Wheat Germ Premium Expression Kit is a starter kit to ascertain if the Wheat Germ Cell-Free System
ENDEXT TM Technology Wheat Germ Premium Expression Kit Ver 1.7 CellFree Sciences Co., Ltd. 1. Purpose Wheat Germ Premium Expression Kit is a starter kit to ascertain if the Wheat Germ Cell-Free System
A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish
 Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
Developmental Cell Supplemental Information A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish Julien Ablain, Ellen M. Durand, Song Yang, Yi Zhou, and Leonard I. Zon % larvae
HiPer RT-PCR Teaching Kit
 HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
Activin-Induced Factors Maintain goosecoid Transcription through a Paired Homeodomain Binding Site
 DEVELOPMENTAL BIOLOGY 204, 172 186 (1998) ARTICLE NO. DB989065 Activin-Induced Factors Maintain goosecoid Transcription through a Paired Homeodomain Binding Site Roslyn McKendry, Richard M. Harland, 1
DEVELOPMENTAL BIOLOGY 204, 172 186 (1998) ARTICLE NO. DB989065 Activin-Induced Factors Maintain goosecoid Transcription through a Paired Homeodomain Binding Site Roslyn McKendry, Richard M. Harland, 1
Cell-Free Protein Expression Kit
 Cell-Free Protein Expression Kit Handbook Version v.01, January 2018 Cat# 507024 (Sigma 70 Master Mix Kit, 24 Rxns) Cat# 507096 (Sigma 70 Master Mix Kit, 96 Rxns) Please refer to this product in your publication
Cell-Free Protein Expression Kit Handbook Version v.01, January 2018 Cat# 507024 (Sigma 70 Master Mix Kit, 24 Rxns) Cat# 507096 (Sigma 70 Master Mix Kit, 96 Rxns) Please refer to this product in your publication
FMF NIRCA PROTOCOL STEP 1.
 FMF NIRCA PROTOCOL STEP 1. After you have isolated patient s DNA and DNA from a healthy donor (wild type), you perform a nested PCR. The primers used to amplify exon 2 and exon 10 of the mefv gene are
FMF NIRCA PROTOCOL STEP 1. After you have isolated patient s DNA and DNA from a healthy donor (wild type), you perform a nested PCR. The primers used to amplify exon 2 and exon 10 of the mefv gene are
ml recombinant E. coli cultures (at a density of A 600 units per ml)
 for purification of plasmid DNA, from bacterial cultures Cat. No. 1 754 777 (50 purifications) Cat. No. 1 754 785 (50 purifications) Principle Alkaline lysis releases plasmid DNA from bacteria and RNase
for purification of plasmid DNA, from bacterial cultures Cat. No. 1 754 777 (50 purifications) Cat. No. 1 754 785 (50 purifications) Principle Alkaline lysis releases plasmid DNA from bacteria and RNase
Understanding embryonic head development. ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute
 Understanding embryonic head development ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute Head malformations among the most common category of congenital malformations in
Understanding embryonic head development ANAT2341 Tennille Sibbritt Embryology Unit Children s Medical Research Institute Head malformations among the most common category of congenital malformations in
Supplementary Information for Structural and Mechanistic Insights into Cooperative Assembly of Dimeric Notch Transcription Complexes
 Supplementary Information for Structural and Mechanistic Insights into Cooperative Assembly of Dimeric Notch Transcription Complexes Kelly L. Arnett 1,2, Matthew Hass 3, Debbie G. McArthur 1,2, Ma. Xenia
Supplementary Information for Structural and Mechanistic Insights into Cooperative Assembly of Dimeric Notch Transcription Complexes Kelly L. Arnett 1,2, Matthew Hass 3, Debbie G. McArthur 1,2, Ma. Xenia
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes
 OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
OriGene GFC-Arrays for High-throughput Overexpression Screening of Human Gene Phenotypes High-throughput Gene Function Validation Tool Introduction sirna screening libraries enable scientists to identify
Rapid amplification of cdna ends (RACE)
 Rapid amplification of cdna ends (RACE) Rapid amplification of cdna ends (RACE) is a technique used in molecular biology to obtain the full length sequence of an RNA transcript found within a cell. RACE
Rapid amplification of cdna ends (RACE) Rapid amplification of cdna ends (RACE) is a technique used in molecular biology to obtain the full length sequence of an RNA transcript found within a cell. RACE
The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity
 Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Promega Notes Magazine Number 62, 1997, p. 02 The GeneEditor TM in vitro Mutagenesis System: Site- Directed Mutagenesis Using Altered Beta-Lactamase Specificity By Christine Andrews and Scott Lesley Promega
Learning Objectives :
 Learning Objectives : Understand the basic differences between genomic and cdna libraries Understand how genomic libraries are constructed Understand the purpose for having overlapping DNA fragments in
Learning Objectives : Understand the basic differences between genomic and cdna libraries Understand how genomic libraries are constructed Understand the purpose for having overlapping DNA fragments in
Supporting Information
 Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
Supporting Information SI Materials and Methods RT-qPCR The 25 µl qrt-pcr reaction mixture included 1 µl of cdna or DNA, 12.5 µl of 2X SYBER Green Master Mix (Applied Biosystems ), 5 µm of primers and
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
pdsipher and pdsipher -GFP shrna Vector User s Guide
 pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
pdsipher and pdsipher -GFP shrna Vector User s Guide NOTE: PLEASE READ THE ENTIRE PROTOCOL CAREFULLY BEFORE USE Page 1. Introduction... 1 2. Vector Overview... 1 3. Vector Maps 2 4. Materials Provided...
EPIGENTEK. EpiQuik Tissue Chromatin Immunoprecipitation Kit. Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE
 EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
EpiQuik Tissue Chromatin Immunoprecipitation Kit Base Catalog # P-2003 PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Chromatin Immunoprecipitation Kit is suitable for combining the specificity
Neural Induction. Steven McLoon Department of Neuroscience University of Minnesota
 Neural Induction Steven McLoon Department of Neuroscience University of Minnesota 1 Course News Coffee Hour (with Dr. Nakagawa) Friday, Sept 22 8:30-9:30am Surdyks Café in Northrop Auditorium Stop by for
Neural Induction Steven McLoon Department of Neuroscience University of Minnesota 1 Course News Coffee Hour (with Dr. Nakagawa) Friday, Sept 22 8:30-9:30am Surdyks Café in Northrop Auditorium Stop by for
High Pure RNA Isolation Kit for isolation of total RNA from 50 samples Cat. No
 for isolation of total RNA from 50 samples Cat. No. 1 88 665 Principle A single reagent lyses the sample lysis and inactivates RNase. In the presence of a chaotropic salt (guanidine HCl), the released
for isolation of total RNA from 50 samples Cat. No. 1 88 665 Principle A single reagent lyses the sample lysis and inactivates RNase. In the presence of a chaotropic salt (guanidine HCl), the released
Supplementary Materials
 Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supplementary Materials Supplementary Methods Supplementary Discussion Supplementary Figure 1 Calculated frequencies of embryo cells bearing bi-allelic alterations. Targeted indel mutations induced by
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
www.sciencemag.org/cgi/content/full/310/5753/1487/dc1 Supporting Online Material for Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors Rongwen Xi and Ting Xie* *To whom correspondence should
Roche Molecular Biochemicals Technical Note No. LC 10/2000
 Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
Roche Molecular Biochemicals Technical Note No. LC 10/2000 LightCycler Overview of LightCycler Quantification Methods 1. General Introduction Introduction Content Definitions This Technical Note will introduce
The Transfection Collection TCF/LEF Transient Pack Wnt / -catenin Signaling Pathway Catalog #: 79273
 Data Sheet The Transfection Collection TCF/LEF Transient Pack Wnt / -catenin Signaling Pathway Catalog #: 79273 Background The Wnt / -catenin signaling pathway controls a large and diverse set of cell
Data Sheet The Transfection Collection TCF/LEF Transient Pack Wnt / -catenin Signaling Pathway Catalog #: 79273 Background The Wnt / -catenin signaling pathway controls a large and diverse set of cell
Bi8 Lecture 19. Review and Practice Questions March 8th 2016
 Bi8 Lecture 19 Review and Practice Questions March 8th 2016 Common Misconceptions Where Words Matter Common Misconceptions DNA vs RNA vs Protein Su(H) vs Su(H) Replication Transcription Transcribed vs
Bi8 Lecture 19 Review and Practice Questions March 8th 2016 Common Misconceptions Where Words Matter Common Misconceptions DNA vs RNA vs Protein Su(H) vs Su(H) Replication Transcription Transcribed vs
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit
 EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE The EpiQuik Tissue Methyl-CpG Binding Domain Protein 2 ChIP Kit is suitable for
SUPPLEMENTARY INFORMATION
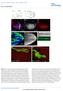 DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
DOI: 10.1038/ncb2806 Supplemental Material: Figure S1 Wnt4 PCP phenotypes are not mediated through canonical Wnt signaling. (a-a ) Illustration of wg, dwnt4 and dwnt6 expression patterns in wing (a) and
Two classes of silencing RNAs move between Caenorhabditis elegans tissues.
 Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Antony M Jose, Giancarlo A Garcia, and Craig P Hunter. Supplementary Figures, Figure Legends, and Tables. Supplementary Figure
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I)
 Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I) We have made considerable progress in our analysis of the gene for
Texas A&M University-Corpus Christi CHEM4402 Biochemistry II Laboratory Laboratory 8: DNA Restriction Digest (II) and DNA Sequencing (I) We have made considerable progress in our analysis of the gene for
Document S1. Supplemental Experimental Procedures and Three Figures (see next page)
 Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
Supplemental Data Document S1. Supplemental Experimental Procedures and Three Figures (see next page) Table S1. List of Candidate Genes Identified from the Screen. Candidate genes, corresponding dsrnas
