Cargo Proteins Facilitate the Formation of Transport Vesicles in the Cytoplasm to Vacuole Targeting Pathway*
|
|
|
- Rhoda Knight
- 6 years ago
- Views:
Transcription
1 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 279, No. 29, Issue of July 16, pp , by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Cargo Proteins Facilitate the Formation of Transport Vesicles in the Cytoplasm to Vacuole Targeting Pathway* Received for publication, April 21, 2004, and in revised form, May 11, 2004 Published, JBC Papers in Press, May 11, 2004, DOI /jbc.M Takahiro Shintani and Daniel J. Klionsky From the Life Sciences Institute and Departments of Molecular, Cellular, and Developmental Biology and Biological Chemistry, University of Michigan, Ann Arbor, Michigan Selective incorporation of cargo proteins into the forming vesicle is an important aspect of protein targeting via vesicular trafficking. Based on the current paradigm of cargo selection in vesicular transport, proteins to be sorted to other organelles are condensed at the vesicle budding site in the donor organelle, a process that is mediated by the interaction between cargo and coat proteins, which constitute part of the vesicle forming machinery. The cytoplasm to vacuole targeting (Cvt) pathway is an unconventional vesicular trafficking pathway in yeast, which is topologically and mechanistically related to autophagy. Aminopeptidase I (Ape1) is the major cargo protein of the Cvt pathway. Unlike the situation in conventional vesicular transport, precursor Ape1, along with its receptor Atg19/Cvt19, is packed into a huge complex, termed a Cvt complex, independent of the vesicle formation machinery. The Cvt complex is subsequently incorporated into the forming Cvt vesicle. The deletion of APE1 or ATG19 compromised the organization of the pre-autophagosomal structure (PAS), a site that is thought to play a critical role in Cvt vesicle/ autophagosome formation. The proper organization of the PAS also required Atg11/Cvt9, a protein that localizes the cargo complex at the PAS. Accordingly, the deletion of APE1, ATG19, or ATG11 affected the formation of Cvt vesicles. These observations suggest a unique concept; in the case of the Cvt pathway, the cargo proteins facilitate receptor recruitment and vesicle formation rather than the situation with most vesicular transport, in which the forming vesicle concentrates the cargo proteins. The localization of many intracellular proteins is dependent upon movement within transient transport vesicles. The precise transport of proteins via vesicular trafficking is guaranteed by the selective incorporation of cargo into the forming vesicles and specific membrane fusion with an acceptor organelle. The cytoplasm to vacuole targeting (Cvt) 1 pathway is * This work was supported by United States Public Health Service Grant GM53396 from the National Institutes of Health (to D. J. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. To whom correspondence should be addressed: Life Sciences Institute, University of Michigan, 210 Washtenaw Ave., Rm. 6036, Ann Arbor, MI Tel.: ; Fax: ; klionsky@umich.edu. 1 The abbreviations used are: Cvt, cytoplasm to vacuole targeting; PAS, pre-autophagosomal structure; GFP, green fluorescent protein; PI, phosphatidylinositol; YFP, yellow fluorescent protein; PE, phosphatidylethanolamine; Ape1, aminopeptidase I; Ams1, -mannosidase; prape1, precursor Ape1. This paper is available on line at an autophagy-related protein targeting pathway in yeast, whereby the resident vacuolar hydrolases, aminopeptidase I (Ape1) and -mannosidase (Ams1), are directly targeted from the cytoplasm to the vacuole. In this pathway, these two hydrolases are selectively enwrapped by double membrane-bound vesicles, termed Cvt vesicles, followed by fusion with the vacuolar membrane and a breakdown of the inner membrane structure to release precursor Ape1 (prape1) and Ams1 into the vacuolar lumen (1, 2). The process of the Cvt pathway and autophagy are topologically and mechanistically similar, even though they are different in their physiological functions (3 5). Autophagy is a cellular process responsible for the non-selective bulk degradation of cytoplasmic components in eukaryotic cells, which is induced in response to environmental cues such as nutrient starvation and hormonal stimuli (6, 7). In yeast, nutrient starvation induces the formation of autophagosomes, which are much larger ( nm in diameter) than Cvt vesicles (150 nm in diameter), for efficient transport of cytoplasm to the vacuole. On the other hand, the Cvt pathway is a constitutive biosynthetic pathway highly specific for prape1 and Ams1. The cargo specificity in the Cvt pathway is conferred by Atg19/Cvt19 that has been characterized as a cargo receptor (8). Recently, we elucidated the mechanism of cargo selection in the Cvt pathway (9). Two independent processes contributed to the selective incorporation of prape1 into the Cvt vesicle: the self-assembly of the prape1 complex, and its recruitment to the perivacuolar vesicle-forming site called the pre-autophagosomal structure (PAS). After synthesis of the prape1 polypeptide, it rapidly forms dodecamers in the cytosol (10), which further assemble into a higher order structure termed the Ape1 complex; assembly of the Ape1 complex is dependent on the prape1 propeptide. The Ape1 complex recruits Atg19 through the interaction between the propeptide of prape1 and a coiled-coil motif of Atg19. Atg11/Cvt9 then binds to the C terminus of Atg19 to target the cargo-receptor complex to the PAS, where the interactions between Atg19 and PAS components ensure the incorporation of the cargo complex into the Cvt vesicle (9). Unlike most receptors that cycle between donor and acceptor membranes, Atg19 is targeted to the vacuole together with cargo proteins and degraded there. The simultaneous binding of another cargo molecule, Ams1, to Atg19 results in an accumulation of Ams1 on the Ape1 complex, allowing an efficient transport of Ams1 to the vacuole. Interestingly, the lack of an Ape1 complex resulted in a dramatic decrease in the turnover of Atg19 as well as a decrease of Ams1 transport to the vacuole (8, 9), suggesting that the Ape1 complex facilitates the incorporation of an Atg19-Ams1 complex into the Cvt vesicles. Because the turnover of Atg19 is dependent on Cvt vesicle formation (8), these results raised a question as to whether the Cvt cargo could also induce Cvt vesicle formation or whether vesicle formation was constitutive and empty vesicles could be formed
2 29890 Cargo Proteins Facilitate Cvt Vesicle Formation TABLE I Yeast strains used in this study Strain Genotype Source SEY6210 MAT his3-200 leu2-3,112 lys2-801 trp1-901 ura3-52 suc2-9 GAL Ref. 34 FRY138 SEY6210; ATG9-YFP::HIS5 S.p. atg1 ::URA3 Ref. 27 TYY014 SEY6210; ATG20-YFP::HIS5 S.p. vps38 ::LEU2 K.l. atg11 ::URA3 K.l. This study YTS135 SEY6210; ATG9-YFP::HIS5 S.p. atg1 ::URA3 atg19 ::LEU2 K.l. This study YTS136 SEY6210; ATG9-YFP::HIS5 S.p. atg1 ::URA3 ape1 ::LEU2 This study YTS150 SEY6210; ATG9-YFP::HIS5 S.p. atg1 ::URA3 atg11 ::LEU2 K.l. This study YTS180 SEY6210; ATG20-YFP::HIS5 S.p. vps38 ::LEU2 K.l. This study YTS184 SEY6210; ATG20-YFP::HIS5 S.p. vps38 ::LEU2 K.l. atg19 ::URA3 K.l. This study YTS185 SEY6210; ATG20-YFP::HIS5 S.p. vps38 ::LEU2 K.l. ape1 ::TRP1 This study YTS187 SEY6210; URA3::GTP-ATG8 This study YTS188 SEY6210; atg1 ::HIS5 S.p. GFP-ATG8:: URA3 This study YTS189 SEY6210; pep4 :: LEU2 GFP-ATG8::URA3 This study YTS190 SEY6210; atg19 ::HIS5 S.p. GFP-ATG8::URA3 This study YTS191 SEY6210; ape1 ::LEU2 GFP-ATG8::URA3 This study YTS192 SEY6210; atg11 ::HIS3 GFP-ATG8::URA3 This study YTS193 SEY6210; atg2 ::HIS5 S.p. [GFP-ATG2 URA3] This study YTS194 SEY6210; atg2 ::HIS5 S.p. atg19 ::LEU2 K.l. [GFP-ATG2 URA3] This study YTS195 SEY6210; atg2 ::HIS5 S.p. ape1 ::LEU2 [GFP-ATG2 URA3] This study YTS196 SEY6210; atg2 ::HIS5 S.p. atg11 ::LEU2 K.l. GFP-ATG2 URA3 This study YTS197 SEY6210 GFP-ATG1 URA3 This study YTS198 SEY6210; atg19 ::HIS5 S.p. GFP-ATG1 URA3 This study YTS199 SEY6210; ape1 ::LEU2 GFP-ATG1 URA3 This study YTS200 SEY6210; atg11 ::HIS3 GFP-ATG1 URA3 This study YTS201 SEY6210; atg1 ::HIS5 S.p. URA3::GFP-ATG8 atg1 ts TRP1 This study YTS202 SEY6210; atg1 ::HIS5 S.p. atg19 ::LEU2 K.l. URA3::GFP-ATG8 atg1 ts TRP1 This study YTS203 SEY6210; atg1 ::HIS5 S.p. ape1 ::LEU2 URA3::GFP-ATG8 atg1 ts TRP1 This study YTS204 SEY6210; atg1 ::HIS5 S.p. atg11 ::LEU2 K.l. URA3::GFP-ATG8 atg1 ts TRP1 This study even without cargo proteins. The role of cargo proteins in facilitating or directing vesicle formation has been difficult to study because in most vesicular trafficking events the full range of cargo molecules have not been defined; in the Cvt pathway, prape1 appears to be the predominant cargo protein, and even Ams1 is a relatively minor constituent. Even though the Ape1 complex is selectively incorporated into autophagosomes under starvation conditions, neither Ape1 nor Atg19 is required for autophagy (8, 11), suggesting that autophagosome formation does not depend on the presence of specific cargo components. In this study, we show that the cargo-receptor complex facilitates the formation of Cvt vesicles but not autophagosomes. EXPERIMENTAL PROCEDURES Strains, Plasmids, and Media The Saccharomyces cerevisiae yeast strains used in this study are listed in Table I. A plasmid expressing GFP-Atg8 was based on prs306 containing the GFP-ATG8 gene with the endogenous ATG8 promoter (12) and used for yeast transformation to integrate GFP-ATG8 at the URA3 locus. The prs316 GFP-APG1 plasmid and YCplac33 GFP-APG2 (pts112) (13) were kind gifts from Dr. Yoshinori Ohsumi (National Institute for Basic Biology, Okazaki, Japan). The atg1 ts allele (12) was cloned on prs414. Yeast cells were grown in SCD medium (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% glucose) supplemented with 0.003% adenine, 0.005% tryptophan, and 0.002% uracil, if necessary. For nitrogen starvation, SD( N) medium (0.17% yeast nitrogen base without ammonium sulfate and amino acids and 2% glucose) was used. Transport of GFP-Atg8 to the Vacuole The strains harboring the atg1 ts allele were grown in SCD medium at 37 C overnight to A The cultures were divided into two tubes, and either rapamycin (0.2 g/ml) or the drug vehicle was added to each tube. After incubation at 37 C for 10 min, the tubes were placed at 30 C to activate the Cvt pathway or autophagy. At various time points, 1 ml of culture was harvested and used to prepare a protein extract. Protein extracts equivalent to A unit of yeast cells were subjected to SDS-PAGE and probed with anti-ape1 antiserum and anti-gfp antibody (Covance Research Products, Berkeley, CA). Fluorescence Microscopy Yeast cells expressing fluorescent proteinfused chimeras were grown in SCD medium to mid-log phase. For nitrogen starvation, the cells grown in SCD medium were washed with water twice and resuspended in SD( N) medium. The cells were observed with a fluorescence microscope as described previously (14). RESULTS Atg8 Can Be Used to Trace Cvt Vesicle Formation Atg19 is a receptor protein for the vacuolar hydrolases Ape1 and Ams1 in the Cvt pathway. It is transported to the vacuole together with cargo proteins and degraded within the vacuole lumen. Previously, we found that the turnover of Atg19 and the transport of Ams1 were severely affected by a lack of Ape1 (9). Because both Atg19 turnover and Ams1 transport are also dependent on the proper function of the Cvt pathway, we asked whether the Ape1 complex could induce the formation of Cvt vesicles or whether empty vesicles could be formed without the Ape1 complex. To analyze the effect of a lack of cargo complex on the formation of Cvt vesicles, we needed to develop a suitable marker protein. Precursor Ape1 is the standard marker used to monitor delivery via the Cvt and autophagy pathways; however, we were unable to use Ape1 as a marker because our goal was to examine vesicle formation in the absence of this specific cargo protein. Among the characterized Atg proteins, only Atg19 and Atg8/Aut7 remain associated with the completed Cvt vesicle/autophagosome. Atg19 was not suitable as a marker protein because it functions in cargo recognition. Accordingly, we used Atg8 as a marker of the Cvt pathway. Atg8 is essential for Cvt vesicle formation and for expansion of the autophagosome membrane (15 17). Similar to Atg19, Atg8 is delivered to the vacuole as a component of the Cvt vesicle/ autophagosome and is degraded after breakdown of the inner vesicle within the vacuole lumen. Accordingly, Cvt vesicle/autophagosome formation can be assessed by measuring the amount of Atg8 protein delivered to the vacuole. Using Atg8 to monitor vesicle formation, however, presented a significant problem for analyzing the Cvt pathway; Atg8 is synthesized at very low levels under vegetative conditions and is only induced upon starvation. Accordingly, it is problematic to monitor delivery of Atg8 under vegetative conditions through a radioactive pulse/chase analysis; the level of protein delivered to the vacuole is too low to allow an accurate quantification, and it is inherently difficult to follow the kinetics of vacuolar delivery by examining loss of a protein.
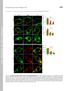
3 Cargo Proteins Facilitate Cvt Vesicle Formation FIG. 1. The Cvt cargo-receptor complex is required for efficient delivery of Atg8 to the vacuole. A, the generation of free GFP from GFP-Atg8 reflects delivery through the Cvt and autophagy pathways. The wild type, atg1, and pep4 cells expressing GFP-Atg8 (YTS187, YTS188, and YTS189, respectively) were grown in SCD (rich medium; growing conditions) to A and starved in SD( N) for 3 h. Cell lysates equivalent to A unit of cells were subjected to immunoblot analysis with anti-ape1 antiserum and anti-gfp antibody. B, delivery of Atg8 to the vacuole was blocked or extensively delayed in the absence of Ape1, Atg19, and Atg11. The atg1 ts, atg1 ts ape1, atg1 ts atg19, and atg1 ts atg11 cells expressing GFP-Atg8 (YTS201, YTS203, YTS202, and YTS204, respectively) were grown in SCD to A at a non-permissive temperature of 37 C. After treatment with or without 0.2 g/ml rapamycin (Rap) for 10 min at 37 C, the cells were transferred to a permissive temperature of 30 C and incubated for the indicated times. Ape1 and GFP-Atg8 were analyzed by immunoblotting as indicated in A. The positions of precursor and mature Ape1 (mape1) and of full-length GFP-Atg8 and free GFP are indicated. The asterisk marks a nonspecific cross-reacting band. Because of these technical difficulties, we designed an alternate strategy to monitor vesicle formation. GFP tagging of Atg8 has been used to trace the process of the Cvt pathway and autophagy (12, 18). Even though Atg8 is subject to degradation upon vacuolar delivery, the relative stability of GFP to the activity of vacuolar hydrolases led us to propose that the GFP moiety would accumulate in the vacuolar lumen as a consequence of the transport of GFP-Atg8 to the vacuole; the accumulated GFP could be easily monitored and would reflect delivery of Atg8. To confirm the suitability of this approach, we performed immunoblot analysis of GFP-Atg8 using anti-gfp antibody. In wild type cells expressing GFP-Atg8 grown in rich medium, we detected protein bands of 40 and 26 kda (Fig. 1A, bottom panel). These molecular masses correspond to the predicted sizes of full-length GFP-Atg8 and free GFP. As controls, we examined atg1 /agp1 and pep4 strains, which are defective in Cvt vesicle/autophagosome formation and the breakdown of intravacuolar vesicles, respectively. In contrast to the result seen in wild type cells, only full-length GFP-Atg8 was seen in the mutant strains (Fig. 1A, bottom panel). These results indicated that the generation of free GFP was dependent on the proper function of the Cvt pathway. When autophagy was induced by incubating the cells in starvation medium, the transport of GFP-Atg8 was enhanced in wild type cells, whereas again no liberation of GFP was observed in atg1 or pep4 mutant cells (Fig. 1A, bottom panel). These results support the idea that an increased level of Atg8 is required to form autophagosomes (17). An analysis of Ape1 from the same protein extracts verified that the mutant strains displayed the expected phenotypes with regard to prape1 processing (Fig. 1A, top panel). To allow a kinetic analysis of GFP-Atg8 transport, we took advantage of a conditional atg1 mutant. Cells harboring a temperature-sensitive allele of ATG1, atg1 ts, and expressing GFP-Atg8 were grown to mid-log phase at non-permissive temperature and then shifted to permissive temperature to activate the Cvt pathway, followed by immunoblot analysis with anti-ape1 antiserum and anti-gfp antibody. After shifting to the permissive temperature, prape1 was converted to the mature form (mape1) within 1 h, indicating that prape1 reached the vacuole immediately after the atg1 ts block was released (Fig. 1B, top left, top panel). Similarly, the conversion of GFP- Atg8 to GFP occurred within 1 h after incubation at 30 C, and accumulation of GFP gradually increased during the time course (Fig. 1B, top left, bottom panel). The addition of rapamycin to the culture, which mimics nutrient starvation conditions, resulted in an enhanced transport of GFP-Atg8 to the vacuole (Fig. 1B, top left, bottom panel). These results confirmed that this system is useful for a kinetic analysis of Cvt vesicle/autophagosome formation. Absence of the Cvt Cargo-Receptor Complex Impairs Cvt Vesicle but Not Autophagosome Formation To investigate whether the Cvt cargo complex is involved in Cvt vesicle formation, we carried out kinetic analyses using an atg1 ts ape1 double mutant strain expressing GFP-Atg8. After growth at 37 C, the cells were shifted to 30 C to activate the Cvt pathway. In the absence of rapamycin treatment, the generation of free GFP was severely delayed up to 4 h after incubation at 30 C, suggesting that prape1 is required for proper or efficient Cvt vesicle formation (Fig. 1B, bottom left, bottom panel). The receptor protein Atg19 is concentrated on the Ape1 complex and behaves like an adaptor protein to connect the Ape1 complex and the vesicle forming machinery; the Atg19-Ape1 complex is termed the Cvt complex. Thus, we hypothesized that Atg19 might be also important for Cvt vesicle formation. To test this, we carried out the same analysis with an atg1 ts atg19 strain. As expected, GFP-Atg8 transport was delayed in this strain and showed kinetics similar to that seen in the atg1 ts ape1 strain (Fig. 1B, top right, bottom panel). These results suggested that the cargo-receptor complex induces the formation of the Cvt vesicle rather than being incorporated within a constitutively forming vesicle. In contrast to the block in Cvt vesicle formation seen in rich medium, the deletion of either APE1 or ATG19 did not affect autophagosome formation when cells were treated with rapamycin to induce autophagy (Fig. 1B, bottom left and top right, bottom panels). This result was consistent with previous observations obtained by using autophagy-dependent activation of alkaline phosphatase activity (8, 11). It is believed that Cvt vesicles and autophagosomes are formed at the PAS, where most of the Atg proteins localize (11, 12, 19). The Cvt complex also localizes to this structure (9, 11). To examine whether proper localization of the cargo-receptor complex to the PAS is also important for vesicle formation, we constructed an atg1 ts atg11 /cvt9 strain expressing GFP-Atg8. Atg11/Cvt9 is a specific factor for the Cvt pathway and pexophagy (20) and is required to localize the cargo-receptor complex to the PAS in the Cvt pathway (9). In this strain, no transport of GFP-Atg8 was observed without rapamycin treatment, whereas with ra-
4 29892 Cargo Proteins Facilitate Cvt Vesicle Formation pamycin the transport of GFP-Atg8 was comparable with that of other strains including the wild type (Fig. 1B). This result suggested that cargo recruitment to the PAS was also important for Cvt vesicle formation but not for autophagosome formation. Examination of Ape1 again confirmed the expected phenotype of the various strains. In particular, prape1 was absent in the ape1 strain, showed normal maturation in the atg1 ts strain at the permissive temperature, accumulated in the absence of Atg19, and was partially matured in the atg11 strain only under starvation conditions (Fig. 1B, top panels). The Cvt Complex Is Important for PAS Organization under Growing Conditions The PAS is thought to be an organizing center for Cvt vesicle/autophagosome formation based on several lines of evidence: (i) most of the Atg proteins are located at the PAS under both growing and starvation conditions (12, 14, 19); (ii) the localization of some Atg proteins at the PAS is dependent on the function of other Atg proteins (12, 13, 18, 21); and (iii) GFP-Atg8 transiently localizes at the PAS and is then transported to the vacuole (12, 14). Accordingly, we decided to examine the effect of depletion of the cargo complex on the organization of the PAS. First, we examined the localization of GFP-Atg8 under growing conditions. In a wild type strain, 25% of the cells contained a single GFP-Atg8 punctate signal or a few GFP-Atg8 punctate signals at the PAS (Fig. 2). In contrast, the PAS localization of GFP-Atg8 was observed in only 6.0% and 2.5% of ape1 and atg19 cells, respectively (Fig. 2). The atg11 strain also showed a severe defect in GFP-Atg8 localization at the PAS; only 3.3% of the cells had punctate signals of GFP-Atg8 (Fig. 2). These results suggested that the cargoreceptor complex and its recruitment to the PAS were required for the proper localization of Atg8 at the PAS under growing conditions. On the other hand, these mutant strains did not show any difference in GFP-Atg8 localization compared with the wild type strain under starvation conditions; most of the cells displayed GFP staining in their vacuoles, and about 75 90% of the cells showed a punctate dot of GFP-Atg8 at the PAS (Fig. 2). This result was consistent with previous observations that Ape1, Atg19, and Atg11 are dispensable for autophagy (8, 11, 20) (Fig. 1) and suggested that the PAS localization of Atg8 occurred independently of the recognition of the Cvt cargo under starvation conditions. It was reported that the PAS localization of Atg8 is dependent on its lipidation state (12). Therefore, we examined the lipidation of Atg8 in ape1, atg19, and atg11 cells and found there was no difference in lipidation between the wild type and these mutant strains (data not shown), suggesting that Atg8 still localized on some membrane structure, although it was not concentrated at the PAS in these mutant cells. Because Atg8 physically interacts with Atg19 (9), we wondered whether only Atg8 lost its localization at the PAS or whether the overall organization of the PAS was compromised in the absence of the Cvt cargo. Recently, it was reported that the PAS localization of Atg2/Apg2 is important for its function in Cvt vesicle/ autophagosome formation (13, 22). Because its localization at the PAS is independent of Atg8, we selected Atg2 as a second PAS marker to examine the organization of the PAS. In rich medium, about 40% of wild type cells showed the PAS localization of GFP-Atg2, whereas it was decreased to 7% in ape1, atg19, and atg11 strains (Fig. 2). In contrast, the PAS localization of GFP-Atg2 in these strains was comparable with that in the wild type strain in a starvation medium (Fig. 2). A similar result was obtained with a GFP-Atg1 construct (Fig. 2); Atg1 is a serine/ threonine kinase that is required for both the Cvt pathway and autophagy (23). These results indicated that the cargo-dependent localization of Atg2 or Atg1 at the PAS was seen only under growing conditions; the deletion of APE1, ATG19, or ATG11 did FIG. 2.Quantification of cells containing pre-autophagosomal structures. The wild type, ape1, atg11, and atg19 cells expressing GFP-Atg8 (YTS187, YTS190, YTS191, and YTS192), GFP-Atg2 (YTS193, YTS194, YTS195, and YTS196), GFP-Atg1 (YTS197, YTS198, YTS199, and YTS200), or Atg20-YFP (YTS180, YTS184, YTS185, and TYY014) were grown in SCD (rich medium; growing conditions) to A and starved in SD( N) for 1 h. For each strain, cells containing fluorescent punctate dots were quantified by fluorescence microscopy. Error bars indicate the S.D. of three independent experiments. The chromosomal VPS38 gene was deleted in the cells expressing Atg20-YFP to eliminate punctate localization at the endosome (see text for details). Punctate localization of GFP-Atg8, GFP-Atg2, GFP- Atg1, and Atg20-YFP to the PAS was greatly diminished in the absence of the Cvt cargo-receptor complex under growing conditions, but not under starvation conditions. not result in any defect in the localization of GFP-Atg2 or GFP- Atg1 under starvation conditions. One of the key components that is thought to play an important role in the initial stage of PAS formation is the phosphatidylinositol (PI) 3-kinase complex that includes Vps34, Vps15, and Vps30/Atg6 (24). Function of the PI 3-kinase complex I, which includes Atg14/Apg14 and acts at the PAS, is required for recruitment of the Cvt pathway-specific factors Atg20/ Cvt20 and Atg24/Cvt13. These proteins localize at the PAS dependent on their phox homology domains that bind to PI 3-phosphate (25). The Atg20 and Atg24 proteins are also implicated in protein retrieval from the early endosome to the late Golgi and primarily localize at the endosome for this function (26). The PAS localization of Atg20 and Atg24 is affected by deleting ATG14 encoding a Cvt/autophagy pathway-specific subunit of PI 3-kinase complex I, whereas their endosomal localization is lost by deletion of VPS38 encoding a carboxypeptidase Y pathway-specific subunit of PI 3-kinase complex II (25, 26). We decided to extend our analysis by examining the role of the Cvt cargo and cargo packaging machinery in the localization of factors dependent on PI 3-kinase function. Accordingly, we decided to use a vps38 strain to analyze the Cvt pathwayspecific localization of Atg20. First, we confirmed that the
5 Cargo Proteins Facilitate Cvt Vesicle Formation FIG. 3.Atg9 cycling requires the Cvt cargo-receptor complex under growing conditions. The atg1, atg1 ape1, and atg1 atg19 cells expressing Atg9-YFP (FRY138, YTS136, and YTS135, respectively) were grown in SCD (rich medium; growing conditions) and then shifted to SD( N) (starvation conditions) and observed by fluorescence microscopy. Atg9 normally transits to the PAS and is retrieved from this site in an Atg1-dependent manner. Delivery to the PAS was defective in the absence of the Cvt cargo-receptor complex. punctate dots of Atg20-YFP observed in vps38 cells colocalized with CFP-Atg8 and CFP-Ape1 as markers of the PAS (data not shown). About 22% of the vps38 cells contained dot structures of Atg20-YFP corresponding to the PAS under growing conditions (Fig. 2). Similar to other PAS proteins, the PAS localization of Atg20-YFP was dramatically decreased in vps38 ape1, vps38 atg19, and vps38 atg11 strains under growing conditions (Fig. 2). Interestingly, Atg20-YFP still localized at the PAS under starvation conditions, even though Atg20 is not required for autophagy; more than 70% of the vps38 cells possessed the PAS dots of Atg20-YFP after incubation in a starvation medium (Fig. 2). The additional deletion of APE1, ATG19, or ATG11 had essentially no affect on the PAS localization of Atg20-YFP under autophagy conditions (Fig. 2). Taken together with the observations for other Atg proteins, these results suggested that the cargo-receptor complex might be important for the overall organization of the PAS rather than for the localization of an individual Atg protein at the PAS under growing conditions. Recently, it has been reported that the Atg1-Atg13/Apg13 protein kinase complex is essential for cycling of the transmembrane protein Atg9/Apg9 between the PAS and other cytosolic punctate structures (27). Atg9 localizes to the PAS and the other cytosolic punctate structures in wild type cells (12), whereas its localization is restricted at the PAS in atg1 or atg13 cells (27), suggesting that the Atg1-Atg13 protein kinase complex regulates the retrieval transport of Atg9 from the PAS. Accordingly, we examined whether this restriction of Atg9 localization at the PAS in atg1 cells was perturbed by a lack of the Cvt complex. In atg1 cells, Atg9-YFP was restricted at the PAS as observed previously (27) (Fig. 3). The additional deletion of APE1 or ATG19 resulted in a scattered localization of Atg9-YFP (Fig. 3), suggesting that the cargoreceptor complex is required for the sorting of Atg9 to the PAS under nutrient-rich conditions. After incubating atg1 ape1 or atg1 atg19 cells in starvation medium for 1 h, Atg9-YFP accumulated as a prominent dot, although several tiny dots were still observed (Fig. 3). DISCUSSION Selective incorporation of cargo proteins into the forming vesicle is an important aspect of protein targeting via vesicular trafficking. According to the paradigm of cargo selection in vesicular transport, the association of cargo proteins with coat proteins (a key component in the vesicle forming machinery) leads to their concentration at the vesicle budding site in the donor organelle (28). We have investigated the mechanism of cargo selection in the Cvt pathway that is topologically and mechanistically similar to autophagy (8, 9). Unlike the situation in conventional vesicular transport, the Cvt cargo proteins are concentrated as a huge complex, the Cvt complex, which is composed of the resident vacuolar hydrolases Ape1 and Ams1 and the receptor protein Atg19, in a process independent of the vesicle formation machinery, before they are incorporated into a double membrane Cvt vesicle (9). In this study, we propose an unusual concept for cargo selection in vesicular transport: the cargo proteins facilitate the process of vesicle formation rather than concentration of the cargo and incorporation into the forming vesicle through the action of the vesicle forming machinery. In wild type cells, at least some population of most of the Atg proteins is present at the pre-autophagosomal structure. In contrast, in the absence of Ape1, Atg19, and Atg11, there is nearly a complete block in PAS formation based on fluorescence microscopy of tagged proteins. The best-characterized marker protein that appears to be a structural component of the PAS and the resulting sequestering vesicle is Atg8 conjugated to phosphatidylethanolamine (PE). In most strains bearing mutations in components of the vesicle forming machinery, Atg8-PE is typically localized to the PAS; in wild type cells, Atg8-PE is additionally delivered to the vacuole and degraded. Atg8 is induced by starvation but is present at relatively low levels under vegetative conditions. Accordingly, it is difficult to quantify Atg8-PE delivery to the vacuole via the Cvt pathway. We developed a method to monitor delivery of Atg8-PE to the vacuole to allow quantification of Cvt vesicle formation. Our approach relied on the stability of GFP and the instability of Atg8 within the vacuole lumen. We used a GFP-Atg8 fusion to monitor release of free GFP after vacuolar delivery. The appearance of free GFP was dependent on vacuolar hydrolase activity and Atg/Cvt proteins, indicating that it reflected the normal Cvt and autophagy pathways (Fig. 1). The assembly of the Cvt complex and its recruitment to the priming site of Cvt vesicle/autophagosome formation led to the convergence of Atg proteins at this site to organize the PAS under growing conditions (Figs. 2 and 3). Therefore, we concluded that the pre-concentrated cargo complex might attract the vesicle forming machinery to it causing it to be surrounded by the Cvt vesicle. This model provides a reasonable explanation as to why the Cvt vesicle completely excludes cytoplasmic material (4); that is, the Cvt vesicle does not randomly engulf cytosol but rather nucleates around the cargo complex. It seems that a membrane structure is also enriched on the cargo complex together with Atg proteins because the sorting of the transmembrane protein Atg9 (29) to the PAS was also dependent on the cargo complex (Fig. 3). Atg8-PE also behaves like an integral membrane protein (16, 30), and its PAS localization is dependent on its lipidation (12, 18). Although the state of Atg8 lipidation in ape1, atg19, oratg11 strains was comparable to that in the wild type strain (data not shown), there was less PAS localization in these mutants, suggesting that Atg8-PE was localized on some membrane structure other than the PAS. This unidentified membranous structure could contain other Atg proteins and become enriched on the cargo complex in a process mediated by Atg11; Atg11 connects the Cvt complex and the Atg machinery (9, 23). With regard to the source of membrane for the Cvt vesicle and autophagosome, there have been arguments for the requirement of the endoplasmic reticulum (1, 31, 32). Recently, it has been reported that the early stage of the secretory pathway (endoplasmic reticulum and/or Golgi complex) is required for both Cvt vesicle and autophagosome formation (33). Overall, these studies suggest that the
6 29894 Cargo Proteins Facilitate Cvt Vesicle Formation Atg/Cvt protein-localizing membrane structure might be supplied from the early secretory pathway. In contrast to the case under growing conditions, the assembly of the Cvt complex is not required for the formation of the autophagosome (8, 11) (Fig. 1) or the proper organization of the PAS under conditions of nutrient starvation (Fig. 2). These results suggest that autophagy uses a mechanism different from the Cvt pathway to organize the PAS, and the details of this mechanism remain to be solved. The deletion of ATG11 resulted in a more severe defect in Cvt vesicle formation than that seen with deletions of APE1 or ATG19 (Fig. 1), presumably because Atg11 functions in another transport pathway other than the Cvt pathway. It is known that excess peroxisomes are selectively degraded by autophagy, or pexophagy, in an Atg11-dependent manner (20). According to its function in the Cvt pathway, Atg11 might act as an adaptor protein for peroxisomes that is required for their selective degradation. Even under normal growth conditions, a basal level of peroxisome degradation could occur to maintain normal levels of peroxisomes or to remove damaged peroxisomes, which might lead to the formation of vesicles containing Atg8 to sequester this organelle. Therefore, the delivery of GFP-Atg8 was completely blocked in atg11 cells in which both the Cvt pathway and pexophagy are impaired (Fig. 1). Autophagy is an inducible pathway, whereas the Cvt pathway is considered to be constitutive, but what does this mean with regard to vesicle formation? In our study, we have focused on the connection between the cargo and the formation of the sequestering vesicle in the Cvt pathway. The Cvt pathway represents a unique type of vesicle-mediated trafficking process in that the cargo forms a complex independent of its receptor, and the cargo-receptor complex induces the formation of the sequestering vesicle. There must be an additional level of control, however, during the vesicle formation process. When precursor Ape1 is overexpressed, the Cvt complex is enlarged, and a portion of the protein accumulates in the cytosol (1, 4); the vesicle forming machinery is apparently unable to completely enwrap the enlarged complex, suggesting that the membrane does not simply wrap around the Cvt complex that acts as a scaffold. It is possible that the limited supply of membrane and/or Atg proteins that is available to the Cvt pathway under vegetative conditions is not sufficient to enwrap the larger Cvt complex. Cvt vesicles are of a carefully defined size relative to autophagosomes. Whereas transport vesicles that bud off from pre-existing organelles are also of strictly defined sizes, this is presumably due to constraints imposed on vesicle curvature by coat components. The action of a transient coat has not yet been established for the Cvt and autophagy pathways. Further work will be needed to determine additional details about the process of sequestering vesicle formation in these pathways. Acknowledgments We thank Dr. Yoshinori Ohsumi for providing plasmid prs316 GFP-Apg1 and pts112 and Drs. Fulvio Reggiori and Per E. Stromhaug for helpful discussions. REFERENCES 1. Klionsky, D. J., Cueva, R., and Yaver, D. S. (1992) J. Cell Biol. 119, Hutchins, M. U., and Klionsky, D. J. (2001) J. Biol. Chem. 276, Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992) J. Cell Biol. 119, Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J., and Ohsumi, Y. (1997) J. Cell Biol. 139, Scott, S. V., Baba, M., Ohsumi, Y., and Klionsky, D. J. (1997) J. Cell Biol. 138, Klionsky, D. J., and Ohsumi, Y. (1999) Annu. Rev. Cell Dev. Biol. 15, Klionsky, D. J., and Emr, S. D. (2000) Science 290, Scott, S. V., Guan, J., Hutchins, M. U., Kim, J., and Klionsky, D. J. (2001) Mol. Cell 7, Shintani, T., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002) Dev. Cell 3, Kim, J., Scott, S. V., Oda, M. N., and Klionsky, D. J. (1997) J. Cell Biol. 137, Suzuki, K., Kamada, Y., and Ohsumi, Y. (2002) Dev. Cell 3, Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001) EMBO J. 20, Shintani, T., Suzuki, K., Kamada, Y., Noda, T., and Ohsumi, Y. (2001) J. Biol. Chem. 276, Kim, J., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002) J. Biol. Chem. 277, Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999) J. Cell Biol. 147, Huang, W.-P., Scott, S. V., Kim, J., and Klionsky, D. J. (2000) J. Biol. Chem. 275, Abeliovich, H., Dunn, W. A., Jr., Kim, J., and Klionsky, D. J. (2000) J. Cell Biol. 151, Kim, J., Huang, W.-P., and Klionsky, D. J. (2001) J. Cell Biol. 152, Noda, T., Suzuki, K., and Ohsumi, Y. (2002) Trends Cell Biol. 12, Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr., and Klionsky, D. J. (2001) J. Cell Biol. 153, Guan, J., Stromhaug, P. E., George, M. D., Habibzadegah-Tari, P., Bevan, A., Dunn, W. A., Jr., and Klionsky, D. J. (2001) Mol. Biol. Cell 12, Wang, C.-W., Kim, J., Huang, W.-P., Abeliovich, H., Stromhaug, P. E., Dunn, W. A., Jr., and Klionsky, D. J. (2001) J. Biol. Chem. 276, Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000) J. Cell Biol. 150, Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001) J. Cell Biol. 152, Nice, D. C., Sato, T. K., Stromhaug, P. E., Emr, S. D., and Klionsky, D. J. (2002) J. Biol. Chem. 277, Hettema, E. H., Lewis, M. J., Black, M. W., and Pelham, H. R.B. (2003) EMBO J. 22, Reggiori, F., Tucker, K. A., Stromhaug, P. E., and Klionsky, D. J. (2004) Dev. Cell 6, Bonifacino, J. S., and Glick, B. S. (2004) Cell 116, Noda, T., Kim, J., Huang, W.-P., Baba, M., Tokunaga, C., Ohsumi, Y., and Klionsky, D. J. (2000) J. Cell Biol. 148, Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Takao, T., Noda, T., and Ohsumi, Y. (2000) J. Cell Biol. 151, Ishihara, N., Hamasaki, M., Yokota, S., Suzuki, K., Kamada, Y., Kihara, A., Yoshimori, T., Noda, T., and Ohsumi, Y. (2001) Mol. Biol. Cell 12, Hamasaki, M., Noda, T., and Ohsumi, Y. (2003) Cell Struct. Funct. 28, Reggiori, F., Wang, C.-W., Nair, U., Shintani, T., Abeliovich, H., and Klionsky, D. J. (2004) Mol. Biol. Cell 15, Robinson, J. S., Klionsky, D. J., Banta, L. M., and Emr, S. D. (1988) Mol. Cell. Biol. 8,
Autophagy induction [h]
![Autophagy induction [h] Autophagy induction [h]](/thumbs/80/80564099.jpg) A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
A SD-N [h]: SD-N [h]: 1 2 4 1 2 4 1 2 4 -GFP 1 2 4 percentage GFP-Atg8 [%] 1 9 8 7 6 5 4 3 2 1 1 2 4 Autophagy induction [h] -13xmyc SD-N [h]: 1 2 4 -GFP -13xmyc 1 prape1 mape1 - - GFP - 13xmyc mape1 [%]
Macroautophagy is a catabolic pathway used
 JCB: Article The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy Wei-Lien Yen, 1,2 Takahiro Shintani, 4 Usha Nair, 1,2 Yang Cao, 1,2 Brian C. Richardson,
JCB: Article The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy Wei-Lien Yen, 1,2 Takahiro Shintani, 4 Usha Nair, 1,2 Yang Cao, 1,2 Brian C. Richardson,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
Supplemental material JCB Tanaka et al., http://www.jcb.org/cgi/content/full/jcb.201402128/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nonselective autophagy is normal in Hrr25-depleted
The yeast two-hybrid assay
 Master program Molecular and Cellular Life Sciences Block 1: Intracellular membrane processes 11th October 2011 The yeast two-hybrid assay Fulvio Reggiori Department of Cell Biology, UMC Utrecht For what
Master program Molecular and Cellular Life Sciences Block 1: Intracellular membrane processes 11th October 2011 The yeast two-hybrid assay Fulvio Reggiori Department of Cell Biology, UMC Utrecht For what
Practice Problems 5. Location of LSA-GFP fluorescence
 Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
Life Sciences 1a Practice Problems 5 1. Soluble proteins that are normally secreted from the cell into the extracellular environment must go through a series of steps referred to as the secretory pathway.
Cytoplasm-to-vacuole targeting and autophagy employ the same
 Proc. Natl. Acad. Sci. USA Vol. 93, pp. 1234-1238, October 1996 Cell Biology Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole (aminopeptidase
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 1234-1238, October 1996 Cell Biology Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole (aminopeptidase
P52A/R67A. The plasmids expressing AD-fused Atg1 variants were constructed as follows: DNA
 SUPPLEMENTAL DATA The Autophagy-Related Protein Kinase Atg1 Interacts with the Ubiquitin-Like Protein Atg8 via the Atg8 Family Interacting Motif to Facilitate Autophagosome Formation Hitoshi Nakatogawa,
SUPPLEMENTAL DATA The Autophagy-Related Protein Kinase Atg1 Interacts with the Ubiquitin-Like Protein Atg8 via the Atg8 Family Interacting Motif to Facilitate Autophagosome Formation Hitoshi Nakatogawa,
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
Supplemental Material THE JOURNAL OF CELL BIOLOGY Yeung et al., http://www.jcb.org/cgi/content/full/jcb.200903020/dc1 Figure S1. Assessment of the surface charge of maturing phagosomes. (A C and F H) RAW
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Karotki et al., http://www.jcb.org/cgi/content/full/jcb.201104040/dc1 Figure S1. Eisosome proteins form highly stable filaments of variable
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Karotki et al., http://www.jcb.org/cgi/content/full/jcb.201104040/dc1 Figure S1. Eisosome proteins form highly stable filaments of variable
PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF
 YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
YFP-PHF1 CFP-PHT1;2 PHT1;2-CFP YFP-PHF + PHT1;2-CFP YFP-PHF + CFP-PHT1;2 Negative control!-gfp Supplemental Figure 1: PHT1;2 accumulation is PHF1 dependent. Immunoblot analysis on total protein extract
supplementary information
 DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
DOI: 10.1038/ncb2007 Figure S1 Temperature sensitivity of DNA ligase I alleles. a, SSL204 (CDC9), SSL612 (cdc9-1) and SSL613 (cdc9-2) cells were spotted in ten-fold serial dilutions on YPD plates and incubated
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
DOI: 10.1038/ncb2386 Figure 1 Src-containing puncta are not focal adhesions, podosomes or endosomes. (a) FAK-/- were stained with anti-py416 Src (green) and either (in red) the focal adhesion protein paxillin,
Chapter One. Construction of a Fluorescent α5 Subunit. Elucidation of the unique contribution of the α5 subunit is complicated by several factors
 4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
4 Chapter One Construction of a Fluorescent α5 Subunit The significance of the α5 containing nachr receptor (α5* receptor) has been a challenging question for researchers since its characterization by
Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product.
 Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Supplementary Information Supplementary Figure S1. Immunodetection of full-length XA21 and the XA21 C-terminal cleavage product. Total protein extracted from Kitaake wild type and rice plants carrying
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
Supplementary Figure 1 Impaired ITS2 processing results in mislocalization of foot-factors to the cytoplasm. (a) Schematic representation of ITS2 processing. The endonuclease Las1 initiates ITS2 processing
7.06 Problem Set #3, 2006
 7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
7.06 Problem Set #3, 2006 1. You are studying the EGF/Ras/MAPK pathway in cultured cells. When the pathway is activated, cells are signaled to proliferate. You generate various mutants described below.
Supporting Information
 Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
Supporting Information Üstün and Börnke, 2015 Figure S1: XopJ does not display acetyltransferase activity. Autoacetylation activity in vitro. Acetylation reactions using MBP, MBP-XopJ, GST and GST-HopZ1a
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Supplemental material Thompson et al., http://www.jcb.org/cgi/content/full/jcb.200909067/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Modification-specific antibodies do not detect unmodified
Regulation of autophagic activity by ζ proteins associated with class III. phosphatidylinositol-3 kinase. Mercedes Pozuelo Rubio
 ONLINE SUPPORTING INFORMATION Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3 kinase Mercedes Pozuelo Rubio entro Andaluz de Biología Molecular y
ONLINE SUPPORTING INFORMATION Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3 kinase Mercedes Pozuelo Rubio entro Andaluz de Biología Molecular y
Stargazin regulates AMPA receptor trafficking through adaptor protein. complexes during long term depression
 Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Supplementary Information Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long term depression Shinji Matsuda, Wataru Kakegawa, Timotheus Budisantoso, Toshihiro Nomura,
Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast
 Manuscript EMBO-2014-89083 Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast Hanghang Huang, Tomoko Kawamata, Tetsuro Horie, Hiroshi Tsugawa, Yasumune Nakayama, Yoshinori Ohsumi, Eiichiro
Manuscript EMBO-2014-89083 Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast Hanghang Huang, Tomoko Kawamata, Tetsuro Horie, Hiroshi Tsugawa, Yasumune Nakayama, Yoshinori Ohsumi, Eiichiro
SUPPLEMENTARY INFORMATION
 Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Supplementary Table 1 Components of the RAD6 damage tolerance pathway in yeast and humans S. cerevisiae Homo sapiens Functions Rad6 HHR6A and HHR6B Ubiquitin conjugating enzyme (E2) Rad18 RAD18 Ubiquitin
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Figure S2. Verification of ihog and boi
 Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Figure S1. Verification of ihog Mutation by Protein Immunoblotting Extracts from S2R+ cells, embryos, and adults were analyzed by immunoprecipitation and immunoblotting with anti-ihog antibody. The Ihog
Supplemental Data. Farmer et al. (2010) Plant Cell /tpc
 Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplemental Figure 1. Amino acid sequence comparison of RAD23 proteins. Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. The sequence of plant Ub is
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
www.sciencesignaling.org/cgi/content/full/9/429/ra54/dc1 Supplementary Materials for Dephosphorylation of the adaptor LAT and phospholipase C by SHP-1 inhibits natural killer cell cytotoxicity Omri Matalon,
Aminoacid change in chromophore. PIN3::PIN3-GFP GFP S65 (no change) (5.9) (Kneen et al., 1998)
 Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Table. Table S1. Fluorophore characteristics of used fluorescent proteins, Related to Figure 2 and 3. Transgenic line Fluprescent marker Aminoacid change in chromophore pk(a) Ref. PIN3::PIN3-GFP
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15
 Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Supplemental Fig. 1. Mcr alleles show defects in tracheal tube size and luminal protein accumulation. (A-F) Confocal projections of living stage 15 embryos expressing GFP and Verm-RFP in tracheal cells
Autophagy/Cytotoxicity Dual Staining Kit
 Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Supplemental Information. Pacer Mediates the Function of Class III PI3K. and HOPS Complexes in Autophagosome. Maturation by Engaging Stx17
 Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Molecular Cell, Volume 65 Supplemental Information Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17 Xiawei Cheng, Xiuling Ma, Xianming Ding,
Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana.
 SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
SUPPLEMENTARY FIGURES Figure S1. Immunoblotting showing proper expression of tagged R and AVR proteins in N. benthamiana. YFP:, :CFP, HA: and (A) and HA, CFP and YFPtagged and AVR1-CO39 (B) were expressed
Coleman et al., Supplementary Figure 1
 Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
Coleman et al., Supplementary Figure 1 BrdU Merge G1 Early S Mid S Supplementary Figure 1. Sequential destruction of CRL4 Cdt2 targets during the G1/S transition. HCT116 cells were synchronized by sequential
ab Autophagy/ Cytotoxicity Dual Staining Kit
 ab133075 Autophagy/ Cytotoxicity Dual Staining Kit Instructions for Use For the study of the regulation of autophagy and cytotoxicity at the cellular level. This product is for research use only and is
ab133075 Autophagy/ Cytotoxicity Dual Staining Kit Instructions for Use For the study of the regulation of autophagy and cytotoxicity at the cellular level. This product is for research use only and is
3.1 The Role of the Enzymatic Activity of Sir2 for Efficient. The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and
 35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
35 3 RESULTS 3.1 The Role of the Enzymatic Activity of Sir2 for Efficient Association of the SIR Complex with DNA The Sir proteins are recruited to DNA by site-specific DNA-binding proteins and subsequently
Supporting information
 Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
Supporting information Construction of strains and plasmids To create ptc67, a PCR product obtained with primers cc2570-162f (gcatgggcaagcttgaggacggcgtcatgt) and cc2570+512f (gaggccgtggtaccatagaggcgggcg),
JCB. Supplemental material THE JOURNAL OF CELL BIOLOGY. Hong et al.,
 Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplemental material JCB Hong et al., http://www.jcb.org/cgi/content/full/jcb.201412127/dc1 THE JOURNAL OF CELL BIOLOGY Figure S1. Analysis of purified proteins by SDS-PAGE and pull-down assays. (A) Coomassie-stained
Supplementary Figure S1. N-terminal fragments of LRRK1 bind to Grb2.
 Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
Myc- HA-Grb2 Mr(K) 105 IP HA 75 25 105 1-1163 1-595 - + - + - + 1164-1989 Blot Myc HA total lysate 75 25 Myc HA Supplementary Figure S1. N-terminal fragments of bind to Grb2. COS7 cells were cotransfected
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr.
 MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
MIT Department of Biology 7.01: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel iv) Would Xba I be useful for cloning? Why or why not?
Confirming the Phenotypes of E. coli Strains
 Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
Confirming the Phenotypes of E. coli Strains INTRODUCTION Before undertaking any experiments, we need to confirm that the phenotypes of the E. coli strains we intend to use in the planned experiments correspond
Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1.
 Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Supplemental Online Material: Materials and Methods: All strains were derivatives of SK1 and are listed in Supplemental Table 1. Constructs: pclb2-3ha-cdc5 and pclb2-3ha-cdc2 strains were constructed by
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
Supplementary Figure 1 Detection of MCM-subunit SUMOylation under normal growth conditions. a. Sumoylated forms of MCM subunits show differential shifts when SUMO is attached to differently sized tags.
7.012 Problem Set 5. Question 1
 Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
Name Section 7.012 Problem Set 5 Question 1 While studying the problem of infertility, you attempt to isolate a hypothetical rabbit gene that accounts for the prolific reproduction of rabbits. After much
SUPPLEMENTARY MATERIALS
 SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
SUPPLEMENTARY MATERIALS SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Condensin is required for condensation of rdna array during starvation (A) Loss of condensin blocks rdna condensation during
Sarker et al. Supplementary Material. Subcellular Fractionation
 Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Supplementary Material Subcellular Fractionation Transfected 293T cells were harvested with phosphate buffered saline (PBS) and centrifuged at 2000 rpm (500g) for 3 min. The pellet was washed, re-centrifuged
Practice Exam A. Briefly describe how IL-25 treatment might be able to help this responder subgroup of liver cancer patients.
 Practice Exam 2007 1. A special JAK-STAT signaling system (JAK5-STAT5) was recently identified in which a gene called TS5 becomes selectively transcribed and expressed in the liver upon induction by a
Practice Exam 2007 1. A special JAK-STAT signaling system (JAK5-STAT5) was recently identified in which a gene called TS5 becomes selectively transcribed and expressed in the liver upon induction by a
supplementary information
 DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
DOI: 10.1038/ncb2172 Figure S1 p53 regulates cellular NADPH and lipid levels via inhibition of G6PD. (a) U2OS cells stably expressing p53 shrna or a control shrna were transfected with control sirna or
Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of
 Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Supplementary data Fig. S1. CrgA intracellular levels in M. tuberculosis. Ten and twenty micrograms of cell free protein lysates from WT M. tuberculosis (Rv) together with various known concentrations
Gene disruption and construction of C-terminally tagged strains
 Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Supplementary information Supplementary Methods Gene disruption and construction of C-terminally tagged strains A PCR-based gene targeting method (Bähler et al., 1998; Sato et al., 2005) was used for constructing
Aoki et al.,
 JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
JCB: SUPPLEMENTAL MATERIAL Aoki et al., http://www.jcb.org/cgi/content/full/jcb.200609017/dc1 Supplemental materials and methods Calculation of the raw number of translocated proteins First, the average
Supplemental Data. Aung et al. (2011). Plant Cell /tpc
 35S pro:pmd1-yfp 10 µm Supplemental Figure 1. -terminal YFP fusion of PMD1 (PMD1-YFP, in green) is localized to the cytosol. grobacterium cells harboring 35S pro :PMD1-YFP were infiltrated into tobacco
35S pro:pmd1-yfp 10 µm Supplemental Figure 1. -terminal YFP fusion of PMD1 (PMD1-YFP, in green) is localized to the cytosol. grobacterium cells harboring 35S pro :PMD1-YFP were infiltrated into tobacco
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the
 Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Supplementary Fig. 1. Schematic structure of TRAIP and RAP80. The prey line below TRAIP indicates bait and the two lines above RAP80 highlight the prey clones identified in the yeast two hybrid screen.
Table S1 Yeast strains used in this study Strain Genotype JSY7452* MAT ade2-1 leu2-3 his3-11,15 trp1-1 ura3-1 can1-100 JSY7453* MAT ade2-1 leu2-3
 Table S1 Yeast strains used in this study Strain Genotype JSY7452* MAT ade2-1 leu2-3 his3-11,15 trp1-1 ura3-1 can1-100 JSY7453* MAT ade2-1 leu2-3 his3-11,15 trp1-1 ura3-1 can1-100 mfb1::his3 JSY8272 MAT
Table S1 Yeast strains used in this study Strain Genotype JSY7452* MAT ade2-1 leu2-3 his3-11,15 trp1-1 ura3-1 can1-100 JSY7453* MAT ade2-1 leu2-3 his3-11,15 trp1-1 ura3-1 can1-100 mfb1::his3 JSY8272 MAT
Autophagy/Cytotoxicity Dual Staining Kit
 Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Autophagy/Cytotoxicity Dual Staining Kit Catalog Number KA1299 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking
 Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking Chi Wai Lee, Jianzhong Han, James R. Bamburg, Liang Han, Rachel Lynn, and James Q. Zheng Supplementary Figures
Saccharomyces cerevisiae. haploid =
 In this lecture we are going to consider experiments on yeast, a very useful organism for genetic study. Yeast is more properly known as Saccharomyces cerevisiae, which is the single-celled microbe used
In this lecture we are going to consider experiments on yeast, a very useful organism for genetic study. Yeast is more properly known as Saccharomyces cerevisiae, which is the single-celled microbe used
Rapid Learning Center Presents. Teach Yourself AP Biology in 24 Hours
 Rapid Learning Center Chemistry :: Biology :: Physics :: Math Rapid Learning Center Presents Teach Yourself AP Biology in 24 Hours 1/35 *AP is a registered trademark of the College Board, which does not
Rapid Learning Center Chemistry :: Biology :: Physics :: Math Rapid Learning Center Presents Teach Yourself AP Biology in 24 Hours 1/35 *AP is a registered trademark of the College Board, which does not
Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction
 Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Journal of Alzheimer s Disease 20 (2010) 1 9 1 IOS Press Supplementary Material Grb2-Mediated Alteration in the Trafficking of AβPP: Insights from Grb2-AICD Interaction Mithu Raychaudhuri and Debashis
Nature Structural & Molecular Biology: doi: /nsmb.1583
 Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Acetylation by GCN5 regulates CDC6 phosphorylation in the S-phase of the cell cycle Roberta Paolinelli 1,2, Ramiro Mendoza-Maldonado 2, Anna Cereseto 1 and Mauro Giacca 2 1 Molecular Biology Laboratory,
Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous
 Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Preparation and purification of polyclonal antibodies Polyclonal ARHGAP25 antibody was prepared from rabbit serum after intracutaneous injections of glutathione S-transferase-ARHGAP25-(509-619) (GST-coiled
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or
 Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure 1: TDP-43 is subject to lysine acetylation within the RNA-binding domain a) QBI-293 cells were transfected with TDP-43 in the presence or absence of the acetyltransferase CBP and acetylated TDP-43
Figure S1 early log mid log late log stat.
 Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which
Figure S1 early log mid log late log stat. C D E F Fig. S1. The chromosome-expressed EI localizes to the cell poles independent of the growth phase and growth media. MG1655 (ptsi-mcherry) cells, which
Supplementary Figure h HO -GFP-Atg h HO -GFP-Atg8. 8 Breaks. 1 Break - GFP -GFP. No Breaks.
 A 1 Break 0 6 12 18 24h HO 8 Breaks 0 6 12 15 18 24h HO - GFP No Breaks C rad9 0 1 2 3 4 h - GFP B mec1δ WT mec1δ tel1δ tel1δ Rapamycin Normalized GFP ratio 1.2 1 0.8 0.6 0.4 0.2 0 D WT rad53δ chk1δ rad53δ
A 1 Break 0 6 12 18 24h HO 8 Breaks 0 6 12 15 18 24h HO - GFP No Breaks C rad9 0 1 2 3 4 h - GFP B mec1δ WT mec1δ tel1δ tel1δ Rapamycin Normalized GFP ratio 1.2 1 0.8 0.6 0.4 0.2 0 D WT rad53δ chk1δ rad53δ
Figure S1, related to Figure 1. Characterization of biosensor behavior in vivo.
 Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
Developmental Cell, Volume 23 Supplemental Information Separase Biosensor Reveals that Cohesin Cleavage Timing Depends on Phosphatase PP2A Cdc55 Regulation Gilad Yaakov, Kurt Thorn, and David O. Morgan
c Nup82-TAP purification
 a Dbp5-TAP Nup159-TAP b Dbp5-TAP Dbp5-TAP/dyn2Δ Nup116 Dbp5 Nup159 Nsp1 Nup82 Nup159 Nup116 Nsp1 Nup82 Dbp5 # # 1 2 1 2 Western c Nup82-TAP purification input flow through pull-down 1 2 3 α-nup82 1 2 3
a Dbp5-TAP Nup159-TAP b Dbp5-TAP Dbp5-TAP/dyn2Δ Nup116 Dbp5 Nup159 Nsp1 Nup82 Nup159 Nup116 Nsp1 Nup82 Dbp5 # # 1 2 1 2 Western c Nup82-TAP purification input flow through pull-down 1 2 3 α-nup82 1 2 3
Section 10. Junaid Malek, M.D.
 Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Section 10 Junaid Malek, M.D. Cell Division Make sure you understand: How do cells know when to divide? (What drives the cell cycle? Why is it important to regulate this?) How is DNA replication regulated?
Figure S1 is related to Figure 1B, showing more details of outer segment of
 Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Information Supplementary Figure legends and Figures Figure S1. Electron microscopic images in Sema4A +/+ and Sema4A / retinas Figure S1 is related to Figure 1B, showing more details of outer
Supplemental Materials and Methods. Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2,
 Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
Supplemental Materials and Methods Yeast strains. Strain genotypes are listed in Table S1. To generate MATainc::LEU2, the LEU2 marker was inserted by PCR-mediated recombination (BRACHMANN et al. 1998)
No wash 2 Washes 2 Days ** ** IgG-bead phagocytosis (%)
 Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Figures Supplementary Figure 1. No wash 2 Washes 2 Days Tat Control ** ** 2 4 6 8 IgG-bead phagocytosis (%) Supplementary Figure 1. Reversibility of phagocytosis inhibition by Tat. Human
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full//59/ra7/dc1 Supplementary Materials for Dok-7 ctivates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation kane Inoue, Kiyoko Setoguchi, Yosuke Matsubara,
www.sciencesignaling.org/cgi/content/full//59/ra7/dc1 Supplementary Materials for Dok-7 ctivates the Muscle Receptor Kinase MuSK and Shapes Synapse Formation kane Inoue, Kiyoko Setoguchi, Yosuke Matsubara,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues.
 Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
Supplemental Figure 1. Alignment of the NbGAPC amino acid sequences with their Arabidopsis homologues. Homologs from N. benthamiana (NbGAPC1, NbGAPC2, NbGAPC3), Arabidopsis (AtGAPC1, AT3G04120; AtGAPC2,
12 Interaction of Genes
 12 Interaction of Genes Yeast genetics has been particularly amenable for identifying and characterizing gene products that directly or indirectly interact with each other, especially when two mutations
12 Interaction of Genes Yeast genetics has been particularly amenable for identifying and characterizing gene products that directly or indirectly interact with each other, especially when two mutations
MCB 421 First Exam October 4, 2004
 1. (10 pts). An E. coli strain (strain A) that lacks an inducing prophage and carries the F factor is heavily irradiated with UV light and then mixed 1:1 with a second E. coli strain (strain B) that carries
1. (10 pts). An E. coli strain (strain A) that lacks an inducing prophage and carries the F factor is heavily irradiated with UV light and then mixed 1:1 with a second E. coli strain (strain B) that carries
Gene Expression: Transcription
 Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Gene Expression: Transcription The majority of genes are expressed as the proteins they encode. The process occurs in two steps: Transcription = DNA RNA Translation = RNA protein Taken together, they make
Chapter 4 - The nucleus controls the functions of life
 Chapter 4 - The nucleus controls the functions of life 4.1 The Function of the Nucleus within the Cell Cell Parts and Organelles Animal Cells Animal cells are equipped with many structures that allow the
Chapter 4 - The nucleus controls the functions of life 4.1 The Function of the Nucleus within the Cell Cell Parts and Organelles Animal Cells Animal cells are equipped with many structures that allow the
Supporting Information
 Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Supporting Information Chen et al. 1.173/pnas.153331112 Fig. S1. The sae2δ-suppressing mre11 alleles: protein level and location. (A) Protein level of Mre11 and Mre11 P11L expressed from the endogenous
Autophagic degradation of peroxisomes in the alkane-assimilating yeast Yarrowia lipolytica
 Autophagic degradation of peroxisomes in the alkane-assimilating yeast Yarrowia lipolytica A dissertation submitted to the Technical University of Dresden For the degree of Doctor rerum naturalium In Biology
Autophagic degradation of peroxisomes in the alkane-assimilating yeast Yarrowia lipolytica A dissertation submitted to the Technical University of Dresden For the degree of Doctor rerum naturalium In Biology
Yeast GFP Clone Collection
 Yeast GFP Clone Collection Catalog nos. 95700, 95701, 95702 Version B 30 September 2008 25-0730 IMPORTANT! Beginning in 2009, all Invitrogen clone manuals will only be available online at www.invitrogen.com
Yeast GFP Clone Collection Catalog nos. 95700, 95701, 95702 Version B 30 September 2008 25-0730 IMPORTANT! Beginning in 2009, all Invitrogen clone manuals will only be available online at www.invitrogen.com
SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION
 APPLICATION NOTE SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION Summary: Interest in using nanoparticles specifically, Surface Enhanced Raman Scattering
APPLICATION NOTE SURFACE ENHANCED RAMAN SCATTERING NANOPARTICLES AS AN ALTERNATIVE TO FLUORESCENT PROBES AN EVALUATION Summary: Interest in using nanoparticles specifically, Surface Enhanced Raman Scattering
DNA Binding Domains: Structural Motifs. Effector Domain. Zinc Fingers. Zinc Fingers, continued. Zif268
 DNA Binding Domains: Structural Motifs Studies of known transcription factors have found several motifs of protein design to allow sequence-specific binding of DNA. We will cover only three of these motifs:
DNA Binding Domains: Structural Motifs Studies of known transcription factors have found several motifs of protein design to allow sequence-specific binding of DNA. We will cover only three of these motifs:
THE JOURNAL OF CELL BIOLOGY
 Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
Supplemental Material THE JOURNL OF CELL BIOLOGY Onischenko et al., http://www.jcb.org/cgi/content/full/jcb.200810030/dc1 Figure S1. Characterization of Ndc1 interactions. () Intracellular distribution
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
Supplementary Figure 1 sirna and shrna mediated depletion of ATP7A results in loss of melanosomal ATP7A staining. a-h, sirna mediated ATP7A depletion. Immunofluorescence microscopy (IFM) analysis of ATP7A
A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole
 The EMBO Journal Vol.17 No.9 pp.2482 2493, 1998 A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole Jennifer J.Vowels and Gregory
The EMBO Journal Vol.17 No.9 pp.2482 2493, 1998 A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole Jennifer J.Vowels and Gregory
HOUR EXAM I BIOLOGY 422 FALL, In the spirit of the honor code, I pledge that I have neither given nor received help on this exam.
 Name First Last (Please Print) PID Number - HOUR EXAM I BIOLOGY 422 FALL, 2011 In the spirit of the honor code, I pledge that I have neither given nor received help on this exam. 1 Signature 2 3 4 5 6
Name First Last (Please Print) PID Number - HOUR EXAM I BIOLOGY 422 FALL, 2011 In the spirit of the honor code, I pledge that I have neither given nor received help on this exam. 1 Signature 2 3 4 5 6
Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes
 Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Supplemental Material for Different Upstream Activators Stimulate Transcription Through Distinct Coactivator Complexes Dong-ki Lee, Soyoun Kim, and John T. Lis Due to be published as a Research Communication
Solution key. c) Consider the following perturbations in different components of this signaling pathway in cells.
 Solution key Question 1 During a summer hike you suddenly spy a grizzly bear. This triggers a fight or flight response activating the signaling pathway that is shown on last Page. a) Circle the best option.
Solution key Question 1 During a summer hike you suddenly spy a grizzly bear. This triggers a fight or flight response activating the signaling pathway that is shown on last Page. a) Circle the best option.
Viral RNAi suppressor reversibly binds sirna to. outcompete Dicer and RISC via multiple-turnover
 Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
Supplementary Data Viral RNAi suppressor reversibly binds sirna to outcompete Dicer and RISC via multiple-turnover Renata A. Rawlings 1,2, Vishalakshi Krishnan 2 and Nils G. Walter 2 * 1 Biophysics and
4 Mutant Hunts - To Select or to Screen (Perhaps Even by Brute Force)
 Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 4 Mutant Hunts - To Select or to Screen
LiveReceptor AMPAR <Endogenous AMPAR Labeling Reagent>
 9-7 Hongo 2-Chome, Bunkyo-Ku Tokyo 113-0033, Japan LiveReceptor AMPAR Product Background Neurotransmitter receptors including glutamate receptors and GABA receptors
9-7 Hongo 2-Chome, Bunkyo-Ku Tokyo 113-0033, Japan LiveReceptor AMPAR Product Background Neurotransmitter receptors including glutamate receptors and GABA receptors
1. An alteration of genetic information is shown below. 5. Part of a molecule found in cells is represented below.
 1. An alteration of genetic information is shown below. 5. Part of a molecule found in cells is represented below. A-G-T-A-C-C-G-A-T A-G-T-G-A-T This type of alteration of the genetic information is an
1. An alteration of genetic information is shown below. 5. Part of a molecule found in cells is represented below. A-G-T-A-C-C-G-A-T A-G-T-G-A-T This type of alteration of the genetic information is an
The Two-Hybrid System
 Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine The Two-Hybrid System Carolina Vollert & Peter Uetz Institut für Genetik Forschungszentrum Karlsruhe PO Box 3640 D-76021 Karlsruhe
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
doi: 10.1038/nature06147 SUPPLEMENTARY INFORMATION Figure S1 The genomic and domain structure of Dscam. The Dscam gene comprises 24 exons, encoding a signal peptide (SP), 10 IgSF domains, 6 fibronectin
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-18 CELL BIOLOGY BIO-5005B Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-18 CELL BIOLOGY BIO-5005B Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
AUTOPHAGY. Industry-leading Tools for Analysis of Autophagic Pathways
 AUTOPHAGY Industry-leading Tools for Analysis of Autophagic Pathways 3Find Your Autophagy Solution COMPREHENSIVE SOLUTIONS FOR AUTOPHAGY Enabling a Clearer Understanding of Autophagy and Disease Through
AUTOPHAGY Industry-leading Tools for Analysis of Autophagic Pathways 3Find Your Autophagy Solution COMPREHENSIVE SOLUTIONS FOR AUTOPHAGY Enabling a Clearer Understanding of Autophagy and Disease Through
Notes on experimental technique: 1. Enzyme activity was measured by noting the increase in absorbance at 341 nm, as NADPH +
 Case 2 Glucose-6-phosphate dehydrogenase activity and cell growth Focus concept The activity of the pentose phosphate pathway enzyme glucose-6-phosphate dehydrogenase has been found to be important in
Case 2 Glucose-6-phosphate dehydrogenase activity and cell growth Focus concept The activity of the pentose phosphate pathway enzyme glucose-6-phosphate dehydrogenase has been found to be important in
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2
 Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
Supplementary Figure 1 Phosphorylated tau accumulates in Nrf2 (-/-) mice. Hippocampal tissues obtained from Nrf2 (-/-) (10 months old, 4 male; 2 female) or wild-type (5 months old, 1 male; 11 months old,
MICB688L/MOCB639 Advanced Cell Biology Exam II
 MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
MICB688L/MOCB639 Advanced Cell Biology Exam II May 10, 2001 Name: 1. Briefly describe the four major classes of cell surface receptors and their modes of action (immediate downstream only) (8) 2. Please
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplemental material Wang et al., http://www.jcb.org/cgi/content/full/jcb.201405026/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Generation and characterization of unc-40 alleles. (A and
Supplementary information
 Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
Supplementary information Supplementary figures Figure S1 Level of mycdet1 protein in DET1 OE-1, OE-2 and OE-3 transgenic lines. Total protein extract from wild type Col0, det1-1 mutant and DET1 OE lines
MCB 421 Exam #1 (A) Fall There are 9 questions and 1 supplement on last page. Answer all 9 questions. Be sure your name is on each page
 MCB 421 Exam #1 (A) Fall 2006 There are 9 questions and 1 supplement on last page. Answer all 9 questions. Be sure your name is on each page 1). (8 points) A Luria-Delbruck fluctuation test was done to
MCB 421 Exam #1 (A) Fall 2006 There are 9 questions and 1 supplement on last page. Answer all 9 questions. Be sure your name is on each page 1). (8 points) A Luria-Delbruck fluctuation test was done to
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Supplemental material Moutin et al., http://www.jcb.org/cgi/content/full/jcb.201110101/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Tagged Homer1a and Homer are functional and display different
Transcription in Eukaryotes
 Transcription in Eukaryotes Biology I Hayder A Giha Transcription Transcription is a DNA-directed synthesis of RNA, which is the first step in gene expression. Gene expression, is transformation of the
Transcription in Eukaryotes Biology I Hayder A Giha Transcription Transcription is a DNA-directed synthesis of RNA, which is the first step in gene expression. Gene expression, is transformation of the
